Highlights
-
•
We determine the mode of interaction between ELMO1 and Hck with direct methods.
-
•
The SH3 domain of Hck interacts specifically with the polyproline motif of ELMO1.
-
•
ELMO1 polyproline affinity for the SH3 of Hck is modulated by Tyr-phosphorylation.
Abbreviations: ELMO, EnguLfment and cell MOtility protein family; DOCK, Downstream Of CrK protein family; GEF, Guanine nucleotide Exchange Factor; ERM, Ezrin–Radixin–Moesin protein family; TAMs, Tyro3, Axl and Mer receptor tyrosine kinase family; Hck, Hematopoietic cell kinase; SH3, Src Homology 3 domain; RBD, Rho-Binding Domain; EID, ELMO Inhibitory Domain; PH, Pleckstrin Homology domain; EAD, ELMO Autoregulatory Domain; PxP, Polyproline motif; GST, Glutathione S-Transferase; GSH, Glutathione (reduced); TEV, Tobacco Etch Virus; FRET, Förster (Fluorescence) resonance energy transfer
Keywords: Hck, ELMO, SH3, Polyproline, Phosphorylation, Phagocytosis
Abstract
Eukaryotic EnguLfment and cell MOtility (ELMO) proteins form an evolutionary conserved family of regulators involved in small GTPase dependent actin remodeling processes that regulates the guanine exchange factor activity of some of the Downstream Of CrK (DOCK) family members. Gathered data strongly suggest that DOCK activation by ELMO and the subsequent signaling result from a subtle balance in the binding of partners to ELMO. Among its putative upward modulators, the Hematopoietic cell kinase (Hck), a member of the Src kinase superfamily, has been identified as a binding partner and a specific tyrosine kinase for ELMO1. Indeed, Hck is implicated in distinct molecular signaling pathways governing phagocytosis, cell adhesion, and migration of hematopoietic cells. Although ELMO1 has been shown to interact with the regulatory Src Homology 3 (SH3) domain of Hck, no direct evidence indicating the mode of interaction between Hck and ELMO1 have been provided in the literature. In the present study, we report convergent pieces of evidence that demonstrate the specific interaction between the SH3 domain of Hck and the polyproline motif of ELMO1. Our results also suggest that the tyrosine-phosphorylation state of ELMO1 tail might act as a putative modulator of Hck kinase activity towards ELMO1 that in turn participates in DOCK180 activation and further triggers subsequent signaling towards actin remodeling.
1. Introduction
The evolutionarily conserved signaling ELMO (EnguLfment and cell MOtility) and DOCK (Downstream Of CrK) proteins interact to promote the Guanine Exchange Factor (GEF) activity of DOCK in a number of actin-dependent cellular processes [1,2]. Indeed, the ELMO/DOCK pathway has been described as an essential signaling cascade that leads to the activation of several Rho GTPase-dependent events triggering the actin cytoskeleton remodeling. Hence, the ELMO/DOCK pathway regulates apoptotic cell engulfment by phagocyte, but also fundamental processes such as neurite outgrowth [3], cell migration [4], cell invasion and proliferation [5], or, for ELMO3, metastasis [6]. DOCK proteins regulate several Rac1- and Cdc42-dependent pathways. Although the ubiquitous, archetype of the family, DOCK180, has been shown to be sufficient to act as a GEF for Rac1 both in vitro and in cellulo [7,8], DOCK180 cooperates with ELMO for an efficient Rac1 signaling in vivo [4,9–12]. Thus, ELMO1 has been shown (i) to relieve the auto-inhibition of DOCK180 through direct binding, (ii) to trigger DOCK180 stabilization of the nucleotide free form of Rac1 [10,13], and (iii) to facilitate the recruitment of DOCK180 to the membrane [14,15]. Furthermore, gathered data strongly suggest that DOCK activation and the subsequent signaling may be the result of a subtle balance between binding of modulators to ELMO and the enhanced accessibility of the GEF domain of DOCK proteins [16,17]. Among ELMO upward modulators, RhoG, in its GTP-bound form, interacts with ELMO1/DOCK180 to form a ternary complex that promotes Rac1 activation [18]; Ezrin–Radixin–Moesin (ERM) family members have been shown to bind ELMO and have been suggested to contribute to the co-localization of ERM proteins and ELMO within active GEF complexes with RhoG [19,20]; and very recently, the receptor tyrosine kinase Axl of the Tyro3, Axl, Mer family (TAMs) has been discovered to participate in ELMO phosphorylation and to promote cell migration and proliferation of breast cancer cells [5].
The Hematopoietic cell kinase (Hck), a member of the Src kinase superfamily, has been identified as an ELMO1 partner [21]. Hck is a tissue specific tyrosine kinase implicated in distinct molecular signaling pathways governing several cellular processes of hematopoietic cells, ranging from phagocytosis, cell adhesion and migration, or lysosome functions [22–24]. Two Hck isoforms have been described: p61Hck expression is restricted to lysosomes, while p59Hck is membrane associated and plays a crucial role among other regulators in actin cytoskeleton reorganization [25]. The kinase activity of p59Hck has been demonstrated to be necessary for Rac1 and Cdc42 activation during Fcγ receptor mediated phagocytosis [26], a process that has been shown to rely on CrkII and DOCK180 signaling [27], thus suggesting a functional interplay between p59Hck and the ELMO/DOCK pathway. Indeed, co-expression of Hck and ELMO1 in mammalian cell models increases phagocytosis efficiency and triggers ELMO1 phosphorylation [21]. These data suggest a possible role for Hck in the control of ELMO/DOCK dependent Rac1 activation and actin remodeling, although its exact mechanism remains uncharacterized.
The primary studies of ELMO1/Hck interaction have shown a direct binding between ELMO1 and the Src Homology 3 (SH3) domain of Hck, suggesting a possible role of the C-terminal polyproline motif of ELMO1 in this interaction [21]. Interestingly, co-precipitation experiments have initially shown that the polyproline motif of ELMO1 could interact with the SH3 domain of DOCK180 [4]. However, both the SH3 domain and the polyproline motif are dispensable for the interaction [8] that is then believed to rely on an extended interaction between the C-terminal region of ELMO including the Pleckstrin Homology (PH) domain with a N-terminal region of DOCK covering the SH3 domain and its flanking α-helices [28]. Our group further demonstrated that the SH3 domain of DOCK180 interacts significantly with ELMO1, regardless of the presence of the polyproline motif, but that the polyproline/SH3 interaction may increase the life-time of the ELMO1/DOCK180 complex, supporting the hypothesis that the polyproline motif of ELMO1 and the SH3 domain of DOCK180 contribute to enhance actin cytoskeleton remodeling [29].
To date, no evidence for the direct binding between the polyproline motif of ELMO1 and the SH3 domain of Hck has been provided and the mode of interaction between Hck and ELMO1 remains to be identified. Here, we report evidence that, in contrast with the SH3 domain of DOCK180, the interaction of the SH3 domain of Hck with the C-terminal domain of ELMO1 depends on the presence of the polyproline motif.
2. Materials and methods
2.1. Cloning
The cDNA encoding the wild-type murine ELMO1 was kindly provided by Dr. Yoshinori Fukui. Full length and truncated forms of ELMO1 were generated by PCR (ELMO1–727, ELMO532–727, ELMO532–707, Fig. 1) and subcloned either using ligation-independent cloning [30] in frame with a Tobacco Etch Virus (TEV) protease cleavable 6-Histine tag for bacterial expression (pPROEXHtb) or by conventional ligation into a modified pEGFP-N1 vector for mammalian expression, in which the EGFP ORF is replaced by a TEV cleavable SNAP tag (New England Biolabs). The cDNA for bacterial expression encoding the SH3 domain of Hck (residues 77–141 of Uniprot Protein Database: P08631; SH3Hck) in fusion with a PreScission (GE-Healthcare) cleavable GST tag has been gifted by S. Feuerstein (pGEX6P2-Hck-SH3). For mammalian expression SH3Hck coding sequence was amplified by PCR (residues 77–141) and cloned in frame with the CLIP tag of the pCLIPf vector (New England Biolabs) The cDNA encoding for the human SH3 domain of DOCK180 (DOCKSH3: residues 9–69 of DOCK180 from Uniprot Protein Database: Q14185) was optimized for bacterial expression, synthesized (Geneart AG) and subcloned into pGEX 4T3 in frame with a TEV cleavable GST tag for bacterial expression.
Fig. 1.
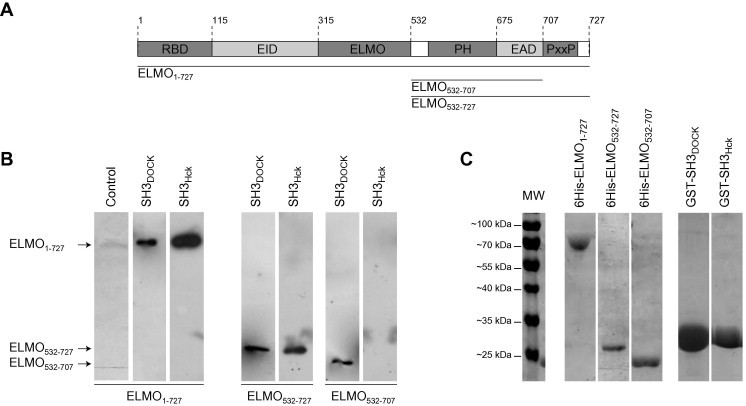
Binding of ELMO1 domains on immobilized SH3 domains of Hck and DOCK180. (A) Schematic representation of the different domains of ELMO1: The wild type ELMO1 protein (ELMO1–727), the C-terminal domain with (ELMO532–727) or without the polyproline motif (ELMO532–707) used in this study are indicated. RBD: Rho-Binding Domain; EID: ELMO Inhibitory Domain; ELMO: ELMO conserved region; PH: Pleckstrin Homology domain; EAD: ELMO Autoregulatory Domain; PxP: Polyproline motif. (B) SH3Hck binding to ELMO1 is in vitro dependent of ELMO1 polyproline motif, SH3DOCK is not: Recombinant 6His-tagged wild type ELMO1, its various deletion mutants and GST-tagged SH3 domains of Hck and DOCK180 were bacterially produced and purified from BL21 (DE3) E. coli. ELMO1 domains were incubated with immobilized either GST-fused SH3DOCK or SH3Hck and after elution, ELMO1 constructs were detected by anti-6His immuno-western blotting. (C) Coomassie-blue stained gel of the purified protein constructs overexpressed in E. Coli: Purity of ELMO and SH3 domains constructs was analyzed after FPLC purification by Coomassie-blue stained polyacrylamide gel electrophoresis in denaturing conditions (12% slab gels). Each construct migrates at its expected apparent molecular weight.
2.2. Purification of recombinant proteins from bacteria
For bacterial production and purification, the recombinant protein constructs were overexpressed in BL21(DE3) Escherichia coli strain using conventional IPTG induction (1 mM) in LB medium for 3 h at 37 °C. For 6-His-tagged constructs, bacteria were lysed using a Microfluidizer® (Microfluidics) in Phosphate-Buffered Saline (PBS), supplemented with DNAse (25 μg/ml), 1 mM MgCl2 and Complete® cocktail (Roche Diagnostics). The lysate was clarified by centrifugation and loaded on a Ni–NTA column (Qiagen). The protein was eluted with an imidazole gradient and further purified by gel filtration using a FPLC system (Purifier Akta – GE Healthcare). For GST fusion constructs, bacteria were lysed in 50 mM Hepes, 100 mM NaCl, 10 mM EDTA, 10% glycerol, 1% v/v Triton ×100, pH = 7.5 supplemented with DNAse (25 μg/ml) and Complete® cocktail (Roche Diagnostic). The lysate was clarified by centrifugation and loaded on glutathione column (Protino-Gluthatione Agarose 4B, Machery-Nagel). For pull-down experiments, GST-SH3Hck or GST-SH3DOCK domain-loaded glutathione columns were stored at 4 °C with Complete® cocktail (Roche diagnostic) and sodium azide. Overexpressed proteins were tested for their purity and integrity by gel electrophoresis and when needed by mass-spectrometry and 1D NMR analysis (quality control NMR and mass spectrometry facilities, IBS).
2.3. Pull Down experiments using bacterial recombinant constructs
The recombinant 6-His-tagged ELMO1 and GST-SH3Hck or GST-SH3DOCK constructs were expressed and purified as described above. The different ELMO1 domain constructs were incubated with 50 μl of immobilized GST-SH3Hck or GST-SH3DOCK resins (1 mg/ml) for one hour at 4 °C using the purification buffer. The resin was then washed 3 times with 5 column volumes of the same buffer. Proteins were eluted in denaturing conditions with 50 μl of electrophoresis loading buffer. The proteins were separated by SDS–PAGE and analyzed by western blot using peroxidase conjugated anti-polyHistidine antibody (Mouse monoclonal HIS1, Sigma) to reveal the presence of captured 6His-ELMO1 constructs.
2.4. Cell culture, labeling and flow cytometry analysis
HEK293T endothelial cells were maintained in DMEM supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin–streptomycin. Cells were transiently transfected using Jet Prime according to the manufacturer’s instruction. Flow cytometry experiments were performed on a MACSQuant VYB flow cytometer (Miltenyi Biotech, M4D cell imaging platform, IBS). For FRET experiments, HEK293T were transiently co-transfected with SNAP-ELMO532–727 or -ELMO532–707 constructs and CLIP-SH3Hck or with CLIP-SH3Hck alone (control). Cells were labeled with SNAP Oregon green (Ex at 490 nm, Em at 514 nm) and CLIP430 (Ex at 430 nm, Em at 484 nm) according to the manufacturer instructions (New England Biolabs™). Cells were eventually fixed with 90% methanol for 5 min at −20 °C and analyzed by flow cytometry. For each sample, a minimum of 100,000 events was collected. The donor fluorescence of CLIP430 was acquired in the V1 channel (Ex 405, Em 450/50 nm) and the acceptor (SNAP Oregon Green) in the B1 channel (Em 488, Exc 525/50 nm). The FRET intensity was measured as the median and standard deviation values of the intensity of the V2 channel (Ex 405 nm, Em 525/50 nm) after gating of the population for transfected cell singlets (FSC-H/A exclusion, FSC-A/SSC-A homogenous cell population and V1 positive cells) using MACSQuantify software. The final gated population represents a minimum of 10,000 cells. The percentile of FRET positive cells is calculated by the ratio of cells with a V2 FRET intensity above the threshold defined as the median value of the ELMO532–727 transfected assay.
2.5. Pull Down of CLIP-SH3Hck by ELMO1 from cellular extracts
HEK293T cells were transiently transfected with SNAP-ELMO1 domains and CLIP-SH3Hck or with only CLIP-SH3Hck for the control well. After 48 h, cells were lysed for 30 min at 4 °C then centrifuged at 14,000 rpm. The cell lysate was diluted in the immobilization buffer (50 mM Tris–HCl, 100 mM NaCl, 1 mM DTT, 0.1% Tween, pH = 7.5), tested for protein expression by anti-SNAP western-blot analysis (Supplementary Fig. S1), then incubated with SNAP capture magnetic beads (New England Biolabs™) for one hour at room temperature. The SNAP-ELMO1–727 constructs retained by beads due to a covalent binding were eluted after protease (TEV) cleavage of the SNAP tag overnight at 4 °C. The beads were recovered by contact with a magnetic rack, and the solubilized fraction is analyzed by western-blot. The presence of CLIP-SH3Hck retained in case of interaction with ELMO1 domains was detected using an anti SNAP antibody (New England Biolabs).
2.6. NMR spectroscopy
The chemically-synthesized peptides corresponding to the 28 C-terminal amino-acids of ELMO1 (ELMO700–727, DLENIQIPDAPPPIPKEPSNYDFVYDCN) and its phosphorylated counterpart on tyrosine 720 (ELMO(p)700–727) were commercially obtained (Proteogenix SAS, France).
For NMR analysis, recombinant 15N labeled GST-SH3Hck was produced in E. coli harvested in 15N-labeled M9 minimal medium containing 1 g/L 15NH4Cl and 2 g/L glucose and purified as described above. Elution of the SH3 domain of Hck is achieved after specific cleavage of the GST tag: the glutathione resin with immobilized GST-SH3Hck was incubated with PreScission protease (GE Healthcare) for 4 h at 4 °C using the standard protocol provided by the manufacturer. The uniformly labeled 15N SH3Hck was then eluted and prepared at a concentration of 1 mg/ml in 50 mM Hepes, 100 mM NaCl, 10 mM EDTA, pH = 7.5. Interactions between ELMO1 peptides (ELMO700–727 or phosphorylated, ELMO(p)700–727) and the SH3 domain were investigated by the addition of aliquots of a concentrated solution of unlabeled chemically synthesized peptide directly to isotope labeled SH3Hck to a final ratio 1:55.
NMR measurements were performed on Agilent VNMRS 600 MHz or 800 MHz (NMR assignment, SH3 titration experiments) systems equipped with cryogenically cooled triple-resonance probes and pulsed z-field gradients. For all 2D-H–N correlation experiments, BEST-TROSY optimized pulse sequences were used. All NMR data were processed using NMRPipe [31] and evaluated using NmrViewJ (One Moon Scientific Inc.).
For the titration experiments, increasing molar ratio of the ELMO700–727 and ELMO(p)700–727 peptides (up to 55) were obtained adding aliquots of a peptide solution at either 3 mM (ligand/protein ratio of 1–5.5) or 30 mM (ligand/protein ratio of 5.5–55) to a 100 μM solution of SH3Hck. Prior to addition, the pH of both peptide solutions was adjusted to 7.6 using a 1 M NaOH solution. Concentration of ELMO1 peptides was determined by UV–visible spectrometry either at 280 nm or 286 nm using a calculated absorption coefficient at 280 nm of 2980 M−1 L−1 for ELMO700–727 and a calculated absorption coefficient at 286 nm of 910 M−1 L−1 for ELMO(p)700–727 [32]. Integrity of the ELMO(p)700–727 peptide was assessed by mass spectrometry (m/z: 3282.4). 1H and 15N chemical shift changes were monitored in 2D [1H,15N]-HSQC spectra at 298 K along the titration. Chemical shift perturbations were calculated as:
Binding constants have been determined from the chemical shift perturbations of the S95, D110, W112, W113 and S129 residues which present the greater variations for both peptides, using in-house modified non-linear least squares fitting procedures with Monte-Carlo error analysis from NMRPipe [31] and formula that take into account the dilution of the protein sample along the titration [33].
3. Results
Primary studies have suggested that the binding of ELMO1 to Hck might occur through the interaction between the only polyproline motif found in the C-terminal tail of ELMO1 and the SH3 domain of Hck [21]. However, the only data allowing this assessment are based on competitive experiments between ELMO1 and a known specific peptide for Src kinases. Moreover, previous work from our group and others demonstrated that ELMO1 interacts in vitro with the SH3 domain of its first identified binder, DOCK180, regardless of the presence of the polyproline-containing C-terminal end. This particularity of the polyproline motif of ELMO1 and the lack of direct evidence of its binding to Hck, led us to investigate by direct methods the interaction of ELMO1 with the isolated SH3 domain of Hck and to compare it to its interaction with the SH3 domain of DOCK.
3.1. Pull-down with bacterially-expressed recombinant constructs
First, we performed pull-down experiments on immobilized GST fused SH3 domains of Hck (SH3Hck) or DOCK180 (SH3DOCK) using different poly-Histidine tagged ELMO1 constructs including or not the polyproline C-terminal extension (Wild type ELMO1–727 and deletion mutants ELMO532–727 and ELMO532–707, Fig. 1A). The overexpressed proteins were controlled for their integrity after FPLC purification (Cf. Section 2 and Fig. 1C). As expected, ELMO1–727 is successfully pulled-down by both SH3 domains, but not in control experiments (mock GSH resin, Fig. 1B). As expected from our previous work, ELMO532–727 and its counterpart lacking the polyproline extension ELMO532–707, are efficiently pulled-down by SH3DOCK. By contrast, ELMO532–727 is specifically pulled-down by SH3Hck but ELMO532–707 is not. These data clearly show that in our experimental conditions, the interaction between the C-terminal domain of ELMO1 and the SH3 of Hck is strictly dependent on the presence of the polyproline motif of ELMO1; a behavior that clearly contrasts with the interaction of the SH3 domain of DOCK180 with ELMO1 lacking the polyproline C-terminal motif.
3.2. In cellulo pull-down and Förster resonance energy transfer (FRET) analysis of ELMO1 and SH3Hck interactions
We next examined if this interaction could occur in the cell context after co-transfection of HEK293T cells. To efficiently pull-down the SH3 domain of Hck, we expressed SNAP-tag and CLIP-tag [34,35] N-terminal fusion constructs of ELMO1 and SH3Hck, respectively. Lysates of HEK293T cells transiently transfected for the co-expression of the SH3 domain of Hck in the presence of wild type ELMO1–727 or the N-terminal deletion mutants ELMO532–727 and ELMO532–707, were checked for correct protein expression by western-blot (Supplementary Fig. S1) and subjected to pull-down experiments using ELMO1 as a bait, taking advantage of the specific and covalent binding of SNAP fusion constructs to the SNAP-capture magnetic beads (New England Biolabs, cf. Section 2). The successful pull-down of SH3Hck was assessed by antibody labeling of the N-terminal CLIP tag of the construct. As shown in Fig. 2 (panel A), CLIP-SH3Hck is revealed when it is co-expressed with ELMO1–727 or its C-terminal domain, ELMO532–727. As expected, when CLIP-SH3Hck is co-expressed with the ELMO1 C-terminal domain lacking its polyproline motif (ELMO532–707), only a residual signal similar to the one observed in the negative control (cells transfected with CLIP-SH3Hck alone) is apparent, despite the correct expression of CLIP-SH3Hck and ELMO532–707 deletion mutant (Supplementary Fig. S1). However, a comparable result was observed when mono-transfected cell lysates were pooled before pull-down (data not shown), raising the possibility that the interaction could also arise after cell lysis and not only in the cell context.
Fig. 2.
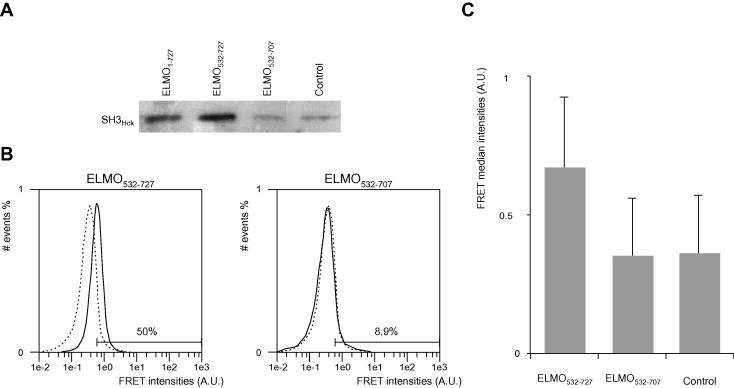
Binding of ELMO1 domains to SH3 domain of Hck in the cellular context. (A) SH3Hck binding to ELMO1 is dependent of ELMO1 polyproline motif in co-transfected HEK293T cell extract: CLIP-SH3Hck was expressed either alone or with SNAP-ELMOWT and its various deletion mutants (ELMO532–727 or ELMO532–707) in HEK293T cells. Cell lysates were then incubated with SNAP-capture magnetic beads. Proteins were eluted after TEV digestion, separated by SDS–PAGE and the presence of retained CLIP-SH3Hck by western-blot using a polyclonal antibody anti-SNAP-tag (which is also specific for CLIP tag). The control well (cells mono-transfected with CLIP-SH3Hck) shows the residual non-specific signal. (B) Flow cytometry distribution of FRET intensities of HEK293T cells co-transfected with SH3Hck and ELMO1 deletion mutants: Superposition of the histograms of the FRET intensities (acceptor fluorescence) from the control cells (transfected with CLIP-SH3Hck, dash line) and cells transfected with CLIP-SH3Hck and either with SNAP-ELMO532–727 or SNAP-ELMO532–707 (plain lines). The FRET signal was measured in the V2 channel Ex 405 nm, Em 525 ± 25 nm. A shift of the maximum of the double positive population is observed for the measured FRET channel for SNAP-ELMO532–727 relative to the negative control, while no difference is noticeable for SNAP-ELMO532–707. The shift is also expressed as the ratio of cells above the threshold in comparison with the total number of cells in the gated region (donor positive cells, see Section 2). The data shown originate from a single experiment representative of three independent experiments. (C) Bar graph of FRET median values: FRET intensities median values of the histograms presented in B. The standard deviation errors of the donor positive population are shown.
To ascertain that this interaction occurs within the cell, we performed FRET experiments using the same constructs transiently expressed in HEK293T epithelial cells. SNAP and CLIP-tag can be efficiently coupled to fluorophores, and have been successfully used to measure FRET signals in living cells [36]. We used flow cytometry to analyze the FRET signals of co-transfected or control HEK293T cell populations. Fig. 2B and C represent a representative set of the FRET data obtained from independent experiments. In brief, we calculated the median value of the FRET intensities of the transfected cells (cf. Section 2 for details). The overlaid histograms of the FRET intensity of control (dash lines) and co-transfected (plain lines) cells are shown in Fig. 2B. The cells co-transfected with SNAP-ELMO532–727 and CLIP-SH3Hck show a significantly higher median FRET intensity compared to control cells, while any significant FRET variation for the cells co-transfected with SNAP-ELMO532–707 was detected (50% and 8.5% of events above the threshold respectively, Fig. 2B). Taken together, these results strongly suggest that the interaction between the C-terminal domain of ELMO1 and the SH3 domain of Hck in the physiological context, requires the presence of the polyproline tail of ELMO1, further confirming our data obtained on purified overexpressed proteins.
3.3. NMR structural characterization
To characterize the specific interaction of the SH3 domain of Hck with the C-terminal polyproline motif of ELMO1, we next investigated their interaction via NMR spectroscopy. In brief, we studied the chemical shift mapping variations of this SH3 domain, which have been 15N labeled, during its titration with increasing amounts of a synthetic peptide encompassing the polyproline motif (ELMO700–727, Fig. 1A). As shown by both 2D 15NH-BEST-TROSY spectra of the 15N-SH3Hck in absence (black) or in presence (red) of ELMO700–727, the titration of SH3Hck resulted in a continuous change of the chemical shifts of several residues revealing fast exchange kinetics on the NMR time scale, which is indicative for a comparatively low binding affinity (Fig. 3A). As illustrated, the majority of induced chemical shifts changes are less than 0.05 ppm, which demonstrates that the overall SH3Hck structure is unchanged. Nonetheless, several residues present more significant variations (>0.05 ppm) of their resonance peaks upon addition of ELMO700–727 peptide (Fig. 3A, B). As expected, the amino-acids of SH3Hck affected by the C-terminal ELMO1 peptide are mainly located in the canonical binding pocket of SH3 domains for polyproline motifs [37], which is formed by the RT- and N-Src-loop as well as the β-strands c and d. Among them, the following residues showed particularly large chemical shift variations: the canonical aspartic acid D95, serine S110, glutamic acid E112, tryptophan W113 and serine S129 (Fig. 3B, C).
Fig. 3.
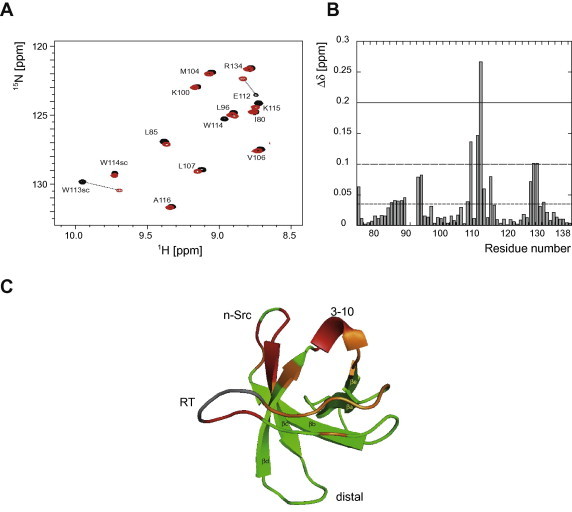
NMR structural characterization of the interaction between of the SH3 domain of Hck and the C-terminal end of ELMO1. (A and B) Chemical shift mapping of the SH3 domain of Hck in presence of ELMO1 polyproline-containing peptide: 2D 15N-HSQC spectra of 100 μM of 15N-labeled SH3Hck alone (in black) and with increased concentrations of ELMO707–727 peptide (in red: 55 equivalent concentration) were acquired using an 800 MHZ NMR spectrometer at 25 °C. Variation in the chemical shifts for different residues are clearly observable. The measured variations (in Δδppm) is also represented as a histogram. (C) SH3 domain of Hck may bind to the polyproline motif of ELMO1 in a canonical manner: the backbone representation of SH3Hck 3D structure (PDB: 4HCK, [37]) is colored according to the variation in chemical shift of each amino-acid (green: Δδppm < 0.05 ppm; orange: 0.05 ppm < Δδppm < 0.1 ppm; red: 0.1 ppm < Δδppm, gray: not observed). The residues of SH3Hck implicated in the interaction are mainly located in the canonical binding pocket of SH3 domains for polyproline motifs, which is formed by the RT- and N-Src-loop as well as the β-strands c and d. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Additionally, the NMR titration experiment allows determining the dissociation constant for the SH3Hck/ELMO700–727 complex formation. The chemical shift changes of the most representative residues, as defined by the extent of their chemical shift variations (D95, S110, E112, W113 and S129), were plotted as a function of the ELMO700–727 concentration (Fig. 4). A global fit of the titration curves yields a dissociation constant Kd = 1.6 ± 0.3 mM (see Supplemental Table S1 for details).
Fig. 4.
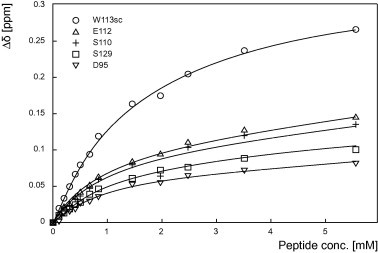
The interaction between the SH3 domain of Hck and the polyproline motif of ELMO1 is of moderate affinity. Titration of the 15N-labeled SH3 domain of Hck with increasing amounts of the ELMO707–727 peptide (up to 55 equivalents). The titration curves result from the fitting of the chemical shift variations of the aspartate 95, serine 110 and 129, tryptophan 113 and the glutamate 112 as a function of the ELMO707–727 peptide concentration. These variations were measured for the five different residues and the dissociation constant was calculated as described in the Section 2.
This structural characterization further confirms that the SH3 domain of Hck binds ELMO1 through the polyproline C-terminal motif, and strongly suggests that the binding may mimic the canonical binding mode of SH3 domains to polyproline motifs.
4. Discussion
Despite the data accumulated over the past decade, the understanding of the ELMO/DOCK pathway regulation in mammalian cells remains highly entangled. A number of identified factors, acting as upstream modulators of the ELMO/DOCK GEF activity have been characterized in a variety of model systems. ELMO1 has been identified as a primary binding partner and a target of the tyrosine kinase Hck in cells of the hematopoietic lineage. In macrophages, Hck has been suggested to be implicated in the regulation of the signaling events that are governed by ELMO/DOCK and lead to actin cytoskeleton regulation [21,38]. It is well established that phagocytosis depends on tyrosine kinase activity. Indeed, the functions of several receptors implicated in phagocytosis or migration, such as TAMs [39] or CD36 [40] have been demonstrated to rely on phosphorylation of tyrosine residues [5,41]. The recruitment of the p130cas–CrkII–DOCK180 complex during apoptotic cell clearance is triggered by a yet unidentified tyrosine kinase in the αvβ5 integrin pathway [42]. Furthermore, phagocytosis mediated by Fcγ receptor is largely regulated by kinases of the Src family [43] and it has also been shown that specific inhibition of the Src family kinase by PP2 decreases phagocytosis efficiency in macrophages [44].
In the present work, we have provided several lines of evidence showing that ELMO1 interacts with the SH3 domain of Hck through its C-terminal polyproline motif. Using purified protein domains we have demonstrated that, in contrast with the binding of the SH3 domain of DOCK180, Hck interaction is strictly dependent on the C-terminal polyproline motif of ELMO1. The characterization of this interaction using a synthetic polyproline peptide by resonance displacements probed by NMR, gave a dissociation constant of 1.6 ± 0.3 mM. The specific residues of the SH3 domain of Hck affected by the binding are highly correlated to the residues implicated in its well characterized binding with the PD1 model polyproline peptide [37], further reinforcing the fact that SH3Hck binding to ELMO1 polyproline is archetypical in nature. Still, the molecular interaction between the C-terminal peptide of ELMO1 and the SH3 domain of Hck appears to be of modest affinity compared to polyproline/SH3 domain bindings commonly-found for Hck. However, preliminary solid-phase enzyme immunoassay data obtained in the presence of ELMO532–727 show that the dissociation rate of SH3Hck is in the ten-micromolar range (data not shown). This observation suggests that in the context of the integer protein, the ELMO1/Hck interaction may be of higher affinity and may involve other regions of ELMO1 in the complex formation. The set of data that we are providing thus implies that the ELMO1 polyproline/SH3Hck interaction likely contributes to the kinase specificity towards ELMO1 and to the recruitment of Hck by ELMO1 or vice versa during DOCK protein regulation and Rac1 activation.
Nevertheless, the physiological consequences of the interaction of the SH3 domain of Hck with ELMO1 polyproline motif remain today an open question. SH3 domains in kinases of the Src family are known to participate in the kinase activity regulation and specificity [45,46]. More than triggering DOCK GEF activity through ELMO1 as suggested before, one possible outcome of Hck/ELMO1 complex formation would be also the activation or activity regulation of Hck upon ELMO1 binding. Indeed, ELMO1 is phosphorylated by Hck on five identified tyrosines (residues 18, 216, 395, 511 and 720) [38] and using non-phosphorylatable mutants, Yokoyama and collaborators reported that phosphorylation is not required for ELMO1/DOCK180 complex formation, although it contributes synergistically to Rac1 activation and increases phagocytosis efficiency and migration of LR73 fibroblasts. The authors showed that tyrosine 511 to phenylalanine mutation drastically reduces phagocytosis efficiency, whereas tyrosine 720 to phenylalanine mutation does not. In contrast, Abu-Thuraia and collaborators recently demonstrated that ELMO2 tyrosine 713 phosphorylation (equivalent to tyrosine 720 in ELMO1) by the receptor tyrosine kinase Axl promotes cell invasion in MDA-MB-231 breast cancer cells [5]. Intriguingly, tyrosine 720 stands within the polyproline motif of ELMO1. We thus addressed the question of the influence of the phosphorylation of tyrosine 720 on ELMO1 polyproline binding to the SH3 domain of Hck. Using the phosphorylated counterpart of the polyproline motif containing peptide ELMO700–727, we measured the chemical shifts variations during the titration of the 15N labeled recombinant SH3 domain of Hck. When comparing the side chains of SH3 affected by the binding of the phosphorylated ELMO(p)700–727 or of the non-phosphorylated form, we observed that the same residues were affected, indicating that the binding site is not specifically altered by tyrosine 720 phosphorylation (data not shown). Hence, the chemical shifts experienced lower variations for the phosphorylated peptide at similar peptide:HckSH3 ratios. A global fit of the variations of the same representative residues as a function of the increased ratio ELMO(p)700–727:HckSH3 yields a higher dissociation constant Kd = 5 ± 1.5 mM (Fig. 5 and Table S1), significantly different from the Kd for the non-phosphorylated peptide (Kd = 1.6 ± 0.3 mM), thus indicating that phosphorylation of tyrosine 720 may weaken the ELMO1/Hck binding. Previously published work by Yokoyama and collaborators shows that tyrosine 720 mutation to phenylalanine results in an apparent overall increased phosphorylation of the remaining target tyrosines of ELMO1 expressed in Cos7 cells [38]. It is well established that the SH3 domain interaction of the kinases of the Src family is implicated in the kinase activity regulation [47]. It is then tempting to speculate that the observed decreased affinity of phosphorylated ELMO1 tail for Hck SH3 domain might in turn participate in the down-regulation of the kinase. Taken together, the present results suggest that the polyproline of ELMO1 might act as a putative modulator of Hck activity towards ELMO1 that in turn participates, by a yet unknown mechanism, to DOCK180 activation and further triggers Rac1 GTP loading. Interestingly, tyrosine 720 of ELMO1 has been recently demonstrated to be phosphorylated by the Mer related receptor tyrosine kinase Axl [5]. This new study and the illustrated role of members of the TAMs family in apoptotic corpses clearance [39,48] suggest that ELMO1 might also be a physiological target of Mer during phagocytosis. These recent results and our present data emphasize the role of ELMO C-terminal tail, and in particular its tyrosine and its phosphorylation, in the modulation of the downstream signaling cascade leading to actin remodeling via DOCK proteins, a role that remains to be further clarified.
Fig. 5.
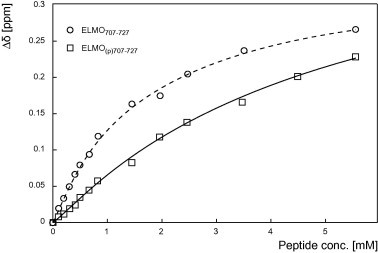
Comparison of the interaction between the SH3 domain of Hck and the phosphorylated or the non-phosphorylated polyproline motif of ELMO1. Titration of the 15N-labeled SH3 domain of Hck with increasing amounts of the phosphorylated ELMO(p)707–727 peptide (open squares) or of the non-phosphorylated ELMO707–727 peptide (open circles). The titration curves of the chemical shift variations of the representative tryptophan 113 side chain are compared as a function of the peptides concentrations (up to 55 equivalents). These variations were measured for at least five different residues and the dissociation constant was calculated as described in the Section 2.
Author contributions
RA performed experiments, analyzed data, wrote the manuscript; IA and AC performed experiments; MS contributed reagents or other essential material; PG performed experiments, analyzed data; PF performed critical reading of the manuscript; JBR analyzed data, wrote the manuscript; JPK planned experiment, wrote the manuscript.
Acknowledgements
This work used the platforms of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEAUJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
We thank Françoise Lacroix and Rose-Laure Revel-Goyet for access to the cell imaging/flow cytometry facility (M4D). We are grateful to Nicole Thielens and Dominique Housset of the “Immune Response to Pathogens and Altered Self” group for supporting this work. We would like to thank the “Viral Infection and Cancer” group for hosting part of RA and JPK experimental work; Dr. Dominique Housset for the critical reading of the manuscript; Joanna Timmins for precious help in editing the manuscript for English; and Dominique Bicout and Timothy Ziman for their help in statistical analysis.
Appendix A. Supplementary data
Western-blot analysis of the cellular extracts of HEK293T cells transfected with ELMO1 deletion mutants or SH3Hck. Lysates of HEK293T cells transfected (control well, left) or cotransfected (ELMO wells) with SNAP-ELMO and CLIP-SH3 constructs, were analyzed by anti-SNAP antibody immunolabeling after polyacrylamide gel electrophoresis and transfer. SNAP antibodies (1/500) reveal both CLIP and SNAP tagged constructs expressed in the distinct HEK293T cell lysates tested (arrows). The cell lysates show that the expressed proteins and domains are present in the extracts at comparable expression levels.
Non-linear least square fitting with Monte Carlo error of the chemical shifts mapping variations. Calculated constants and errors from the chemical shifts mapping variations of the 5 most representative residues during titration of the SH3 domain of Hck with increasing peptide concentrations of the polyproline motif of ELMO under its phosphorylated (ELMO(p)707–727) or non-phosphorylated form (ELMO707–727).
References
- 1.Lu M., Ravichandran K.S. Dock180-ELMO cooperation in Rac activation. Methods Enzymol. 2006;406:388–402. doi: 10.1016/S0076-6879(06)06028-9. [DOI] [PubMed] [Google Scholar]
- 2.Gadea G., Blangy A. Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 2014;93(10–12):466–477. doi: 10.1016/j.ejcb.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Namekata K., Kimura A., Kawamura K., Harada C., Harada T. Dock GEFs and their therapeutic potential: Neuroprotection and axon regeneration. Prog. Retin. Eye Res. 2014;43:1–16. doi: 10.1016/j.preteyeres.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Gumienny T.L., Brugnera E., Tosello-Trampont A.C., Kinchen J.M., Haney L.B., Nishiwaki K., Walk S.F., Nemergut M.E., Macara I.G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M.O., Ravichandran K.S. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 5.Abu-Thuraia A., Gauthier R., Chidiac R., Fukui Y., Screaton R.A., Gratton J.-P., Côté J.-F. Axl phosphorylates Elmo scaffold proteins to promote Rac activation and cell invasion. Mol. Cell Biol. 2015;35(1):76–87. doi: 10.1128/MCB.00764-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen L.S., Søes S., Hansen L.L. ELMO3: a direct driver of cancer metastasis? Cell Cycle. 2014;13(16):2483–2484. doi: 10.4161/15384101.2014.947228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Côté J.-F., Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 8.Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., Côté J.-F. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell. 2008;19:4837–4851. doi: 10.1091/mbc.E08-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brugnera E., Haney L., Grimsley C., Lu M., Walk S.F., Tosello-Trampont A.-C., Macara I.G., Madhani H., Fink G.R., Ravichandran K.S. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 10.Lu M., Kinchen J., Rossman K., Grimsley C.M., deBakker C., Brugnera E., Tosello-Trampont A., Haney L., Klingele D., Sondek J., Hengartner M., Ravichandran K. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struc. Mol. Biol. 2004;11(8):756–762. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y., Tsai M., Cheng L., Chou C., Weng N. C. elegans CED-12 acts in the conserved CrkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z., Hartwieg E., Horvitz H.R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C-elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 13.Lu M., Kinchen J., Rossman K., Grimsley C.M., Hall M., Sondek J., Hengartner M., Yajnik V., Ravichandran K. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005;15:371–377. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 14.deBakker C., Haney L., Kinchen J., Grimsley C.M., Lu M., Klingele D., Hsu P., Chou B., Cheng L., Blangy A., Sondek J., Hengartner M., Wu Y., Ravichandran K. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr. Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 15.Grimsley C.M., Kinchen J., Tosello-Trampont A., Brugnera E., Haney L., Lu M., Chen Q., Klingele D., Hengartner M., Ravichandran K. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- 16.Côté J.-F., Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel M., Pelletier A., Côté J.-F. Opening up on ELMO regulation: new insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases. 2011;2:268–275. doi: 10.4161/sgtp.2.5.17716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh H., Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–464. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- 19.D’Angelo R., Aresta S., Blangy A., Del Maestro L., Louvard D., Arpin M. Interaction of ezrin with the novel guanine nucleotide exchange factor PLEKHG6 promotes RhoG-dependent apical cytoskeleton rearrangements in epithelial cells. Mol. Biol. Cell. 2007;18:4780–4793. doi: 10.1091/mbc.E06-12-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimsley C.M., Lu M., Haney L.B., Kinchen J.M., Ravichandran K.S. Characterization of a novel interaction between ELMO1 and ERM proteins. J. Biol. Chem. 2006;281:5928–5937. doi: 10.1074/jbc.M510647200. [DOI] [PubMed] [Google Scholar]
- 21.Scott M.P., Zappacosta F., Kim E.Y., Annan R.S., Miller W.T. Identification of novel SH3 domain ligands for the Src family kinase Hck. Wiskott–Aldrich syndrome protein (WASP), WASP-interacting protein (WIP), and ELMO1. J. Biochem. 2002;277:28238–28246. doi: 10.1074/jbc.M202783200. [DOI] [PubMed] [Google Scholar]
- 22.Fitzer-Attas C.J., Lowry M., Crowley M.T. Fcγ Receptor-Mediated Phagocytosis in Macrophages Lacking the Src Family Tyrosine Kinases Hck, Fgr, and Lyn. J. Exp. Med. 2000;191(4):669–682. doi: 10.1084/jem.191.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler S.F., Marth J.D., Lewis D.B., Perlmutter R.M. Novel protein-tyrosine kinase gene (Hck) preferentially expressed in cells of hematopoietic origin. Mol. Cell. Biol. 1987;7:2276–2285. doi: 10.1128/mcb.7.6.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suen P.W., Ilic D., Caveggion E., Berton G., Damsky C.H., Lowell C.A. Impaired integrin-mediated signal transduction, altered cytoskeletal structure and reduced motility in Hck/Fgr deficient macrophages. J. Cell Sci. 1999;112(Pt 22):4067–4078. doi: 10.1242/jcs.112.22.4067. [DOI] [PubMed] [Google Scholar]
- 25.Carreno S., Gouze M.E., Schaak S., Emorine L.J., Maridonneau-Parini I. Lack of palmitoylation redirects p59(Hck) from the plasma membrane to p61(Hck)-positive lysosomes. J. Biochem. 2000;275:36223–36229. doi: 10.1074/jbc.M003901200. [DOI] [PubMed] [Google Scholar]
- 26.Carreno S., Caron E., Cougoule C., Emorine L., Maridonneau-Parini I. P59Hck isoform induces F-actin reorganization to form protrusions of the plasma membrane in a Cdc42- and Rac-dependent manner. J. Biochem. 2002;277:21007–21016. doi: 10.1074/jbc.M201212200. [DOI] [PubMed] [Google Scholar]
- 27.Lee W., Cosio G., Ireton K., Grinstein S. Role of CrkII in FCgamma receptor-mediated phagocytosis. J. Biol. Chem. 2007;282(15):11135–11143. doi: 10.1074/jbc.M700823200. [DOI] [PubMed] [Google Scholar]
- 28.Hanawa-Suetsugu K., Kukimoto-Niino M., Mishima-Tsumagari C., Akasaka R., Ohsawa N., Sekine S.-I., Ito T., Tochio N., Koshiba S., Kigawa T., Terada T., Shirouzu M., Nishikimi A., Uruno T., Katakai T., Kinashi T., Kohda D., Fukui Y., Yokoyama S. Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. Proc. Natl. Acad. Sci. U.S.A. 2012;109(9):3305–3310. doi: 10.1073/pnas.1113512109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sévajol M., Reiser J.B., Chouquet A., Perard J., Ayala I., Gans P., Kleman J.-P., Housset D. The C-terminal polyproline-containing region of ELMO contributes to an increase in the life-time of the ELMO-DOCK complex. Biochimie. 2012;94:823–828. doi: 10.1016/j.biochi.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Aslanidis C., de Jong P. Ligation-independent cloning of PCR products (LIC-PCR) Nucl. Aci. Res. 1990;18(20):6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Apostol I., Kuciel R., Wasylewska E., Ostrowski W.S. Phosphotyrosine as a substrate of acid and alkaline phosphatases. Acta Biochim. Pol. 1985;32:187–197. [PubMed] [Google Scholar]
- 33.Lecoq L., Bougault C., Hugonnet J.-E., Veckerlé C., Pessey O., Arthur M., Simorre J.-P. Dynamics induced by β-lactam antibiotics in the active site of Bacillus subtilis l, d-transpeptidase. Structure. 2012;20:850–861. doi: 10.1016/j.str.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Gautier A., Juillerat A., Heinis C., Corrêa I.R., Kindermann M., Beaufils F., Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 36.Maurel D., Comps-Agrar L., Brock C., Rives M.-L., Bourrier E., Ayoub M.A., Bazin H., Tinel N., Durroux T., Prézeau L., Trinquet E., Pin J.-P. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat. Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt H., Hoffmann S., Tran T., Stoldt M., Stangler T., Wiesehan K., Willbold D. Solution structure of a Hck SH3 domain ligand complex reveals novel interaction modes. J. Mol. Biol. 2007;365:1517–1532. doi: 10.1016/j.jmb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama N., deBakker C.D., Zappacosta F., Huddleston M.J., Annan R.S., Ravichandran K.S., Miller W.T. Identification of tyrosine residues on ELMO1 that are phosphorylated by the Src-family kinase Hck. Biochemistry. 2005;44:8841–8849. doi: 10.1021/bi0500832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seitz H.M., Camenisch T.D., Lemke G., Earp H.S., Matsushima G.K. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J. Immunol. 2007;178:5635–5642. doi: 10.4049/jimmunol.178.9.5635. [DOI] [PubMed] [Google Scholar]
- 40.Moore K.J., Khoury El J., Medeiros L.A., Terada K., Geula C., Luster A.D., Freeman M.W. A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. J. Biol. Chem. 2002;277:47373–47379. doi: 10.1074/jbc.M208788200. [DOI] [PubMed] [Google Scholar]
- 41.Scott R., McMahon E., Pop S., Reap E., Caricchio R., Cohen P., Earp H., Matsushima G. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411:207–211. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- 42.Albert M., Kim J., Birge R. Alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat. Cell Biol. 2000;2:899–905. doi: 10.1038/35046549. [DOI] [PubMed] [Google Scholar]
- 43.Strzelecka A., Kwiatkowska K., Sobota A. Tyrosine phosphorylation and Fcγ receptor-mediated phagocytosis. FEBS Lett. 1997;400(1):11–14. doi: 10.1016/s0014-5793(96)01359-2. [DOI] [PubMed] [Google Scholar]
- 44.Hu B., Punturieri A., Todt J., Sonstein J., Polak T., Curtis J.L. Recognition and phagocytosis of apoptotic T cells by resident murine tissue macrophages require multiple signal transduction events. J. Leukoc. Biol. 2002;71:881–889. [PMC free article] [PubMed] [Google Scholar]
- 45.Kato J.Y., Takeya T., Grandori C., Iba H., Levy J.B., Hanafusa H. Amino acid substitutions sufficient to convert the nontransforming p60c-src protein to a transforming protein. Mol. Cell. Biol. 1986;6:4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Superti-Furga G., Courtneidge S.A. Structure-function relationships in Src family and related protein tyrosine kinases. BioEssays. 1995;17:321–330. doi: 10.1002/bies.950170408. [DOI] [PubMed] [Google Scholar]
- 47.Yadav S.S., Miller W.T. Cooperative activation of Src family kinases by SH3 and SH2 ligands. Cancer Lett. 2007;257:116–123. doi: 10.1016/j.canlet.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y., Singh S., Georgescu M., Birge R. A role for Mer tyrosine kinase in alphavbeta5 integrin-mediated phagocytosis of apoptotic cells. J. Cell Sci. 2005;118:539–553. doi: 10.1242/jcs.01632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western-blot analysis of the cellular extracts of HEK293T cells transfected with ELMO1 deletion mutants or SH3Hck. Lysates of HEK293T cells transfected (control well, left) or cotransfected (ELMO wells) with SNAP-ELMO and CLIP-SH3 constructs, were analyzed by anti-SNAP antibody immunolabeling after polyacrylamide gel electrophoresis and transfer. SNAP antibodies (1/500) reveal both CLIP and SNAP tagged constructs expressed in the distinct HEK293T cell lysates tested (arrows). The cell lysates show that the expressed proteins and domains are present in the extracts at comparable expression levels.
Non-linear least square fitting with Monte Carlo error of the chemical shifts mapping variations. Calculated constants and errors from the chemical shifts mapping variations of the 5 most representative residues during titration of the SH3 domain of Hck with increasing peptide concentrations of the polyproline motif of ELMO under its phosphorylated (ELMO(p)707–727) or non-phosphorylated form (ELMO707–727).


