Abstract
For the past several decades, due to technical limitations, the field of transcriptomics has focused on population-level measurements that can mask significant differences between individual cells. With the advent of single-cell RNA-Seq, it is now possible to profile the responses of individual cells at unprecedented depth and thereby uncover, transcriptome-wide, the heterogeneity that exists within these populations. Here, we describe a method that merges several important technologies to produce, in high-throughput, single-cell RNA-Seq libraries. Complementary DNA (cDNA) is made from full-length mRNA transcripts using a reverse transcriptase that has terminal transferase activity. This, when combined with a second “template-switch” primer, allows for cDNAs to be constructed that have two universal priming sequences. Following preamplification from these common sequences, Nextera XT is used to prepare a pool of 96 uniquely indexed samples ready for Illumina sequencing.
Keywords: SMART-Seq, Single-Cell RNA-Seq, Transcriptomics, cell type, Single-cell, low input RNA-Seq, SMART-Seq2, Next-generation sequencing, template-switching
INTRODUCTION
The development of single-cell RNA-Seq affords new opportunities to study complex cellular systems at unprecedented resolution. While previous RNA-Seq methods were only capable of capturing the average behavior of a population, single-cell methods now enable studies of cellular heterogeneity, as well as the discovery of novel cell types and states. These methods employ a variety of techniques to address the technical challenges posed by single cells – most notably, the ultra-low input quantity of RNA.
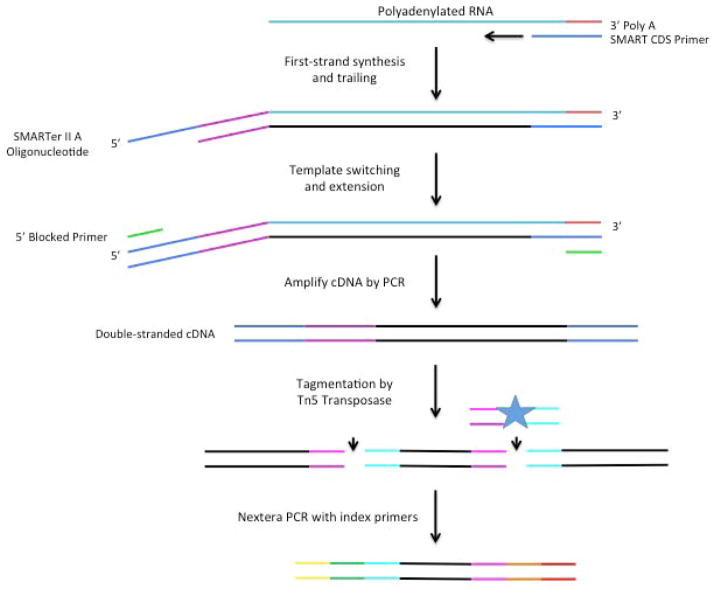
Here, we exploit the terminal transferase activity of the SMARTScribe reverse transcriptase in conjunction with a “template-switch” primer to make cDNAs that have PCR priming sites on both ends directly from full length mRNA (Figure 1). First, single cells are lysed in a guanidine thiocyanate solution. Second, a bead cleanup is used to isolate RNA. Third, cDNA is synthesized from the full-length transcripts. Fourth, these cDNA products are amplified by PCR and normalized. Fifth, the amplified cDNA is incubated with a Tn5 transposase to fragment full-length transcripts and append adapters on each molecule. Sixth, each single-cell library is individually barcoded by PCR with index primers, and the sample set is pooled. And finally, the resulting pool is purified by a bead cleanup and is then sent for Illumina sequencing.
Figure 1.
Single-cell RNA-Seq library construction. Illustrated are the steps required to convert mRNA to cDNA with sequencing adapters.
Materials
Microseal® ‘F’ Foil (Bio-Rad Laboratories, Inc., cat. no. MSF-1001)
Microseal® ‘B’ Adhesive Seals (Bio-Rad Laboratories, Inc., cat. no. MSB-1001)
2-mercaptoethanol, ≥ 99.0%
Buffer TCL (Qiagen, cat. no. 1031576)
Eppendorf twin.tec® PCR 96-well plate, skirted, (Eppendorf, cat. no. 951020401)
Dry ice
RNeasy Micro Kit
Agencourt RNAClean XP SPRI beads (Beckman Coulter, cat. no. A63987)
RNaseZap® RNAse Decontamination Solution (Life Technologies, cat. no. AM9780)
RNase-free water
200 proof ethanol
-
SMARTer® Ultra™ Low Input RNA Kit for Illumina® Sequencing-HV, 96 reactions (Clontech, cat. no. 634828) containing:
Dilution Buffer
3′ SMART CDS Primer II A (12 μM)
RNase Inhibitor (40 U/μl)
5X First-Strand Buffer (RNase-free)
Dithiothreitol (DTT; 100mM)
SMARTer dNTP Mix (dATP, dCTP, dGTP, and dTTP, each at 20mM)
SMARTer II A Oligonucleotide (12 μM)
SMARTScribe Reverse Transcriptase (100 U/μl)
Nuclease-Free Water
-
IS PCR Primer (12 μM)
Components of kit not used:
Purification Buffer (10mM Tris-Cl, pH 8.5)
Control Total RNA (1 μg/μl)
-
Advantage® 2 PCR Kit, 100 reactions (Clontech, cat. no. 639206) containing:
10X Advantage® 2 PCR Buffer
50X dNTP Mix (dATP, dCTP, dGTP, and dTTP, each at 10mM)
50X Advantage® 2 Polymerase Mix
-
PCR-Grade Water
Components of kit not used:
10X Advantage® 2 SA PCR Buffer
Control DNA Template (100 ng/μl)
Control Primer Mix, 5′ primer and 3′ primer (10 μM each)
Agencourt AMPure XP SPRI beads (Beckman Coulter, cat. no. A63881)
TE Buffer, 1x (see recipe)
Agilent High Sensitivity DNA Kit (Agilent Technologies, cat. no. 5067-4626)
Qubit® dsDNA HS assay kit, 100 reactions (Life Technologies, Inc., cat. no. Q32851)
Qubit® Assay Tubes (Life Technologies, Inc., cat. no. Q32856)
Taqman® Fast Advanced Master Mix, 100 reactions (Life Technologies, Inc., cat. no. 4444557)
Taqman® Probe set (Choose housekeeping genes that are appropriate for your system)
-
Nextera XT DNA Sample Preparation Kit, 96 samples (Illumina, cat. no. FC-131-1096) containing:
Amplicon Tagment Mix (ATM)
Tagment DNA Buffer (TD)
Nextera PCR Master Mix (NPM)
Neutralize Tagment Buffer (NT)
-
Components of kit not used:
Resuspension Buffer
Library Normalization Additives 1
Library Normalization Wash 1
Hybridization Buffer
Library Normalization Beads 1
Library Normalization Storage Buffer 1
-
Nextera XT Index Kit, 96 indices, 384 samples (Illumina, cat. no. FC-131-1002) containing:
Index Primers, S501 to S508 (8 tubes)
Index Primers, N701 to N712 (12 tubes)
TruSeq Index Plate Fixture Kit (Illumina, cat. no. FC-130-1005)
Plate centrifuge
Vortex
DynaMag™-96 side skirted magnet (Life Technologies, cat. no. 12027)
RNAse and DNAse-free 1.5ml tubes
Thermal cycler
Qubit® 2.0 Fluorometer (Life Technologies, Inc., cat. no. Q32866)
2100 Electrophoresis Bioanalyzer Instrument (Agilent Technologies, cat. no. G2939AA)
DynaMag™-2 Magnet (Life Technologies, Inc., cat. no. 12321D)
Oligonucleotide Primer Sequences
3′ SMART CDS Primer IIA: 5′ AAGCAGTGGTATCAACGCAGAGTACT(30)VN
SMARTer II A Oligonucleotide: 5′ AAGCAGTGGTATCAACGCAGAGTACATrGrGrG
IS PCR Primer: 5′ AAGCAGTGGTATCAACGCAGAGT
TSO: 5′ AAGCAGTGGTATCAACGCAGAGTACATrGrG+G
Stage I: Preparation of single-cell lysates
Note: In this protocol, we use Microseal F to seal for long-term storage and Microseal B when thermal cycling.
-
1
Prepare a solution of 1% 2-mercaptoethanol (by volume) in TCL buffer and distribute 5 μL of this solution into each well of a skirted-side 96-well PCR plate, and 350 μL into a 1.5 mL RNase-free centrifuge tube. Cover this plate with Microseal F and keep at room temperature until ready for single-cell isolation.
-
2
Prepare a cell suspension in complete media and use a FACS machine to sort a single cell into each well of the abovementioned 96-well plate containing Buffer TCL, and an additional sample of ≥10,000 cells into the 1.5 mL centrifuge tube to use as a population control. Once sorting is completed seal plate with Microseal F and centrifuge (800g, 1 min). Immediately freeze plate and population control on dry ice and keep at −80°C until ready for lysate cleanup.
Note: To improve yield, we recommend sorting single cells on both the presence of a positive viability indicator (e.g., Calcein AM, Life Technologies) and the absence of a cell death marker (e.g., the membrane-impermeant DNA stain EthD-1, Life Technologies).
Stage II: Lysate cleanup and reverse transcription of mRNA species
Note: The following steps can be performed inside a biosafety cabinet or a RNA workstation (if available); otherwise, they can be carefully performed on a standard benchtop.
-
3
Purify RNA from the population control sample using the RNEasy Plus Micro Kit (Qiagen) according to the manufacturers recommendations. Keep purified RNA at 4°C until step 18 (−80°C for long term storage).
-
4
Bring RNA-SPRI beads (Agencourt RNAClean XP SPRI beads) to room temperature (allow 30 minutes) and use RNAseZap to clean workbench and all equipment used to process RNA.
-
5
Thaw lysate plate on ice, and then centrifuge (800g, 1 min).
-
6
Optional: Add 10 μL RNase-free H2O to lysate to reduce the difficulty of performing the following bead-cleanup steps.
-
7
Thoroughly vortex RNA-SPRI beads to ensure a uniform suspension. Add 2.2 volumes RNA-SPRI beads to each well of lysate (2.2 × 5 μL cell lysate = 11 μL of beads are added, and 2.2 × 15 μL = 33 μL) and mix well by pipetting 10 times. If each solution is not in the bottom of its respective well, centrifuge plate (300×g, 1 min).
Note A: The following steps are done at room temperature
Note B: Do not centrifuge plates containing SPRI beads at greater than 300 × g. Beads will adhere to the well sidewall and impair RNA recovery.
-
8
Incubate lysate and bead suspension for 10 minutes on bench.
Note: If this procedure is performed on a standard benchtop, the plate should be kept covered (we use the lid from a fresh box of pipette tips, placed slightly ajar) to prevent dust and debris from falling into samples during this and subsequent incubation steps.
-
9
Place plate on 96-well plate magnet (Life Technologies DynaMag™-96 side skirted magnet, Cat No. 12027) and incubate for 5 min. Remove supernatant from each column of the plate, being careful not to aspirate the beads collected on the side of each well.
Note: The magnet described draws the beads to alternating sides of each column of the plate. It is therefore recommended to use an eight-channel pipette to remove the supernatant.
-
10
Wash beads by adding 75 μL of 80% ethanol (prepared same day with nuclease-free water) to each well. Wait approximately 30 seconds. Aspirate ethanol and repeat wash two more times.
-
11
Aspirate final ethanol wash. Leave the plate on the magnet and allow beads to dry at room temperature for approximately 10 minutes. Keep plate loosely covered with the lid from a fresh box of pipette tips, placed slightly ajar.
Note: Allowing beads to dry for too long will impair recovery of RNA. Immediately proceed with protocol when visible cracks appear in bead pellet.
-
12
Once bead pellet has dried, remove plate from magnet and elute RNA from beads by resuspending dried beads in 4.5 μL of the following mix, using reagents from the SMARTer Ultra Low Input RNA Kit (amounts per sample):
2.375 μL Dilution Buffer
1 μL RNase-free H2O
1 μL 24 μM 3′ SMART CDS Primer II A
-
0.125 μL RNase Inhibitor
Note A: Do not remove eluent from original plate!
Note B: For population control, substitute 1 μL purified RNA in place of H2O.
-
1
Seal and incubate for 1 minute at room temperature.
-
2
Centrifuge (800g, 1 min).
-
3
Incubate for 3 min at 72°C to anneal 3′ SMART CDS Primer II A.
-
4
Place plate on ice immediately following incubation.
-
5
Remove seal and add the following as a mastermix (amounts per sample):
2 μL First Strand Buffer
0.25 μL 100 mM DTT
1 μL 10 mM dNTP Mix
1 μL 12 uM SMARTer II A Oligo
0.25 μL RNase Inhibitor
1 μL SMARTScribe Reverse Transcriptase
Mix well by pipetting, reseal with a new Microseal B, and centrifuge (800g, 1 min).
-
3
Carry out the following Reverse Transcription (RT) in a thermal cycler using the following conditions:
Initial step: 90 min 42°C Inactivate: 10 min 72°C Cool: Hold 4°C. Note A: Following this step, the product can be stored at 4°C overnight.
Note B: Residual SPRI beads remain in each well with the libraries until step 28.
Note C: The protocol provided with the SMARTer kit describes a SPRI cleanup at this stage of the protocol, which we omit here. It is important to avoid losing material before amplification and it has been found that the cleanup is not essential to generating a high-quality sequencing library (Shalek et al., 2013; Picelli et al., 2013).
Stage III: Performing whole transcriptome amplification) and post-PCR cleanup
-
6
Centrifuge the plate (800g, 1 min) and add the following as a mastermix (amounts per sample):
5 μL 10X Advantage 2 PCR Buffer
2 μL 10 mM dNTP mix
2 μL 12 uM IS PCR Primer (from the Clontech Ultra Low input RNA Kit)
2 μL 50X Advantage 2 Polymerase Mix
29 μL H2O
Mix well by pipetting, seal, and centrifuge (800g, 1 min).
-
7
Carry out the whole transcriptome amplification (WTA) in a thermal cycler using the following conditions:
Initial step: 1 min 95°C (initial denaturation) 5 cycles: 20 sec 95°C (denaturation) 4 min 58°C (annealing) 6 min 68°C (extension) 9 cycles: 20 sec 95°C (denaturation) 30 sec 64°C (annealing) 6 min 68°C (extension) 7 cycles: 20 sec 95°C (denaturation) 30 sec 64°C (annealing) 7 min 68°C (extension) 1 cycle: 10 min 72°C (final extension) Cool: Hold 4°C Note A: Following this step, the product can be stored at 4°C overnight
Note B: For this amplification, we have made a few modifications to the thermal cycling steps described in the original SMARTer protocol. We have found that these modifications improve annealing, denaturation, and extension for the diverse transcript sets with which we work.
-
8
Bring DNA SPRI beads (Agencourt AMPure XP) to room temperature.
-
9
Centrifuge WTA plate (800g, 1 min).
-
10
Unseal and add 0.8 volumes of DNA SPRI beads to each well and mix well by pipetting.
-
11
Cover with a clean lid from a pipette tip box and incubate bead suspension for 5 min on bench.
-
12
Place plate on 96-well plate magnet and incubate for another 5 minutes.
-
13
Wash beads by adding 100 μL 80% ethanol to each well and move the plate left and right on the magnet to move the beads from side to side in each well. Continue shifting plate on magnet for approximately 30 sec. Aspirate ethanol and repeat wash two more times.
-
14
Aspirate final ethanol wash. Leave the plate on the magnet and allow beads to dry at room temperature for approximately 10 minutes. Keep plate covered with the lid of a pipette tip box (left slightly ajar) to prevent dust and debris from falling into samples.
-
15
Once bead pellet has dried, remove plate from magnet and elute DNA from beads by resuspending dried beads with 20 μL TE buffer. Transfer eluent to a new 96-well plate.
Note: Following this step, the new product plate can be sealed and stored at 4°C overnight or at −20°C for months.
-
29
Use 1 μL of the purified PCR product to measure the fragment size distribution using the Agilent HS DNA BioAnalyzer or similar instrumentation (as per the manufacturer’s recommendations) and 1 μL to estimate the library concentration using the Qubit® dsDNA HS Assay kit with Qubit® assay tubes and the Qubit® fluorometer or similar instrumentation (as per the manufacturer’s recommendations).
-
30
Use 1 μL of the purified PCR product to measure single-cell viability with the Taqman qPCR assay.
Note: In most single-cell sample sets, a small subset of libraries will have low complexity and will lack expression of highly expressed housekeeping genes. Since there are, in most systems, a set of housekeeping genes that should be expressed in every cell (Shalek et al., 2013), we use qPCR of two housekeeping genes (usually ACTB and B2M) to identify viable samples. Samples with low or no expression of these genes can be removed from Nextera library construction to reduce cost.
-
31
For each sample, dilute the purified PCR product to a concentration of between 0.1 and 0.2 ng/μL with buffer TE in preparation for Nextera XT library construction.
Stage IV: Nextera XT sequencing-library construction
-
32
Before beginning library construction, thaw index primers and mix by brief vortexing followed by centrifugation. Arrange Nextera index tubes in the TruSeq Index Plate Fixture, such each slot in the fixture holds one index tube. The 12 index 1 (i7) primers, which have orange caps, should be arranged in order horizontally, such that each tube corresponds to a column (Figure 2). The column 1 slot should hold N701 and the column 12 slot should hold N712. Similarly, the index 2 (i5) primers should be arranged in order vertically, such that S501 is in the slot for row A and S508 is in the slot for row H.
Using a multichannel pipette, distribute 10 μL of each index into the corresponding row/column. (i7 primers will be distributed down rows, while i5 primers will be distributed across columns.)
Seal, vortex mildly to mix, and then centrifuge (800g, 1 min). These mixed index primers can be used immediately, or stored long-term at −20°C.
There is enough combined index primer solution in each well to generate 8 × 96 single-cell libraries.
-
33
Using a new 96-well plate, add to each well:
2.5 μL TD Buffer
1.25 μL ATM
1.25 μL Diluted PCR product from SMART RT-PCR
Mix well by pipetting, seal, and centrifuge (800g, 1 min).
-
34
Carry out tagmentation using the following conditions:
Initial step: 10 min 55°C Cool: Hold 10°C -
35
Unseal and immediately add 1.25 μL NT Buffer to each well and mix well by pipetting. Wait 5 minutes at room temperature.
-
36
Add 3.75 μL NPM and 2.5 μL combined Index primer solution to each well. Mix well by pipetting, seal, and centrifuge (800g, 1 min).
-
37
Carry out the amplification in a thermal cycler using the following conditions:
Initial step: 3 min 72°C (initial annealing) 1 cycle: 30 sec 95°C (denaturation) 12 cycles: 10 sec 95°C 30 sec 50°C 60 sec 72°C 1 cycle: 5 min 72°C Cool: Hold 4°C Note: Following this step, the product can be stored at 4°C overnight.
Figure 2.
Layout to prepare the combined dual-index primer plate that is used in Nextera XT library construction.
Stage V: Pooling and DNA SPRI bead cleanup
-
38
Bring DNA SPRI beads (Agencourt AMPure XP) to room temperature.
-
39
Spin down plate (800g, 1 min), unseal, and pool 2.5 μL from each well into a single 1.5 mL tube. Measure total volume of resulting pool and add 0.9 volumes SPRI beads to tube and mix well by pipetting.
-
40
Incubate bead suspension for 5 min on bench.
-
41
Place tube on Dynamag™-2 magnet, and incubate for 5 min.
-
42
Wash beads by adding 500 μL 80% ethanol to the tube and rotate it on the magnet to move the beads from front to back across the tube. Aspirate ethanol and repeat wash once.
-
43
Aspirate final ethanol wash. Leave the tube on the magnet and allow beads to dry at room temperature for approximately 10 minutes. Keep the tube covered to prevent dust and debris from falling into your sample.
-
44
Once bead pellet has dried, remove the tube from the magnet and elute DNA from beads using 30 μL TE buffer (consider smaller volumes if working with fewer than 96 samples). Transfer eluent to a new 1.5 mL tube and repeat 0.9X SPRI bead cleanup once.
-
45
Use 1 μL of the purified PCR product to measure the fragment size distribution using the Agilent HS DNA BioAnalyzer or similar instrumentation (as per the manufacturer’s recommendations) and 1 μL to estimate the library concentration using the Qubit® dsDNA HS Assay kit with Qubit® assay tubes and the Qubit® fluorometer or similar instrumentation (as per the manufacturer’s recommendations).
Stage VI: Sequencing
-
46
Sequence libraries on an Illumina sequencer using paired-end reads.
Note A: We typical run 2x25bp for expression quantification.
Note B: Expression estimates will saturate around ~ 1 million aligned reads, so one lane of HiSeq should be sufficient for 96 single cells.
Alternate Protocol: Smart-seq2 single-cell RNA-Seq modified method
In single-cell RNA-Seq, small amounts of sample loss during a number of steps can lead to significant decreases in transcript detection sensitivity. A decrease in assay sensitivity results in data that is only accurate and reproducible for highly expressed genes, limiting the scope and confidence of gene expression analyses. Further complications in assay sensitivity arise from an uneven distribution of sequencing reads along a transcript; usually, in SMARTer, there is a bias towards more reads at the 3′ end of the transcript. Even coverage along a transcript improves the accuracy of analytical tools used to quantify gene expression and transcript isoform abundance. A method published by Picelli et al (Nature Methods, 2013) modified the traditional SMARTer protocol to address this by improving transcript detection, coverage, accuracy, yield, and cost. Following the same strategy as SMARTer library construction, Smart-seq2 uses several alternative reagents to generate whole-transcriptome full-length cDNA libraries.
Avoiding small-volume, bead-based SPRI cleanups of each sample is an effective way of reducing loss and increasing assay sensitivity. Lysing single cells in a guanidine thiocyanate buffer necessitates SPRI cleanup due to the protein denaturing effects of the compound, which will affect downstream reactions, like reverse transcription. Multiple alternative lysis buffers exist that address this. The Ambion Single Cell Lysis buffer (Life technologies, #4458235), often used for single-cell RT-PCR, only requires the addition of a stop solution to inactivate its lytic activity before subsequent reactions. A hypotonic lysis buffer with small amounts of RNase-inhibitor and surfactant, as described in Smart-seq2, is the preferred buffer due to the lack of a need for a post-lysis cleanup or the addition of a stop solution prior to reverse transcription. However, the optimal lysis strategy will depend on the experimental system being analyzed.
Smart-seq2 takes additional steps to minimize sample loss during library construction. The reverse transcription is improved by the addition of betaine and additional magnesium chloride to the reaction mix and by the use of a template-switch oligonucleotide with one locked nucleic acid (LNA) riboguanosine base. These improvements assist in the hybridization between the template-switch oligonucleotide and the cDNA product, thereby increasing the probability of successfully introducing a second PCR adapter onto the cDNA product (see Figure 1). A second key improvement was made in the preamplification PCR step, which can be heavily biased against either long transcripts or those containing regions with high G/C content. Picelli et al found that the preamplification PCR is improved by using the KAPA HiFi HotStart ReadyMix, which dramatically improved coverage and sensitivity, particularly for GC-rich transcripts.
Additional materials
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
SUPERase-In RNase-Inhibitor, 40 U/μL (Life Technologies, Inc., cat. no. AM2694)
10 μM Reverse transcription DNA oligonucleotide primer (custom synthesized by Integrated DNA Technologies, see table for sequence)
dNTP Mix (dATP, dCTP, dGTP, and dTTP, each at 10mM) (Thermo-Fisher, cat. no. R0192)
-
SuperScript™ II Reverse Transcriptase Kit (Life Technologies, Inc., cat. no. 18064-014) containing:
5X First-Strand Buffer
100 mM DTT
SuperScript™ II Reverse Transcriptase
5 M betaine
100 mM MgCl2
10 μM SMARTer TSO primer (custom synthesized by Exiqon, see table for sequence)
RNase-free water
KAPA HiFi HotStart PCR ReadyMix (KAPA Biosystems, cat. no. KK2601)
10 μM PCR oligonucleotide primer (custom synthesized by Integrated DNA Technologies, see table for sequence)
8. Reagents and Solutions
TE Buffer, 1x
10 mM Tris-HCl, pH 8.0
1 mM EDTA
Stage I: Preparation of single-cell lysates
-
1
Prepare a mild hypotonic lysis buffer of 0.2% Triton X-100 and 2 U/μL RNase-Inhibitor and distribute 4 μL of this solution to each well of a 96-well plate as described above.
-
2
Sort single cells with a FACS machine as described above.
Stage II: RT of mRNA species
-
3
Prepare work space and thaw plate as above. Add 1 μL of 10 μM anchored oligo-dT RT primer and 1 μL of 10 mM dNTP mix to each lysate well, seal plate with Microseal ‘B’ and centrifuge (800g, 1 min).
-
4
Incubate for 3 minutes at 72 °C and immediately place on ice.
-
5
Centrifuge plate (800g, 1 min), unseal, and add the following as a mastermix:
2 μL SuperScriptII first strand buffer
0.25 μL 100 mM DTT
2 μL 5M betaine
0.9 μL 100 mM MgCl2
1 μL 10 μM template switching oligonucleotide
0.1 μL H2O
0.25 μL 40 U/μL RNase-Inhibitor
0.5 μL 200 U/μL SuperScriptII RT
Mix well by pipetting, seal, and centrifuge (800g, 1 min).
-
6
Carry out the following RT in a thermal cycler using the following conditions:
Initial step: 90 min 42°C 10 cycles: 2 min 50°C 2 min 42°C Inactivation: 15 min 70°C Cool: Hold 4 °C
Stage III: Performing WTA and Post-PCR Cleanup
-
7
Add the following as a mastermix:
13 μL KAPA HiFi HotStart ReadyMix
1 μL 10 μM IS PCR primer
-
8
Carry out the amplification in a thermal cycler using the following conditions:
Initial step: 3 min 98°C 20 cycles: 15 sec 98°C 20 sec 67°C 6 min 72°C Extension: 5 min 72°C Cool: Hold 4°C Note: Following this step, the product can be stored at 4°C overnight
-
9
Carry out DNA SPRI bead cleanup of preamplification product as described above (Stage III, #16-24)
Stage IV: Nextera XT sequencing-library construction
-
10
Carry out Nextera XT library construction as described above (Stage IV, 25-30)
Stage V: Pooling and SPRI bead cleanup
-
11
Carry out pooling and SPRI bead cleanup as described above (Stage IV, 31-38)
Support Protocol: Single-cell RNA-Seq expression analysis
Following sequencing of the cDNA libraries on an Illumina sequencer, data is generated as a series of files in the FASTQ format. For each unique sample specified in the sequencing sample sheet, four files are generated: one containing the “left-hand” read data (one end of the paired-end reads), one containing the “right-hand” read data (the other end of the pair), one containing the “left-hand” Nextera indexing read data, and one containing the “right-hand” Nextera indexing read data. RNA-Seq analysis uses computational tools to match each read pair, align the read pair to the genome sequence, and quantify the number of reads that align within each annotated gene.
The GenomeSpace web portal was developed to assist researchers with minimal computational analysis experience. Using its drag-and-drop interface, data sets and modules of pre-built analytic tools can be organized into customizable pipelines for numerous applications. Despite its ease of use, GenomeSpace uses cloud storage and computing power, making it less efficient for a large number of sequencing analyses or if a researcher has access to higher computing power at their own institution; as an alternative strategy for higher throughput, we also provide a Unix-based workflow.
Using GenomeSpace for expression analysis
Create an account at http://www.genomespace.org/
Upload each of the raw FASTQ files from the sequencing run into the home directory of the GenomeSpace interface via drag-and-drop onto the GenomeSpace interface.
Under the “Recipes” drop-down menu on the GenomeSpace interface, select “Analyzing data with GenomeSpace tools”.
Select the appropriate application for which the data will be analyzed, and follow the instructions to generate an analysis pipeline using the tools available through GenomeSpace.
Using Unix command line for expression analysis
-
Ensure that the following programs are installed and ready to use on the computer or server that will run the analysis:
TopHat – http://tophat.cbcb.umd.edu/
Bowtie (or Bowtie2) – http://bowtie-bio.sourceforge.net/
Samtools – http://samtools.sourceforge.net/
Picard tools – http://picard.sourceforge.net/
Integrative Genomics Viewer (IGV) – http://www.broadinstitute.org/igv/
Cufflinks – http://cufflinks.cbcb.umd.edu/
-
Run the program TopHat to match each of the paired-end reads with its mate and align the reads to the desired reference genome.
Files required:
- Reference genome index(download at ftp://ftp.ccb.jhu.edu/pub/data/bowtie_indexes/)
- Transcriptome reference annotation file (.GTF)(see http://tophat.cbcb.umd.edu/igenomes.shtml for up-to-date reference annotations)
Left reads FASTQ file (either compressed or uncompressed)
- Right reads FASTQ file (either compressed or uncompressed
% tophat [options] [genome index base] [~/LeftReads.fastq.gz] [~/RightReads.fastq.gz] For full list of [options], see http://tophat.cbcb.umd.edu/manual.shtml
TopHat will create several files, including ~/accepted_hits.bam. The data in this file contains the alignment and pairing information for all reads that successfully paired and aligned in the BAM file format
-
The Picard suite of command-line tools contains programs that provide important metrics regarding the sequencing data. Run various tools on the ~/accepted_hits.bam file to analyze the sequencing quality. Helpful tools include the following:
CollectAlignmentSummaryMetrics.jar – view statistics on the number of reads that correctly align to the reference genome
- CollectInsertSizeMetrics.jar – view statistics on the lengths of the sequenced cDNA fragments
% java –jar ~/picard/CollectAlignmentSummaryMetrics.jar [options] I=~/accepted_hits.bam O=~/AlignmentMetrics.txt % java –jar ~/picard/CollectInsertSizeMetrics.jar [options] H=~/InsertSizeHistogramChart.txt I=~/accepted_hits/bam -
For full list of tools and [options], see
- Sort the ~/accepted_hits.bam file using the Picard tool SortSam.jar, which organizes the BAM file data based on the aligned reads’ locations in the reference genome.
% java –jar ~/picard/SortSam.jar I=~/accepted_hits.bam O=~/accepted_hits.sort.bam SO=coordinate - Index the sorted BAM file using the Samtools suite of command-line tools.
% samtools index ~/accepted_hits.bam For graphical visualization of the data, load the aligned sequencing reads into the program IGV (Integrative Genomics Viewer). Open the IGV program, select the appropriate reference genome, and load the ~/accepted_hits.sort.bam file. This will display a graph for each file loaded with the location within the genome on the horizontal axis and the number of reads on the vertical axis.
-
For a quantitative measure of gene or transcript abundance, run the program Cufflinks on the ~/accepted_hits.sort.bam file. The two primary output files generated by Cufflinks represent quantified expression level estimates at the gene and transcript isoform level. Each of these presents the expression level estimates as a table relating each gene or transcript to its relative abundance in fragments per kilobase of exon per million mapped fragments (FPKM).
-
Files required:
BAM data file
-
Transcriptome reference annotation file (.GTF)
(see http://cufflinks.cbcb.umd.edu/igenomes.html for up-to-date reference annotations)% cufflinks [options] ~/accepted_hits.sort.bam
-
For full list of [options], see http://cufflinks.cbcb.umd.edu/manual.html
9. Commentary
Background Information
It has long been known that population-level observations of cellular transcription do not always accurately represent transcriptional patterns across the single cells that comprise them (Eberwine et al. 1992; Sandberg 2014; Shapiro et al. 2013). Earlier methods developed to explore this transcriptional heterogeneity in cellular populations first interrogated individual genes of interest in single cells (Eberwine et al. 1992), and later allowed for highly multiplexed qRT-PCR of a up to 96 genes simultaneously across 96 single cells (see, for example, Sanchez-Freire et al. 2012).
Recent technical advances that make high-throughput construction of next-generation sequencing libraries possible from picogram quantities of RNA facilitate studies of whole-transcriptome expression profiles in single cells. Relative to population measurements, single-cell RNA-Seq is able to deconvolute transcriptional profiles of cellular subpopulations and reveal transcriptional profiles of rare cellular subtypes that would be hidden below the detection threshold in population-level analysis.
A key tool that makes the construction of single-cell RNA-Seq cDNA libraries possible is the introduction of uniform adapter sequences to the transcripts of interest, allowing for the amplification and sequencing of all mRNA transcripts. A number of strategies have been developed to efficiently add these adapters. Pioneering work on mRNA-Seq utilized poly(A) tailing of reverse transcribed products followed by poly(T)-primed PCR amplification (Tang et al. 2009). CEL-Seq, a more recently published method, relies on in vitro transcription (IVT) to linearly amplify reverse transcribed products, followed by ligation of adapter sequences to the 3′ end of amplified RNA (Hashimshony et al. 2012). Presented here, the SMARTer protocol leverages the terminal transferase activity of a M-MLV-derived reverse transcriptase to reverse transcribe mRNA and then, with a template-switch primer, add an adapter sequence in a single reaction (Zhu et al. 2001). Each method has its own distinct advantages, disadvantages, and biases specific to the biochemical reactions underlying each protocol. For example, CEL-Seq avoids biases introduced by PCR amplification of reverse transcription products by linearly amplifying its reverse transcription products with IVT; this, however, necessitates a cleanup of both reverse transcription products and IVT amplification products prior to subsequent reactions (Hashimshony et al. 2012). With varying levels and tolerances for biases arising in specific experimental contexts, selecting the appropriate method for a given study depends on the system being assayed.
As the throughput and power of single-cell transcriptome profiling increases, these methodologies can be applied to increasingly complex biological questions. Whole-transcriptome sequencing across cell types can reveal transcriptional profiles specific to each type, as well as the heterogeneity within them. Identification of cell types from a population of cells of unknown composition based solely on transcriptional profiles is now possible and is increasingly easy as more single-cell RNA-Seq profiling data is added to the knowledge base (Shapiro et al. 2013). Meanwhile, primary cells, or rare subtypes of cells, desirable for study, may not appear in extracted populations in sufficient frequency to conduct RNA-Seq at a population level, necessitating single-cell methods. The transcriptional profiles of a patient’s tumor cells may elucidate the origin, development, and function of various cell lineages within a tumor. Other questions about cellular response to stimuli (Shalek et al. 2013), transcriptional fluctuation across time (Shapiro et al. 2013), and differential gene expression in disease or pathological tissue (Sandberg 2014) are now being explored; we envision that many more topics will be fruitfully investigated with single-cell RNA-Seq as it becomes a more widely used tool.
Critical Parameters and Troubleshooting
Proper sample preparation is key when preparing viable single-cell RNA-Seq libraries, as degradation and contamination of input material are the most common sources of error in downstream steps. Prior to sorting, sample integrity can be improved by minimizing the time spent preparing cells for sorting, and then freezing lysate plates no more than 10 minutes after sorting has concluded. When preparing for cDNA synthesis, it is vitally important to ensure that all workspaces, equipment, and reagents are RNase free by using products such as RNase-ZAP and RNase inhibitors. Cross contamination is also of great concern throughout this protocol, and care should be taken to avoid sharing tips between samples. Additionally, samples should be covered whenever possible to prevent airborne contamination.
Because sample integrity is a common source of error, and because small quantities of starting material are frequently used in this protocol, several parameters must be assayed following cDNA synthesis, amplification and cleanup to ensure library quality. Average fragment size, cDNA concentration, and the presence of transcripts for housekeeping genes are effective measures of library quality. Measuring, via qPCR, at least two well-established housekeeping genes is particularly important at this stage. In virtually any set of singe cell libraries, there will be samples of low complexity that lack expression of key housekeeping genes, and the results of qPCR measurements can be used to eliminate these samples.
The Tn5 transposase used in Nextera XT library construction fragments dsDNA at a rate proportional to [dsDNA] in your library. Thus, it is crucial to normalize the concentration of your SMARTer single-cell amplified cDNAs prior to Nextera XT library construction. This ensures that dsDNA from each single cell sample will yield tagmentation products with similar distributions of fragment length, and further that after pooling, cleanup, and sequencing the distribution of reads across each sample will be unbiased.
Anticipated Results
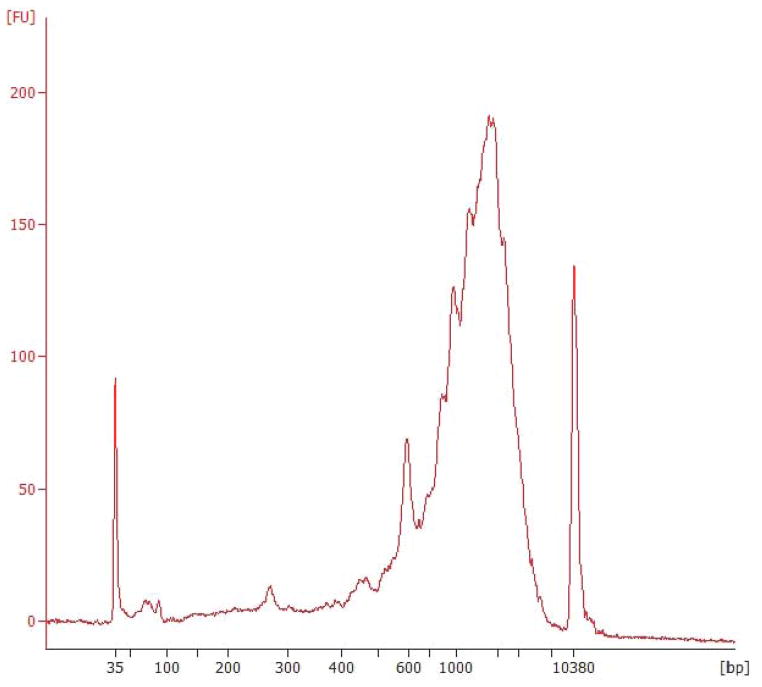
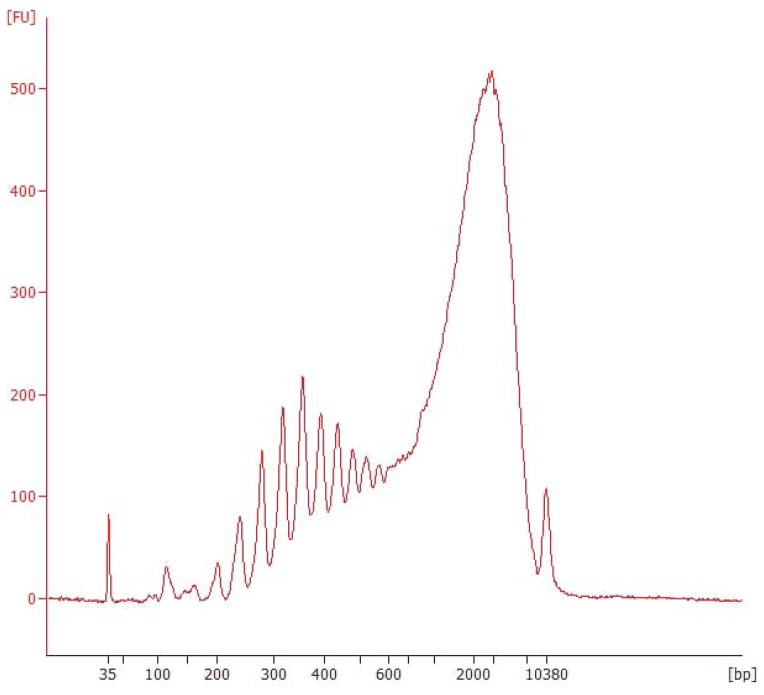
Following RT, PCR, and SPRI cleanup, successful single-cell libraries can have concentrations ranging from 0.1–50 ng/ μL. At this stage in the protocol, each library should have an average fragment size of approximately 2000 bp, with few or no species smaller than 400 bp as shown in Figure 3. When running qPCR, genes in viable libraries will amplify in fewer than ~26 cycles. Contaminated samples may yield libraries that align poorly to a corresponding reference genome, which can often be identified by regularly-spaced spiky peaks between 100 and 600 bp on a Bioanalyzer trace, as shown in Figure 4.
Figure 3.
Typical best-case results following RT-PCR and cleanup. The ideal average WTA product is around 2000 bp.
Figure 4.
Regularly-spaced spiked peaks indicate sample contamination
After a Nextera library has been pooled and cleaned up its concentration can range from 2–100 ng/μL, depending largely on the volume used for elution after the final SPRI cleanup. These libraries will have average fragment sizes of 350–550 bp with a somewhat flat peak as shown in Figure 5.
Figure 5.
Typical best-case results following Nextera library construction, pooling, and normalization. The ideal average fragment size for a Nextera library is 350–500 bp.
Time Considerations
The SMARTer library construction and normalization take 2–3 days, and Nextera XT library construction and pooling takes 1 day.
Table 1.
Troubleshooting for Single-Cell RNA-Seq Library Preparation
| Problem | Likely Cause | Solution | |
|---|---|---|---|
| Little or no WTA yield | RNA degradation | Start with high-quality RNA; use RNA with RNA integrity number >8; begin with a single RNA sample and work quickly; work in an RNase-free environment | |
| RNA-SPRI failure | Make sure beads have been fully resuspended prior to use; make sure beads are never exposed to RNases. Use freshly prepared ethanol. Don’t overdry beads. | ||
| Reverse transcription failure | Make sure all primers are added to the correct reaction | ||
| Too few PCR cycles | Increase nul/exZc mber of PCR cycles | ||
| PCR failure | Poor library generation, perhaps due to inactive enzymes/reagents | ||
| PCR-SPRI failure | Make sure beads have been fully resuspended prior to use | ||
| Cell sorting failure | Make sure cell sorter dispenses cells accurately to center of each well | ||
| Presence of WTA products that are longer than expected (>4 kb) | Too many PCR cycles | Reduce number of PCR cycles | |
| Presence of WTA products that are shorter than expected (<500 bp) | RNA degradation | Start with high-quality RNA; use RNA with RNA integrity number >8; begin with a single RNA sample and work quickly; work in an RNase-free environment | |
| PCR-SPRI failure or ineffective cleanup | Make sure beads have been fully resuspended prior to use; repeat 0.8X SPRI cleanup | ||
| Regularly-spaced spiky peaks on WTA Bioanalyzer trace | Contamination | Make sure to prepare samples and reactions in a clean and sterile environment | |
| Small fragment size after Nextera | Tagmentation reaction ran too long | Reduce reaction time of Nextera tagmentation | |
| Low tagmentation input cDNA concentration | Make sure tagmentation input cDNA is at the correct concentration; make sure the cDNA Bioanalyzer traces show little primer contamination (<500 bp peaks) after Nextera | ||
| Low alignment rate of reads to genome | Contamination | Make sure to prepare samples and reactions in a clean and sterile environment | |
| Low library complexity/number of genes detected | Too many PCR cycles | Reduce number of PCR cycles | |
| Loss of material prior to PCR | Make sure loss of material is limited during the initial RNA-SPRI cleanup | ||
Acknowledgments
Work was supported by an NIH Postdoctoral Fellowship (1F32HD075541-01, RS), an NIH CEGS Award (1P50HG006193-01, AR), NIH Pioneer Awards (DP1OD003958-01 to AR), the Broad Institute (AR), HHMI (AR), and the Klarman Cell Observatory at the Broad Institute (AR).
10. Literature Cited
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA. 1992;89(7):3010–4. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing Subgroup. 2009. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 1000;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell-Reports. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Picelli S, Björklund Å, Faridani O, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Sanchez-Freire V, Ebert A, Kalisky T, Quake S, Wu J. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nature Protocols. 2012;7(5):829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- Shalek A, Satija R, Adiconis X, Gertner R, Gaublomme J, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta J, Gennert D, Gnirke A, Goren A, Hacohen N, Levin J, Park H, Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Reviews Genetics. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch B, Siddiqui A, Lao K, Surani M. mRNA-Seq whole transcriptome analysis of a single cell. Nature Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg L. TopHat: discovering splice junctions with RNA-Seq. Bioninformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D, Pimentel H, Salzberg S, Rinn, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Machleder E, Chenchik A, Li R, Siebert P. Reverse Transcriptase Template Switching: a SMART Approach for Full-Length cDNA Library Construction. Biotechniques. 2001;30(4):892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]