Abstract
Chemical modifications of transfer RNA (tRNA) molecules are evolutionarily well conserved and critical for translation and tRNA structure. Little is known how these nucleoside modifications respond to physiological stress. Using mass spectrometry and complementary methods, we defined tRNA modification levels in six yeast species in response to elevated temperatures. We show that 2-thiolation of uridine at position 34 (s2U34) is impaired at temperatures exceeding 30°C in the commonly used Saccharomyces cerevisiae laboratory strains S288C and W303, and in Saccharomyces bayanus. Upon stress relief, thiolation levels recover and we find no evidence that modified tRNA or s2U34 nucleosides are actively removed. Our results suggest that loss of 2-thiolation follows accumulation of newly synthesized tRNA that lack s2U34 modification due to temperature sensitivity of the URM1 pathway in S. cerevisiae and S. bayanus. Furthermore, our analysis of the tRNA modification pattern in selected yeast species revealed two alternative phenotypes. Most strains moderately increase their tRNA modification levels in response to heat, possibly constituting a common adaptation to high temperatures. However, an overall reduction of nucleoside modifications was observed exclusively in S288C. This surprising finding emphasizes the importance of studies that utilize the power of evolutionary biology, and highlights the need for future systematic studies on tRNA modifications in additional model organisms.
Keywords: yeast, tRNA modification, URM1, temperature stress, evolution
INTRODUCTION
Transfer RNA (tRNA) is critical for translation by linking the codon of the messenger RNA (mRNA) to the appropriate amino acid (Crick et al. 1957). Interestingly, all tRNA molecules feature post-transcriptional chemical modifications, some of which are highly conserved throughout all domains of life (Grosjean 2009; Grosjean et al. 2014). To date, ∼110 RNA modifications have been reported, and among the 42 tRNA species found in yeast, 25 modifications occur at 36 individual nucleotide positions (Phizicky and Hopper 2010). The greatest diversity of modified nucleotides is found at the anticodon loop positions 34 and 37—two nucleotides that are critical for decoding and maintenance of the translational reading frame (Helm and Alfonzo 2014). Furthermore, some modifications at the tRNA core affect the stability of specific tRNA species, whereas only minor effects are seen on other tRNAs (Phizicky and Alfonzo 2010).
Little is known about the regulation of tRNA modifications and how they respond to external factors. Certain nucleoside methylations were reported to increase upon chemical stress, which might indicate a need for tighter structural rigidity (Chan et al. 2010, 2012). Moreover, it has been proposed that tRNA modification changes might affect translation in response to stress (Begley et al. 2007; Chan et al. 2010, 2012; Bauer et al. 2012; Laxman et al. 2013). An interesting example is uridine at position 34 (U34), which pairs with the wobble base of the codon. In isoacceptors tEUUC, tKUUU, and tQUUG, U34 is modified to 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U). The mcm5s2U modification is generated through the concerted action of the elongator (ELP) pathway, which is required for formation of the mcm5 side chain, and the URM1 pathway that is essential for 2-thiolation (s2) (Huang et al. 2005, 2008; Björk et al. 2007; Nakai et al. 2008; Schlieker et al. 2008; Leidel et al. 2009; Noma et al. 2009). The physiological role of mcm5s2U is not well established, although it is thought to stabilize codon–anticodon interactions (Grosjean 2009; Rodriguez-Hernandez et al. 2013), and yeast strains lacking U34 modifications are sensitive to high temperatures and chemical stress (Esberg et al. 2006; Leidel et al. 2009).
Studies that connect tRNA modifications to stress generally apply harsh chemical treatments that are often incompatible with cellular viability. This complicates the evaluation of such a relationship and it is therefore desirable to analyze physiological stress conditions. Moderate temperature stress constitutes a key example of such a physiologically relevant experimental setup. The effect of elevated temperatures is well established on the transcriptional level in Saccharomyces cerevisiae, where >1000 genes respond to temperature shifts (Gasch et al. 2000). However, little attention has been paid to genes that are required for tRNA modification. Furthermore, temperature-induced changes in the tRNA modification landscape have not been extensively analyzed in eukaryotes, although studies have been made in bacteria and archaea (Agris et al. 1973; Watanabe et al. 1976; Kumagai et al. 1980; Kowalak et al. 1994). This is surprising, as growth at high temperature is associated with pathogenicity of yeast (McCusker et al. 1994), and a detailed study of tRNA modifications could potentially reveal mechanisms that facilitate temperature tolerance.
Finally, the majority of studies in yeast utilize a few common laboratory strains of S. cerevisiae. These strains have been domesticated and evolved in an environment that differs markedly from their natural habitat (Liti et al. 2009). Therefore, it is an open question whether such strains might respond to environmental stress differently than yeasts that have not evolved in the laboratory for thousands of generations.
To elucidate how cells respond to stress, we set out to analyze how high temperatures affect tRNA modifications in yeast. To this end, we quantitatively characterized nucleoside modification levels in six yeast species—S. cerevisiae, Saccharomyces bayanus, Saccharomyces mikatae, Saccharomyces paradoxus, Candida glabrata, and Schizosaccharomyces pombe—at ambient and elevated temperatures. We used reversed-phase ultraperformance liquid chromatography–mass spectrometry (RP-UPLC–MS), affinity gel electrophoresis, and Northern blots. This allowed us to establish the first comparative inventory of tRNA modifications in several yeast species. To our knowledge, this is the first comprehensive evolutionary analysis of how tRNA modifications respond to temperature stress. We found that high temperatures induced profound changes in the tRNA modification landscape. Surprisingly, all strains but one responded by globally up-regulating modification levels. We observed the opposite effect only in S288C. When specifically analyzing mcm5s2U34, we detected a significant decrease in 2-thiolation at high temperature in S. cerevisiae and S. bayanus. This reduction is reversible and recovers fully upon removal of the stress. Using polymerase inhibitors and a temperature-sensitive mutant strain of RNA polymerase III, we found no evidence that thiolation is actively down-regulated. In contrast, our results suggest that the URM1 pathway is inactive at high temperatures, and that loss of thiolation occurs as a result of diluting modified tRNA by newly transcribed unmodified species. Moreover, mcm5U34 modification levels are not negatively affected. Taken together, these findings suggest that the URM1 pathway is temperature sensitive in S. cerevisiae and S. bayanus, potentially reflecting a domestication event.
RESULTS
The S. cerevisiae S288C and W303 strains are capable of growth at elevated temperatures
We sought to quantitatively characterize tRNA modification landscapes of yeast in response to a relevant physiological stress. In most natural environments yeasts experience high temperatures, making it a very appropriate experimental paradigm. Furthermore, some tRNA modification mutants confer temperature sensitivity, implying a connection between temperature stress and tRNA modification (Esberg et al. 2006; Phizicky and Hopper 2010). We first determined the upper temperature limit of two commonly used laboratory strains—S288C and W303—by serial dilution spotting at temperatures ranging between 25°C and 43°C (Fig. 1). Both colony morphology and growth were normal at temperatures of up to 37°C. Interestingly, S288C is capable of growing at 39°C, whereas W303 is significantly impaired at this temperature. Neither of the strains was able to grow at temperatures exceeding 40°C (Fig. 1).
FIGURE 1.

Saccharomyces cerevisiae tolerates elevated temperatures. Serial dilutions (1:5) of S. cerevisiae S288C and W303 were spotted onto rich growth medium (YPD) plates and incubated for 3 d at the indicated temperatures. Note the absence of growth for both strains at 41°C and 43°C.
Mass spectrometry analysis of tRNA modification levels reveals unexpected variation
To quantitatively characterize tRNA modifications, we grew yeast at different temperatures in liquid culture and isolated their tRNA. The total tRNA was enzymatically converted into mononucleosides, which we analyzed by RP-UPLC–MS (Fig. 2A). Overlapping nucleosides were differentiated based on at least two of the following criteria: the isotopologue profile, the reported mass/charge (m/z) ratio, or the expected neutral loss—i.e., the mass change induced by dissociation of the ribosyl moiety from the nucleobase during in-source fragmentation. If the nucleobase is methylated, loss of the ribose yields a mass change of 132.04, whereas methylation of the ribose leads to a neutral loss of 146.06 (Fig. 2B). In addition, lack of neutral loss serves as a distinguishing feature to discriminate pseudouridine (Ψ) from uridine (Dudley et al. 2005). To compare the relative change in tRNA modification levels as a function of temperature, we adapted pyQms in a newly developed pipeline system for the analysis of nucleoside modifications. PyQms is a Python module for high-throughput quantification of MS experiments (Barth et al. 2014). Briefly, the area under the curve was determined for each modification from the MS1 spectra and normalized against adenosine. The normalized values were then compared with the respective value at the reference temperature (30°C) to reveal relative changes in modification levels. The feasibility of this approach, as well as the technical reproducibility and sensitivity of the LC–MS instrument, was validated by a set of 18 chemically synthesized nucleoside modifications (Supplemental Fig. S1A).
FIGURE 2.
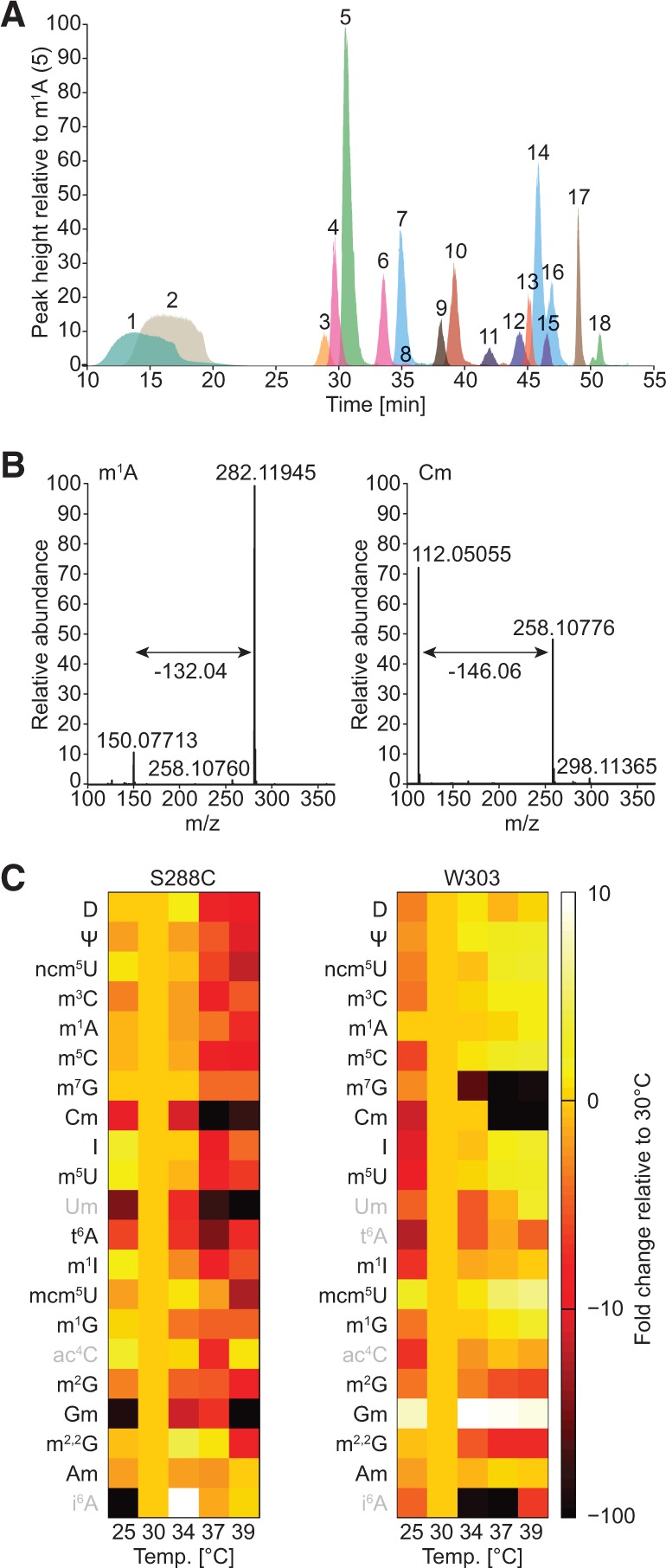
Quantitative liquid chromatography mass spectrometry analysis reveals differences in tRNA modification levels following temperature stress. (A) Superimposed extracted-ion chromatograms (XICs) of the 18 most prominent nucleoside modifications found in yeast. The average retention time (n = 3) for each modification in this chromatographic setup is indicated in minutes: 1, D (13.8); 2, Ψ (17.37); 3, ncm5U (29.03); 4, m3C (30.05); 5, m1A (31.78); 6, m5C (34.57); 7, m7G (36.24); 8, Cm (36.75); 9, I (38.28); 10, m5U (39.32); 11, t6A (43.44); 12, m1I (44.82); 13, mcm5U (45.37); 14, m1G (45.75); 15, ac4C (46.53); 16, m2G (46.63); 17, m2,2G (48.83); 18, Am (51.65). In addition, we detect yW (26.68), mcm5s2U (39.43), Um (42.06), ct6A (44.44), Gm (46.93), ncm5Um (47.92), and i6A (55.50). The canonical bases are omitted for clarity. (B) Full mass spectrum depicting the neutral loss occurring upon ionization-induced fragmentation of a nucleoside with a native (m1A) or methylated (Cm) ribosyl group, respectively. (C) Heat map depicting the relative comparison of normalized modification levels at various temperatures for 21 nucleosides detected by pyQms in samples from S. cerevisiae S288C and W303. The scale bar indicates the fold change in modification levels compared with 30°C (10-fold increase shown as white, no change as pale orange, and 100-fold decrease as black). Modifications that yield a weak MS signal are labeled in gray.
Manual analysis of the MS data for S288C at 30°C revealed 25 chemical modifications, of which pyQms reproducibly detected and quantified 21 (Fig. 2C; Supplemental Fig. S1B). We observed a clear temperature-dependent decrease in modification levels for S288C. At 25°C and 34°C, most nucleoside modifications are 1.2- to 1.8-fold less abundant than at 30°C, whereas dihydrouridine (D) and 7-methylguanosine (m7G) remain largely unaffected (Fig. 2C; Supplemental Table S1). However, a significant reduction occurs at 37°C and 39°C where all modifications are on average approximately fourfold down-regulated, apart from 2′-O-methyladenosine (Am) that remains largely unaltered (Fig. 2C; Supplemental Table S1). Interestingly, at elevated temperatures Ψ levels decrease noticeably (∼10-fold reduction at 39°C), whereas modifications associated with the structural stability of tRNA molecules, such as 1-methylguanosine (m1G) and 2-methylguanosine (m2G), are less affected (∼3.0-fold). Indeed, the relative abundance of m1G and m2G at 37°C and 39°C increases by ∼38% and ∼12%, respectively (Fig. 2C; Supplemental Table S1). In contrast, wobble position modifications, including 2′-O-methylcytidine (Cm) and 2′-O-methylguanosine (Gm) decrease strongly (Fig. 2C).
The W303 and S288C strains are evolutionarily closely related (Liti et al. 2009). Surprisingly, their modification landscape differs markedly (Fig. 2C). A general reduction in modification levels following temperature stress could not be seen for W303. Most nucleoside modifications remain at relatively high levels and rise, as temperature increases (Fig. 2C). The 1-methyladenosine (m1A) and Ψ modifications increase 1.4- and 1.9-fold, respectively, at 39°C (Supplemental Table S1). The same trend is observed for many other modifications, with m7G, Cm, and m2G being notable exceptions (Fig. 2C). In particular, m7G decreases to nearly nondetectable levels at 37°C and 39°C. Furthermore, when comparing the overall modification landscape of W303 to S288C, it appears that W303 is hypomodified at ambient temperatures but becomes hypermodified at 39°C (W303: ∼1.4-fold reduction at 25°C, ∼1.5-fold increase at 39°C; S288C: ∼1.2-fold reduction versus ∼4.0-fold reduction). This shows that although S288C and W303 may have a similar genotype, their tRNA modification phenotypes differ substantially.
2-Thiolation of tRNA decreases as a response to temperature stress
This perplexing difference in tRNA modifications observed in our MS analysis prompted us to perform an in-depth analysis of one specific modification. We chose mcm5s2U34 for several reasons. First, we did not detect this modification by MS, but it can be accurately quantified by ([N-acryloylamino]phenyl)mercuric chloride (APM)-affinity gel electrophoresis and Northern blotting with radiolabeled probes. In this assay, thiolated tRNAs are specifically retarded in a polyacrylamide gel (Igloi 1988). Second, mcm5s2U34 only occurs on three cytoplasmic tRNAs—tEUUC, tKUUU, and tQUUG—allowing us to quantify all modification targets in parallel. Third, this wobble uridine modification has been implicated in translational control (Grosjean 2009; Rodriguez-Hernandez et al. 2013).
Strikingly, we found a significant reduction in mcm5s2U34 levels at elevated temperatures (Fig. 3). For S288C, ∼60% of tKUUU is modified at 25°C and 30°C, whereas modification levels drop below 20% at 39°C (Fig. 3A). A slight decrease to ∼50% is seen at 34°C, suggesting that this temperature constitutes the upper boundary for the thermal adaptation of the strain. A similar trend is observed for tEUUC as well, albeit the overall 2-thiolation levels are slightly lower at all temperatures. Interestingly, tQUUG shows even lower 2-thiolation levels (∼35%) at 25°C–34°C, and an almost complete loss of 2-thiolation at 37°C and 39°C (Fig. 3A). This indicates that the specificity or affinity of the enzymes toward the individual isoacceptors differ substantially in S288C. In W303, 2-thiolation levels for all three isoacceptors are more similar, suggesting that formation of the mcm5s2U34 modification proceeds with a lower isoacceptor preference (Fig. 3B). In contrast to most other modifications and as observed in S288C, tRNA thiolation levels clearly decrease at elevated temperatures in W303.
FIGURE 3.
2-Thiolation levels in S. cerevisiae respond to elevated temperatures. (A,B) Representative autoradiogram of tRNA extracted at different growth temperatures from (A) S288C and (B) W303 following ([N-acryloylamino]phenyl)mercuric chloride (APM)-affinity gel electrophoresis and Northern blot with the tKUUU probe. The horizontal arrows in this and subsequent Northern blot images mark thiolated (black) and non-thiolated (gray) tRNA species. 2-Thiolation levels were quantified for all three tRNA isoacceptors (n = 3, apart from W303 samples grown at 25°C where n = 2). (C) Superimposed UV-chromatograms from UPLC analysis of enzymatically digested tRNA of S288C. The position of the canonical bases is indicated, and synthetic standards of mcm5U (20.3 min) and mcm5s2U (31.5 min; boxed) were used to determine the average retention times (n = 3). Note the decreasing height and area of the mcm5s2U peak at elevated temperatures. The relative change in peak area for mcm5s2U is depicted (normalized to 30°C).
We independently verified our results for the S288C samples by UPLC. As seen in the chromatogram overlay, the intensity of the mcm5s2U34 signal decreased as temperature increased (∼0.61% of the total signal at 30°C, ∼0.28% at 34°C, ∼0.071% at 37°C, and ∼0.021% at 39°C) (Fig. 3C). This analysis confirmed that 2-thiolation levels are reduced at higher temperatures, although it did not provide information regarding possible isoacceptor preferences.
2-Thiolation levels are reduced in ELP-complex deficient strains (Björk et al. 2007; Leidel et al. 2009). Such a deficiency could account for the reduction in 2-thiolation that we observed. However, careful analysis of MS data speaks against this. In W303 cells, mcm5U34 levels increase ∼1.2-fold at 34°C to ∼3.8-fold at 39°C, whereas mcm5s2U34 levels decrease approximately threefold to fivefold depending on the isoacceptor (Supplemental Table S1; Figs. 2C, 3B). This suggests that the ELP pathway remains active. In S288C, the picture is more complicated due to the overall reduction of most modifications at high temperatures. However, ncm5U34 levels can act as a proxy for ELP pathway activity that is independent of s2 levels (Björk et al. 2007). The ratio of mcm5U34 to ncm5U34 increases in S288C from ∼1.3-fold at 34°C to ∼2.5-fold at 37°C, and in W303 from ∼1.4-fold at 34°C to ∼1.8-fold at 37°C and 39°C, respectively. This indicates that the ELP pathway is active and relative increases in mcm5U34 levels are observed due to lack of 2-thiolation. Thus, the observed loss of mcm5s2U34 modification at high temperatures is not due to a defect in the ELP pathway, but rather due to temperature sensitivity of one or more components in the URM1 pathway.
2-Thiolation recovers upon removal of temperature stress
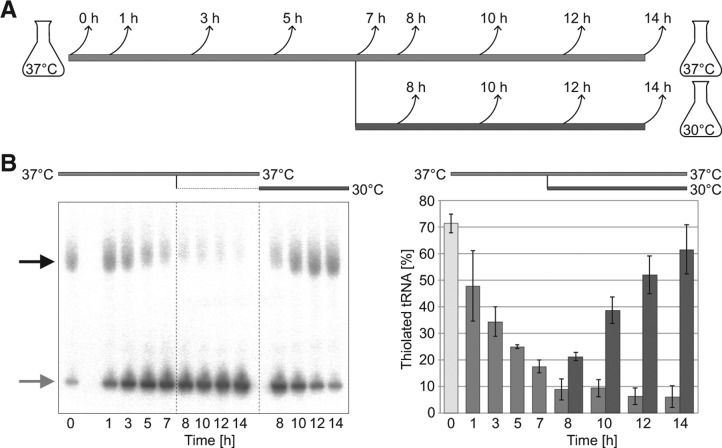
To the best of our knowledge, temperature sensitivity of a cytoplasmic U34 modification has not been described before. Therefore, we wanted to better understand the kinetics of this inactivation and to test whether 2-thiolation levels can recover. To address this question, we maintained S288C cells in logarithmic growth at 37°C for 7 h, at which point the cultures were split into two. One culture was kept at 37°C and the other shifted to 30°C, and growth was continued for additional 7 h (Fig. 4A). We found that the mcm5s2U34 modification levels decreased at 37°C in an exponential manner, reaching the half point after ∼3 h (Fig. 4B; Supplemental Fig. S2). However, in cells that were shifted to 30°C, 2-thiolation levels recovered within a few hours. This shows that cells maintain the ability to recover.
FIGURE 4.
Temperature-induced reduction of tRNA 2-thiolation is reversible. (A) Scheme of the time course assay. The 37°C culture is shown as a light gray line; the culture at 30°C is depicted by a dark gray line (for this and all subsequent figures). Arrows indicate time points for sample collection. (B) Representative autoradiogram following APM-affinity gel electrophoresis and Northern blot with tKUUU probe of tRNA collected at the indicated time points. 2-Thiolation levels were quantified by image densitometry (n = 3).
Inhibition of tRNA synthesis stalls 2-thiolation at elevated temperatures
We have demonstrated that loss of mcm5s2U34 modification is reversible in wild-type yeast. However, different mechanisms could underlie the loss of 2-thiolation at higher temperatures: 2-thiolated tRNA molecules might be actively degraded, the modification itself might be actively removed, or newly transcribed tRNA might not acquire the modification at 37°C, thereby diluting the modified tRNA.
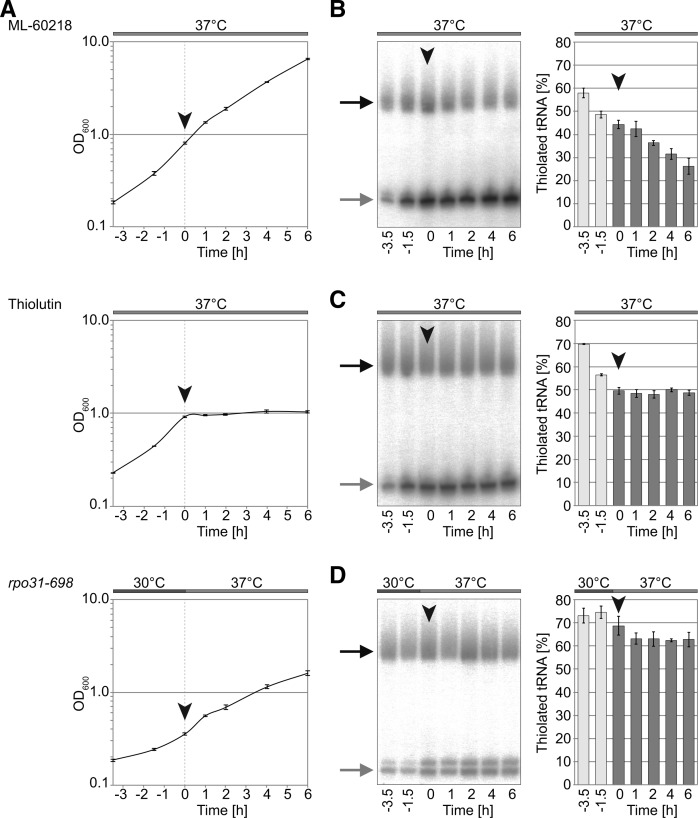
To address this question, we decided to inhibit RNA synthesis. First, we used ML-60218, an indazolo-sulfonamide compound that specifically inhibits RNA polymerase III (Wu et al. 2003). ML-60218 does not completely inhibit tRNA synthesis, but a leakiness of 20%–30% is expected (Iwasaki et al. 2010). This hypomorphic phenotype allowed us to reduce tRNA synthesis while only slightly reducing growth rates during the first hours of the experiment (Fig. 5A, top panel). Importantly, the loss of tRNA modification occurred slower relative to the situation without the inhibitor (Figs. 4B, 5B; Supplemental Figs. S2, S3A). This suggests that tRNA synthesis rather than active removal of thiolated tRNA or thiolated nucleosides affect thiolation levels at high temperatures.
FIGURE 5.
Inhibition of tRNA synthesis stalls 2-thiolation. (A) Growth curve measurements of S288C cells in logarithmic growth at 37°C supplemented with RNA polymerase inhibitor ML-60218 (top panel) or thiolutin (middle panel) at the 0 h time point. rpo31-698 was grown at 30°C and shifted to 37°C at the 0 h time point (bottom panel). (B) Representative autoradiogram of tRNA extracted at indicated time points from ML-60218 treated cells following APM-affinity gel electrophoresis and Northern blot with tEUUC probe. Arrowheads highlight the 0 h time point at which the inhibitor was added. Thiolation levels were quantified by image densitometry (n = 3). (C) As in B for thiolutin. (D) As in B for rpo31-698, where inhibition of tRNA synthesis is induced by the temperature shift to 37°C.
To verify this hypothesis, we used thiolutin to completely block transcription of RNA polymerases I–III (Tipper 1973). Addition of thiolutin led to cessation of growth within 1 h (Fig. 5A, middle panel). Similarly, the mcm5s2U34 modification levels remained constant throughout the experiment (Fig. 5C; Supplemental Fig. S3B). To exclude that this is an indirect effect caused by inactivation of RNA polymerases I and II, we utilized S. cerevisiae rpo31-698, a temperature-sensitive mutant strain of RNA polymerase III (Li et al. 2011). When shifted to the restrictive temperature at 37°C, growth slowed down (as seen for ML-60218) (Fig. 5A) and 2-thiolation levels remained constant following an initial decrease during the first hour after shift (Fig. 5D; Supplemental Fig. S3C). These experiments suggest that the tRNA isoacceptors and the modification per se remain stable and are not actively turned over. It is unlikely that addition of the inhibitors directly affect 2-thiolation levels, as we observed no change in mcm5s2U34 levels in cells grown at 30°C in the presence of ML-60218 or thiolutin (Supplemental Fig. S4A,B). Furthermore, cells grown at 37°C and shifted to 30°C upon addition of thiolutin did not increase 2-thiolation levels (Supplemental Fig. S4C). Taken together, our experiments suggest that tRNA synthesized before exposure to heat retain their modifications whereas tRNA synthesized at high temperatures is not 2-thiolated.
Temperature tolerance and 2-thiolation vary markedly in other yeast species
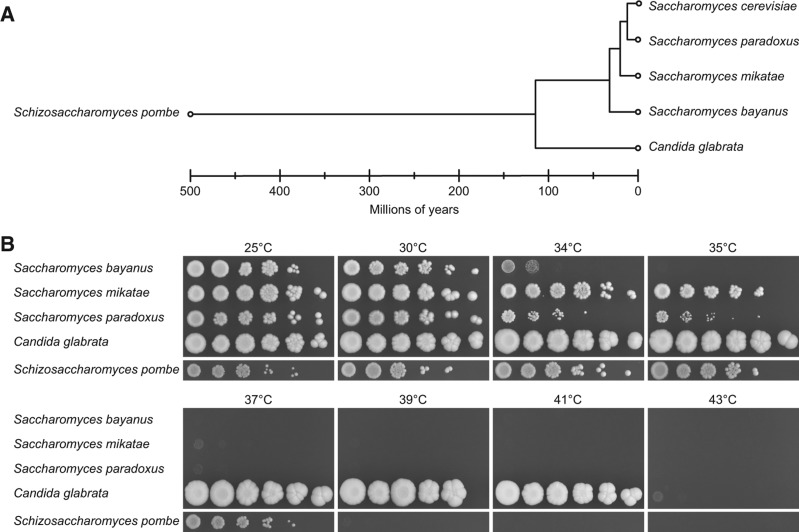
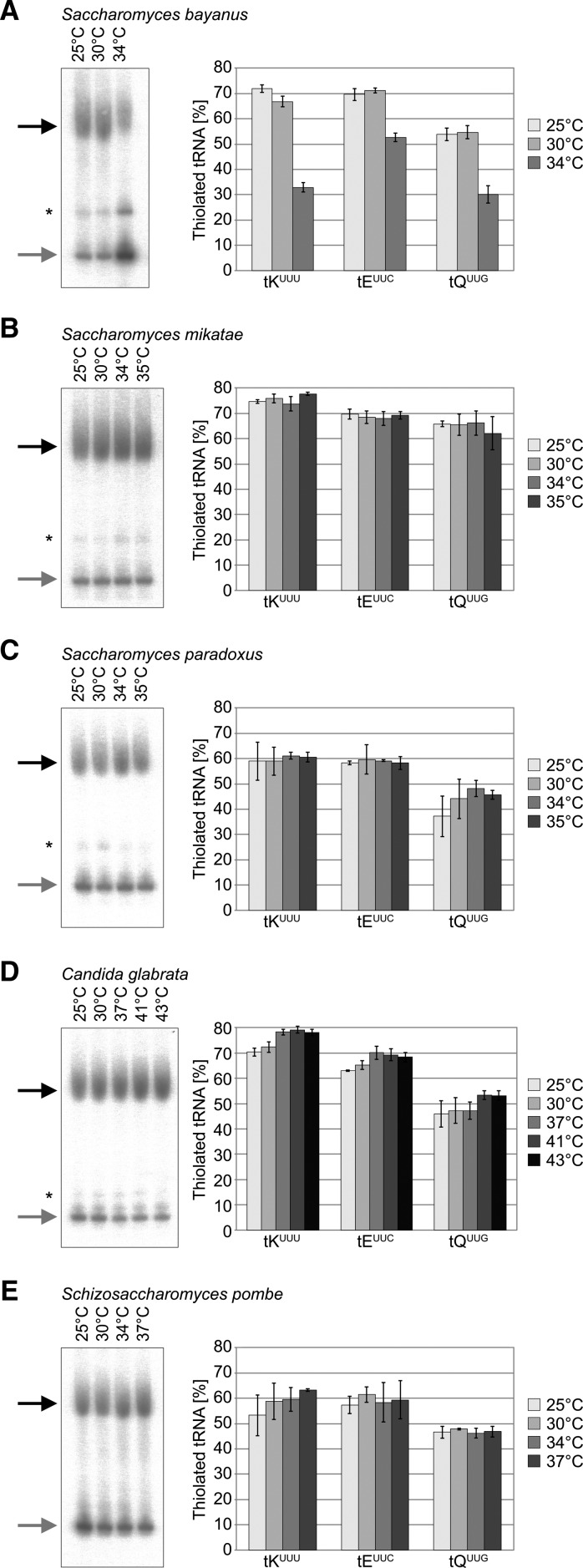
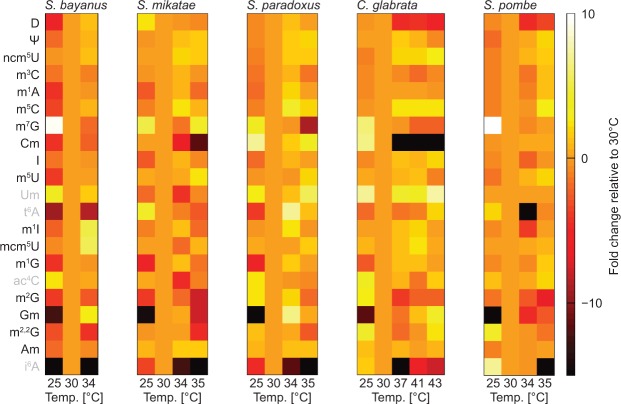
As we found reduced s2U34 levels at elevated temperature in S. cerevisiae, we wondered whether this trait is evolutionarily conserved and constitutes a common adaptation of yeast to high temperature. To this end, we analyzed representative strains of five evolutionarily divergent yeast species; S. bayanus, S. mikatae, S. paradoxus, C. glabrata, and S. pombe (Fig. 6A), and performed serial dilution spottings at the same temperature range as for S. cerevisiae (Figs. 1, 6B). Candida glabrata grows very well at 41°C and slight growth can be seen at 43°C. In contrast, S. bayanus did not grow above 34°C, whereas S. mikatae and S. paradoxus grew at 35°C. S. pombe is also capable of growth at 37°C (Fig. 6B). Based on these results we isolated tRNA from liquid cultures for subsequent analysis by Northern blot and MS. As expected, all yeast species contained 2-thiolated tRNA. However, the levels varied between 50% and 60% for the different isoacceptors in S. pombe to ∼70% in S. mikatae. Furthermore, the phenomenon that tQUUG is less 2-thiolated than tEUUC and tKUUU appeared to be evolutionarily conserved. Similar to S. cerevisiae, S. bayanus is the only yeast that showed decreased thiolation levels in response to elevated temperatures (Figs. 3, 7A). In S. mikatae, S. paradoxus, and S. pombe, we did not detect obvious changes in 2-thiolation levels in response to heat (Fig. 7B,C,E). However, in C. glabrata, which is a pathogenic yeast species capable of growth at high temperature, 2-thiolation levels rose slightly in response to increased temperature (Fig. 7D). Thus, loss of 2-thiolation does not constitute a conserved response to high temperatures in yeast.
FIGURE 6.
Evolutionary divergent yeasts have different temperature preferences. (A) Phylogenetic relationship and evolutionary distance of all yeast species included in this study. (Data adapted from Rhind et al. 2011.) (B) Serial dilutions (1:5) of five yeast species were spotted onto rich growth medium (YPD or YES) plates and incubated for 3 d at the indicated temperatures.
FIGURE 7.
2-Thiolation levels are not affected by temperature in most other yeast species. Representative autoradiogram images of tRNA extracted at different growth temperatures from (A) S. bayanus, (B) S. mikatae, (C) S. paradoxus, (D) C. glabrata, and (E) S. pombe following APM-affinity gel electrophoresis and Northern blot with the tKUUU probe. 2-Thiolation levels were quantified by image densitometry (n = 3, apart from samples grown at 25°C where n = 2). Asterisks (*) show unspecific binding of the probe.
Furthermore, MS analysis of these representative yeast species revealed tRNA modification landscapes that share remarkably similar features (Fig. 8; Supplemental Fig. S5). In contrast to S288C, all other strains share the trend of up-regulating modification levels at higher temperatures (Figs. 2C, 8). In particular, S. bayanus is very similar to W303 as many modifications respond comparably with increased temperatures (Figs. 2C, 8). Candida glabrata surpasses its modification levels at 30°C by an average of ∼12% at 39°C–43°C. With the exception of S288C, Ψ modification levels increase ∼1.2- to 2.0-fold at high temperatures in all other strains analyzed (Fig. 8; Supplemental Table S1). Methylations associated with the structural stability of the tRNA molecule, such as m5C and m1G, show clear temperature dependance in all species and become more prevalent as temperature increases (Fig. 8). More divergence is observed for position 34 modifications, as mcm5U and Am remain stable throughout all temperatures in all species, and Cm decreases significantly in S. mikatae and C. glabrata but not in S. pombe (Fig. 8; Supplemental Table S1). In addition, m7G is detected at slightly higher intensity only at ambient temperature in S. bayanus and S. pombe. This surprising similarity between species implies that neither evolutionary distance nor natural environments have forced a radiation of modification phenotypes.
FIGURE 8.
tRNA modification landscapes for divergent yeast species show surprising similarity. Heat map of the relative change in nucleoside modification levels compared with 30°C in S. bayanus, S. mikatae, S. paradoxus, C. glabrata, and S. pombe using pyQms. The scale bar shows a 10-fold increase in modification levels as white, no change as pale orange, and a 31.6-fold decrease as black. Modifications that yield a weak MS signal are labeled in gray.
DISCUSSION
We conducted a comprehensive evolutionary study to quantitatively characterize the effects of temperature stress on tRNA modification levels in yeast. Importantly, we established that elevated temperatures lead to a reduction of the mcm5s2U34 level in the commonly used S. cerevisiae laboratory strains S288C and W303. However, this reduction is reversible, as 2-thiolation levels return to normal within a few hours after relief of the stress. This is an intriguing finding, as modification of U34 in tEUUC has been proposed to dynamically regulate the translation of key damage response proteins (Begley et al. 2007). A decrease of s2U34 might similarly redirect the translational program of cells at high temperature by disfavoring recognition of the cognate codons for tEUUC, tKUUU, and tQUUG. However, it is unlikely that loss of 2-thiolation in response to high temperatures might be an active regulatory process, which is exploited by the cell. First, the decrease in 2-thiolation levels correlates with cellular growth rates, suggesting that newly synthesized, unmodified tRNA dilutes the pool of modified tRNA. Similarly, recovery of thiolation upon shift to ambient temperatures is slow compared with the immediate response that follows environmental stress (Gasch et al. 2000). Thus, a translationally regulated response would be slow and follow the transcriptional rewiring of the cell rather than preceding it. Second, reducing or blocking the synthesis of nascent tRNA slows down or prevents loss of 2-thiolation. If s2U34 would be actively degraded, it would likely be uncoupled from tRNA synthesis. Third, loss of U34 modification is likely to affect the translation of a large number of proteins, thereby causing cellular stress, which is corroborated by the up-regulation of Gcn4p upon U34 hypomodification (Zinshteyn and Gilbert 2013). Therefore, it is unlikely that S. cerevisiae would gain fitness benefits for growth at high temperatures by actively reducing 2-thiolation. Finally, our analysis of other yeasts at high temperature reveals that s2U34 levels are reduced only in S. cerevisiae and S. bayanus. In contrast, their close relatives, as well as S. pombe, which constitutes the phylogenetic outgroup in our study, do not exhibit the same phenotype. Thus, a decrease of 2-thiolation in response to high temperatures is unlikely to constitute an ancient cellular program. Taken together, our results suggest that the reduction of s2U34 levels at high temperatures is likely not an active regulatory process used to dynamically rewire the translational program of the cells. Rather, it may reflect the temperature sensitivity of one or more components of the URM1 pathway.
It is puzzling that loss of 2-thiolation may have been independently acquired in S. cerevisiae and S. bayanus. However, both were originally used in fermentation. It is possible that constant growth at ambient temperatures in rich media selected for modifications of cellular metabolism, which decrease fitness of yeast in its natural environment. Therefore, it will be interesting to extend the analysis to yeast strains from different ecological niches, and to test whether loss of 2-thiolation at high temperature is a general behavior of S. cerevisiae.
Importantly, our analysis of 2-thiolation reveals enzymatic features of the URM1 pathway. We found that isoacceptors are not fully modified and that relative 2-thiolation levels generally follow the order tKUUU > tEUUC > tQUUG. This raises the question, whether functional constraints limit the pathway to fully modify all tRNA molecules or whether some isoacceptors are less important (Phizicky and Alfonzo 2010). The latter is supported by tRNA overexpression experiments in S. cerevisiae and S. pombe, where overexpression of tKUUU and tQUUG, but not tEUUC, leads to the rescue of different stress sensitivities in mcm5s2U34 deficient strains (Esberg et al. 2006; Björk et al. 2007; Leidel et al. 2009; Fernández-Vázquez et al. 2013). Furthermore, 2-thiolation is not temperature sensitive in all yeasts. Thus, it is feasible to complement mutants in S. cerevisiae with the genes of other species that form s2U34 at high temperature to identify the genes that cause temperature sensitivity of 2-thiolation.
We sought to define the tRNA modification landscapes in a range of yeast species in response to stress. By choosing representative strains of S. cerevisiae, S. bayanus, S. mikatae, S. paradoxus, and C. glabrata, we analyzed several prominent models in the Saccharomyces clade, including one pathogenic strain. Furthermore, S. pombe is separated from the other species due to a whole-genome duplication event (Vivancos et al. 2006). Surprisingly, all strains except S288C showed a fairly similar pattern—most tRNA modifications increase modestly if temperature rises >30°C. This may either reflect a coordinated response to temperature stress or a passive increase in pathway activities due to more favorable reaction conditions at higher temperatures. Our global analysis also did not address whether all tRNA modifications occur at relevant positions. Theoretically, the increase could reflect unspecific modifications through decreased selectivity toward the natural modification targets. However, examples in archaea show that tRNA modifications are more abundant at high temperature (Noon et al. 2003; Shigi et al. 2006). Similar findings have not been reported in a eukaryotic system. When looking at the modification landscapes more closely we find that Ψ, which is one of the most abundant nucleoside modifications, remains prominent at elevated temperatures in all strains analyzed, apart from S. cerevisiae S288C where it decreases. Clearly, methylations implicated in structural stability, such as m5C and m1G, become more prevalent at high temperatures, emphasizing a need for tighter conformational control of the tRNA molecule. Indeed, these observations support the notion that high temperature warrants increased tRNA modification levels also in eukaryotic cells.
Upon comparison to all other strains, it is surprising that the commonly used laboratory strain S288C reacts in such a divergent manner to increasing temperatures, especially as the response is so different from the closely related strain W303. It is important to keep this in mind. Even though S288C offers numerous experimental advantages, we need to be cautious when basing our conclusions on results obtained in a single wild-type strain. The selective pressures in the laboratory are unusual, and not every conclusion may stand the test in a natural environment. This emphasizes the need to perform more studies that make use of the great power of the evolutionary biology of yeasts.
MATERIALS AND METHODS
Yeast strains and growth conditions
Haploid mating type a strains of S. cerevisiae and S. pombe were used and diploid strains of S. bayanus, S. mikatae, S. paradoxus, and C. glabrata. Refer to Supplemental Table S2 for further information about these strains.
All experiments were conducted in rich growth medium (YPD; Formedium), except for S. pombe, which was grown in YES. The solid growth media were supplemented with 2% agar. Overnight (o/n) starter cultures were grown at 30°C, 200 rpm. The starter culture was inoculated into prewarmed medium to yield an OD600 = 0.2, grown at the indicated temperatures to logarithmic growth phase (OD600 = 0.8–1.0), and harvested by centrifugation for 3 min at 5000g.
For time-course experiments, an o/n starter culture of S288C was used to inoculate cultures at 37°C. The cultures were kept at logarithmic growth by regular dilution with fresh prewarmed medium. At 7 h post-inoculation, the cultures were split into two, one being further incubated at 37°C and the other shifted to 30°C. Samples were collected at the indicated time points for tRNA isolation.
Isolation and purification of tRNA
tRNA was isolated essentially as previously described (Emilsson and Kurland 1990; Björk et al. 2001). In brief, cells were resuspended in 4 mL of 0.9% NaCl and lysed in an equal volume of acidic phenol (pH 4.3) supplemented with 0.1 vol. of 1-bromo-3-chloropropane (BCP) and glass beads. The mixture was vortexed for 5–10 min followed by centrifugation for 15 min at 10,000g. The RNA containing aqueous phase was transferred to a fresh centrifugation tube containing 2 mL acidic phenol and 0.1 vol. of BCP. The suspension was vortexed for 30 sec and centrifuged for 10 min at 10,000g. This step was repeated twice. Subsequently, the total RNA containing aqueous phase was applied onto Nucleobond AX-100 columns (Macherey-Nagel) that were preequilibrated with 10 mL of buffer EQ (10 mM Tris, pH 6.3, 15% ethanol, 200 mM KCl, 0.15% Triton X-100). The columns were washed twice with 12 mL of buffer WA (10 mM Tris, pH 6.3, 15% ethanol, 300 mM KCl), and the tRNA was eluted with 10 mL buffer EL (10 mM Tris, pH 6.3, 15% ethanol, 650 mM KCl) into 2.5 vol. of 99.6% ethanol. The tRNA was precipitated o/n at −20°C and pelleted by centrifugation for 20 min at 10,000g. Residual salt was removed by washing the pellet three times with 80% ethanol. The tRNA pellet was dried at room temperature for ∼10 min and resuspended into 50 µL of RNase-free water.
Electrophoretic fractionation and Northern hybridization
Northern blot analysis was performed essentially as previously described (Emilsson and Kurland 1990; Björk et al. 2001). For DNA oligonucleotides refer to Supplemental Table S3. Signals were collected onto BAS-IP MS 2025-E image plates and scanned using a Fuji FLA 7000 phosphorimager (Fujifilm). Digital image analysis (densitometry) was performed using Fiji-ImageJ2 (The Fiji Project), measuring the band intensities using the Analyze/Gels tool.
Inhibition of tRNA synthesis
To assess turnover of nucleoside modifications, tRNA synthesis was inhibited by the addition of RNA polymerase inhibitors. S288C cultures at 37°C were grown to OD600 = 0.6 as described above, at which point the cultures were supplemented with either 5 µg/mL thiolutin (Sigma-Aldrich) or 50 µg/mL N1-[3-(5-chloro-3-methylbenzo[b]thiophen-2-yl)-1-methyl-1H-pyrazol-5-yl]-2-chlorobenzene-1-sulfonamide (ML-60218; Calbiochem). The rpo31-698 (Li et al. 2011) culture was grown to OD600 = 0.6 at 30°C and subsequently shifted to 37°C. The cultures were grown for an additional 6 h and samples for total RNA isolation were collected at indicated time points. Total RNA isolation was performed as described above and precipitated by adding 2.5 vol. of 99.6% ethanol to the final aqueous phase. The total RNA was precipitated o/n at −20°C and washed with 80% ethanol.
Enzymatic digestion of tRNA and nucleoside purification
Dephosphorylated mononucleosides were generated by enzymatic digestion of 25 µg tRNA using 80 milliunits of Nuclease P1 from Penicillium citrinum (Sigma-Aldrich) in the presence of 0.2 unit of Shrimp Alkaline Phosphatase (Fermentas) in 30 µL reactions at 37°C containing 2 mg/mL ZnCl2 and 1× NEB3 buffer. Following 1.5 h incubation, the reaction mixture was supplemented with 15 µL 0.5 M ammonium bicarbonate and incubation was resumed at 37°C for 1 h. The reaction was terminated by adding 5.0% trifluoroacetic acid (TFA) in water to a final concentration of 1.0%. Nucleosides were purified with HyperSep Hypercarb Spin Tips (Thermo Scientific). Briefly, the spin tips were equilibrated three times with 50 µL 0.1% TFA in H2O. The digested samples were applied and washed three times with 50 µL 0.1% TFA in H2O. The nucleosides were eluted twice with 50 µL 0.1% TFA in 80% acetonitrile. All centrifugation steps were carried out in a microcentrifuge for 30 sec to 1 min at 845g. The samples were dried to completion in a Savant SpeedVac concentrator (Thermo Scientific) and resuspended in either 0.1% formic acid (for chromatography analysis) or 5 mM sodium acetate (for mass spectrometry).
Reverse-phase high-performance liquid chromatography of nucleosides
Analysis of nucleosides was performed on a Hypercarb 3 µm 2.1 × 150 mm porous graphitic carbon column (Thermo Scientific) using a Knauer PLATINblue UPLC system with a PDA-1 photodiode array detector (Dr. Ing. Herbert Knauer GmbH). The column was equilibrated in 0.1% formic acid with 2% acetonitrile at a flow rate of 0.75 mL/min until a stable baseline was achieved. Twenty-five micrograms of nucleoside digest was loaded onto the column and washed for 5 column volumes (CV) with 2% acetonitrile. Separation was obtained by applying a linear gradient from 10% to 40% acetonitrile at 0.75 mL/min for 40 CV, followed by a wash at 98% acetonitrile and regeneration at 2% acetonitrile for 5 CV, respectively. Absorption at 260 nm was recorded.
The average retention times for all nucleotide modifications were obtained by analyzing synthetic nucleoside standards (Carbosynth Ltd.). mcm5U, mcm5s2U, ncm5U, and ncm5s2U were a generous gift by Andrzej Małkiewicz. Nucleoside modification levels were determined using automated peak analysis in ChromGate 3.3.2 (Dr. Ing. Herbert Knauer GmbH) by normalization of the signal intensity to adenosine in each run.
Liquid chromatography mass spectrometry
Nucleosides were resuspended in 25 µL of 5 mM sodium acetate buffer pH 5.3 and subjected to LC–MS analysis using a Proxeon EASY nLC online coupled via a C18 reversed-phase column (Synergi 4u Hydro-RP 80A 0.3 × 150 mm, Phenomenex Ltd.) and a stainless steel emitter (Proxeon, 0.03 × 40 mm) to a Q Exactive mass spectrometer (Thermo Finnigan). The column was attached to a nano ESI source from Proxeon. Injection volume was 5 µL. The buffer system consisted of 5 mM sodium acetate pH 5.3 (Buffer A) and 40% acetonitrile (Buffer B). Nucleosides were separated using a multistep gradient (0%–1% B in 5 min; 1%–2% B in 4:20 min; 2%–3% B in 1:40 min; 3%–5% B in 2 min; 5%–25% B in 19 min; 25%–50% in 6 min; 50%–75% B in 5:30 min; hold at 75% for 1:30 min; 75%–99% B in 5 min; hold at 99% for 7 min) at a flow rate of 1 µL/min. The mass spectrometer was run in the positive mode at a resolution of 70,000. Full MS spectra were recorded in profile mode in the scan range from m/z 100–700, with the AGC target value set to 1 × 105 and the maximum fill time to 200 msec. Only single charged ions were allowed (Nuc+H+), excluding uncharged compounds and compounds with z ≥ 2. In source, CID fragmentation was enabled (5.0 eV) to allow for incomplete fragmentation of most nucleosides at the glycosidic bond between the ribose and the corresponding base. Loss of the sugar is reflected by a mass difference of 132.04 (ribosyl) or 146.06 (methylated ribosyl) between the intact nucleoside and remaining base, for both of which signals are present in the MS spectrum. This mass loss is characteristic for ribonucleosides and simplifies identification of the nucleoside spectra in complex mixtures. Average retention times were obtained as described above.
Automated quantification of nucleoside modifications
Identification and quantification of nucleoside modifications was performed using pyQms (Barth et al. 2014). In brief, pyQms generates high accuracy isotopologues from chemical formulas and matches those onto MS1 spectra using a weighted similar match score (Gower 1971). A manually curated list of all potentially expected nucleosides was used as input for pyQms (data obtained from MODOMICS) (Machnicka et al. 2013). The isotopologues were grouped into five classes that share the same nitrogenous base and differ only by one of the following neutral loss masses: ribose (C5O4H8), methylated ribose (C6O4H10), ribophosphate (C5O7H9P1), methylated ribophosphate (C6O7H11P1), or phosphate (H1O3P1). When insource fragmentation is applied, multiple members of one class can be detected in the same MS1 scan. This allows us to distinguish molecules that share the same chemical formula, yet differ in their chemical compositions after neutral loss.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank Prof. Andrzej Małkiewicz for synthesis of modified nucleosides, and Dr. Tamara Bar-Magen Numhauser for skillful assistance in MS data analysis. This work was supported by grants from the Max Planck Society (Max-Planck-Gesellschaft), the North Rhine-Westphalian Ministry for Innovation, Science and Research [314-400 010 09], and the European Research Council [ERC-2012-StG 310489-tRNAmodi] (to S.A.L.). L.P.S. is a Sigrid Jusélius Fellow (2012–2014). F.A. is supported by the International Max Planck Research School—Molecular Biomedicine.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048199.114.
Freely available online through the RNA Open Access option.
REFERENCES
- Agris PF, Koh H, Söll D 1973. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch Biochem Biophys 154:277–282. [DOI] [PubMed] [Google Scholar]
- Barth J, Bergner SV, Jaeger D, Niehues A, Schulze S, Scholz M, Fufezan C 2014. The interplay of light and oxygen in the reactive oxygen stress response of Chlamydomonas reinhardtii dissected by quantitative mass spectrometry. Mol Cell Proteomics 13:969–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D 2012. Translational control of cell division by Elongator. Cell Rep 1:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 28:860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, Persson OP 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Huang B, Persson OP, Byström AS 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13:1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ 2010. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6:e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH, Griffith JS, Orgel LE 1957. Codes without commas. Proc Natl Acad Sci 43:416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E, Tuytten R, Bond A, Lemière F, Brenton AG, Esmans EL, Newton RP 2005. Study of the mass spectrometric fragmentation of pseudouridine: comparison of fragmentation data obtained by matrix-assisted laser desorption/ionisation post-source decay, electrospray ion trap multistage mass spectrometry, and by a method utilising electrospray quadrupole time-of-flight tandem mass spectrometry and in-source fragmentation. Rapid Commun Mass Spectrom 19:3075–3085. [DOI] [PubMed] [Google Scholar]
- Emilsson V, Kurland CG 1990. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J 9:4359–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Byström AS 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24:139–148. [DOI] [PubMed] [Google Scholar]
- Fernández-Vázquez J, Vargas-Pérez I, Sansó M, Buhne K, Carmona M, Paulo E, Hermand D, Rodriguez-Gabriel M, Ayté J, Leidel S, et al. 2013. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 9:e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower JC 1971. A general coefficient of similarity and some of its properties. Biometrics 27:857–871. [Google Scholar]
- Grosjean H 2009. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. Landes Bioscience, Austin, TX. [Google Scholar]
- Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barre A, Yoshizawa S, Fourmy D, de Crecy-Lagard V, et al. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of Mollicutes. PLoS Genet 10:e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Alfonzo JD 2014. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 21:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Byström AS 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Lu J, Byström AS 2008. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14:2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL 1988. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27:3842–3849. [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33:7869–7876. [DOI] [PubMed] [Google Scholar]
- Kumagai I, Watanabe K, Oshima T 1980. Thermally induced biosynthesis of 2′-O-methylguanosine in tRNA from an extreme thermophile, Thermus thermophilus HB27. Proc Natl Acad Sci 77:1922–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP 2013. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154:416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458:228–232. [DOI] [PubMed] [Google Scholar]
- Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, et al. 2011. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol 29:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker JH, Clemons KV, Stevens DA, Davis RW 1994. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42°C and form pseudohyphae. Infect Immun 62:5447–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Hayashi H 2008. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 283:27469–27476. [DOI] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T 2009. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37:1335–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon KR, Guymon R, Crain PF, McCloskey JA, Thomm M, Lim J, Cavicchioli R 2003. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C). J Bacteriol 185:5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Alfonzo JD 2010. Do all modifications benefit all tRNAs? FEBS Lett 584:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK 2010. tRNA biology charges to the front. Genes Dev 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Hernandez A, Spears JL, Gaston KW, Limbach PA, Gamper H, Hou YM, Kaiser R, Agris PF, Perona JJ 2013. Structural and mechanistic basis for enhanced translational efficiency by 2-thiouridine at the tRNA anticodon wobble position. J Mol Biol 425:3888–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL 2008. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci 105:18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N, Sakaguchi Y, Suzuki T, Watanabe K 2006. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J Biol Chem 281:14296–14306. [DOI] [PubMed] [Google Scholar]
- Tipper DJ 1973. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J Bacteriol 116:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos AP, Jara M, Zuin A, Sanso M, Hidalgo E 2006. Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol Genet Genomics 276:495–502. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Shinma M, Oshima T, Nishimura S 1976. Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem Biophys Res Commun 72:1137–1144. [DOI] [PubMed] [Google Scholar]
- Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, Bulawa CE, Gould AE, Ocain TD, Dick LR, Errada P, et al. 2003. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell 2:256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 9:e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.