Abstract
Na+/H+ exchanger isoform 1 (NHE1) has been reported to be hyperactive in 4.1R-null erythrocytes (Rivera A et al., Am J Physiol Cell Physiol, 291, C880–886, 2006), supporting a functional interaction between NHE1 and 4.1R. Here we demonstrate that 4.1R binds directly to the cytoplasmic domain of NHE1 (NHE1cd) through the interaction of an EED motif in 4.1R FERM (Four.one/Ezrin/Radixin/Moesin) domain with two clusters of basic amino acids, K519R and R556FNKKYVKK, in NHE1cd, previously shown to mediate phosphatidylinositol 4,5-bisphosphate (PIP2) binding (Aharonovitz et al. J. Cell. Biol., 150, 213–224, 2000). The affinity of this interaction (Kd=100–200nM) is reduced in hypertonic and acidic conditions, demonstrating that this interaction is of electrostatic nature. The binding affinity is also reduced upon binding of Ca2+-saturated calmodulin (Ca2+/CaM) to 4.1R FERM domain. We propose that 4.1R regulates NHE1 activity through a direct protein-protein interaction that can be modulated by intracellular pH and Na+ and Ca2+ concentrations.
Keywords: protein 4.1R, NHE1, sodium-proton exchange, cytoskeleton, calmodulin
INTRODUCTION
Na+/H+ exchanger (NHE) 1 is the most ubiquitously expressed member of the NHE family and is therefore often considered the ”housekeeping” NHE. NHE1 participates in the regulation of intracellular volume and pH in all cell types, including erythrocytes [1]. It is also the major isoform expressed in mammalian erythrocytes [1–6]. Highly relevant to this study, NHE1 is the only resident plasma membrane NHE isoform in erythrocytes. There is now a large body of evidence for the membrane skeleton being involved in regulating the basal rate of NHE1-mediated ion transport and in the transmission of signals that modulate NHE1 activity in response to physical stimuli [1–7]. Additionally, a recent study showed that β-adducin (Add2) null mouse erythrocytes, which adopt a spherocytic shape, lack α-adducin and SLC9A1/NHE1 [8]. Protein 4.1R-null mouse erythrocytes exhibit a dramatic NHE1 phenotype characterized by cell dehydration and high intracellular Na+ resulting from hyperactivity of NHE1 [9,10]. This latter observation demonstrates unequivocally a role for 4.1R80 in modulating NHE1 activity. The aim of the present study is to extend these findings by performing a detailed characterization of 4.1R80-NHE1 interaction and of its regulation.
Protein 4.1R is a key membrane skeletal protein in erythrocytes. The erythrocyte isoform, 4.1R80, is comprised of four major chymotryptic domains, a N-terminal 30kDa domain referred to as “FERM” domain, a 16kDa domain, a 10kDa domain and a C-terminal 24kDa domain [11,12]. The FERM domain mediates interaction with various transmembrane proteins, including the anion exchanger band 3 [13–15], glycophorin C (GPC) [16], CD44 [17], and with the membrane-associated protein, p55 [18,19]. The 10kDa domain of 4.1R80 binds to spectrin and actin filaments [11,12]. Through these multiple and dynamic interactions, 4.1R80 participates in the maintenance of the mechanical stability of human erythrocytes.
Calmodulin (CaM), in association with Ca2+, regulates 4.1R80 binding to membrane proteins [12,20,21] and to the spectrin/actin complex [20] in human erythrocytes. As a result, it destabilizes the erythrocyte membrane mechanical stability [12,22]. We have previously shown that the binding affinity of 4.1R FERM domain for band 3 and GPC decreases when Ca2+/CaM binds simultaneously to two regions of 4.1R FERM domain: the A264KKLWKVCVEHHTFFRL peptide (pep 11) that mediates Ca2+-independent CaM binding and the A181KKLSMYGVD-LHKAKDL peptide that is responsible for Ca2+-sensitive CaM binding [23]. The Ser185 residue in the second peptide sequence is critical for Ca2+-dependent regulation of 4.1R FERM domain binding to membrane proteins by CaM, mutation of Ser185 resulting in a loss of Ca2+/CaM regulatory activity [23,24].
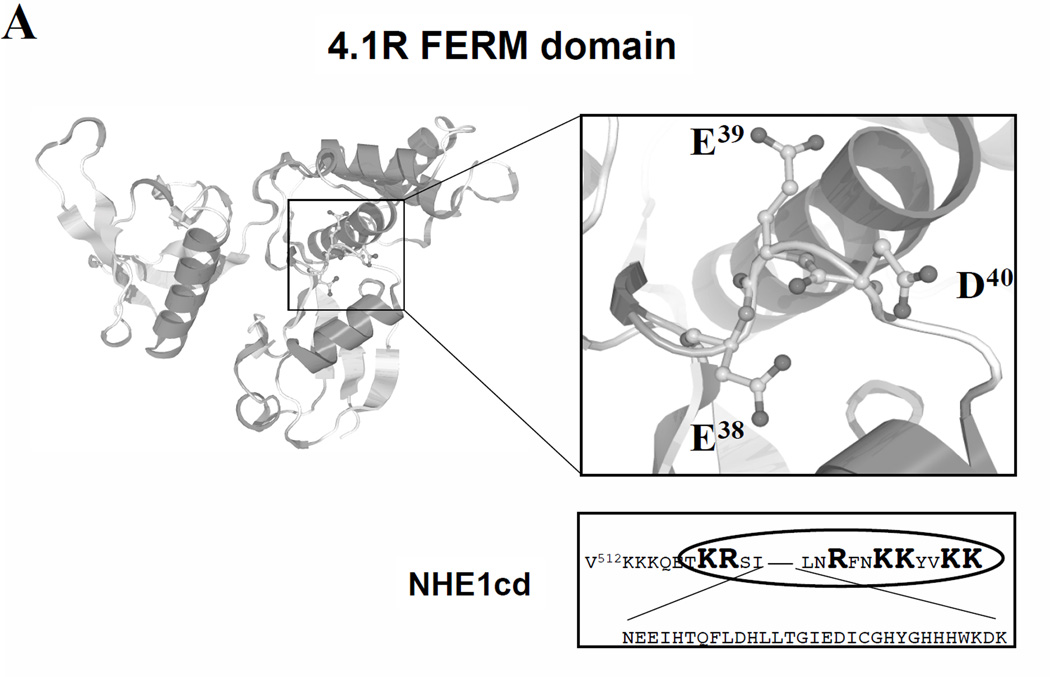
Based on sequence homologies with the motif in the cytoplasmic domain of anion exchanger isoform 1 (AE1 or band 3), referred to as band 3cd, that we previously showed mediates the interaction of band 3 with 4.1R80 [15], we identified positively charged candidate binding sequences in the juxta-membrane region of NHE1 cytoplasmic domain (NHE1cd) that may mediate NHE1 interaction with 4.1R80 (Figure 1A). Strikingly, these basic amino acids sequences in NHE1cd, K513KKQETKR and R556FNKKYVKK, were previously reported to play a key role in increasing NHE1 activity through their interaction with phosphatidylinositol 4,5-bisphosphate (PIP2) [25]. We identified the E38ED sequence in 4.1R80 FERM domain and two clusters of basic amino acids in NHE1cd as critical for 4.1R80-NHE1 interaction. The interaction between 4.1R80 and NHE1 differed from that previously described between 4.1R80 and band 3, not only because of the type and number of motifs it involved but also because the binding of 4.1R80 to NHE1 was more sensitive to tonicity and pH than 4.1R80 binding to band 3. Furthermore, the interaction between 4.1R80and NHE1cd differed from that previously described for ERM proteins and NHE1cd [26] because the binding of 4.1R80 required both basic sequences. In addition, because Ca2+/CaM reduced the binding affinity of 4.1R80 FERM domain for NHE1cd,we hypothesize that Ca2+/CaM reduces the affinity of 4.1R80 binding to NHE1, thus favoring PIP2 binding to NHE1, and that sustained binding of PIP2 to NHE1 may account for the hyper activation of NHE1 observed in 4.1R-null mouse erythrocytes [25]. This study provides novel insights into the complexity and the versatility of the structural and functional interactions between the cytoskeleton and ion exchangers.
Figure 1. Alignment of AE1 (band 3) and NHE1 cytoplasmic domain juxta-membrane regions and of 4.1R exon 5-encoded wild type and EED mutant peptides and pull down assay showing interaction of 4.1R80 with the cytoplasmic domain of sodium proton exchanger, NHE1cd.
(A)The motif responsible for anion exchanger AE1 (also known as band 3, accession No.P23562) interaction with 4.1R80 has been identified as a positively charged RRR cluster (shown in bold and larger font) in the juxta membrane region of the cytoplasmic domain [4]. This cluster interacts primarily with a negatively charged EED motif located in 4.1R FERM domain exon 5-encoded peptide (underlined characters). Strikingly, a positively charged amino acid cluster KKK, very similar to the RRR band 3 motif involved in 4.1R80 interaction, is present in the juxta-membrane region of the cytoplasmic domain of NHE1 (shown in bold and larger font). This cluster is part of the previously defined M1 motif (V512KKKQETKR), another cluster of positively charged residues further downstream being referred to as M2 motif (R556FNKKYVKK), (B). GST-fusion proteins encoding the C-terminal cytoplasmic domains of NHE1 (amino acids 501–815; lane 1) or of a variant of NHE1 cytoplasmic domain lacking amino acids 501–637 in its juxta-membrane region (lane 2) or GST alone (lane 3; to assess non-specific binding) were incubated with [35S]-methionine labeled in vitro-translated human 4.1R80. The pool of 4.1R80 bound to GST-NHE fusion proteins was separated by SDS-PAGE and detected by autoradiography (C). Symbols, + and – represent as positive and negative binding to 4.1R80, respectively.
EXPERIMENTAL
Cloning of cytoplasmic domain of transmembrane proteins
cDNAs encoding full length (P501-Q815) or truncated (N638-Q815) cytoplasmic domains of wild type rat (Rattus norvegicus) NHE1 (NHE1cd, accession No. NP_036784), or mutant M1 (K513KKQETKR→ AAAQETAA), mutant M2 (R556FNKKYVKK→AFNAAYVAA), and double mutant M1+M2 rat NHE1 cytoplasmic domain N-terminal region (L506-Q575) were cloned into the glutathione S-transferase (GST) fusion protein vector pGEX-2T (GE Healthcare UK Ltd., Buckinghamshire, England) (Figure 1A and 1B). A less extensively mutated version of the M1 mutant (K513KK→AAA), referred to as M3 mutant in the following, was generated from the wild type construct by site directed mutagenesis following the manufacturer's recommendations (Stratagene Inc. La Jolla, CA, USA). Preparation of 43kDa cytoplasmic domain of human (band 3cd) has been previously described [23].
Preparation of recombinant full length 4.1R80 and its polypeptide fragments
cDNAs encoding full length human 4.1R80 (accession No.P11171, expressing exon 16-encoded region but lacking exon 17b-encoded region) was cloned 5’-NsiI-XhoI-3’ into the Histidine-tagged pET31b(+) vector (Novagen Inc., Madison, WI, USA). Histidine-tagged 4.1R80 was expressed in BL21 Gold (DE3) E. Coli (Stratagene, La Jolla, CA, USA) and purified according to the manufacturer’s instructions (Novagen Inc., Madison, WI, USA).
Recombinant 4.1R FERM domain was cloned into the pGEX-KG vector and expressed as GST-fusion protein in BL21 bacteria [27]. Following sonication, the bacterial lysate was loaded onto a glutathione affinity column to purify and the recombinant GST fusion protein was eluted from the column after cleavage of the GST tag with thrombin, as previously described [27]. After desalting, the protein was further purified on a heparin-Sepharose column to remove contaminants and breakdown products. Finally, 4.1R FERM domain was loaded onto a Sephacryl S-100 size exclusion chromatography equilibrated with 50 mM Tris-HCl, pH7.5, 0.5 M NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 1 mM benzamidine, 1% glycerol and 2 mM NaF (Figures 2A and 2B). Preparation of exon 5 region deleted FERM domain (FERM ΔExon5) and E38ED mutagenesis to A38AA (FERM Exon 5 mut.) have been previously described [28]. A cDNA encoding exon 5-encoded human 4.1R80 polypeptide was cloned 5’-EcoRI-XhoI-3’ into the GST fusion protein vector pGEX4T-2 vector (GE Healthcare UK Ltd., Buckinghamshire, England). Domains of 4.1R80, including the 16kDa-, 10kDa- and 22kDa C-terminal domains were also expressed as GST fusion protein as previously described [23]. All constructs were checked by DNA sequencing (Elim BioPharmaceuticals Inc., Hayward, CA, USA) prior to expression in bacteria. After affinity purification, recombinant proteins were dialyzed extensively against 10mM Na2HPO4/NaH2PO4, pH 7.4 containing 0.15M NaCl (PBS) prior to use in IAsys binding assays. Protein purity was assessed by SDS-PAGE after protein separation on a 12.5% polyacrylamide gel and staining with Gelcode Blue® (Pierce Inc., Rockford, IL, USA). FERM domain concentration was determined by absorbance at 280nm, the E1% corresponding to 14 at molar concentration for tyrosine (ε=1340), tryptophan (ε=5550) and cycteine (ε=200) [29].
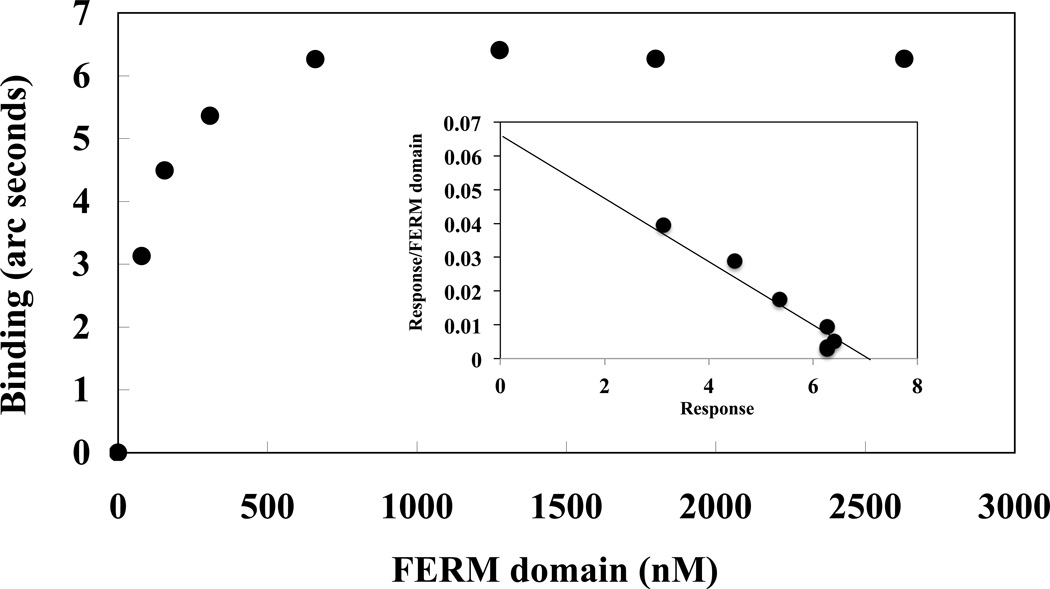
Figure 2. Analysis of 4.1R FERM domain binding to NHE1cd.
4.1R FERM domain binding to NHE1cd immobilized on aminosilane cuvette was measured using the IAsys system. The Bmax (arc seconds) at each 4.1R FERM domain concentration [FERM] was calculated using the software package Fastfit® and Bmax was plotted against the Bmax / [FERM] ratio. The maximum binding (Bmax) at the crossing point on the X-axis was 6.4. The half maximum binding occurred at 87nM [FERM], an affinity similar to the Kd value calculated from the kinetic binding analysis (44nM, Table II).
Preparation of recombinant CaM
A cDNA encoding human calmodulin (CaM) with four extra N-terminal amino acids, Gly-Ser-His-Met, was a kind gift from Ms. Marilyn Parra (Lawrence Berkeley National Laboratory, University of California at Berkeley, CA, USA). Human CaM was cloned into the histidine-tag pET15b(+) vector after restriction of the NdeI cloning site. Recombinant CaM was purified sequentially on a Ni+ column, a phenyl Sepharose column and a Sephacryl S-100 column equilibrated with 50 mM Tris-HCl pH7.5 containing 500 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 2 mM NaF and 1% glycerol. Protein retention time was recorded on an Akta Prime™ Plus (GE Healthcare UK Ltd., Buckinghamshire, England). The purity of recombinant CaM was assessed by SDS-PAGE and Western blot analysis. CaM concentration was calculated based on the absorbance at 280 nm and an E1% of 1.6 for CaM [27].
Preparation of PC and PIP2/PC liposomes and liposome binding to NHE1cd
Phosphatidylcholine (PC) liposomes containing 20% PIP2 or liposomes containing only PC (used as control) were prepared as previously described [30,31]. Liposomes were resuspended at 1mg/ml in 20mM MES, pH6.0 or pH6.8 or 50mM Tris-HCl, pH7.5, 0.1M NaCl and 1mM EDTA with or without 1.5mM CaCl2. PIP2/PC or PC liposome binding to NHE1cd immobilized on an aminosilane cuvette was evaluated by measurement of maximal binding as described below and in our previous report [32].
Pull down assays
Pull down assays were performed as previously described [26,33]. Briefly, GST or GST fusion proteins encoding full length or truncated cytoplasmic domains of NHE1 (P501-Q815, N638-Q815) were purified from BL21 cells induced at 28°C for 2h with 0.25 mM isopropyl-β-D-thiogalactopyranoside. Proteins were extracted from bacteria by sonication (4 × 15sec) on ice in PBS supplemented with protease inhibitors followed by incubation for 1h in presence of 1% Triton X-100. Glutathione-coupled beads were pre-equilibrated in binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Nonidiet P-40) for 1h. [35S]-methionine labeled 4.1R80, synthesized for 1h at 30°C using the TNT coupled in vitro transcription/translation system (Promega Corporation, Madison, WI), was incubated for 1h with 2mg of either GST, GST-full length NHE1 cytoplasmic domain (NHE1cd) or GST-truncated NHE1cd fusion protein. After extensive washes in binding buffer, beads were denatured by boiling for 3 min in 1 volume of 2xSDS buffer and samples were separated by SDS-PAGE. The fraction of 4.1R80 bound to GST-fusion proteins was detected by autoradiography of dried gels.
Resonant mirror detection binding assays
Kinetic analysis
Interactions of 4.1R FERM domain with NHE1cd were examined using the IAsys® resonant mirror detection system following the manufacturer’s instructions (Affinity Sensors, Cambridge, UK) [34]. In the following, the protein immobilized on the cuvette is referred to as the "ligand" while the one added in solution to the cuvette is referred to as "analyte". GST-fusion protein of NHE1cd was immobilized on aminosilane cuvettes as previously described [17,24]. 4.1R FERM domain, dissolved in 50 mM Tris-HCl, pH 7.5, 0.1 M NaCl, 1 mM EDTA and 1mM 2-mercaptoethanol(Buffer A), was added at concentrations ranging from 50 nM to 1 µM. All binding assays were conducted at 25°C with constant stirring. Kinetic analysis of analyte binding to ligand was conducted as previously reported [12,17,23]. Dissociation constants at equilibrium (Kd) were calculated using equation (1):
| (1) |
where ka is the association rate constant and kd the dissociation rate constant. Kd was obtained from the means of 3–5 measurements for ka and kd and was confirmed by Scatchard plotting using maximum binding (Bmax) and molar concentrations of analyte [23,24,35]. Bmax was calculated from binding characteristics using the software package FASTfit® version 2.1.
Maximum binding
The maximum response detected upon addition of 4.1R FERM domain to GST-NHE1cd, “Beq” (expressed in “arc second”), was estimated from the binding profile using the software package FASTfit® version 2.1. The stoichiometry of 4.1R FERM domain binding to GST-NHE1cd was calculated from the equation: (Bmax 4.1R FERM domain/32,428) : (GST-NHE1cd/30,381), where 32,428 and 30,381 are apparent molecular weights (in Da) for 4.1R FERM domain and GST-NHE1cd, respectively) as described in the Method Guide of the IAsys® system. Cuvettes were reused after cleaning with 20mM HCl. The original binding curves could be replicated after HCl washing, indicating that the washing had not denatured the bound ligands.
NaCl-dependent FERM domain binding to NHE1cd and band 3cd
4.1R FERM domain binding to NHE1 was evaluated in the presence of increasing NaCl concentrations (0.1M~0.5M) in 50mM Tris-HCl, pH7.5, 1mM EDTA, 1mM 2-mercaptoethanol, using Beq, as described above.
pH-dependent FERM domain binding to NHE1cd and band 3cd
FERM domain was dissolved in various buffers: 0.1M NaCl, 1mM EDTA and 1mM 2-mercaptoethnol: 50mM CH3COOH/CH3COONa buffered at either pH 4.0, pH 5.0, pH 5.5 or pH 6.0; 25mM Na2HPO4/NaH2PO4 buffered at either pH 6.0, pH 6.5, pH6.8, pH 7.0, pH 7.2, pH 7.5 or pH 8.0; 50mM Tris-HCl, buffered at either pH 7.5, pH 8.0, pH 8.5; 50mM NaHCO3/Na2CO3 buffered at pH9.0 and kinetic binding analysis to NHE1cd of band 3cd was carried out.
Binding ratio determination using the quartz crystal micro-balance
Single-Q® (Scinics Ltd., Tokyo, Japan) based on quartz crystal micro-balance was used to determine the stoichiometry of 4.1R FERM domain binding to NHE1cd. NHE1cd was immobilized on the sensor chip according to Affinix® instructions and changes in mass of immobilized NHE1cd following the addition (binding) of 4.1R FERM domain were recorded [36]. The maximal binding value (Bmax) of 4.1R FERM domain to NHE1cd immobilized on the sensor chip was determined.
Representation of 4.1R FERM domain 3D structure
Ribbon and surface model 3D structures of 4.1R FERM domain (PDB accession No. 1GG3) were generated using the MolFeat Ver. 4.6 (FiatLux Corp., Tokyo, Japan).
RESULTS
Direct interaction of 4.1R80 with NHE1in vitro
Our previous finding that NHE1 is hyperactive in 4.1R-null mouse erythrocytes [10] suggests a functional interaction between 4.1R and NHE1, In testing this prediction, we now show that this phenotype reveals an actual physical interaction between 4.1R80 and NHE1. A GST fusion protein encoding the cytoplasmic domain of NHE1 (NHE1cd; aa 501–815) was able to pull down in vitro translated human 4.1R80 (Figure 1C, lane 1). In contrast, a GST-NHE1cd construct lacking the juxta membrane region of NHE1cd (amino acids 501–637; Figure 1C, lane 2), or GST alone (Figure 1C, lane 3), showed a markedly reduced interaction with 4.1R80. This observation documented that the NHE1 peptide encompassing P501–N637 mediated NHE1 interaction with 4.1R80 (Figure 1B). IAsys-based in vitro binding assays enabled us to further identify the minimal region in NHE1cd interacting with 4.1R80 to the L502-Q572 peptide (Table I). Of particular note, the binding affinity of 4.1R80 for NHE1 was very similar (Kd ~100–200 nM; Table I) to those previously reported for 4.1R80 binding to its two major transmembrane binding partners in erythrocytes, GPC and band 3 [19,23].
Table I.
In vitro interactions of 4.1R80 with NHE1cd
| Analyte | NHE1 | ka(M−1 s−1) | kd(s−1) | Kd(nM) |
|---|---|---|---|---|
| 4.1R80 | wild | 1.0 ± 0.20 × 105 | 9.6 ± 0.11 × 10−3 | 99 ± 23 |
| FERM | wild | 4.3 ± 0.19 × 104 | 9.5 ± 0.18 × 10−3 | 214 ± 17 |
| Exon 5 | wild | 4.8 ± 0.25 × 104 | 5.1 ± 0.10 × 10−3 | 106 ± 6 |
| FERM /ΔExon 5 | wild | No binding | No binding | No binding |
| FERM EED mut. | wild | 1.2 ± 0.14 × 104 | 1.7 ± 0.15 × 10−2 | 1442 ± 211 |
| FERM | M1 | No binding | No binding | No binding |
| Exon5 | M1 | 2.5 ± 0.25 × 102 | 1.2 ± 0.10 × 10−3 | 4791 ± 652 |
| FERM | M2 | No binding | No binding | No binding |
| Exon 5 | M2 | 2.4 ± 0.25 × 102 | 6.2 ± 0.10 × 10−3 | 26060 ± 2465 |
| FERM | M3 | 1.2 ± 0.10 × 105 | 6.5 ± 0.16 × 10−2 | 545 ± 47 |
| FERM | M1+M2 | No binding | No binding | No binding |
Kd values for interactions of 4.1R80, several versions of 4.1R FERM domain or 4.1R FERM domain exon 5-encoded peptide with immobilized NHE1cd are shown. Wild type 4.1R FERM domain (FERM) or a FERM domain either lacking exon 5 (FERM/ΔExon5) or bearing a mutation of the EED motif (FERM EED mut.) were probed. The sequences for wild type NHE1 (wild), or M1, M2, M3 and M1+M2 NHE1 mutants are shown in Figure 1. Each analyte (50nM to 1 µM) was incubated with the immobilized ligand as described in the Methods section. Kd values were determined from the binding curves obtained by the resonant mirror detection method using the software package FAST-Fit ™.
Mapping of the motifs in 4.1R FERM domain and NHE1cd responsible for their interaction
Having confirmed a direct interaction of 4.1R80 with NHE1, we mapped the critical motifs in both 4.1R80 and NHE1 responsible for this interaction. The use of various 4.1R80 recombinant proteins enabled us to show that 4.1R FERM domain, and more specifically a 35 amino acid peptide encoded by alternative exon 5 within this domain, mediated the 4.1R80 interaction with NHE1 (Table I). Although 4.1R FERM domain bound to NHE1cd with a similar affinity as full-length 4.1R80, a variant 4.1R FERM domain lacking the exon 5-encoded peptide failed to interact with NHE1 (Table I). Mutation of the EED motif within the exon 5-encoded peptide, a motif previously reported to participate in 4.1R80 interaction with band 3 (Figure 1A) [28], resulted in a significant decrease in 4.1R80 binding affinity for NHE1cd (Kd~1400nM; Table I). The other domains of 4.1R80, 16kDa, 10kDa spectrin/actin binding domain and C-terminal 24kDa domain did not bind to NHE1cd (data not shown).
Alignment of the amino acid sequences of rat anion exchanger 1 (AE1)/band 3and rat NHE1 juxta-membrane regions of their cytoplasmic domains revealed the presence of a common cluster of positively charged residues, i.e., RRR and KKK, respectively (Figure 1A). Such a positively charged cluster in NHE1 was an obvious candidate for interaction with the negatively charged EED motif in 4.1R80. However, mutation of this K513KK→AAA cluster (referred to as M3 motif) induced only a slightly decreased affinity for NHE1 and 4.1R80 (FERM domain) binding (Kd = 545nM, Table I). In contrast, a more extensive mutation of this region K513KKQETKR→AAAQETAA, previously characterized as the M1 motif (Figure 1A), mutation of another more distal cluster of positively charged residues, R556FNKKYVKK→AFNAAYVAA, previously characterized as the M2 motif (Figure 1A) [25], and simultaneous mutation of both M1 and M2 motifs resulted in a dramatic decrease in 4.1R80 binding affinity for NHE1 (Kd = 4791nM, 26060nM and no binding, respectively; Table I). These data led us to conclude that both M1 and M2 sequences are necessary to mediate NHE1 interaction with 4.1R80. Of particular note, these two motifs are absent in the same region of band 3 cytoplasmic domain (Figure 1A), which strongly suggests a different type of interaction between 4.1R80 and band 3 and 4.1R80 and NHE1.
IAsys binding analysis estimated the binding ratio of 4.1R FERM domain to NHE1 as approximately 1:1 (Figure 2). Binding affinity was calculated by Scatchard plot analysis (Figure 2, insert), with maximal binding being observed in the 10−7 M range. Another method, based upon the use of a quartz crystal micro-balance biosensor, confirmed the binding ratio and binding affinity values (data not shown). These results indicate that one molecule of 4.1R FERM domain interacts simultaneously with the two NHE1cd binding sites described above.
Influence of pH on 4.1R FERM domain-NHE1cd interaction
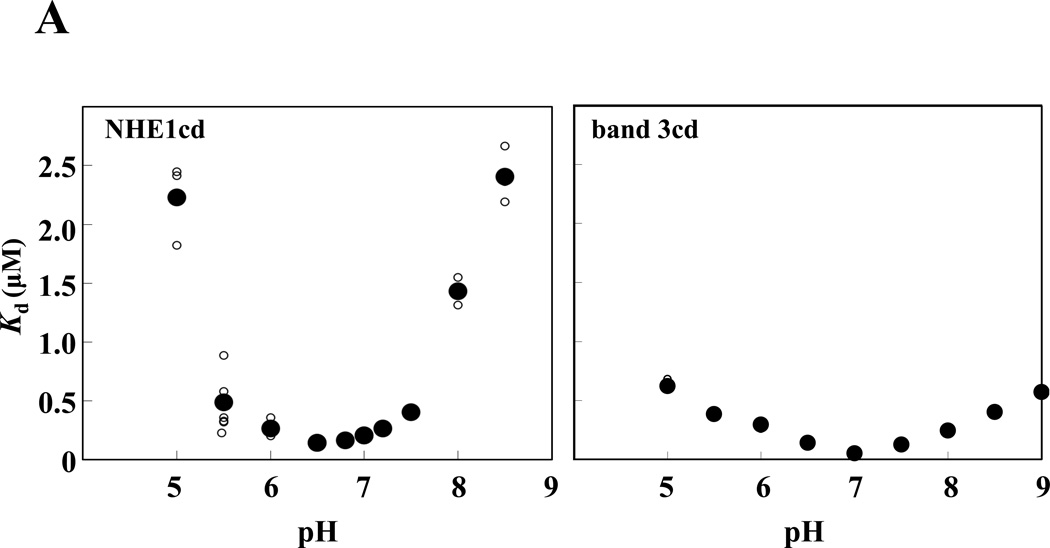
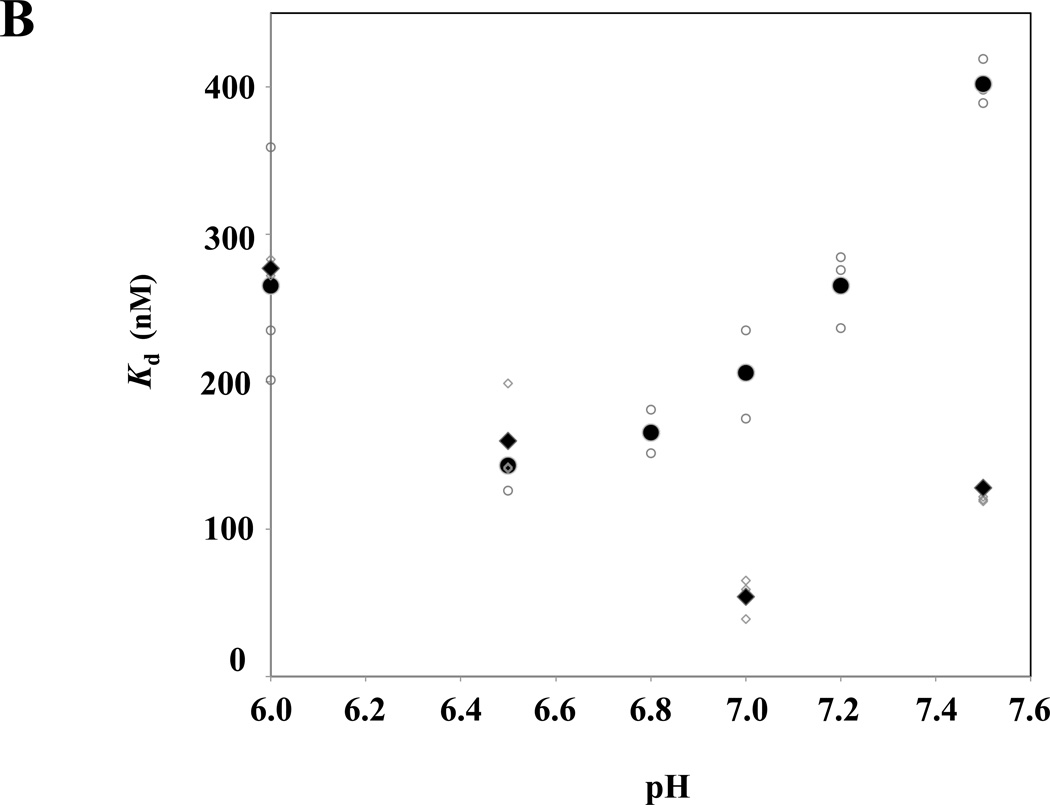
Because NHE1 plays a key role in intracellular pH homeostasis, we investigated whether 4.1R FERM domain binding to NHE1cd is regulated by pH. As shown in Figure 3A, 4.1R FERM domain binding to NHE1cd as a function of pH adopted a deep parabola shape, the maximum binding (minimum Kd ~ 150nM) being observed at pH 6.5. The affinity of 4.1R FERM domain binding to NHE1cd was similar between pH 6.0 and 7.2. In contrast, binding affinities decreased dramatically at more basic (~pH8.0) or acidic pH (~pH5.5); the dissociation constants (Kds) increased more than 20 times at pH 5.5 and 8.5 (Supplementary Fig. S1). In contrast, Kds for 4.1R FERM domain binding to band 3 were not significantly different over a large range of pH (5.0–8.5). In the more physiological range of pH 6.5 to 7.5, the Kd of 4.1R FERM domain binding to NHE1 was significantly higher than to the affinity for binding to band 3cd (Figure 3B). Furthermore, maximum binding affinity (Kd ~ 50nM) for 4.1R FERM interaction with band 3 was observed at pH 7.0, which further supports different characteristics for 4.1R80 interaction with NHE1 and band 3.
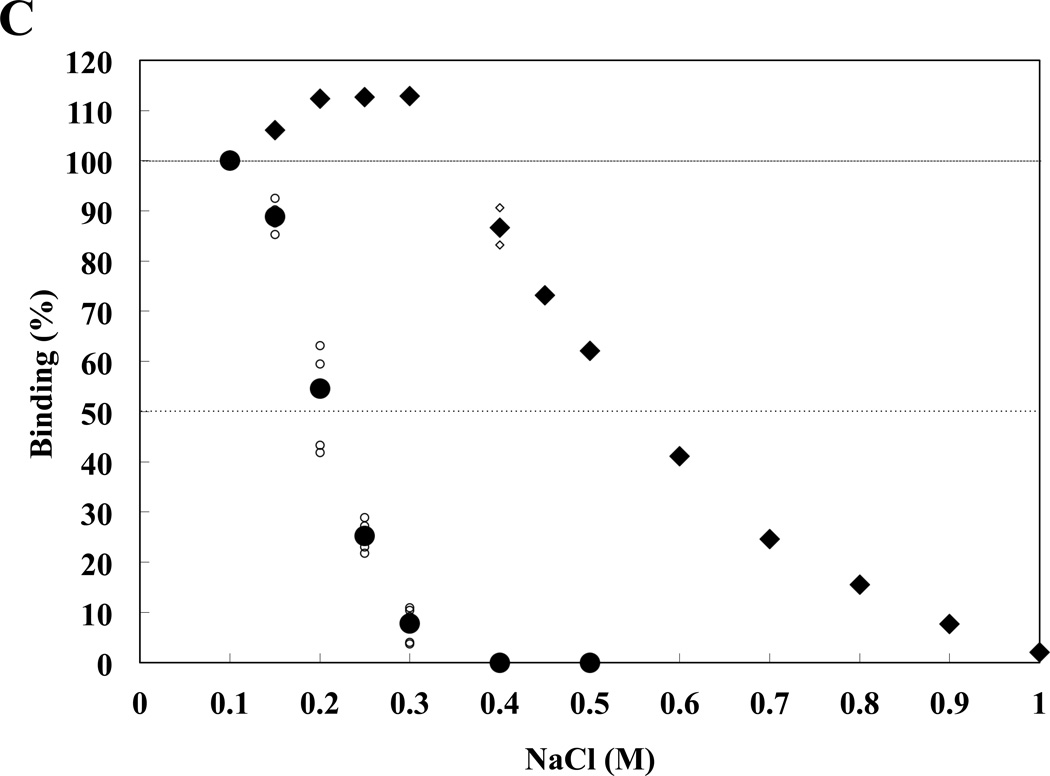
Figure 3. Relative pH- and Na+-dependence of 4.1R FERM domain binding to NHE1cd and band 3cd.
(A) 4.1R FERM domain binding to NHE1cd or band 3cd immobilized on aminosilane cuvette was measured at various pHs using the IAsys system. Small and large circles represent raw data from 3 independent determinations and mean values, respectively. (B) A magnification of the data shown in (A) in the 6.5–7.5 pH range is shown. Circles ( ) and diamonds (※) indicate 4.1R FERM domain binding to NHE1 and band 3 respectively. Open and closed symbols correspond to individual and averaged raw data, respectively. (C) Circles and diamonds represent binding data for NHE1cd and band 3cd, respectively. Small circles and diamonds and large circles and diamonds represent individual and averaged raw data, respectively. The "binding (%)" at each concentration of NaCl is relative to the maximum (100%) binding detected at equilibrium in 0.1M NaCl.
) and diamonds (※) indicate 4.1R FERM domain binding to NHE1 and band 3 respectively. Open and closed symbols correspond to individual and averaged raw data, respectively. (C) Circles and diamonds represent binding data for NHE1cd and band 3cd, respectively. Small circles and diamonds and large circles and diamonds represent individual and averaged raw data, respectively. The "binding (%)" at each concentration of NaCl is relative to the maximum (100%) binding detected at equilibrium in 0.1M NaCl.
Influence of Na+ concentration on 4.1R FERM domain-NHE1cd interaction
In addition to pH-dependent differences between4.1R80 binding to NHE1cd and band 3cd, we also found that 4.1R FERM domain interaction with NHE1cd but not band 3cd is more sensitive to Na+. The binding of 4.1R FERM domain to NHE1cd at pH 7.5 was already reduced by 50% at NaCl concentrations as low as 0.2M, the binding being totally inhibited at 0.3M NaCl (Figure 3B). In contrast, 4.1R FERM domain binding to band 3 was unaffected up to 0.5M NaCl, the binding being ablated at 1.0M NaCl. 4.1R FERM domain binding to NHE1 was therefore much more sensitive to variations in NaCl concentration than that to band 3cd, in particular in the physiological range of 0.1M to 0.2M NaCl. These observations supported once again the different characteristics for 4.1R80 interaction with NHE1 and band 3.
Regulation of 4.1R FERM domain-NHE1cd interaction by Ca2+/CaM
We previously showed that CaM binds to 4.1R FERM domain in the absence of Ca2+ and decreases binding affinity of 4.1R FERM domain for its binding partners in a Ca2+-dependent manner [12,23,37]. We therefore asked whether there is a Ca2+/CaM-dependent regulation of 4.1R FERM domain binding to aNHE1cd construct lacking the two CaM binding sites, i.e. R636-A656 and N664-L684 [38,39]. In the absence of CaM, the Kd observed for 4.1R80-NHE1 interaction at physiological pH 7.5 was similar to that of previously characterized binding partners (Table II). At pH 6.0, 4.1R FERM domain could still bind to NHE1cd (Table II) and to Ca2+/CaM with a Kd of ~ 100nM (ka=6.7 × 10−2 s and kd=7.0×104 s · M−1). Ca2+/CaM binding to 4.1R FERM domain reduced the binding affinity of 4.1R FERM domain for NHE1cd (Table II), the inhibitory effect of Ca2+/CaM being more pronounced at pH 7.5 than at pH 6.0. These results indicate that the 4.1R FERM domain-NHE1 interaction is regulated by CaM in a Ca2+-dependent manner and that this regulation is likely most dramatic at physiological pH values.
Table II.
Ca2+/CaM-mediated down-regulation of 4.1R FERM domain interaction with NHE1cd
| pH7.5 | ||||
| CaM | ka(M−1 s−1) | kd(s−1) | Kd(nM) | |
| − | EDTA | 1.8 ± 0.12 × 104 | 3.4 ± 0.24 × 10−3 | 182 ± 10 |
| − | Ca2+ | 1.7 ± 0.23 × 104 | 7.1 ± 0.17 × 10−3 | 380 ± 29 |
| + | EDTA | 1.3 ± 0.11 × 104 | 5.4 ± 0.18 × 10−3 | 396 ± 9 |
| + | Ca2+ | 1.4 ± 0.13 × 103 | 9.3 ± 0.20 × 10−3 | 6362 ± 634 |
| pH6.0 | ||||
| CaM | ka(M−1 s−1) | kd(s−1) | Kd(nM) | |
| − | EDTA | 7.8 ± 0.18 × 104 | 2.1 ± 0.04 × 10−2 | 265 ± 67 |
| − | Ca2+ | 3.4 ± 0.07 × 104 | 9.5 ± 0.13 × 10−3 | 276 ± 26 |
| + | EDTA | 2.1 ± 0.10 × 104 | 1.0 ± 0.11 × 10−2 | 344 ± 24 |
| + | Ca2+ | 1.1 ± 0.17 × 104 | 1.6 ± 0.20 × 10−2 | 1413 ± 58 |
Kd values for 4.1R FERM domain interactions with immobilized NHE1cd either in the presence (Ca2+) or absence of Ca2+ (EDTA) and CaM are shown. Each analyte (50nM to 1 µM) was incubated with the identified immobilized ligand as described in the Methods section. Kd values (mean ± S.D) were determined from the binding curves obtained by the resonant mirror detection method in 3–5 independent experiments using the software package FAST-Fit™.
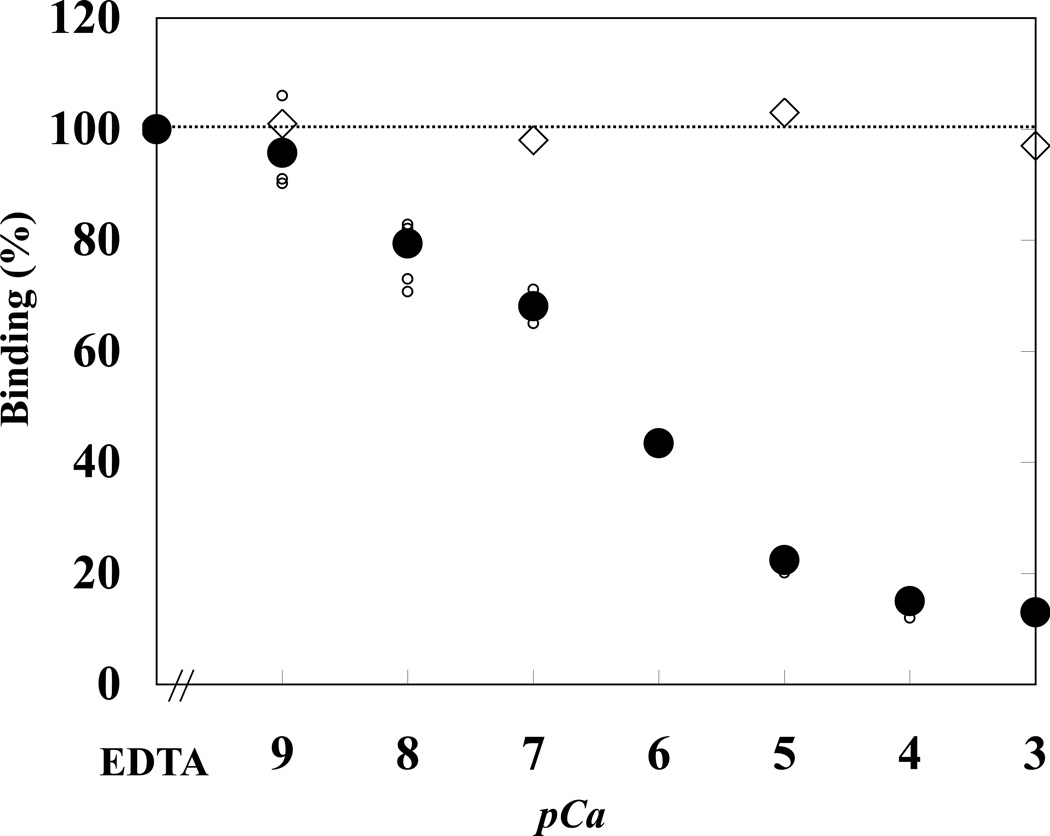
We then investigated the Ca2+ dependence of the CaM-modulated interaction of 4.1R FERM domain with NHE1cd at pH 7.4 (Figure 4). At Ca2+ concentrations greater than 0.01 µM (pCa=8), the extent of 4.1R binding to NHE1 started to decline, the maximal inhibition being reached at Ca2+ concentrations of 100 µM and higher (pCa=4). Half-maximal inhibition was observed at a Ca2+ concentration of ~0.1 µM (pCa=7). In the absence of CaM, Ca2+ had no effect on the 4.1R80-NHE1 binding affinity (Figure 4). At pH 6.0, binding of 4.1R FERM domain to NHE1cd was no longer dependent on Ca2+ (data not shown).
Figure 4. Ca2+-dependence of 4.1R FERM domain binding to NHE1cd.
4.1R FERM domain binding to NHE1cd was measured at various concentrations of Ca2+ either in the presence ( ) or absence (★) of 5 µM CaM. Small and large circles represent individual and averaged raw data, respectively. Ca2+ concentrations (pCa) were adjusted with a Ca2+/EGTA buffer system. 4.1R FERM domain binding to NHE1cd was plotted as a function of Ca2+ concentration. The maximal extent of binding under different experimental conditions was quantitated as described under “Materials and Methods.” Maximal binding in the presence of EGTA was used to normalize the extent of binding under different experimental conditions. In the absence of CaM, there was no change in the binding of 4.1R FERM domain to NHE1cd regardless of Ca2+ concentrations (★).
) or absence (★) of 5 µM CaM. Small and large circles represent individual and averaged raw data, respectively. Ca2+ concentrations (pCa) were adjusted with a Ca2+/EGTA buffer system. 4.1R FERM domain binding to NHE1cd was plotted as a function of Ca2+ concentration. The maximal extent of binding under different experimental conditions was quantitated as described under “Materials and Methods.” Maximal binding in the presence of EGTA was used to normalize the extent of binding under different experimental conditions. In the absence of CaM, there was no change in the binding of 4.1R FERM domain to NHE1cd regardless of Ca2+ concentrations (★).
PIP2/PC liposome binding to NHE1cd at different pHs in the presence and absence of Ca2+
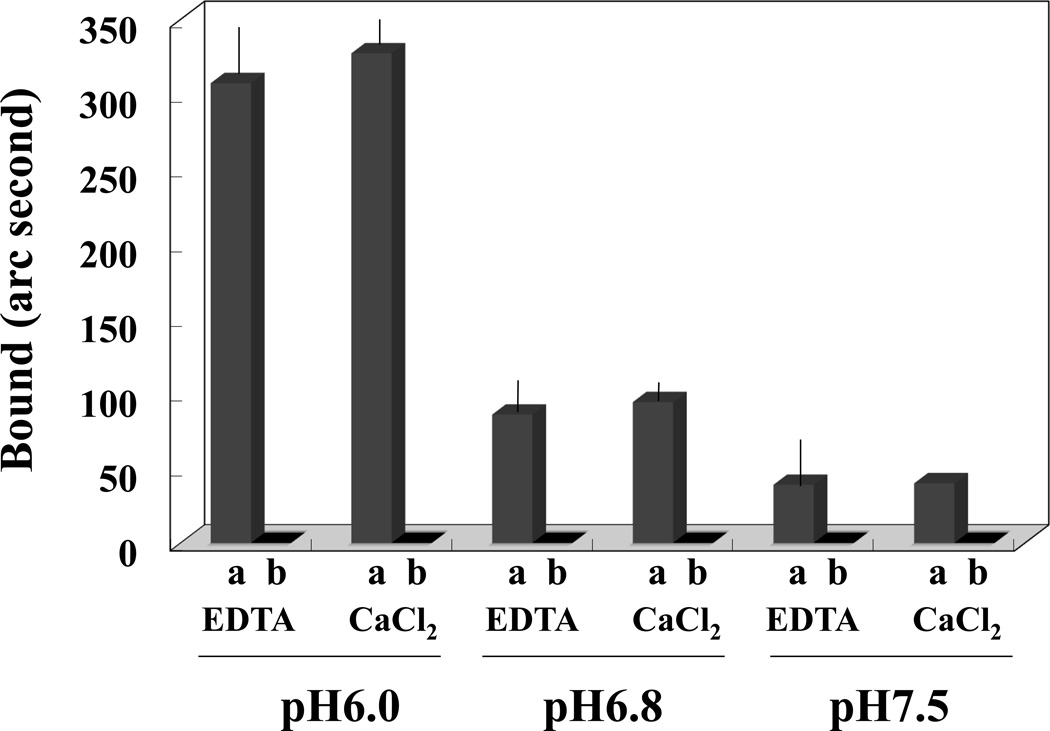
We also compared PIP2 binding to NHE1cd at different pH values and in the presence or absence of Ca2+. Binding of PIP2/PC liposomes to NHE1cd immobilized on a cuvette was measured using the IAsys system. Consistent with a previous report [25], PIP2/PC liposomes bound more strongly to NHE1cd at pH 6.0 than at pH 6.8 or pH 7.5 (Figure 5). Furthermore, we found that this binding was not significantly affected by Ca2+ (Figure 5). PC liposomes, used as a negative control, failed to bind to NHE1cd.
Figure 5. PIP2/PC liposome binding to NHE1 as a function of pH.
Liposomes containing either 20% PIP2-80% PC (columns a) or 100% PC (columns b) were resuspended at 1mg/ml of a medium buffered at pH 6.0, pH 6.8 or pH 7.5 and tested for binding to immobilized NHE1cd in the presence (CaCl2) or absence of Ca2+(EDTA) as described in the Methods section. PIP2-PC liposomes bound to NHE1cd whereas PC liposomes did not in any of the conditions tested. Moreover, PIP2-PC liposomes binding to NHE1cd was higher at more acidic pH (6.0) than at more neutral pHs (6.8 and 7.5).
DISCUSSION
Mice with engineered deletions of the four members of the protein 4.1 family (4.1R, 4.1G, 4.1N and 4.1B) have been the subject of an increasing number of studies over the last 10 years [11,21]. Relevant to our current study, erythrocytes from 4.1R-null mice exhibit hyperactivity of NHE1 compared with erythrocytes from wild type mice [10]. We now show that 4.1R directly binds to NHE1 in vitro and identify the motifs in 4.1R and NHE1 mediating this interaction. We also reveal that the 4.1R80-NHE1 interaction is modulated by changes in pH and by concentrations of Na+ and Ca2+/CaM. Our in vitro data clearly demonstrate that at acidic pH, 4.1R dissociates from NHE1cd but binding of PIP2 to NHE1cd is increased. This distinct behavior may be heightened by variations in intracellular CaM and Ca2+ concentrations as the regulatory effect of Ca2+/CaM on 4.1R80-NHE1 interaction contrasts dramatically with its inability to regulate the PIP2-NHE1 interaction. We hypothesize that the antagonistic effects of 4.1R80 and PIP2 on NHE1 activity [10,25] play an important role in the regulation of NHE1 activity and that, in absence of 4.1R80, sustained binding of PIP2 to NHE1cd facilitates increased NHE1 activity. This phenotype may be heightened in erythrocytes because their PIP2 content is higher than in other cells [40].
The L37EEDY sequence that is shown here to mediate 4.1R80 interaction with NHE1 has been reported to enable 4.1R80 interaction with band 3 [14]. Analysis of the 3D structure of 4.1R FERM domain reveals that the EED motif is located in a loop structure [41] (Figure 6A). The side chains of each amino acid of the EED motif adopt completely different directions, conferring upon this motif a “T-like” shape (Figure 6A). Another important finding of our study is that NHE1 interaction with 4.1R80 requires simultaneously the M1 and M2 motifs in the NHE1 cytoplasmic domain and, as a correlate, that two motifs in the FERM domain are likely involved in this interaction as well. Our predicted model is supported by our finding that 4.1R FERM domain binds to NHE1cd with a 1:1 stoichiometry. The observation that 4.1R FERM domain cannot bind to a double (M1+M2) mutant NHE1cd supports that 4.1R FERM domain binds simultaneously to the M1 and M2 motifs in NHE1cd.
Figure 6. Tridimensional schematic representation of 4.1R80 binding site for NHE1cd and proposed model for regulation of NHE1 activity by 4.1R80 in erythrocytes.
(A) A representation of 4.1R FERM domain (PDB No. 1GG3, [45]) 3D structure was obtained using NCBI 3D-structure viewer (see Methods). Key residues responsible for predicted interactions are displayed in both NHE1cd and 4.1R FERM domain. These interactions include electrostatic interactions between 4.1R80 E38 and NHE1 K513, 4.1R80 D40 and NHE1 K515 and 4.1R80 R29 and NHE1 E517. A hydrophobic interaction between 4.1R80 L36 and NHE1 A511 is also predicted. 4.1R80 E39 is not predicted to play a role in 4.1R80 interaction with NHE1. (B) Schematic representation of the lipid bilayer and NHE1-4.1R80 interaction, involving the motifs M1 and M2 in NHE1 cytoplasmic domain and he EED motif in 4.1R FERM domain. Competition between 4.1R and PIP2 for binding to NHE1 as a function of Ca2+/CaM concentration is also shown.
The 3D structure of the cytoplasmic domain of NHE1 has not been resolved except for the CaM binding sites and the calcineurin B homologous protein (CHP) domain [42,43]. A recent study reported that the distal part of the carboxyl cytoplasmic domain of NHE1 is intrinsically disordered but that it contains conserved regions of transient α-helicity that play an important role in NHE1 trafficking [44]. We predict that the spacing of the two basic residue clusters in NHE1cd, K513KKQETKR and R556FNKKYVKK, would enable wrapping of the 4.1R EED motif despite the presence of Phe and Val hydrophobic residues. In that respect, 4.1R FERM domain-NHE1 interaction may resemble a key-keyhole structure. Crystal structure studies will enable further characterization of these interactions at the atomic level.
The 4.1R80-NHE1 interaction differs from the 4.1R80-band 3 interaction in which only one motif in band 3cd has been shown to interact with 4.1R80 [14]. The distinction is not only structural as we also show that, while 4.1R FERM domain interaction with NHE1cd is highly sensitive to variations in Na+concentration and at a lesser extent to variations in pH, that of 4.1R FERM domain with band 3cd is not. We hypothesize that these differences may result from a different type of interaction, 4.1R80 interaction with band 3 likely involving hydrophobic residues, such as Leu37 and Tyr40. These striking observations lead us to propose that upon changes in intracellular pH and Na+ concentration4.1R80 would rapidly dissociate from NHE1 but would still be bound to band 3. We predict that the high sensitivity of 4.1R-NHE1 interaction to serum NaCl concentrations could be highly relevant in pathological conditions of hyponatremia (<135mEq/L) or hypernatremia (>145mEq/L) that result primarily from an excess and loss of free water, respectively, and that have been both associated with seizures due to either brain swelling or shrinkage. Indeed, an increase in 4.1R binding to NHE1in hyponatremic conditions (Figure 3C) would result in a decrease in NHE1 activity [10] and therefore in a decrease in influx of Na+ in cells in an attempt to avoid a further decrease in serum Na+ levels whereas the opposite would be predicted to occur in hypernatremic conditions.
We showed that the functional 4.1R FERM domain-NHE1 complex is potentially regulated by Ca2+ through CaM binding to 4.1R FERM domain in erythrocytes, the inhibitory effect of Ca2+/CaM on 4.1R80-NHE1 interaction being maximal around physiological pH (pH 7.5). Under physiological conditions, CaM is mostly unsaturated (Ca2+-free) in erythrocytes [27]. Given an estimated 3×105 molecules of CaM and 2×105 molecules of 4.1R80 per erythrocyte [27], one can predict that all 4.1R80 molecules are potentially complexed with CaM in erythrocytes. We have shown that the recruitment of CaM may confer stability on 4.1R80 structure [27]. Upon increase in intracellular Ca2+, Ca2+/CaM complex formation would result in the dissociation of 4.1R80 from NHE1, thus promoting NHE1 activity. This hypothesis is consistent with previous reports that show that an increase in intracellular Ca2+ is accompanied by an increase in NHE1 activity [45,46], this effect requiring CaM binding to NHE1 [38,39].
Based on the present results, we propose a model for NHE1 regulation by protein 4.1R80 in erythrocytes (Figure 6B), in which an increase in intracellular [Na+] mediated by NHE1 activity resulting in a rapid dissociation of 4.1R80 from NHE1. In contrast, 4.1R80 interaction with band 3cd is not affected by such changes in pH or [Na+], possibly because of the hydrophobic nature of 4.1R80-band 3 interaction.
The functional consequences of these structural differences are also true for cytoskeletal proteins. Thus, ezrin, a member of the protein 4.1 superfamily of cytoskeletal proteins that is not expressed in erythrocytes, also binds to NHE1cd, but only through the M2 motif [4,6,47]. The poor conservation of 4.1R and ezrin FERM domains (31% identity, Supplementary Fig. S2) could well explain the different properties of 4.1R and ezrin binding to NHE1cd. Relevant to our study, the motif in the exon 5-encoded peptide involved in interaction with NHE1 differs greatly in 4.1R (L37EEDY) and ezrin (L39REVWY). Our comparative analysis of the binding properties of the various members of the protein 4.1R family (4.1R, 4.1G. 4.1N and 4.1B) shows that all 4.1 proteins bind to NHE1cd with the same affinity although 4.1G, 4.1N and 4.1B bear a LEKDY motif instead of a LEEDY motif, indicating that the middle E residue in the LEEDY motif is not essential for FERM domain binding to NHE1 (manuscript in preparation).
A recent study showed that β-adducin (Add2)-null mouse erythrocytes, which adopt a spherocytic shape, lack α-adducin and SLC9A1/NHE1 [7]. Although there is no evidence for a direct interaction between NHE1 and adducin, the observations in adducin-null mouse erythrocytes suggest that adducin interacts either directly with NHE1 or indirectly through 4.1R binding partners, such as p55 [10,11,20]. One major junctional complex results from the association of the transmembrane protein glycophorin C, 4.1R80, the spectrin-actin network and other proteins, such as adducin. Alterations in NHE1 expression and/or activity, as a result of the lack of expression of various components of this complex, such as adducin or 4.1R, are therefore not surprising.
This study illustrates the complexity of the structural and functional interactions between the cytoskeleton and plasma membrane ion exchangers, in particular sodium-proton exchangers, and highlight a “sensor” role for these scaffolding proteins.
Supplementary Material
Changes in Kd resulted primarily from changes in ka, although both kd and ka changed significantly between pH 7.0 and 7.5.
The residues E38ED in human 4.1R FERM domain and the corresponding residues, R40EWV, in human ezrin are depicted using a Stick-ball model. The PDB accession ID is 1gg3 and 1ni2 for human 4.1R and ezrin FERM domains, respectively.
ACKNOWLEDGMENTS
We thank Dr. Shotaro Tanaka, Department of Biochemistry, Tokyo Women’s Medical University, Tokyo, Japan, for bacterial expression of calmodulin. We thank Ms. Marilyn Parra, Lawrence Berkeley National Laboratory, University of California at Berkeley, CA, USA, for providing us with a cDNA encoding human calmodulin. We also thank Dr. Sergio Grinstein, Hospital for Sick Children Research Institute, Toronto, Canada, for providing us with wild type and mutant rat NHE1 cDNAs.
FUNDING
This work was supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sport, Science and Technology of Japan 15570123to WN and by National Institutes of Health grants GM58642 to DLB and DK56355 to PG.
Abbreviations
- 4.1R80
erythrocyte 80kDa protein 4.1R
- AE1
anion exchanger isoform 1
- band 3cd
cytoplasmic domain of anion exchanger band 3
- CaM
calmodulin
- Ca2+/CaM
Ca2+ saturated CaM
- FERM
Four.one/Ezrin/Radixin/Moesin
- GPC
glycophorin C
- GST
glutathione-S-transferase
- NHE1
Na+/H+ exchanger isoform 1
- PC, NHE1cd
cytoplasmic domain of NHE1; phosphatidylcholine
- PIP2
phosphatidylinositol-4,5 bisphosphate
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Footnotes
AUTHOR CONTRIBUTION
Wataru Nunomura designed, performed, analyzed and interpreted the binding assays and wrote the manuscript. Sheryl Denker performed in vitro pull down assays and Diane Barber contributed guidance and expertise on NHE1 and ERM proteins. Yuichi Takakuwa contributed to interpretation of the data. Philippe Gascard conceived the overall study, cloned some of the constructs and edited the manuscript.
REFERENCES
- 1.Sarangarajan R, Dhabia N, Soleimani M, Baird N, Joiner C. NHE-1 is the sodium-hydrogen exchanger isoform present in erythroid cells. Biochim. Biophys. Acta. 1998;1374:56–62. doi: 10.1016/s0005-2736(98)00130-8. [DOI] [PubMed] [Google Scholar]
- 2.Slepkov E, Fliegel L. Structure and function of the NHE1 isoform of the Na+/H+ exchanger. Biochem. Cell Biol. 2002;80:499–508. doi: 10.1139/o02-151. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi H, Szaszi K, Grinstein S. Multiple modes of regulation of Na+/H+ exchangers. Ann. N. Y. Acad. Sci. 2002;976:248–258. doi: 10.1111/j.1749-6632.2002.tb04747.x. [DOI] [PubMed] [Google Scholar]
- 4.Meima ME, Mackley JR, Barber DL. Beyond ion translocation: structural functions of the sodium-hydrogen exchanger isoform-1. Curr. Opin. Nephrol. Hypertens. 2007;16:365–372. doi: 10.1097/MNH.0b013e3281bd888d. [DOI] [PubMed] [Google Scholar]
- 5.Stock C, Schwab A. Role of the Na+/H+ exchanger NHE1 in cell migration. Acta Physiol (Oxf) 2006;187:149–157. doi: 10.1111/j.1748-1716.2006.01543.x. [DOI] [PubMed] [Google Scholar]
- 6.Slepkov ER, Rainey JK, Sykes BD, Fliegel L. Structural and functional analysis of the Na+/H+ exchanger. Biochem. J. 2007;401:623–633. doi: 10.1042/BJ20061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denker SP, Barber DL. Ion transport proteins anchor and regulate the cytoskeleton. Curr Opin Cell Biol. 2002;14:214–220. doi: 10.1016/s0955-0674(02)00304-6. 2002. Review. [DOI] [PubMed] [Google Scholar]
- 8.Wooden JM, Finney GL, Rynes E, Maccoss MJ, Lambert AJ, Robledo RF, Peters LL, Gilligan DM. Comparative proteomics reveals deficiency of SLC9A1 (sodium/hydrogen exchanger NHE1) in beta-adducin null red cells. Br. J. Haematol. 2011;154:492–501. doi: 10.1111/j.1365-2141.2011.08612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi ZT, Afzal V, Coller B, Patel D, Chasis JA, Parra M, Lee G, Paszty C, Stevens M, Walensky L, Peters LL, Mohandas N, Rubin E, Conboy JG. Protein 4.1R-deficient mice are viable but have erythroid membrane skeleton abnormalities. J. Clin. Invest. 1999;103:331–340. doi: 10.1172/JCI3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera A, De Franceschi L, Peters LL, Gascard P, Mohandas N, Brugnara C. Effect of complete protein 4.1R deficiency on ion transport properties of murine erythrocytes. Am. J. Physiol. Cell Physiol. 2006;291:C880–C886. doi: 10.1152/ajpcell.00436.2005. [DOI] [PubMed] [Google Scholar]
- 11.Diakowski W, Grzybek M, Sikorski AF. Protein 4.1, a component of the erythrocyte membrane skeleton and its related homologue proteins forming the protein 4.1/FERM superfamily. Folia Histochemica et Cytobiologica. 2006;44:231–248. [PubMed] [Google Scholar]
- 12.Nunomura W, Takakuwa Y. Regulation of protein 4.1R interactions with membrane proteins by Ca2+ and calmodulin. Front BioSci. 2006;11:1522–1539. doi: 10.2741/1901. [DOI] [PubMed] [Google Scholar]
- 13.Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. An alternative binding site for an element of the membrane skeleton. J. Biol. Chem. 1985;260:3676–3683. [PubMed] [Google Scholar]
- 14.Jöns T, Dreckahn D. Identification of the binding interface involved in linkage of cytoskeletal protein 4.1 to the erythrocyte anion exchanger. EMBO J. 1992;11:2863–2867. doi: 10.1002/j.1460-2075.1992.tb05354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombardo CR, Willardson BM, Low PS. Localization of the protein 4.1-binding site on the cytoplasmic domain of erythrocyte membrane band 3. J. Biol. Chem. 1992;267:9540–9546. [PubMed] [Google Scholar]
- 16.Anderson RA, Lovrien RE. Glycophorin is linked by band 4.1 protein to the human erythrocyte membrane skeleton. Nature. 1984;307(5952):655–658. doi: 10.1038/307655a0. [DOI] [PubMed] [Google Scholar]
- 17.Nunomura W, Takakuwa Y, Tokimitsu R, Kawashima M, Krauss S, Mohandas N. Regulation of CD44-protein 4.1 interaction by Ca2+ and calmodulin: implications for modulation of CD44-ankyrin interaction. J. Biol. Chem. 1997;272:30322–30328. doi: 10.1074/jbc.272.48.30322. [DOI] [PubMed] [Google Scholar]
- 18.Marfatia SM, Morais-Cabral JH, Kim AC, Byron O, Chishti AH. The PDZ domain of human erythrocyte p55 mediates its binding to the cytoplasmic carboxyl terminus of glycophorin C. Analysis of the binding interface by in vitro mutagenesis. J. Biol. Chem. 1997;272:24191-2417. doi: 10.1074/jbc.272.39.24191. [DOI] [PubMed] [Google Scholar]
- 19.Nunomura W, Takakuwa Y, Parra M, Conboy J, Mohandas N. Regulation of protein 4.1R, p55, and glycophorin C ternary complex in human erythrocyte membrane. J. Biol. Chem. 2000;275:24540–24546. doi: 10.1074/jbc.M002492200. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T, Kadowaki K, Lazarides E, Sobue K. Ca2+-dependent regulation of the spectrin/actin interaction by calmodulin and protein 4.1. J. Biol. Chem. 1991;266:1134–1140. [PubMed] [Google Scholar]
- 21.Takakuwa Y, Mohandas N. Modulation of erythrocyte membrane material properties by Ca2+ and calmodulin. Implications for their role in regulation of skeletal protein interactions. J. Clin. Invest. 1998;82:394–400. doi: 10.1172/JCI113611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunomura W, Gascard P, Takakuwa Y. Insights into the function of the unstructured N-Terminal domain of proteins 4.1R and 4.1G in Erythropoiesis. Int. J. Cell Biol. 2011;2011:943272. doi: 10.1155/2011/943272. (2011), 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunomura W, Takakuwa Y, Parra M, Conboy J, Mohandas N. Ca2+-dependent and Ca2+-independent calmodulin binding sites in erythrocyte protein 4.1. Implications for regulation of protein 4.1 interactions with transmembrane proteins. J. Biol. Chem. 2000;275:6360–6367. doi: 10.1074/jbc.275.9.6360. [DOI] [PubMed] [Google Scholar]
- 24.Nunomura W, Takakuwa Y, Cherr GN, Murata K. Characterization of Protein 4.1R in erythrocytes of zebrafish (Danio rerio: Unique binding properties with transmembrane proteins and calmodulin. Comp. Biochem. Physiol. Part B. 2007;148:124–138. doi: 10.1016/j.cbpb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Aharonovitz O, Zaun HC, Balla T, York JD, Orlowski J, Grinstein S. Intracellular pH regulation by Na+/H+ exchange requires phosphatidylinositol 4,5-bisphosphate. J. Cell. Biol. 2000;150:213–224. doi: 10.1083/jcb.150.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na+/H+ exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell. 2000;6:1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 27.Nunomura W, Sasakura D, Shiba K, Nakamura S, Kidokoro SI, Takakuwa Y. Structural stabilization of protein 4.1R FERM domain upon binding to apo-calmodulin: novel insights into the biological significance of the calcium-independent binding of calmodulin to protein 4.1R. Biochem. J. 2011;440:367–374. doi: 10.1042/BJ20110676. [DOI] [PubMed] [Google Scholar]
- 28.Gascard P, Nunomura W, Lee G, Walensky LD, Krauss SW, Takakuwa Y, Chasis JA, Mohandas N, Conboy JG. Deciphering the nuclear import pathway for the cytoskeletal red cell protein 4.1R. Mol. Biol. Cell. 1999;10:1783–1798. doi: 10.1091/mbc.10.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wetlaufer DB. Ultraviolet spectra of proteins and amino acids. Adv. Protein Chem. 1962;17:303–391. [Google Scholar]
- 30.Niggli V, Andréoli C, Roy C, Mangeat P. Identification of a phosphatidyl-inositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 1995;376:172–176. doi: 10.1016/0014-5793(95)01270-1. [DOI] [PubMed] [Google Scholar]
- 31.An X, Zhang X, Debnath G, Baines AJ, Mohandas N. Phosphatidylinositol-4,5-biphosphate (PIP2) differentially regulates the interaction of human erythrocyte protein 4.1 (4.1R) with membrane proteins. Biochemistry. 2006;45:5725–3572. doi: 10.1021/bi060015v. [DOI] [PubMed] [Google Scholar]
- 32.Omoe K, Nunomura W, Kato H, Li ZJ, Igarashi O, Araake M, Sano K, Ono HK, Abe Y, Hu DL, Nakane A, Kiyono H, Takakuwa Y, Shinagawa K, Uchiyama T, Imanishi K. High affinity of interaction between super- antigen and T cell receptor Vbeta molecules induces a high level and prolonged expansion of superantigen-reactive CD4+ T cells. J. Biol. Chem. 2010;285:30427–30435. doi: 10.1074/jbc.M110.140871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denker SP, Barber DL. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 2002;159:1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cush R, Cronin JM, Stewart WJ, Maule CH, Molloy J, Goddard NJ. The resonant mirror: a novel optical biosensor for direct sensing of biomolecular interactions Part I: Principle of operation and associated instrumentation. Biosensors & Bioelectronics. 1993;8:347–353. [Google Scholar]
- 35.Nunomura W, Kinoshita K, Parra M, Gascard P, An X, Mohandas N, Takakuwa Y. Similarities and differences in the structure and function of 4.1G and 4.1R135, two protein 4.1 paralogs expressed in erythroid cells. Biochem. J. 2010;432:407–446. doi: 10.1042/BJ20100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunomura W, Parra M, Hebiguchi H, Sawada K, Mohandas N, Takakuwa Y. Marked difference in membrane-protein-binding properties of the two isoforms of protein 4.1R expressed at early and late stages of erythroid differentiation. Biochem. J. 2009;417:141–148. doi: 10.1042/BJ20081372. [DOI] [PubMed] [Google Scholar]
- 37.Gascard P, Pawelczyk T, Lowenstein JM, Cohen CM. The role of inositol phospholipids in the association of band 4.1 with the human erythrocyte membrane. Eur. J. Biochem. 1993;211:671–681. doi: 10.1111/j.1432-1033.1993.tb17595.x. [DOI] [PubMed] [Google Scholar]
- 38.Wakabayashi S, Bertrand B, Ikeda T, Pouysségur J, Shigekawa M. Mutation of calmodulin-binding site renders the Na+/H+ exchanger (NHEI) highly H+-sensitive and Ca2+ regulation-defective. J. Biol. Chem. 1994;269:13710–13715. [PubMed] [Google Scholar]
- 39.Bertrand B, Wakabayashi S, Ikeda T, Pouyseégur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHEl) is a novel member of the calmodulin-binding proteins: identification and characterization of calmodulin-binding sites. J. Biol. Chem. 1994;269:13703–13709. [PubMed] [Google Scholar]
- 40.Quist E, Powell P. Polyphosphoinositides and the shape of mammalian erythrocytes. Lipids. 1985;20:433–438. doi: 10.1007/BF02534234. [DOI] [PubMed] [Google Scholar]
- 41.Han BG, Nunomura W, Takakuwa Y, Mohandas N, Jap JK. Protein 4.1R core domain structure and insights into regulation of cytoskeletal organization. Nat. Struct. Biol. 2000;7:871–875. doi: 10.1038/82819. [DOI] [PubMed] [Google Scholar]
- 42.Ammar YB, Takeda S, Hisamitsu T, Mori H, Wakabayashi S. Crystal structure of CHP2 complexed with NHE1-cytosolic region and an implication for pH regulation. EMBO J. 2006;25:2315–2325. doi: 10.1038/sj.emboj.7601145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köster S, Pavkov-Keller T, Kuhlbraandt W, Yildiz Ö. Structure of human Na+/H+ exchanger NHE1 regulatory region in complex with CaM and Ca2+ J. Biol. Chem. 2011;286:40954–40961. doi: 10.1074/jbc.M111.286906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nørholm AB, Hendus-Altenburger R, Bjerre G, Kjaergaard M, Pedersen SF, Kragelund BB. The intracellular distal tail of the Na+/H+ exchanger NHE1 is intrinsically disordered: implications for NHE1 trafficking. Biochemistry. 2011;50:3469–3480. doi: 10.1021/bi1019989. [DOI] [PubMed] [Google Scholar]
- 45.Berridge MJ, Bootman MD, Roderick HL. Calcium signaling: dynamics, homeostasis and remodeling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 46.Yi YH, Ho PY, Chen TW, Lin WJ, Gukassyan V, Tsai TH, Wang DW, Lew TS, Tang CY, Lo SJ, Chen TY, Kao FJ, Lin CH. Membrane targeting and coupling of NHE1-integrin alphaII bbeta3-NCX1 by lipid rafts following integrin-ligand interactions trigger Ca2+ oscillations. J. Biol Chem. 2009;284:3855–3864. doi: 10.1074/jbc.M804334200. [DOI] [PubMed] [Google Scholar]
- 47.Malo ME, Fliegel L. Physiological role and regulation of the Na+/H+ exchanger. Can. J. Physiol. Pharmacol. 2006;84:1081–1095. doi: 10.1139/y06-065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Changes in Kd resulted primarily from changes in ka, although both kd and ka changed significantly between pH 7.0 and 7.5.
The residues E38ED in human 4.1R FERM domain and the corresponding residues, R40EWV, in human ezrin are depicted using a Stick-ball model. The PDB accession ID is 1gg3 and 1ni2 for human 4.1R and ezrin FERM domains, respectively.