Abstract
The freeware Java tool PAJ, created to perform 3D point analysis, was tested in an independent laboratory setting. The input data consisted of images of the hippocampal perforant pathway from serial immunocytochemical localizations of the rat brain in multiple views at different resolutions. The low magnification set (2× objective) comprised the entire perforant pathway, while the high magnification set (100× objective) allowed the identification of individual fibers. A preliminary stereological study revealed a striking linear relationship between the fiber count at high magnification and the optical density at low magnification. PAJ enabled fast analysis for down-sampled data sets and a friendly interface with automated plot drawings. Noted strengths included the multi-platform support as well as the free availability of the source code, conducive to a broad user base and maximum flexibility for ad hoc requirements. PAJ has great potential to extend its usability by (a) improving its graphical user interface, (b) increasing its input size limit, (c) improving response time for large data sets, and (d) potentially being integrated with other Java graphical tools such as ImageJ.
Introduction
Image analysis is increasingly recognized as a crucial step in the quantitative characterization of neuroanatomical structures. New visualization techniques from the macroscopic to the microscopic levels create more and more graphical options about the type and content of information available about the nervous system. In this context, analytical tools can be particularly valuable if they can be applied to imaging modalities across multiple scales. The software presented in this issue (Condron 2008), PAJ (Point Analysis in Java), was originally developed to quantify synaptic distributions, but is in principle suitable for extension to other domains. As described in the related News Item, analyses available in PAJ include rank, density map, lacunarity, and Voronoi distributions. Additionally, a Monte Carlo (MC) option is included to further investigate spatial attributes of the analyzed data. The MC process generates 100 randomized data sets with summary statistics that are extracted from the input data. The resulting plots can be used to determine whether the input data is regularly distributed, randomly distributed, or clustered, based on their comparisons with the MC results.
We were eager to test this new tool on experimental data other than the exemples provided by the PAJ author. To this aim, we used a histological tract tracing preparation available in our laboratory. Tract tracing is a fundamental experimental technique that underlies much of the effort to construct connectional maps of the mammalian brain at multiple scales (Ascoli and Scorcioni 2006). In a recent research project, we explored the possibility of characterizing the perforant pathway projection that emanates from the entorhinal cortex and invades the dentate gyrus and Ammon’s horn regions. This projection pathway constitutes a major extrinsic input to the hippocampus, and provides both granule and pyramidal cells with direct connections from neocortical principal cells. The anatomical and topological organization of the perforant pathway may shape and constrain functional information processing, and is subject of intense investigation (van Groen 2001; van Groen et al. 2003; Witter 2007).
The preliminary stereological investigation of this histological material revealed a striking relation across imaging scales and resolutions. In particular, the optical density (grayscale level) recorded at a sufficently low magnification to image the entire perforant pathway was linearly correlated with the microscopic count of single fiber identified in the same regions of interest at high resolution. This observation provided the motivation to further quantify the correspondence between these two different anatomical scales. In particular, we were interested in determining what structural features would either change or remain consistent from the system to the cellular level. We thus welcomed the opportunity to use these data sets to road-test the new software PAJ. In this report we present a user’s account of this application to the analysis of our perforant pathway histological images at both low and high resolution.
Histological Procedures
Tract tracing experiments were performed following established procedures (e.g. Wouterlood and Jorritsma-Byham 1993, Reiner et al. 2000). Briefly, three adult male Sprague-Dawley rats (280 – 450 g) were anesthetized (ketamine, xylazine, and acepromozine; 0.08 ml/ 100 g) and the entorhinal cortex was injected stereotaxically with the anterograde tracer biotinylated dextran amine (BDA;10% in 0.01 M sodium phosphate buffer). Tracer was delivered over 5 minutes through a Hamilton microliter syringe (Cases 1 and 2, 1000 nL) or a nanoliter injector (Case 3, 600 nL) using coordinates from the atlas of Paxinos and Watson (6.3 mm caudal to Bregma, 4.0 mm lateral to the midline, and 8.2 mm ventral from the skull with the toothbar at 3.3 mm below the intra-aural plane). Following 3 to 7 days survival, the animals were deeply anesthetized with pentobarbital and perfused transcardially with paraformaldehyde (4% in 0.1 M sodium phosphate buffer). Each brain was removed, postfixed for 2 days, and serially sectioned in the sagittal plane at 50 (Case 3) or 75 µm (Cases 1 and 2) using a Leica Vibratome. After phosphate buffered saline rinsing (3×5 minutes), BDA was detected using the Vectastain Elite avidin biotin (ABC) kit (Vector Laboratories) following the manufacturer's procedure. Sections were mounted serially on gelatin-coated slides, air-dried, dehydrated in ethanol, cleared in xylenes, and coverslipped using Cytoseal 60 (Richard Allen Scientific).
Image acquisition and preliminary analysis
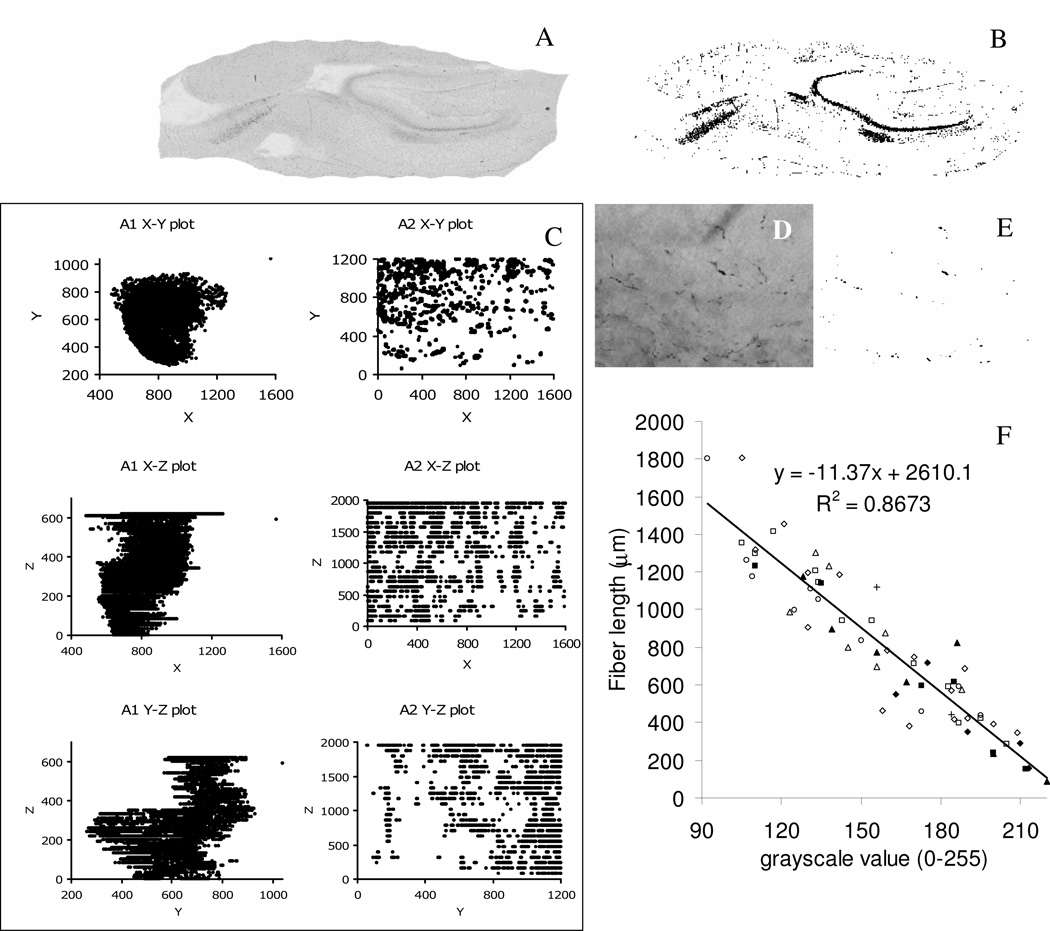
The first set of data, A1, was generated by digitizing case 2 at low resolution. All of the serial images of sections from this case were acquired with an Olympus Provis AX70 microscope and a 2× dry objective. Images were captured with a Microfire camera (Optronics; Goleta, CA) at the native resolution of 1600×1200 with the Neurolucida software package (MicroBrightfield; Burlington, VT) as a mosaics collection of multiple combined frames. Subsequently, they were aligned based on blood vessels. Non-hippocampal regions and any area that was not involved with the injection site were excluded from the analysis (Figure 1A). The second set, A2, was a high-resolution representation generated from a single section of case 2. This set was composed of 26 single frame images (encompassing the full thickness of the section) captured from selected regions of interest with a 100× oil immersed objective at a resolution of 1600×1200 (Figure 1C).
Figure 1.
(A) Grayscale single sagittal section of set A1 and its thresholded image (B); (C) grayscale single sagittal section of set A2 and its thresholded image (D); (E) stereological correlation between the 2× grayscale value and fiber length computed at 100× in the same region of interest, for 65 unique locations. Different symbols represent different sections (filled and empty symbols correspond to experiments 1 and 3, respectively). (F) 2D X-Y-Z plots (all units are in pixels) of down sampled sets A1 and A2 (XY corresponds to the sagittal plane for both sets).
To link the two levels of detail, a preliminary stereological fiber count was performed and plotted against grayscale levels. The study involved visually selecting a total of 65 locations on 8 sections from cases 1, 2, and 3. Each location was first captured using the 2× objective and its grayscale level recorded. On the same location, the 100× oil immersion objective was used for the stereological analysis with the Stereo Investigator software package (MicroBrightfield; Burlington, VT). The area of interest was highlighted in the high magnification image and selected in such a way that the stereology software only generated a single point of collection, which was recorded with a hemi-sphere of 9 µm radius. Once all fibers that intercepted the hemi-sphere were manually identified, Stereo Investigator computed the total amount of fibers contained in the area of interest. Each single data point in Figure 1E represents the grayscale value recorded at 2× versus the fiber count at 100× lens for the same individual location. The trend line over all 65 locations illustrates the high correlation value (R2=0.87).
Point analysis in Java
PAJ was downloaded from http://faculty.virginia.edu/PAJ and executed as instructed (Condron 2008). PAJ is capable of analyzing a maximum of 10,000 points distributed in 3-dimensional (3D) space. In order to convert the grayscale datasets in compatible format, we developed a Java application, named Threshold, which takes a series of image files, each representing a unique Z value, and outputs a set of 3D coordinates compatible with PAJ. In particular, this application lists all points below a given grayscale level (the source code is freely available from http://krasnow.gmu.edu/cn3/ by selecting the Threshold link under Data/Tools++). Grayscale thresholds were manually chosen to optimize the signal/noise ratio. The value of 190 used for set A1 generated 774,535 3D points (Figure 1B represents the thresholded image of panel A). Set A2 yielded 133,387 3D points with a grayscale threshold of 60 (Figure 1D represents the thresholded image of panel C). To reduce both sets below the 10,000 points limit a sampling rate of 1/78 was applied to set A1 while a rate of 1/14 was applied to set A2. The XY, XZ and YZ plots of both re-sampled sets are shown in Figure 1F. For both sets, the XY plane represents the sagittal plane. Set A1 highlights the macroscopic aspect of the overall path of the fibers, while set A2 highlights the microscopic aspect of the fiber path.
Two different platforms were used to test PAJ: a Windows XP pro computer P4 3.0GHz, 2GB RAM, 320GB disk, java 1.6.0_02-b06; and a Linux Fedora 8, Intel Quad Core 2.4 Ghz, 8GB RAM, 73GB disk, Java 1.7.0_b21. PAJ requires a Java runtime environment which can be freely downloaded from http://java.sun.com. Having previously installed the Java environment, only a small number of user friendly steps were required to perform the point analysis, namely (a) starting PAJ, (b) cutting and pasting the lists of 3D coordinates, and (c) clicking the "Run All" button. The analysis took about 6 minutes (355 seconds) on the Windows computer and slightly more than 5 minutes (312 seconds) in Linux. The Monte Carlo analysis took approximately 13 hours under Windows and just below 6 hours on the Linux machine.
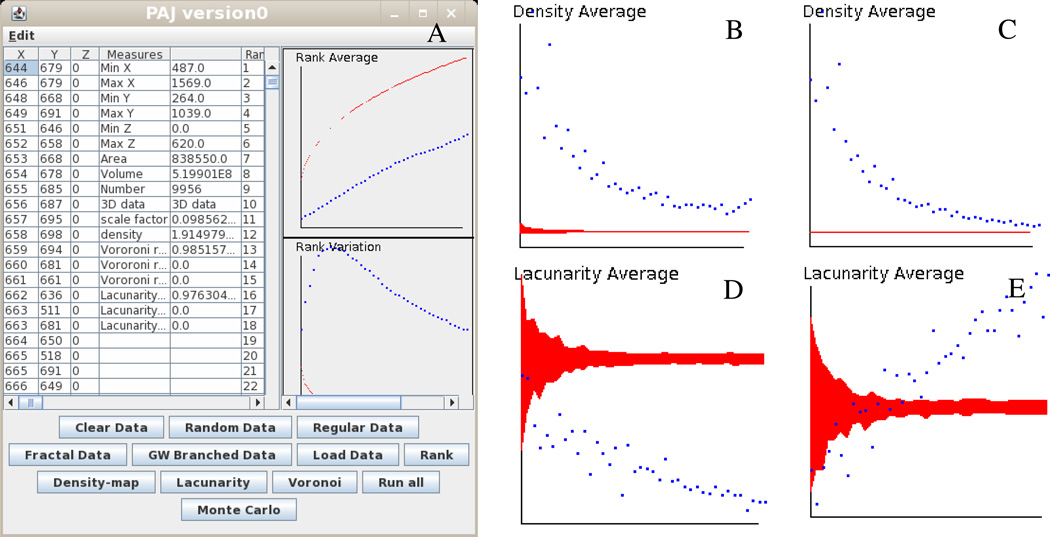
A screenshot of the PAJ graphical user interface (GUI) loaded with set A2 is shown in Figure 2A. Figures 2B and 2C represent Voronoi distributions for sets A1 and A2, respectively. Both distributions and associated MC simulations present similar patterns. In fact, in both cases the dotted lines corresponding to the analysis results are above the MC simulations, represented by dark region or thin black line. This indicates that sets A1 and A2 are regularly distributed with respect to average density. Figures 2D and 2E represent Lacunarity Average for sets A1 and A2, respectively. This scenario illustrates different behaviors for the two sets. Set A1 is clustered since the dotted line is below the MC simulation area, while set A2 starts as clustered and then quickly moves above the MC area becoming regularly distributed. All plots of Figure 2 are unit-less and were taken directly from the PAJ GUI.
Figure 2.
(A) PAJ graphical interface; (B) Density Average for set A1; (C) Density Average for set A2; (D) Lacunarity Average for set A1; (E) Lacunarity Average for set A2. Dotted lines correspond to the measured quantity; gray areas are associated with the Monte Carlo simulations.
As preliminary finding, these results suggest that some structural features of the perforant pathway are indeed conserved across scales. In particular, regular distribution of density averages at both system and cellular level present the same trend, as shown in Figure 2B and 2C. In contrast, other aspects clearly differ between the two magnifications, such as lacunarity averages which show opposite trends in Figure 2D and 2E. However, it is important to explicitly state that these results are obtained from heavily down sampled data. The analysis was performed at the maximum capacity of PAJ (10,000 points in both cases), and yet included only 1.3% and 7.1% of sets A1 and A2, respectively. Even if the sampling process is deterministically reproducible, the analyzed collection of points only represents a coarse caricature of the original images. We therefore caution potential users that for applications to histological preparations the 10,000 point limit may constitute a serious filter to the kinds of scientific question that can be addressed.
As the PAJ author reported in the ReadMe file, the graphical output behaved erratically. Plots are visualized only after resizing the window. In addition, any window resizing or handlebar movement triggers an automated inversion of the y-axis on all plots. This action results in two orthogonal views; the correct one can only be identified by utilizing the provided Excel file, named Excel_analysis_of_PAJ.xls. There is an opportunity to improve the graphical layout in terms of both optimal space usage and manual adjustments so that users could adapt the layout to their preferences (as an example, Figure 2A represents an optimally edited view of PAJ). PAJ allows “cut and paste” operations from its outputs, an essential function to generate the correct plots with the provided Excel file. We recommend adding column labels, which would allow users to easily manipulate data within their preferred statistical/graphical software. The Excel file generates corrected plots with appropriate units, except for the Lacunarity Average plot (mislabeled “Lacunarity Variation”).
PAJ allows a user-friendly point analysis of 3D data. Its strength relies on the ability to generate a multitude of plots that summarize the 3D characteristics of the input data. Until now, no other tools have provided this amount of functionality in a single package that is both freely available and can be executed on multiple platforms. In addition, this tool allows ad hoc modifications, since its source code is available from the website. For example, it was straightforward to increase the 10,000 point limit to 140,000, which allowed us to load the whole set A2 in PAJ. In this extended case, however, the data analysis without MC required a significantly longer time to complete (more than 20 hours in Windows). PAJ functionality could be further improved by optimizing the graphical user interface behavior/layout, by extending the 10,000 points limit, and by potentially including it as a plug-in for other scientific packages, such as ImageJ (Abramoff, 2004). PAJ integration within the ImageJ package would provide an ideal opportunity to directly analyze image stacks with a threshold parameter that the user can specify. Moreover, the availability of PAJ source code could allow either the author or other users to extend and improve its functionality. For example, it could be possible to implement a multi-thread version of PAJ which could speed up the MC simulation significantly on multi-core CPU computers.
Acknowledgements
This work was supported by NIH R01 grants NS39600 and NS24288.
Footnotes
Information Sharing Statement
All software code described in this report is freely available to the general public. PAJ can be downloaded from http://faculty.virginia.edu/PAJ, while Threshold can be downloaded from http://krasnow.gmu.edu/cn3/ (select the Threshold link under Data/Tools++).
References
- Ascoli GA, Scorcioni R. Neuron and network modeling. In: Zaborszky L, Wouterlood Lanciego J, editors. Tract Tracing Methods. 3rd ed. New York NY: Springer; 2006. pp. 604–630. [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;7:36–42. [Google Scholar]
- Condron B. A freeware Java tool for the spatial point analysis of neuronal structures. Neuroinformatics. 2008 doi: 10.1007/s12021-008-9008-z. (in press) [DOI] [PubMed] [Google Scholar]
- Reiner A, Veenman CL, Medina L, Jiao Y, Del Mar N, Honig MG. Pathway tracing using biotinylated dextran amines. Journal of Neuroscience Methods. 2000;103:23–37. doi: 10.1016/s0165-0270(00)00293-4. [DOI] [PubMed] [Google Scholar]
- Van Groen T, Miettinen P, Kadish I. The entorhinal cortex of the mouse: organization of the projection to the hippocampal formation. Hippocampus. 2003;13:8133–8149. doi: 10.1002/hipo.10037. [DOI] [PubMed] [Google Scholar]
- Van Groen T. Entorhinal cortex of the mouse: cytoarchitectonical organization. Hippocampus. 2001;11:397–407. doi: 10.1002/hipo.1054. [DOI] [PubMed] [Google Scholar]
- Witter MP. The perforant path: projections from the entorhinal cortex to the dentate gyrus. Progress in Brain Research. 2007;163:43–61. doi: 10.1016/S0079-6123(07)63003-9. [DOI] [PubMed] [Google Scholar]
- Wouterlood FG, Jorritsma-Byham B. The anterograde neuroanatomical tracer biotinylated dextran-amine: comparison with the tracer Phaseolus vulgaris-leucoagglutinin in preparations for electron microscopy. Journal of Neuroscience Methods. 1993;48:75–87. doi: 10.1016/s0165-0270(05)80009-3. [DOI] [PubMed] [Google Scholar]