Abstract
RNA chaperones are ubiquitous, heterogeneous proteins essential for RNA structural biogenesis and function. We investigated the mechanism of chaperone-mediated RNA folding by following the time-resolved dimerization of the packaging domain of a retroviral RNA at nucleotide resolution. In the absence of the nucleocapsid (NC) chaperone, dimerization proceeded via multiple, slow-folding intermediates. In the presence of NC, dimerization occurred rapidly via a single structural intermediate. The RNA binding domain of hnRNP A1 protein (UP1), a structurally unrelated chaperone, also accelerated dimerization. Both chaperones interacted primarily with guanosine residues. Replacing guanosine with more weakly pairing inosine yielded an RNA that folded rapidly without a facilitating chaperone. These results show RNA chaperones can simplify RNA folding landscapes by weakening intramolecular interactions involving guanosine and explain many RNA chaperone activities.
Outside the cellular environment or in the absence of chaperone proteins, most RNAs fold via complex pathways involving multiple, long-lived intermediates. RNA chaperone proteins with non- or semi-specific RNA binding activities accelerate adoption of the thermodynamically most stable RNA structure by lowering the energetic barriers between RNA states and by facilitating rearrangement of misfolded states (1–4). Retroviruses package two RNA genomes in each virus particle (5). These genomes dimerize near their 5′ ends, and dimerization is catalyzed by an RNA chaperone, the nucleocapsid (NC), which is derived from the retroviral Gag protein that co-assembles with the viral RNA to generate replication competent virus (2, 6, 7). By following the dimerization of a region of the Moloney murine leukemia virus (MuLV) genomic RNA at single-nucleotide resolution, we uncovered a simple mechanism for how a retroviral nucleocapsid chaperone protein functions.
We studied an RNA construct spanning the 170-nt MuLV dimerization region (8–10) and including 5′ and 3′ flanking sequences of 46 and 115 nucleotides, respectively. This RNA dimerizes under physiological-like conditions in vitro and has a structure similar to that of genomic RNA isolated from virions (11, 12). Point mutations in this region of the MuLV genome eliminate its selective packaging into virions (10). We followed dimerization at single-nucleotide resolution using time-resolved, selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (13, 14). A fast-acting reagent, benzoyl cyanide (BzCN), that either reacts to form a 2′-O-adduct at conformationally flexible nucleotides or undergoes rapid self-inactivation by hydrolysis (with a 0.25-sec half-life) was used (14). Each time point, obtained over reactions spanning tens of minutes, thus yields a structural snapshot of ~1 second duration.
SHAPE profiles for the initial monomer and final dimer forms agree well with accepted structures for the MuLV dimerization domain (Fig. S1 and text S1). Five key regions underwent large-scale structural changes during dimerization (Fig. 1). The loops of hairpins SL1 and SL2 (positions 329-332 and 363-366) were reactive in the monomer and became unreactive during dimerization (within 7 sec), consistent with formation of a stable intermolecular loop-loop kissing interaction (15). Two palindromic sequences, PAL1 (positions 210-219) and PAL2 (positions 283-298), were initially reactive but became unreactive due to intermolecular duplex formation in the dimer. Conversely, two regions that form the ‘anchoring helix’ (positions 231-251 and 290-315) in the monomer became more reactive upon dimer formation (Figs. 1 and S1).
Figure 1. Time-resolved SHAPE analysis of MuLV RNA dimerization.
SHAPE reactivities are shown for monomer (no magnesium), dimer (60 min), and representative time points in which specific structural intermediates predominate. Key structural interactions that change during dimerization are highlighted within sets of dashed blue lines.
We obtained SHAPE data for every nucleotide within the 170-nt MuLV domain in sixteen one-second snapshots yielding over 2,700 structural data points. We grouped nucleotides with similar kinetic behaviors by k-means clustering (16). In the presence of 5 mM Mg2+ and without a protein chaperone, there were seven distinct kinetic behaviors involving four net rates (Fig. 2A). Rates were identical, within error, over a three-fold change in RNA concentration (Fig. S2) indicating that most conformational changes reflect pseudo-unimolecular transitions between two interacting RNAs. The fastest rate of ≥5 min−1 (Fig. 2A; cluster 1a, in orange on bottom structures) occurred at nucleotides at the apexes of SL1 and SL2, suggesting formation of a complex between two RNAs before the first time point. PAL1 nucleotides became less reactive at a net rate of 1.6 ± 0.4 min−1 (Fig. 2A, cluster 1b, green on structures). Anchoring helix and PAL2 nucleotides demonstrated opposing kinetic behaviors (rates of 0.30 ± 0.03 min−1, Fig. 2A, clusters 2 and 3, in red), suggestive of a single process involving both structures. Positions in a large flexible domain (positions 251-282) showed slower kinetic behavior with a net rate of 0.11 ± 0.02 min−1 (Fig. 2A, cluster 4, in black on structures). Finally, nucleotides in clusters 5 and 6 showed biphasic kinetic behavior in which the SHAPE reactivity first increased and then decreased over time, or vice versa, with rates of 1.6 and 0.1 min−1. Time-resolved SHAPE analysis of the MuLV domain thus reveals that dimerization is complex, slow, and characterized by multiple structurally distinct transitions and intermediates.
Figure 2. Model-free clustering of nucleotide-resolution kinetic profiles for dimerization.
SHAPE reactivities at 600 nM MuLV RNA (A) in the absence of and (B) in the presence of 8 μM NC and (C) for an RNA containing inosine in place of guanosine. Each data point is shown on a scale (black to red) corresponding to its SHAPE reactivity (see Fig. 1). Y-axis shows every nucleotide (170 positions) in the MuLV dimerization domain RNA in an order determined by k-means clustering rather than linear sequence. Major kinetic clusters are labeled and representative kinetic profiles and observed net rates are shown for each cluster. Rates are reported as the mean for all nucleotides in each cluster ± the standard deviation. Positions of nucleotides in each cluster are shown in structural cartoons below each kinetic profile, colored by rate: orange > green > red > black. For clarity, only one strand of the dimer is colored.
We next performed an analogous set of experiments initiating dimerization by simultaneous addition of magnesium ion and the MuLV NC protein. With the addition of chaperone, clustering of the SHAPE data revealed that the NC protein collapsed dimerization into a single kinetic process that occurred at a net rate of 1.6 ± 0.4 min−1 (Fig. 2B). There was no evidence of the slow and multi-rate processes that characterized the RNA-only reaction.
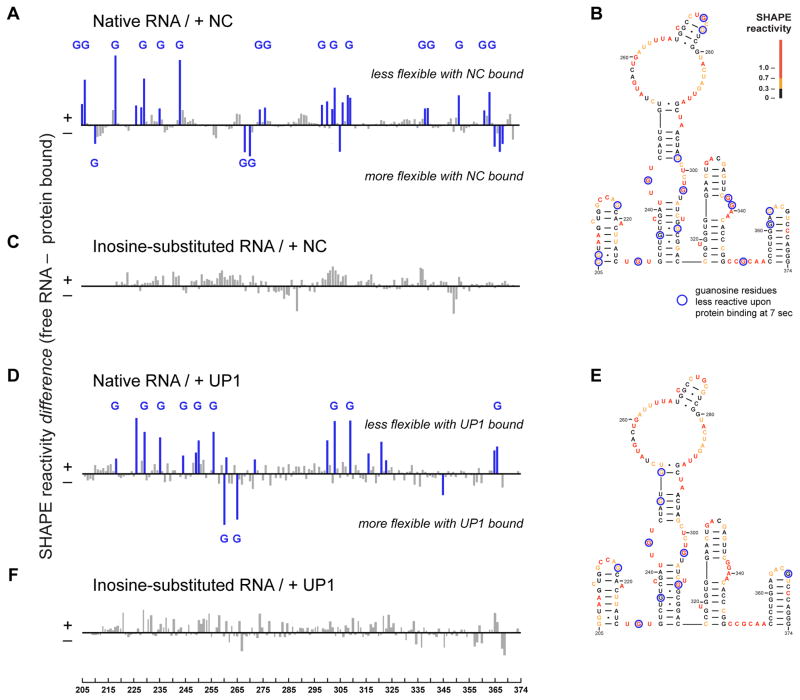
Initial binding interactions between the NC chaperone and RNA monomer were readily detected in a difference analysis in which the SHAPE profile immediately after NC binding was subtracted from that of reactivity profile of the free RNA (Fig. 3A). Of the 29 nucleotides with the largest changes in SHAPE reactivity, 19 (or 66%) are guanosine residues (Figs. 3A and Fig. 3B and text S2), consistent with studies showing that NC contains a cleft that binds guanosine (17). Sites of protection (positive peaks) likely correspond to sites of stable binding by NC during the 1-second window of the time-resolved SHAPE experiment; the smaller number of guanosine residues with higher reactivity in the presence of NC (negative peaks) likely reflect either a rapid binding and release or NC-induced conformational changes.
Figure 3. Initial interactions between NC and UP1 with the MuLV monomer.
SHAPE difference plots illustrating the effect of (A) NC or (D) UP1 binding to the native dimerization domain 7 sec after protein addition. Sites of strongest interaction, corresponding to SHAPE differences greater than 20%, are highlighted blue; those that occur at guanosine residues are labeled with a G. Superposition of strongest initial interaction sites for (B) NC and (E) UP1 on a MuLV dimerization domain secondary structure model. Structures are colored by SHAPE reactivity prior to protein binding. SHAPE difference plots illustrating the lack of an effect of (C) NC or (F) UP1 binding to the inosine-substituted RNA.
The preference of NC to interact at guanosine residues prompted us to consider whether NC exerts its RNA chaperone activity by destabilizing interactions between guanosine and other nucleotides. We explored the dimerization reaction using an RNA in which all guanosine residues were replaced by inosine, in essence removing a single amine group from each guanosine position. Inosine-cytosine pairs are iso-structural with guanosine-cytosine pairs, but are ~1 kcal/mol less stable (Fig. 4A); inosine also pairs more weakly with uridine than guanosine (18). The guanosine-to-inosine substitution will thus reduce both the strength and promiscuity of alternative base pairs during the RNA folding reaction.
Figure 4. Role of guanosine in RNA structure and mechanism of chaperone-mediated RNA folding.
(A) Average stabilities (ΔG°37) of nearest neighbor base pair combinations involving one G-C, I-C, or A-U pair and one Watson-Crick pair in 1 M NaCl (18, 22) with representative structures. Standard deviations for the nearest-neighbors combinations are shown with lines. Correlations between SHAPE reactivity profiles of (B) native and inosine-substituted dimers (obtained at 30 min dimerization time points), (C) native and inosine-substituted monomers (obtained just prior to addition of Mg2+), and (D) the native RNA after a 7-second interaction with NC versus the protein-free inosine-substituted monomer (both in the presence of Mg2+). The inosine dimer and native dimer in the presence of NC (not shown) also show a strong correlation (R2 = 0.89) reflecting that NC binds at relatively few sites in the native dimer (Fig. S6). Structures of (E) NC (17) and (F) UP1 (23) chaperones, emphasizing that both have a guanosine-binding pocket and that flanking nucleotides interact in an extended conformation. NC and UP1 bind guanosine in distinct ways involving anti and syn nucleotide conformations, respectively. (G) RNA-only (top) and chaperone-catalyzed (bottom) MuLV genome assembly mechanisms. Net rates are reported for each step. The overall reaction proceeds sequentially as indicated by (i) the change in reaction order (from second to first, yielding a large increase in effective RNA concentration) upon formation of the initial SL1-SL2 kissing interaction in the first step and (ii) the observation of biphasic profiles (Fig. 2) that include both the 1.6 and 0.1 min−1 processes. Evidence for a specific order of the 0.3 min−1 process is less strong, and this step may occur in parallel with the 0.1 min−1 conformational change.
The inosine-substituted RNA formed essentially the same final dimer structure as the guanosine-containing MuLV domain as indicated by SHAPE-directed modeling (19) (Fig. S3A), and individual nucleotide SHAPE reactivities for the inosine and native MuLV domain dimers are strongly correlated (R2 = 0.88, Fig. 4B). Although the overall secondary structures for inosine and native RNAs in the monomer states are similar (Figs. S3B and S3C), SHAPE reactivities correlate poorly (R2 = 0.26, Fig. 4C). However, adding NC to the guanosine-containing monomer converts this RNA to a structure that has a SHAPE profile highly similar to that of the inosine RNA monomer (R2 = 0.87; Fig. 4D). The inosine-substituted RNA is thus a good model both for the NC-destabilized native RNA in the monomer state and for the final dimer.
Time-resolved SHAPE analysis of dimerization of the inosine-substituted RNA in the absence of NC revealed a single, fast kinetic step involving similar nucleotides as NC-mediated dimerization of the native sequence RNA (compare Figs. 2B and 2C). The dimerization rate of the inosine RNA was accelerated by 7-fold relative to that of the free native RNA. The NC protein does not affect the structure of the inosine-substituted RNA (Fig. 3C). A non-denaturing gel-based analysis confirmed that addition of NC protein had no effect on the rate of formation of the final dimer state for the inosine-substituted RNA (Fig. S4). Replacement of guanosine with inosine thus both abrogates most of the need for the RNA chaperone activity of the NC protein and converts the RNA into a form that folds via a simple and direct pathway (compare Fig. 2A and 2C).
The unwinding domain of the hnRNP A1 protein (UP1) contains an arginine-rich RNA recognition motif and has potent RNA chaperone activity (1, 20). UP1 has no structural similarity with NC except that both proteins contain clefts that bind guanosine (Figs. 4E and 4F). As in the presence of NC, UP1-mediated dimerization of the MuLV domain proceeded in a single, fast kinetic step (Fig. S5) accelerated by ~20-fold (kobs> 2 min−1) relative to the RNA alone. Of the initial interaction sites (at ~7 sec) for UP1 on the native monomer RNA, 52% were guanosine residues (Fig. 3D and text S2). The set of guanosines contacted most strongly by UP1 included some but not all of the guanosines contacted by NC (compare Figs. 3B and 3E). UP1 had no effect on the dimerization rate of the inosine-substituted RNA (Figs. 3F and S4). UP1 is not known to play a role in structure rearrangements for the MuLV RNA genome yet is a potent facilitator of RNA dimerization of the MuLV domain and does so by a mechanism similar to the cognate NC chaperone.
Our data support a model of MuLV genomic dimerization in which two MuLV monomers initially associate rapidly via loop-loop interactions; subsequent steps for RNA-only folding are complex, involve multiple intermediates, and proceed slowly (Fig. 4G). In the presence of the chaperone, RNA dimerization was accelerated by more than ten-fold and appeared to occur in a single kinetic step, indicating that chaperone function accelerated multiple classes of slow RNA conformational changes (Fig. 4G). Our data indicate that RNA chaperones NC and UP1 both act by binding to exposed guanosine residues in RNA, thereby destabilizing stronger base pairings and creating a simplified folding pathway (text S3). The two proteins contact distinct, partially overlapping sets of guanosine residues in their initial interactions with RNA; thus, many possible guanosine-binding activities may support RNA chaperone function. The NC and UP1 chaperones also bind to the final native sequence dimer, in patterns that are distinct from their initial interactions with the monomer state (Fig. S6). These data suggest that chaperone binding does not discriminate between folded and misfolded RNA states per se, but that guanosine nucleotides are ultimately arranged in the final structure in such a way that chaperone binding (or inosine substitution) does not overly destabilize the final RNA structure. In this way, a guanosine-centric mechanism for RNA chaperone function is analogous to the mechanism of some chaperones that facilitate protein folding that destabilize interactions involving hydrophobic amino acid residues (21). In these cases, both RNA and protein chaperones simply interact with residues especially prone to forming stable intermediate and non-native states.
Supplementary Material
Acknowledgments
We are indebted to David Grawoig for a critical review of the manuscript and to Donald Johnson and Catherine Hixson for assistance in preparing MuLV NC protein. This work was supported by the US National Institutes of Health (GM064803 to K.M.W., GM072518 to J.D.L., and GM031819 to Jack Griffith and B.D.B) and by the National Cancer Institute (National Institutes of Health, under contract HHSN261200800001E with SAIC-Frederick, Inc. to R.J.G.). Datasets of representative clustered kinetic data are provided in the Supporting Online Material.
References
- 1.Herschlag D. J Biol Chem. 1995;270:20871. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 2.Rein A, Henderson LE, Levin JG. Trends Biochem Sci. 1998;23:297. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 3.Rajkowitsch L, et al. RNA Biol. 2007;4:118. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 4.Woodson SA. RNA Biol. 2010;7:677. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- 6.Rein A. RNA Biol. 2010;7:700. doi: 10.4161/rna.7.6.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas JA, Gorelick RJ. Virus Res. 2008;134:39. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adam MA, Miller AD. J Virol. 1988;62:3802. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hibbert CS, Mirro J, Rein A. J Virol. 2004;78:10927. doi: 10.1128/JVI.78.20.10927-10938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherghe C, et al. Proc Natl Acad Sci USA. 2010;107:19248. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badorrek CS, Weeks KM. Biochemistry. 2006;45:12664. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- 12.Gherghe C, Leonard CW, Gorelick RJ, Weeks KM. J Virol. 2010;84:898. doi: 10.1128/JVI.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortimer SA, Weeks KM. J Am Chem Soc. 2008;130:16178. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 14.Mortimer SA, Weeks KM. Nature Protoc. 2009;4:1413. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li PT, Bustamante C, Tinoco I., Jr Proc Natl Acad Sci USA. 2006;103:15847. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey A, York D, Smalls-Mantey A, Summers MF. Biochemistry. 2005;44:3735. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 18.Wright DJ, Rice JL, Yanker DM, Znosko BM. Biochemistry. 2007;46:4625. doi: 10.1021/bi0616910. [DOI] [PubMed] [Google Scholar]
- 19.Deigan KE, Li TW, Mathews DH, Weeks KM. Proc Natl Acad Sci USA. 2009;106:97. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portman DS, Dreyfuss G. EMBO J. 1994;13:213. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartl FU, Hayer-Hartl M. Nature Struct Mol Biol. 2009;16:574. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 22.Xia T, et al. Biochemistry. 1998;37:14719. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 23.Ding J, et al. Genes Dev. 1999;13:1102. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.