Abstract
On average, every fifth residue in secretory proteins carries either a positive or a negative charge. In a bacterium such as Escherichia coli, charged residues are exposed to an electric field as they transit through the inner membrane, which should generate a fluctuating electric force on a translocating nascent chain. Here, we have used translational arrest peptides as in vivo force sensors to measure this electric force during co-translational chain translocation through the SecYEG translocon. We find that charged residues experience a biphasic electric force as they move across the membrane, including an early component with a maximum when they are 47-49 residues away from the ribosomal P-site, followed by a more slowly varying component. The early component is generated by the transmembrane electric potential while the second may reflect interactions between charged residues and the periplasmic membrane surface.
Keywords: membrane protein; Sec translocon; SecM, translational arrest; charged amino acids; membrane potential
Introduction
In both prokaryotic and eukaryotic cells, proteins are translocated across lipid membranes with the aid of protein-conducting channels (translocons). In their natural context, translocons such as the bacterial SecYEG complex do not conduct ions, so as not to compromise biologically important ion gradients 1, but nevertheless act as non-selective ion channels in the sense that they conduct charged residues in the proteins being transported. In membranes that support a membrane potential, strong electric forces should be expected to act on charged residues during chain translocation, reflecting the local electric potential in the translocon channel. We reasoned that, by measuring such forces at defined positions within the translocon channel we might be able to chart the electric field experienced by the nascent chain, much as electrophysiological studies have been instrumental in developing a detailed picture of the electric environment in classical ion channels, and thereby be able to clarify how membrane electrostatics contribute to the dynamics of chain translocation.
Results
We have recently shown that translational arrest peptides (APs) can be used as transplantable in vivo force sensors to measure forces acting on nascent polypeptide chains during co-translational processes such as membrane-protein biogenesis 2-4. APs are short stretches of polypeptide that can induce ribosomal stalling when translated 5. The family of bacterial SecM APs is particularly interesting in this regard, as the degree of stalling induced by these APs depends on the tension in the nascent polypeptide chain at the precise point when the most C-terminal codon of the AP is in the ribosomal A-site: the higher the tension, the lower the degree of stalling 3,6,7.
To measure forces acting on charged residues during co-translational chain translocation through the E. coli SecYEG translocon, we used an engineered version of the inner membrane protein LepB as a reporter, Fig. 1a. The construct has two transmembrane segments near the N-terminus (TM1, TM2) that serve to target the ribosome-nascent chain complex to SecYEG, and a large C-terminal periplasmic domain that contains a stretch of n charged or uncharged test residues (X) placed L residues upstream of a SecM AP (see Supplementary Fig. 1 and Supplementary Table 1 for sequences of all constructs). The SecM AP is followed by a 23-residue C-terminal tail to ensure that arrested and full-length LepB chains can be easily separated by SDS-PAGE.
Figure 1.
Negatively charged residues experience an electric pulling force during passage through the SecYEG translocon. (a) Design of LepB constructs. A schematic picture of a translating ribosome bound to the SecYEG translocon and the sequence surrounding the nX stretch are shown below. Two engineered Asn-Ser-Thr acceptor sites for N-linked glycosylation (G1, G2) are shown in red; these are not present in the constructs expressed in E. coli but are used in the experiment reported in panel d. (b) Pulse labeling of 5D constructs. “AP” and “mut” indicates, respectively, constructs with a functional SecM(Ms) AP and a non-functional mutant AP with the last Pro residue mutated to Ala. (c) Fraction of full-length protein (fFL) plotted as a function of L+n for constructs with nX = 5N (yellow), 5Q (green), 5K (blue), and 5D (purple) and the SecM(Ms) AP. (d) Determination of the location of the 5D segment in the [5D, L+n = 48] construct by glycosylation mapping. The right panel compares the results obtained for the [5D, L+n = 48] construct with previously determined glycosylation distances (see main text). (e) fFL measured for a 5D construct with L+n = 48 residues (blue bars), and for a construct with L = 39 residues in which the nX stretch is replaced by a hydrophobic transmembrane segment of composition [6L,13A] (gray bars), at increasing concentrations of indole in the growth medium. The amount of non-processed pro-OmpA protein is also shown (red bars). Standard errors (s.e.m.) are indicated.
The basic experiment is simple: a plasmid encoding a particular version of the LepB construct (characterized by the number of test residues n, the identity of the test residues X, the number of linker residues L between the nX stretch and the last residue of the AP, and the particular SecM AP used) is transformed into E. coli, expression of the construct is induced, cells are briefly labeled with [35S]-Met, lysed, subjected to immunoprecipitation with a LepB antiserum, and analyzed by SDS-PAGE, Fig. 1b (see also Supplementary Data Set 1). Constructs in which there is a strong external pulling force F on the nascent chain at the point when the ribosome reaches the last residue in the AP will yield mostly full-length protein, whereas if F is low there will be efficient translational arrest at the AP, yielding mostly the shorter, arrested form of the protein. The fraction of full-length protein, fFL, can therefore serve as a proxy for F3, and force profiles representing the instantaneous pulling force acting on the nascent chain during translocation through the SecYEG channel can be derived by measuring fFL for a series of constructs with different values of L. As force-sensors, we used the eight-residue AP HAPIRGSP from Mannheimia succiniciproducens (Ms) SecM8 and a mutated version, HPPIRGSP (Ms-Sup1; substitution underlined), that requires a stronger tension in the nascent chain to reach a given fFL-value 3,8,9.
Charged residues experience an electric pulling force
Force profiles obtained with the SecM(Ms) AP and nX = 5N, 5Q, 5K, and 5D are shown in Fig. 1c (all experimental data is summarized in Supplementary Table 1). For 5N, 5Q, and 5K, fFL is low for L+n < 50 residues, and then increases slowly. The profile for 5D is dramatically different, with a very rapid increase from L+n = 45 residues to a first peak at L+n = 47–49 residues, followed by a second peak at L+n = 53 residues and a slow decline. At L+n > 60 residues, all profiles merge. That the strong pulling force is seen only with the negatively charged 5D test segment suggests that it is electric in nature and is generated by the electrical component (ΔΨ) of the proton-motive force (pmf). We do not understand the reason for the relatively high fFL values seen for all constructs at L+n > 60 residues, but it is clearly not related to the charge characteristics of the nX stretch.
Unfortunately, there is no good method available to precisely map the location of the 5D stretch in the bacterial SecYEG channel at different linker lengths; instead, we applied the method of glycosylation mapping 10-12 to the [5D, L+n = 48] construct translated in vitro in a rabbit reticulocyte lysate in the presence of dog pancreas rough microsomes derived from the endoplasmic reticulum (ER). In this case there is no membrane potential that can act on charged residues in the nascent chain, but the overall structure of the ribosome-translocon complex and the geometry and relevant distances characterizing the nascent chain ribosome-translocon conduit are highly conserved between bacteria and mammals 13,14.
The method rests on the observation that the Asn residue in an Asn-X-(Thr, Ser) acceptor site for N-linked glycosylation (site G2 in Fig. 1a) in an extended nascent polypeptide chain must be ~65 residues away from the ribosomal P-site (or ~15 residues away from the N-terminal end of a membrane-integrated transmembrane segment) to reach the lumenal active site of the oligosaccharyl transferase in the ER and become half-maximally glycosylated10-12. Two glycosylation acceptor sites (G1, G2) were engineered into the [5D, L+n = 48] construct, as shown in Fig.1a. mRNA truncated at the last Pro codon in the AP was translated in vitro in the presence of dog pancreas rough microsomes and [35S] Met to generate a stalled ribosome-nascent chain complex that spans the membrane as indicated in Fig. 1a, proteins were separated by SDS-PAGE, and the glycosylation status of the G1 and G2 sites were determined from the intensities of the bands representing singly and doubly glycosylated product. The G1 site was kept in a fixed location and is always glycosylated, while the G2 site was moved stepwise relative to the 5D stretch in order to determine the location at which it is half-maximally glycosylated.
As shown in Fig. 1d, we found that the N-terminal end of the 5D stretch in the [5D, L+n = 48] construct is located 28 residues away from the G2-site Asn and therefore that the total chain length between the P-site and the Asn is 28+48 = 76 residues at half-maximal glycosylation, similar to results obtained previously for a highly charged 14K stretch engineered into a secretory mammalian protein 15. This places the 5D stretch in or near the cytoplasmic entrance to the translocon channel, and further implies that the linker region is not fully extended at this point during translocation, and may be in a looped-out conformation as seen in a recent cryo-EM structure 16. The simplest interpretation, also suggested for the 14K stretch 15, is that the ring of hydrophobic residues that defines the narrowest part of the translocon channel 17 presents a barrier for the translocation of charged residues that is only overcome as the nascent chain keeps elongating. Our working hypothesis is therefore that, in E. coli, the 5D stretch is held transiently on the cytoplasmic side of the pore ring and then translocates en bloc when the nascent chain length reaches L+n ≈ 46 residues. To probe the pmf-dependence of fFL for the 5D construct, we grew cells in increasing concentrations of the uncoupler indole, a treatment that has been shown to reduce the pmf from its unperturbed value to essentially zero over a range of 1–5 mM indole (and to block cell division at > 3mM indole) 18. At a fixed L+n = 48 residues, fFL was reduced from its maximal value to background levels over the same range of indole concentrations, Fig. 1e. Translocation of the SecA- and SecYEG-dependent outer membrane protein OmpA was only adversely affected when the indole concentration was > 3 mM, as assayed by the accumulation of the uncleaved precursor form (pro-OmpA). As a further control we tested the effect of indole on a LepB construct in which the nX segment is replaced by a hydrophobic transmembrane segment of composition [6L,13A] that we have previously shown generates a strong pulling force at L = 39 residues, i.e., at the point when it is expected to integrate into the inner membrane 3. In this case, there was no effect of indole on fFL for concentrations ≤ 3 mM. We conclude that the peak in the 5D force profile at L+n = 47–49 residues reflects an electric interaction between the negatively charged residues in the 5D stretch and the transmembrane potential across the SecYEG channel.
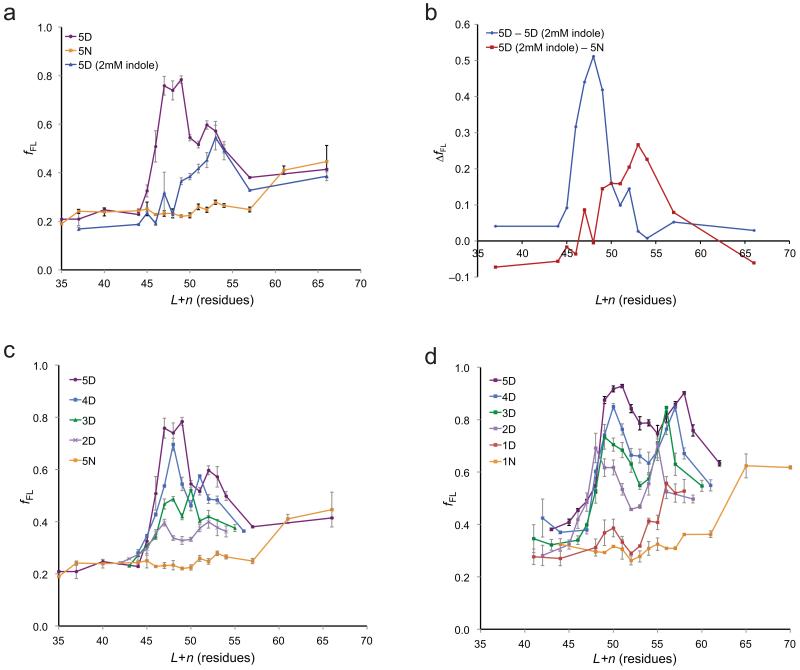
The force acting on the 5D test segment at L+n = 47–49 residues is strong enough to cause fFL to saturate at ~1.0, Fig. 1c. In order to increase the sensitivity of the assay in this region, we obtained force profiles for the 5D and 5N series of LepB constructs using the stronger SecM(Ms-Sup1) AP, Fig. 2a. The overall shapes of the two force profiles are similar to those obtained with the SecM(Ms) AP, but the peak at L+n = 47–49 residues is now better defined and clearly distinct from the second peak at L+n = 52–53 residues. Also shown in Fig. 2a is a 5D-SecM(Ms-Sup1) force profile obtained in the presence of 2 mM indole, i.e., at a concentration where there is no effect on pro-OmpA processing but a strong effect on fFL for the 5D-SecM(Ms) construct (Fig. 1e). Remarkably, the peak at L+n = 47–49 residues is completely obliterated by indole, whereas the peak at L+n = 52–53 residues is hardly affected. This is even more clearly seen in Fig. 2b, where we have isolated the effect indole by subtracting the force profile of the 5D-SecM(Ms-Sup1) constructs obtained in the presence of indole from that obtained in its absence, and have subtracted the 5N-SecM(Ms-Sup1) force profile from that of 5D-SecM(Ms-Sup1) obtained in the presence of indole. The pmf-dependent peak is strong and nearly symmetric around L+n = 48 residues, whereas the pmf-independent peak is of smaller magnitude and extends over the range L+n ≈ 50–60 residues.
Figure 2.
The electric force has both pmf-dependent and pmf-independent components and scales with the number of negatively charged residues in the nD stretch. (a) fFL plotted as a function of L+n for 5D constructs containing the SecM(Ms-Sup1) AP, measured in the absence (purple) and presence (blue) of 2 mM indole in the growth medium. fFL for 5N constructs obtained in the absence of indole is also shown (yellow). (b) Difference plots of the fFL profiles in panel a, representing the pmf-dependent component (blue) obtained by subtracting the 5D (+indole) profile from the 5D (-indole) profile and the pmf-independent component (red) obtained by subtracting the 5N profile from the 5D (+indole) profile. (c) fFL plotted as a function of L+n for nD and 5N constructs containing the SecM(Ms-Sup1) AP. (d) fFL plotted as a function of L+n for nD and 5N constructs containing the SecM(Ms-Sup1) AP and with 10-residue long (SG)5 segments flanking the nX stretch. Standard errors (s.e.m.) are indicated.
How does the force profile vary with the number of Asp residues in the nD stretch? Results obtained with the SecM(Ms-Sup1) AP for n = 2–5 are shown in Fig. 2c, and for n = 10 in Supplementary Fig. 2a. The magnitudes of both the L+n ≈ 48 and L+n ≈ 53 peaks scale approximately with n, as does the initial slope of the pmf-dependent L+n ≈ 48 peak, Supplementary Fig. 3. As for the 5D stretch, 2 mM indole totally suppresses the first peak in the 10D force profile but reduces the second peak only marginally, Supplementary Fig. 2b.
In an attempt to further isolate the effect on fFL of the nX stretch from influences of the surrounding sequence, we measured force profiles for nD stretches (n = 1–5) and a control 1N stretch embedded in an uncharged (SG)5-nX-(SG)5 segment, using the weaker SecM(Ms) AP, Fig. 2d. Overall, the force profiles are similar to those in Fig. 2c, with a clear two-peak appearance. Again, the first peak is pmf-dependent and the second is not, Supplementary Fig. 2c, d. The profiles are shifted by about 2 residues towards higher L+n values, implying that the (SG)5 linker attains a somewhat more compact conformation or makes a longer excursion in the space between the ribosome and the translocon 16 than the linker used in Fig. 2c.
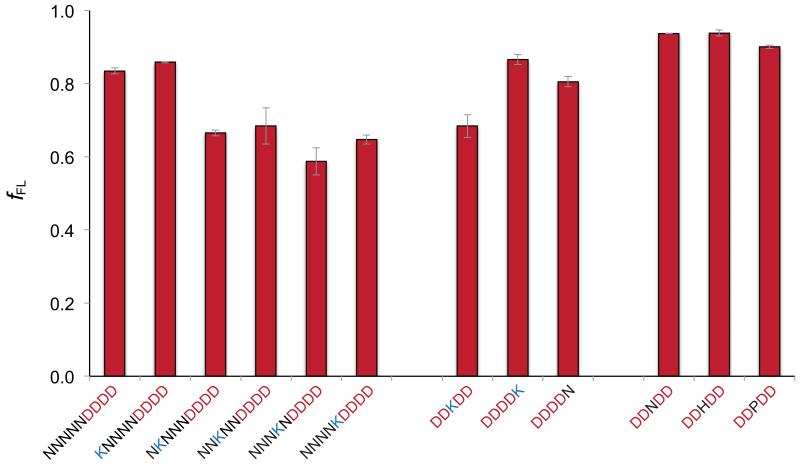
Finally, we analyzed the effect on fFL of a single positively charged Lys residue placed in different positions relative to a 4D stretch. For this experiment, we used the SecM(Ms) AP and fixed L at 43 residues. As seen in Fig. 3, a single Lys residue placed five residues N-terminal to the 4D stretch (thus entering the translocon ahead of the negatively charged residues) has no effect on fFL. In contrast, when the Lys residue is four, three, two, or one residue upstream of the 4D stretch, fFL is reduced from ~0.85 to ~0.60. Placing the Lys residue in the middle of the 4D stretch reduces fFL to ~0.70, but when located just C-terminal to the 4D stretch, the Lys residue has no effect on fFL. Placing either Asn, His, or Pro in the middle of the 4D stretch also has no effect on fFL. This is the pattern expected if the long, positively charged Lys side chain is pushed back by the membrane potential towards the 4D stretch as it enters the SecYEG channel. That the neutralizing effect is seen already when the Lys residue is four residues upstream of the 4D stretch indicates that the NKNNN stretch has a compact conformation as it passes through the SecYEG channel, in contrast to the following 4D stretch that likely is in an extended conformation, as indicated by the lack of an effect on fFL by the helix-breaking Pro mutation.
Figure 3.
A positively charged lysine residue placed N-terminal to a 4D stretch reduces the electric pulling force. fFL values are shown for 4D-SecM(Ms) constructs with Asn, Lys, His, and Pro residues included in the indicated positions. In all cases, the number of residues between the indicated stretch of residues and the P-site is L = 43 residues. Standard errors (s.e.m.) are indicated.
A physical model of chain translocation
As a charged residue translocates through the ribosome and the translocon channel, it will minimally be subjected to interactions with the ribosomal tunnel (which is at a negative potential of −10 to −20 mV relative to the cytoplasm 19), with the transmembrane electric potential ΔΨ, with charged and polar residues in the translocon, and with the membrane surface potential Vs. It will also need to pass through the relatively non-polar interior of the translocon channel, and especially the hydrophobic “pore ring”. Since our constructs were specifically designed to probe electric forces acting on the nascent chain during its passage through the translocon, we asked whether our results could be explained by a physical model including only ΔΨ and Vs (see Supplementary Note); indeed, as shown in Fig. 4b and Supplementary Fig. 4, the experimental fFL profiles can be reproduced with reasonable accuracy by the simple model depicted in Fig. 4a, with parameters optimized over the experimental data. The first peak (at L+n ≈ 50 residues) in the nD fFL profiles is generated by the ΔΨ, and the second peak (at L+n ≈ 55 residues) is generated by the surface potential on the periplasmic side of the membrane. The model also correctly reproduces the increasingly steep rise in the fFL profiles at L+n ≈ 47 for increasing n values, but fails in the region L+n > 60 residues since we did not attempt to model the non-electrostatic forces apparently acting in this region (c.f. above). Additional work probing both electrostatic and hydrophobic interactions between the nascent chain and the ribosome–translocon conduit will be required to substantiate and refine the model to a point where it can accurately predict force profiles for arbitrary nascent chain sequences.
Figure 4.
Physical model for translocation of charged residues (see Supplementary Note for details). (a) The total electric potential (black) experienced by a charged residue has two components: the transmembrane potential ΔΨ (blue) and the membrane surface potential Vs (red). The electric field strength (which is proportional to the force F acting on a point charge) is shown as a dashed line. The nascent polypeptide chain is modeled using the wormlike chain model. Approximate lengths of the ribosomal tunnel (yellow) and the translocon channel (gray) are indicated. (b) Approximate values for the 13 parameters describing the model were obtained from the literature, and a local parameter optimization around 6 of these values was done to simultaneously fit the experimental fFL profiles for the (SG)5-nX-(SG)5-SecM(Ms) constructs and the nX-SecM(Ms-Sup1) constructs (see Supplementary Note). Results for the (SG)5-nX-(SG)5 set are shown (experimental profiles in blue, calculated in red); results for the other data sets are in Supplementary Fig. 4. (c) Experimental fFL values plotted vs. the predicted pulling force F for the (SG)5-nX-(SG)5-SecM(Ms) constructs. The solid line is a sigmoidal fit fFL = (1+A e−F/σ)−1, with A = 5.2 and σ = 7.0 pN. (d) As in c, but for the SecM(Ms-Sup1) AP. The solid line is a sigmoidal fit fFL = (1+A e−F/σ)−1, with A = 5.2 and σ = 11.8 pN.
The relation between the total electric force predicted by the model and the experimentally observed fFL values for the (SG)5-nX-(SG)5 constructs with the SecM(Ms) AP, Fig. 4c, allows us to estimate forces (in pN) from measured fFL values, not only for the nX constructs analyzed here, but in general. A similar relation holds for the stronger SecM(Ms-Sup1) AP, Fig. 4d. The typical forces from the model (up to ~40 pN for the 5D stretch, Fig. 4d) compare favorably with the forces previously measured for, e.g., unfolding of titin domains (~100 pN) 20, the force exerted by a viral packaging motor (~50 pN) 21, or the force generated by the ribosome on the mRNA during the translocation step (~13 pN) 22.
Discussion
Our findings highlight a hitherto unappreciated aspect of membrane translocation dynamics: a strong electric force acting on the nascent polypeptide chain during its passage through the SecYEG translocon. Although our studies are confined to co-translational translocation, there is no reason why post-translationally translocating chains should not experience the same kind of force. Fine-tuning of chain translocation dynamics by charged residues may have implications for, e.g., co-translational protein folding, membrane protein biogenesis, and regulatory processes involving translational arrest peptides 23.
Online Methods
Enzymes and chemicals
All enzymes were obtained from Thermo Scientific (Waltham, MA, USA) and New England Biolabs (Ipswich, MA, USA). Oligonucleotides were from Eurofins MWG Operon (Ebersberg, Germany). [35S]-Met was from PerkinElmer (Waltham, MA, USA). All other reagents were from Sigma-Aldrich (St. Louis, MO, USA).
DNA manipulations
All constructs were generated from the previously described pING1 plasmid carrying the lepB gene containing a [6L,13A] H segment insert and the eight-residue SecM arrest peptide, HAPIRGSP, from M. succiniciproducens under the control of an arabinose-inducible promoter 3. The plasmid was digested with SpeI and KpnI to release the [6L,13A] segment, and oligonucleotides corresponding to the various test residues (GG-nX-GG) were ligated in its place. Shorter linker lengths, L, between the test segments and the arrest peptide were generated by PCR using forward and reverse primers complementary to regions denoted by the right- or left-facing arrows in Supplementary Fig. 1a. In order to generate constructs with the stronger HPPIRGSP and the non-functional HAPIRGSA arrest peptides, QuikChange site-directed mutagenesis was performed to convert the Ala to Pro and Pro to Ala, respectively (underlined).
Pulse-labeling analysis
E. coli MC1061 cells bearing the respective plasmids were grown overnight at 37°C in M9 minimal media supplemented with 19 natural amino acids (1 μg ml−1; no Met), 100 μg ml−1 thiamine, 0.1 mM CaCl2, 2 mM MgSO4, 0.4% (w/v) fructose, and 100 μg ml−1 ampicillin. Cultures were back-diluted to OD600 = 0.1 and grown to OD600 = 0.35. LepB expression was induced with 0.2% (w/v) arabinose for 5 min. For cultures which were treated with indole, appropriate volumes of 1 M indole (in EtOH) were added to the cultures to final concentrations of 1, 2, 3, 4, and 5 mM indole 1 min before pulse-labeling. EtOH was added to 0.5% (v/v) for mock-treated samples. Cells were then pulsed-labeled with [35S]-Met for 2 min at 37°C before being added to an equal volume of 20% trichloroacetic acid (TCA). The samples were incubated on ice for 30 min and spun for 5 min at 20,800 g at 4°C. The pellet was washed with cold acetone, spun again for 5 min at 4°C, and subsequently solubilized in Tris-SDS solution (10 mM Tris-Cl, pH 7.5, 2% SDS) at 95°C for 10 min. The samples were spun for 5 min at room temperature and the lysate was used for immunoprecipitation using LepB and OmpA antisera. The samples were resolved by SDS-PAGE and the gel visualized using a Fuji FLA-3000 phosphoimager and the ImageGauge V4.23 software. Quantification of protein bands was performed using the QtiPlot 0.9.7.10 software. All experiments were repeated three times using independent culture incubations, and standard errors (s.e.m.) were calculated. Original images of autoradiographs used in this study can be found in Supplementary Data Set 1.
In vitro transcription and translation
In vitro transcription was performed with SP6 RNA polymerase according to the manufacturer’s protocol (Promega) using PCR products as templates for the generation of truncated nascent chains. RNA obtained was purified using RNeasy Mini Kit (Qiagen). Translations were performed in a rabbit reticulocyte lysate system as described (Promega) for 15 min at 30 °C in the presence of 0.5 μl of dog pancreas rough microsomes and 1 μl of [35S]-Met (5 μCi). The reaction was stopped by the addition of an equal volume of sample buffer and treated with RNase A (200 μg ml−1) for 15 min at 30 °C before the samples were resolved by SDS-PAGE. All experiments were repeated three times using independent incubations, and standard errors (s.e.m.) were calculated.
Supplementary Material
Acknowledgements
This work was supported by grants from the Swedish Foundation for Strategic Research, the European Research Council (ERC-2008-AdG 232648), the Swedish Cancer Foundation, the Swedish Research Council, and the Knut and Alice Wallenberg Foundation to GvH, and by a grant from the Wenner-Gren Foundation to ML.
References
- 1.Park E, Rapoport TA. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature. 2011;473:239–242. doi: 10.1038/nature10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cymer F, von Heijne G. Cotranslational folding of membrane proteins probed by arrest-peptide-mediated force measurements. Proc Natl Acad Sci U S A. 2013;110:14640–14645. doi: 10.1073/pnas.1306787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ismail N, Hedman R, Schiller N, von Heijne G. A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nature Struct Molec Biol. 2012;19:1018–1022. doi: 10.1038/nsmb.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cymer F, Ismail N, von Heijne G. Weak pulling forces exerted on Nin-orientated transmembrane segments during co-translational insertion into the inner membrane of Escherichia coli. FEBS Lett. 2014;588:1930–1934. doi: 10.1016/j.febslet.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 5.Ito K, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Comm. 2010;393:1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butkus ME, Prundeanu LB, Oliver DB. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J Bacteriol. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gumbart J, Schreiner E, Wilson DN, Beckmann R, Schulten K. Mechanisms of SecM-mediated stalling in the ribosome. Biophys J. 2012;103:331–341. doi: 10.1016/j.bpj.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap MN, Bernstein HD. The translational regulatory function of SecM requires the precise timing of membrane targeting. Mol Microbiol. 2011;81:540–553. doi: 10.1111/j.1365-2958.2011.07713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson I, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 11.Nilsson I, Whitley P, von Heijne G. The C-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J Cell Biol. 1994;126:1127–1132. doi: 10.1083/jcb.126.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson A, Armulik A, Nilsson I, von Heijne G, Johansson S. Determination of N- and C-terminal borders of the transmembrane domain of integrin subunits. J Biol Chem. 2004;279:21200–21205. doi: 10.1074/jbc.M400771200. [DOI] [PubMed] [Google Scholar]
- 13.Frauenfeld J, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita H, Yamagishi M, Kida Y, Sakaguchi M. Positive charges on the translocating polypeptide chain arrest movement through the translocon. J Cell Sci. 2011;124:4184–4193. doi: 10.1242/jcs.086850. [DOI] [PubMed] [Google Scholar]
- 16.Park E, et al. Structure of the SecY channel during initiation of protein translocation. Nature. 2014;506:102–106. doi: 10.1038/nature12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 18.Chimerel C, Field CM, Pinero-Fernandez S, Keyser UF, Summers DK. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim Biophys Acta. 2012;1818:1590–1594. doi: 10.1016/j.bbamem.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 20.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 21.Smith DE, et al. The bacteriophage ⍰29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, et al. Direct measurement of the mechanical work during translocation by the ribosome. eLife. 2014;3:e03406. doi: 10.7554/eLife.03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamori K, Chiba S, Ito K. Identification of a SecM segment required for export-coupled release from elongation arrest. FEBS Lett. 2014;588:3098–3103. doi: 10.1016/j.febslet.2014.06.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.