SUMMARY
MeCP2 is a nuclear protein with an affinity for methylated DNA that can recruit histone deacetylases. Deficiency or excess of MeCP2 causes severe neurological problems, suggesting that the number of molecules per cell must be precisely regulated. We quantified MeCP2 in neuronal nuclei and found that it is nearly as abundant as the histone octamer. Despite this high abundance, MeCP2 associates preferentially with methylated regions and high-throughput sequencing showed that its genome-wide binding tracks methyl-CpG density. MeCP2 deficiency results in global changes in neuronal chromatin structure, including elevated histone acetylation and a doubling of histone H1. Neither change is detectable in glia, where MeCP2 occurs at lower levels. The mutant brain also shows elevated transcription of repetitive elements. Our data argue that MeCP2 may not act as a gene-specific transcriptional repressor in neurons, but might instead dampen transcriptional noise genome-wide in a DNA methylation-dependent manner.
INTRODUCTION
Mammalian genomes exhibit tissue-specific patterns of DNA methylation, predominantly at the dinucleotide sequence CpG. In most tissues, the bulk genome has a relatively low CpG density, but is heavily (~70%) methylated (Ehrlich et al., 1982). This global distribution is interrupted by regions of high CpG density, termed CpG islands (CGIs), which are typically unmethylated and associated with gene promoters (Bird et al., 1985). The effects of methylated and non-methylated CpGs on chromatin metabolism are mediated by proteins that are either attracted or repelled by that modification state. MeCP2 is one of several proteins that bind methylated DNA in vivo and in vitro and is therefore a potential interpreter of the DNA methylation pattern (Lewis et al., 1992). MeCP2 is most highly expressed in the brain, specifically in neurons (Kishi and Macklis, 2004), where neuronal expression rises postnatally as the final stages of neurogenesis are completed (Balmer et al., 2003).
Mutations in the X-linked MECP2 gene are the primary cause of the autism spectrum disorder Rett syndrome (Amir et al., 1999). In Rett patients, apparently normal development gives way to regression after 6-12 months with loss of acquired skills including speech and mobility (Armstrong, 2002). The Mecp2-null mouse recapitulates several features of Rett syndrome, showing a late-onset neurodevelopmental phenotype leading to death at ~12 weeks (Chen et al., 2001; Guy et al., 2001). This link between MeCP2 and a neurological disorder suggests that brain expression is of key functional importance. Accordingly, brain-specific depletion of MeCP2 produced mice that are indistinguishable from Mecp2-null animals (Chen et al., 2001; Guy et al., 2001). Furthermore, a mouse model expressing MeCP2 under the Tau promoter, which is primarily neuron-specific, rescued the null animals (Luikenhuis et al., 2004). These observations highlight the importance of MeCP2 function in neurons.
Transfection experiments showed that MeCP2, when recruited to a promoter via a GAL4 binding domain, could repress transcription and that this repression was in part mediated by an interaction with histone deacetylase (HDAC) containing complexes (Nan et al., 1998). These findings suggested that MeCP2 is a transcriptional repressor targeted to specific genes via DNA methylation. While subsequent studies have identified reproducible gene expression changes between wildtype and Mecp2-null mouse brains, these are generally rather subtle (Jordan et al., 2007; Nuber et al., 2005), querying the view that MeCP2 acts as a classical transcriptional repressor. In contrast to the original model, recent studies have proposed that MeCP2 modulates alternative splicing (Young et al., 2005) and acts as a transcriptional activator by recruiting the transcription factor CREB to specific genes (Chahrour et al., 2008). In addition, there have been reports that MeCP2 binding to DNA may not depend on methylation, as studies using in vitro chromatin assembly suggested that MeCP2 can bind to both methylated and non-methylated DNA and mediate nucleosomal compaction (Georgel et al., 2003; Nikitina et al., 2006). Support for this view has come from chromatin immunoprecipitation-microarray (ChIP-chip) experiments using a neuronal cultured cell line. The report claimed that MeCP2 does not selectively bind methylated promoters, but instead is bound at many unmethylated promoters and has a similar binding pattern to RNA polymerase II (Yasui et al., 2007). These findings call for a re-evaluation of the conventional view that MeCP2 recruits HDACs to methylated sites.
To distinguish these alternative hypotheses for MeCP2 function, we measured its abundance and distribution in the mature mouse brain. An additional motivation was to explain why MeCP2 abundance is so critical for its function. For example, expression of MeCP2 from transgenes in mice only brings about phenotypic rescue if the level of protein is close to that in wildtype brain; over-expression by ~2-fold is severely detrimental (Collins et al., 2004; Luikenhuis et al., 2004). These findings are mirrored in humans where MECP2 gene duplication events give rise to developmental delay and mental retardation (Lubs et al., 1999). We developed a technique to purify neuronal nuclei from the mature mouse brain and found that the absolute abundance of MeCP2 approaches the number of nucleosomes and methyl-CpG moieties in a diploid genome. Consistent with its high abundance, ChIP analysis showed MeCP2 to be globally distributed and to track the density of methyl-CpG in the genome. Bisulfite sequencing of the immunoprecipitated DNA confirmed highly selective binding to methylated DNA sequences. In the absence of MeCP2, histone H3 acetylation levels were globally elevated and histone H1 levels were doubled, suggesting that MeCP2 alters the global chromatin state. These changes were not seen in glia or other tissues where MeCP2 is much less abundant. We propose that a key neuron-specific role of MeCP2 is to impact the entire genome, rather than to act as a gene-specific regulator.
RESULTS
MeCP2 is enriched in neuronal nuclei and almost as abundant as nucleosomes
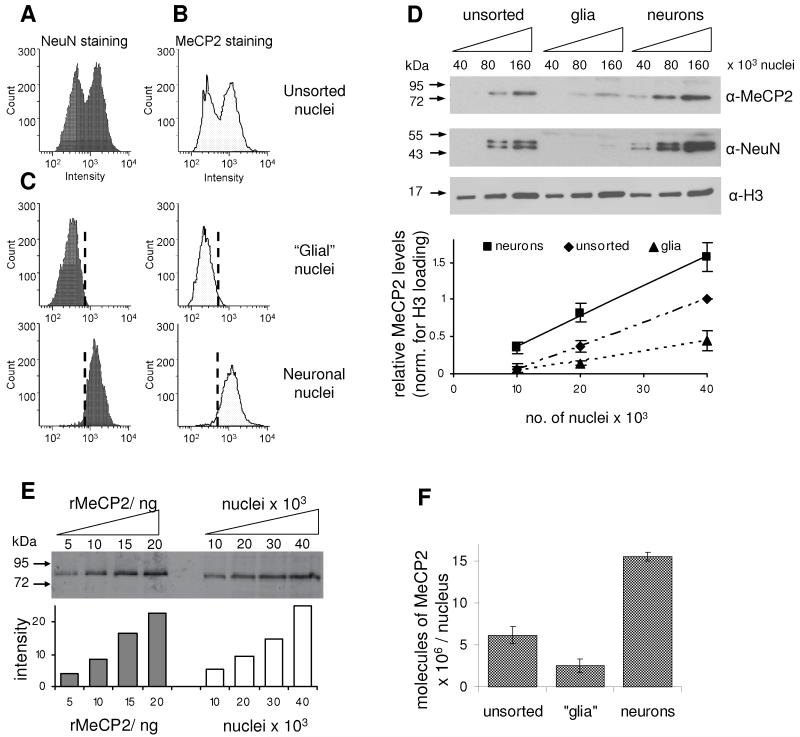
As the correct expression level seems to be crucial to MeCP2 function (Collins et al., 2004; Guy et al., 2001; Luikenhuis et al., 2004), we wanted to determine the absolute abundance of MeCP2 in the mature brain. We first asked when during postnatal development MeCP2 reaches its maximum level by quantitative western blotting using mice of different ages (Fig S1A). MeCP2 was weakly detectable in neonatal brain, but increased rapidly to reach a maximum at ~5 weeks of age, after which total levels remained constant. We decided to use 6-8 week old mice for all subsequent experiments. Immunofluorescence data have indicated that MeCP2 is predominantly expressed in neuronal nuclei, with much less being seen in glia (Kishi and Macklis, 2004). In order to focus on neurons, a procedure was developed to sort neuronal from glial nuclei using fluorescence activated cell sorting (FACS). The vast majority of neuronal nuclei are positive for staining by the neuronal nuclear marker NeuN, whereas glial nuclei are negative (Mullen et al., 1992). NeuN staining of total brain nuclei gave a bimodal intensity distribution comprising 50% NeuN positive nuclei (neurons) and 50% negative nuclei (predominantly glia; Fig 1A). Staining of brain nuclei for MeCP2 gave a similar bimodal distribution: 56% high and 44% low MeCP2 staining (Fig 1B), with an approximately 6-fold difference in staining intensity. Sorting of nuclei on the basis of the NeuN staining purified a neuronal population and a predominantly glial population of nuclei (Fig 1C). As expected, NeuN-positive nuclei co-sorted with the high MeCP2 expressing nuclei.
Fig 1.
The abundance of MeCP2 in purified neuronal nuclei approaches that of the histone octamer. Nuclei were isolated from wildtype whole mouse brain and then FACS-stained for (A) NeuN and (B) MeCP2. (C) Nuclei sorted for NeuN-negative and NeuN-positive staining co-sorted with low-MeCP2 and high-MeCP2 stained nuclei, respectively. (D) Infra-red western blotting for NeuN, MeCP2 and histone H3 was performed on unsorted and FACS sorted nuclei. The graph indicates quantification of western signals normalised for H3 loading. (E) The level of MeCP2 in unsorted brain nuclei was quantified against recombinant MeCP2 by infra-red western blotting. The graph below indicates densitometric analysis using Licor Odyssey (F) Densitometric analysis of western blots to determine absolute abundance of MeCP2 in the different nuclei populations. The positions of molecular weight markers (kDa) are indicated. Error bars indicate the mean +/− SEM. See also Fig S1.
Quantitative western blotting using infra-red imaging confirmed that both NeuN and MeCP2 levels were increased in the neuronal population and depleted in the predominantly glial population relative to unsorted brain nuclei (Fig 1D). We detected approximately 7-fold more MeCP2 in NeuN-positive versus NeuN-negative populations, similar to the difference in the staining intensities recorded by FACS (about 6-fold). Quantification of recombinant MeCP2 against a BSA standard (Fig S1B) allowed us to determine the absolute abundance of MeCP2 in unsorted nuclei isolated from mature mouse brain as 6 × 106 molecules per nucleus (Fig 1E and S1C). This result was verified using an independent MeCP2 antibody (Fig S1D). The amount of MeCP2 in sorted neuronal nuclei (NeuN positive) was 16 × 106 molecules per nucleus, compared with ~2 × 106 molecules per nucleus in the predominantly glial nuclear fraction (Fig 1F). For comparison, the abundance of MeCP2 in liver nuclei was 0.5 × 106 molecules per nucleus (Fig S1E). FACS staining suggested there may be some heterogeneity within the populations, indicating that these average values may not apply to each cell, as previously suggested (LaSalle et al., 2001). To relate MeCP2 levels to the core components of chromatin, we determined the abundance of histone H4 as 64 × 106 molecules per brain nucleus (Fig S1F), which corresponds to 32 × 106 nucleosomes in total, or one nucleosome per 165 base pairs of genomic DNA. The estimated number of methyl CpGs per diploid nucleus is 40 × 106 methylated CpG sites. Therefore, the number of MeCP2 molecules in the nucleus of a mature neuron approaches the number of histone octamers and methyl-CpG sites and may be sufficient to almost “saturate” the genome.
MeCP2 binds globally across the genome in the mouse brain
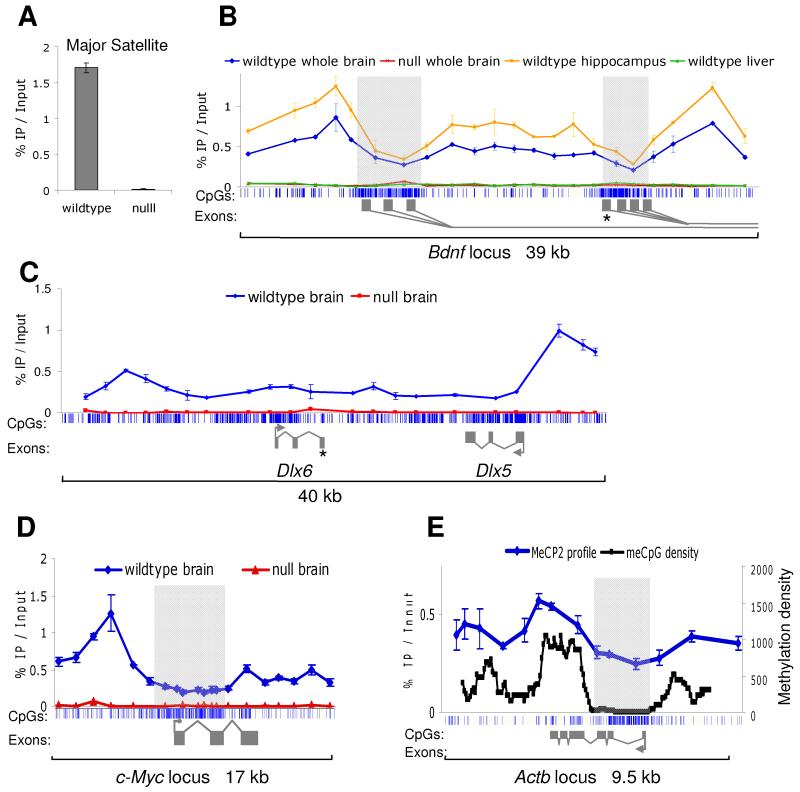
Previous reports using cultured neurons from embryonic or immature mice have suggested that MeCP2 binds at discrete genomic sites, such as at the Brain derived neurotrophic factor gene locus (Bdnf) (Chen et al., 2003) and at the Dlx5/6 locus (Horike et al., 2005). As MeCP2 levels are low in immature mouse brain compared to the high abundance later on (Fig S1), the MeCP2 binding pattern in mature mice was determined. We used whole brain nuclei for MeCP2 chromatin immunoprecipitation (ChIP) as approximately ~89% of brain MeCP2 is derived from neuronal nuclei. We first tested the specificity of the 674 MeCP2 antibody (Nan et al., 1998) using the 234 base pair mouse major satellite repeat, a known MeCP2 binding sequence, as a ChIP probe (Nan et al., 1996). Despite the MeCP2 antibody displaying relatively low efficiency (1.7% IP/Input), it was highly specific, exhibiting a 130-fold enrichment in wildtype compared to Mecp2-null mouse brain (Fig 2A). We concluded that this degree of specificity was sufficient for analysis of MeCP2 binding in the genome.
Fig 2.
MeCP2 shows widespread binding across gene loci in mature brain. Mouse tissues (brain, hippocampus, liver) were dissected and ChIP performed using an antibody against MeCP2. Immunoprecipitated DNA was analysed by real time PCR using a panel of primers. (A) ChIP of the major satellite repeat shows 130-fold enrichment in wildtype brain compared to Mecp2-null brain, indicating that the antibody is highly specific. (B) MeCP2 binding profile across 39 kb of the promoter region of Bdnf. The blue vertical lines below the graph indicate CpG sites, with CGIs shaded in light grey. Alternative Bdnf exons 1 are indicated with dark grey rectangles. The asterisk marks the discreet binding site identified using cultured embryonic cortical neurons (Chen et al, 2003). ChIP was performed on various tissues as indicated. (C) MeCP2 ChIP profile across a 40 kb region encompassing the Dlx5/6 locus. The asterisk marks the key site identified using brains of 1 day old mice. Wildtype whole brain is shown in blue and Mecp2-null whole brain is shown in red. (D) MeCP2 binding profile across the c-Myc locus in wildtype (blue) and Mecp2-null (red) brains. (E) MeCP2 binding profile across the Actb locus follows the meCpG density. MeCP2 ChIP was performed on wildtype brain (blue). Total brain genomic DNA was subjected to bisulfite sequencing for a continuous run of 262 CpG sites across ~9 kb. The methylation density is plotted based on a window size of 650 bp and step of 50 bp (shown in black). Error bars indicate mean +/− SEM.
Initially, we examined binding to the mouse Bdnf locus, which has 8 alternative first exons, most of which are located in two large CGIs, with a large intervening intron. A panel of 22 ChIP primers was designed across a 39 kb region spanning the CGIs and the MeCP2 binding profile was determined in mature mouse brain using ChIP and quantitative PCR. We observed significant enrichment across the entire region compared with the Mecp2-null brain, suggesting that MeCP2 is broadly distributed at the locus (Fig 2B). Interestingly, MeCP2 binding over the Bdnf CGIs is reduced relative to the surrounding regions, raising the possibility that these active promoters fail to attract MeCP2 due to their lack of DNA methylation. The intervening regions are CpG deficient by comparison with CGIs, but limited bisulfite DNA sequencing confirmed that they are largely methylated (data not shown). MeCP2 ChIP over the same region in wildtype liver gave a profile indistinguishable from the Mecp2-null brain, suggesting this profile is a result of the high abundance in neurons. It was possible that the MeCP2 binding pattern in the whole brain is an aggregate of a large number of distinct patterns from individual regions and neuronal subtypes. To address this, the ChIP was repeated using a specific brain region - the hippocampus, which represents a relatively restricted subset of neuronal subtypes. The resulting MeCP2 binding profile was very similar to that of the wildtype whole brain, with MeCP2 bound over the entire locus but reduced over the CGIs (Fig 2B). This finding makes it likely that the ChIP results represent a consistent pattern across many types of neuron.
A similar approach was used to look at the MeCP2 binding across a 40 kb region encompassing the Dlx5/6 locus. ChIP using mature mouse brain gave a reproducibly different pattern from that previously reported in neonatal mice by (Horike et al., 2005), with MeCP2 bound across the entire region (Fig 2C). ChIP signal over the same region in Mecp2-null brain was minimal, verifying the significance of the data. We next chose to examine MeCP2 occupancy over two housekeeping genes, c-Myc and Actb, which have unmethylated CGIs flanked by methylated DNA that is comparatively CpG deficient. ChIP across the c-Myc locus showed MeCP2 bound to the methylated regions flanking the CGI, but, as in the case of Bdnf, MeCP2 binding was depleted across the CGI (Fig 2D). Analysis of 9.5 kb surrounding the Actb locus, showed a similar pattern with higher MeCP2 occupancy across the flanks, but depleted binding over the CGI (Fig 2E). In order to relate this result to methyl-CpG density, we performed bisulfite sequencing to unambiguously determine modification of all 262 CpG sites across the Actb region (Fig 2E). The results confirm that the CGI is DNA methylation-free, whereas the flanking region is predominantly methylated. Due to the variable CpG density, the methylation level varies dramatically across the locus. Significantly, the MeCP2 binding profile mirrors the methylation density through the region. Taken together, the results of these ChIP experiments fit with the hypothesis that MeCP2 binding tracks methyl-CpG density.
MeCP2 tracks the meCpG density of the genome
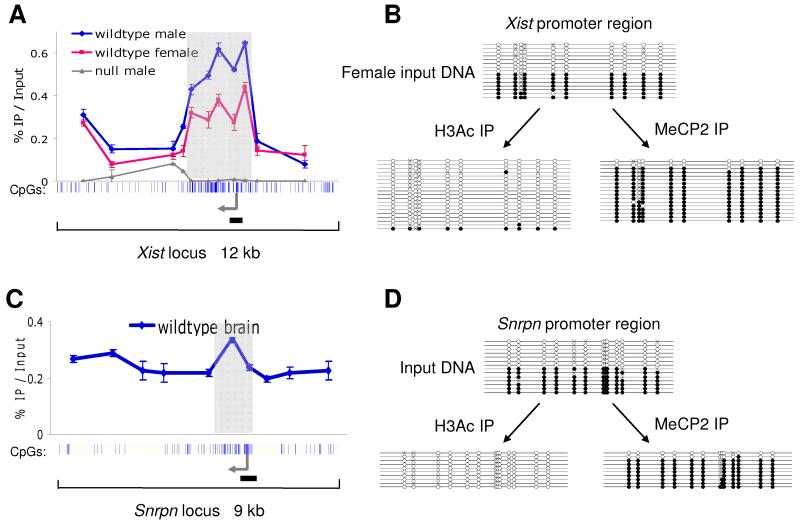
In the light of its high abundance and potentially global binding, we tested whether MeCP2 acts as a methyl-CpG binding protein in neurons or whether it interacts with chromatin regardless of DNA methylation as proposed by in vitro chromatin assembly experiments (Georgel et al., 2003; Nikitina et al., 2006). We selected genomic regions that occur in both methylated and non-methylated states within each cell and asked which of the two forms bound to MeCP2. First, we characterised binding over 12 kb of the X-linked Xist locus, comparing male to female mouse brains (Fig 3A). In male mice there is a single methylated copy of the Xist promoter CGI, whereas in females there are two copies, one methylated and one non-methylated (Hendrich et al., 1993). In the male brain, MeCP2 occupies the entire region, peaking over the densely methylated CGI. The finding that MeCP2 occupancy drops over non-methylated CGIs (Fig 2), but peaks over methylated CGIs implies that MeCP2 is tracking the density of methyl-CpG.
Fig 3.
Brain MeCP2 binds selectively to methylated DNA in vivo. Whole mouse brains were dissected and ChIP performed using antibodies against MeCP2 and acetylated histone H3. Input DNA and immunoprecipitated DNA were analysed by real time PCR and subjected to bisulfite sequencing. (A) MeCP2 binding profile across 12 kb of the Xist locus in wildtype male brain (blue), wildtype female brain (pink) and Mecp2-null brain (grey). The blue vertical lines below the graph indicate CpG sites, with the CGI shaded in light grey. The transcription start site is indicated with an arrow. The horizontal black line marks the region amplified for bisulfite sequencing. (B) ChIP was performed on the wildtype female brain for MeCP2 and histone H3 acetylation. Recovered DNA was used for bisulfite sequencing of a region of the Xist promoter. Each line represents a single clone. Open and filled circles indicate non-methylated and methylated CpG sites, respectively. Crosses indicate uncharacterised CpG sites. (C) MeCP2 ChIP profile across 9 kb of the imprinted Snrpn locus in wildtype brain. (D) ChIP was performed on the wildtype brain for MeCP2 and histone H3 acetylation. The resulting DNA was used for bisulfite sequencing of a region of the Snrpn promoter. Error bars indicate mean +/− SEM.
In females, MeCP2 occupancy flanking the Xist CGI is indistinguishable from the male profile. Over the Xist CGI, however, MeCP2 binding is reduced by half relative to the male brain (Fig 3A). This result is consistent with the notion that MeCP2 is only bound to one of the two Xist CGI alleles in the female. To test this more rigorously, we combined the ChIP protocol with bisulfite sequencing to determine the methylation status of the immunoprecipitated DNA. Input DNA from female brain gave approximately equal numbers of non-methylated and methylated clones, as expected, confirming unbiased PCR amplification (Fig 3B). As a control, we bisulfite sequenced Xist DNA immunoprecipitated by an antibody against acetylated histone H3. This histone mark is specific for the active Xist allele and duly recovered the non-methylated allele in 97% of clones (n=39 clones). Following ChIP with anti-MeCP2 antibody, however, 88% of the recovered clones were methylated (n=58 clones). A similar result was obtained by analysing 9 kb of the imprinted Snrpn locus, which has a paternal unmethylated allele and a maternal methylated allele (Sutcliffe et al., 1994). MeCP2 is bound through the entire region, peaking over the imprinted CGI (Fig 3C). Using the ChIP-bisulfite protocol, the input DNA gave approximately equal numbers of non-methylated and methylated clones (Fig 3D). As before, acetylated histone H3 ChIP recovered 100% non-methylated clones (n=23), whereas 89% of the clones obtained by MeCP2 ChIP were methylated (n=19). We conclude that MeCP2 preferentially binds to the methylated allele in each case.
High throughput sequencing confirms genome wide binding
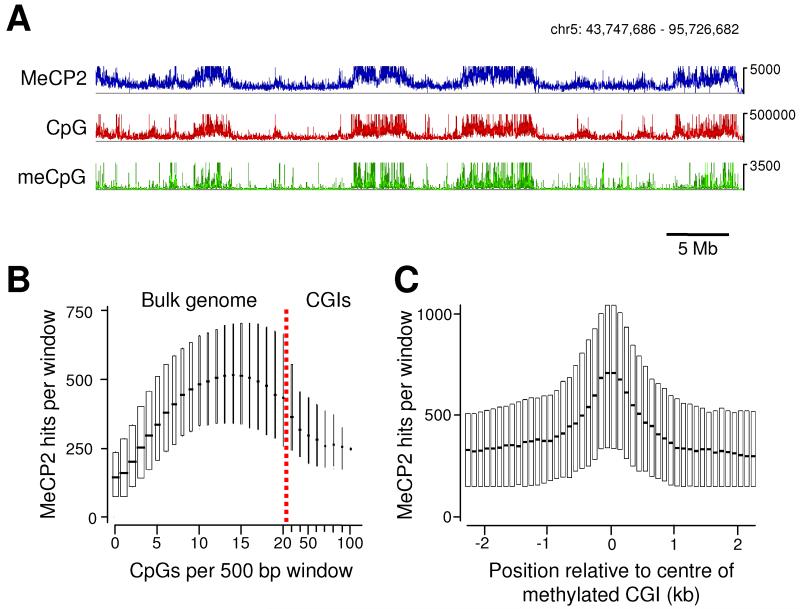
Analysis of candidate loci across ~150 kb of the mouse brain genome suggests that MeCP2 is widely distributed. To view MeCP2 occupancy genome-wide, we performed high throughput DNA sequencing of total MeCP2-bound chromatin. In all, 2.9 × 109 bases were sequenced, of which 1.3 × 109 were uniquely mappable. Despite relatively extensive sequencing, clear peaks were not identified. Instead sequencing reads were dispersed throughout the genome, covering 56% of the mouse genome (2.5 × 109 bp of which 1.8 × 109 is repeat masked). This result fits with our analysis of candidate regions by ChIP-PCR. Since MeCP2 binding is genome-wide, it would be necessary to sequence the entire genome several-fold to obtain a comprehensive high-resolution profile. With 56% genome coverage, however, we were able to robustly test for a relationship with DNA methylation by comparing the profile of the CpG sequence in a sliding 5 kb window with that of MeCP2. The match was striking (Fig 4A), suggesting that MeCP2 binding coincides with this dinucleotide sequence, which is ~70% methylated in brain DNA (Ehrlich et al., 1982). As a more direct test, we determined the distribution of meCpG by fractionating whole genomic DNA based on its affinity for an immobilized methyl-CpG binding domain (Cross et al., 1994; Illingworth et al., 2008). Retained sequences were analyzed by high-throughput sequencing. An earlier comparison between this method of meCpG mapping and use of an anti-meC antibody (MeDIP) indicated that both are equally effective at measuring meCpG density (Zhang et al., 2006). We conclude that in vivo-bound MeCP2 mirrors meCpG over long regions of chromosomal DNA (Fig 4A).
Fig 4.
High throughput sequencing of immunoprecipitated chromatin shows MeCP2 globally distributed and tracking the meCpG density. (A) Profiles of MeCP2-bound sequences (blue), CpG density (red) and parallel sequencing from methyl-CpG-rich sequences (green) were analysed using a sliding window (5 kb window; 1 kb step). A 52 Mb region of chromosome 5 is shown. The vertical axis represents sequencing hits or number of CpGs per window. (B) The genome was scanned using a 500 bp window (100 bp step) and the number of MeCP2 hits per window plotted against the number of CpGs per window. Boxplots are shown with the horizontal line indicating the median and the surrounding box showing the interquartile range. The width of the box is proportional to the fraction of the genome that corresponds with that CpG density. The majority of CGIs contain greater than 5 CpG sites/100 bp as indicated by red line, whereas the bulk genome has a lower CpG density. (C) Methylated CGIs were indentified by methyl-CpG affinity chromatography. The surrounding 5 kb of genomic DNA was analyzed for MeCP2 binding using a 500 bp window (100 bp step). Results are presented as for (B). See Fig S2.
In a second test for the relationship between MeCP2 binding and meCpG, we asked whether the depression of MeCP2 binding seen at non-methylated CGIs associated with the c-Myc, Dlx5/6, Actb and Bdnf loci applied genome-wide. We binned the mouse genome sequence according to CpG density in 500 bp windows and determined the level of MeCP2 binding in each bin. The results (Fig 4B) show that MeCP2 binding initially increases with base composition as predicted by Fig 4A, but drops steeply at densities above 25 CpGs per 500 bp. This density predominantly reflects non-methylated CGIs (Fig S2). Thirdly, we examined the binding of MeCP2 to methylated CGIs, as analysis of the Xist and Snrpn loci suggested increased binding (Fig 3). The results show a peak of MeCP2 binding centred over methylated CGIs (Fig 4C). These findings match closely the predictions based on candidate regions of the genome (Fig 2) and therefore reinforce the conclusion that MeCP2 tracks DNA methylation genome-wide.
MeCP2 globally alters the chromatin state
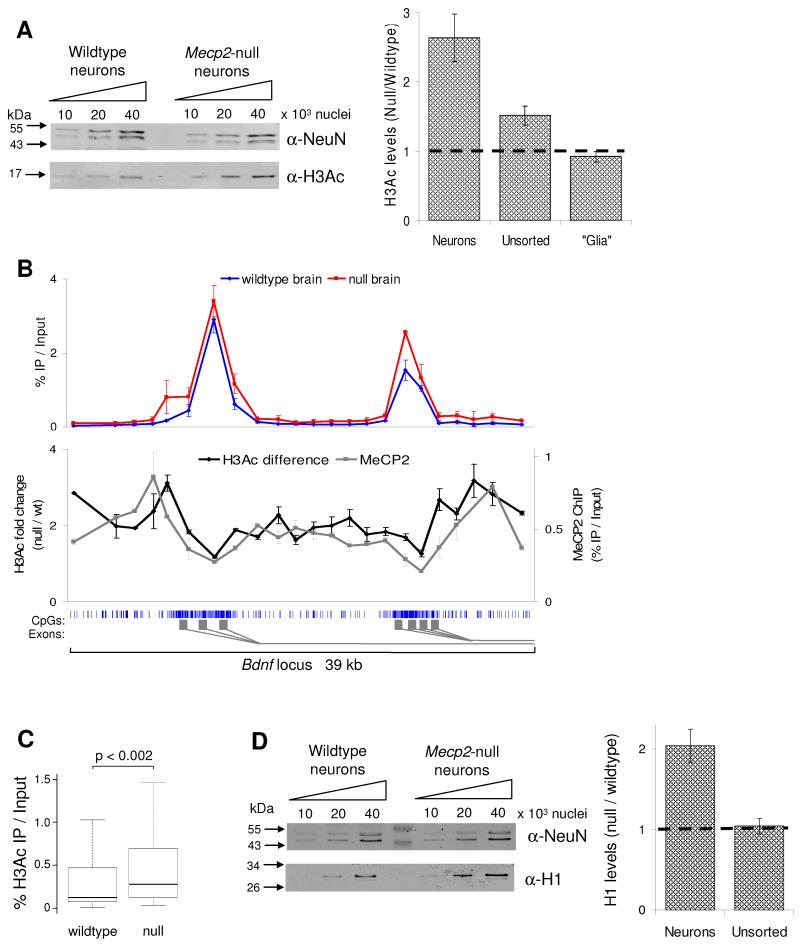
The high abundance of MeCP2 in neurons suggests that it may influence chromatin structure globally. It was previously reported that in the Mecp2-mutant mouse brain levels of histone H3 acetylation (H3Ac) are elevated (Shahbazian et al., 2002). As approximately 89% of MeCP2 is in the neurons, we hypothesised that any differences in chromatin state between wildtype and Mecp2-null brain would be restricted to the neurons, but be absent from glia. To test this, we compared H3Ac levels in unsorted nuclei with purified neuronal and glial nuclei by western blotting (Fig 5A). Unsorted Mecp2-null brain nuclei displayed a 1.5-fold increase in H3Ac compared to wildtype. This difference increased significantly to 2.6-fold in sorted neuronal nuclei (Kolmogorov-Smirnov [KS] test p≤0.01), whereas glial nuclei consistently showed no significant increase in H3Ac (0.9 fold decrease). We conclude that the small elevation of H3Ac seen in whole brain is entirely due to neurons where MeCP2 is highly abundant. Our findings do not corroborate a recent report that histone H3 acetylation is increased in Mecp2-null glia (Ballas et al., 2009).
Fig 5.
MeCP2-deficiency affects the global chromatin state by elevating levels of histone H3 acetylation (H3Ac) and the linker histone H1. (A) H3Ac levels were determined by western blotting of unsorted brain nuclei and FACS purified nuclei from both wildtype and Mecp2-null brain. The western blot shows H3Ac levels in the neuronal nuclei plus histones from the same samples resolved and stained by coomassie blue as a loading control (lower panel). The graph indicates densitometric analysis of H3Ac levels in the different nuclei populations, normalised for differences in loading. The horizontal line represents no change between wildtype and Mecp2-null. (B) The upper graph shows the H3Ac profile across the promoter region of the Bdnf locus in wildtype (blue) and Mecp2-null (red) brains. The lower graph indicates the H3Ac fold difference between wildtype and MeCP2-null brain (pink); the MeCP2 ChIP profile is also shown (black). The blue vertical lines below the graph indicate CpG sites, with the CGI shaded in light grey. The gene structure is indicated by dark grey rectangles below. This data represents the average of 3 independent experiments. (C) Boxplot showing all H3Ac ChIP results for wildtype and Mecp2-null brain (n=100). (D) Quantitative western blotting for histone H1 and NeuN (as a loading control) using FACS purified neuronal nuclei from wildtype and Mecp2-null mice. The graph indicates densitometric analysis of H1 levels in the different nuclei populations. Error bars indicate mean +/− SEM and the KS test was used to determine statistical significance. See also Fig S3.
Widespread chromosome binding by MeCP2 might be expected to modulate chromatin structure globally rather than at specific sites. We asked whether ChIP using an anti-acetyl H3 antibody could detect alterations across specific chromatin domains in the MeCP2-deficient brain. Due to low recovery of neuronal nuclei via FACS, whole brain was used, although this probably leads to an under-estimate of changes in neuronal chromatin. Across the 39 kb Bdnf region, H3Ac peaked over the active promoter CGI regions in both wildtype and null (Fig 5B), but was low in flanking regions. Comparing wildtype and mutant brain, levels of immunoprecipitated DNA in the mutant were elevated by 2-fold relative to input throughout the region (KS-test p<0.004). Interestingly, the magnitude of the effect mirrored the pattern of MeCP2 binding (Fig 5B), being lowest where MeCP2 binds least (the CGIs), but highest where most MeCP2 is relatively concentrated. This is compatible with a simple causal relationship between binding of MeCP2 and levels of histone deacetylation. A 1.4-fold elevation in histone acetylation was also seen in regions of the male Xist locus flanking the CGI (Fig S3; KS-test p=0.03). This difference was notably absent over the Xist CGI itself, implying that repression of histone acetylation at this DNA promoter sequence is not solely dependent on MeCP2. Altogether we examined 100 loci covering 90 kb of genomic DNA by quantitative PCR and found an average 1.4-fold elevation in H3Ac (KS-test: p<0.002). These data indicate that the global chromosomal association of MeCP2 imposes a reduction in histone acetylation levels across the genome (Fig 5C).
Histone H1 is present in most cell types at an approximate stoichiometry of one molecule per nucleosome (Woodcock et al., 2006), but uniquely in neurons this is reduced to one molecule every two nucleosomes (Allan et al., 1984; Pearson et al., 1984). Chromatin reconstitution experiments showed that MeCP2 can compete with histone H1 for binding to methylated chromatin and may function as a substitute linker histone (Ishibashi et al., 2008; Nan et al., 1997). As MeCP2 occurs at approximately one molecule per two nucleosomes in neurons, we asked whether the abundance of histone H1 was affected by MeCP2 depletion. Using FACS purified neuronal nuclei, we found that histone H1 is elevated ~2-fold in the Mecp2-null compared to the wildtype (Fig 5D; KS-test p<0.05). No difference was seen in unsorted nuclei from wildtype and mutant whole brain, suggesting that this effect is specific to neurons where MeCP2 is highly abundant. The level of histone H1 present in MeCP2-deficient neurons appears to be close to one molecule per nucleosome, as seen in other cell types, suggesting that the reduced level in neurons may reflect substitution of H1 by MeCP2.
MeCP2 suppresses spurious transcription of repetitive elements
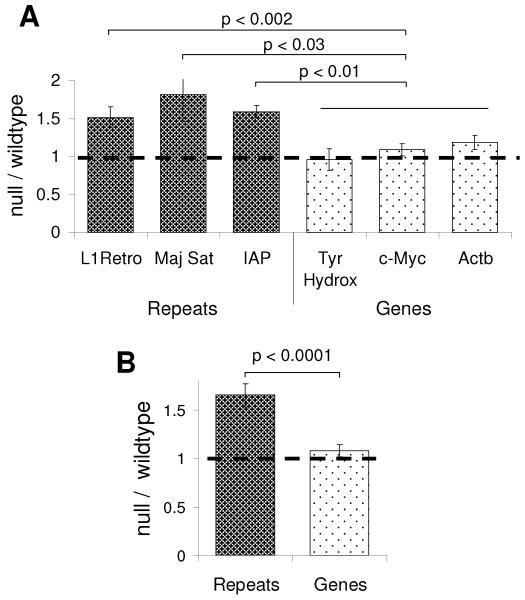
Our results implicate MeCP2 as a global regulator of neuronal chromatin structure. A possible outcome of MeCP2 depletion is that increased histone acetylation could lead to inappropriate transcription from the bulk genome, including dispersed repetitive elements. To test this possibility, we studied expression of L1 retrotransposons, intracisternal A particles and tandem repetitive units of the mouse major satellite, which are methylated and bound by MeCP2 (Fig S4A). RNA was extracted from the whole brain and RT-PCR used to determine the expression levels of these elements. No difference was observed between wildtype and Mecp2-null brain (Fig S4B). We considered, however, that spurious transcripts might be degraded by the exosome and therefore not survive as stable components of cytoplasmic RNA. To reduce the opportunities for transcript degradation, RNA was extracted from isolated nuclei and analyzed by RT-PCR. The same repetitive regions now showed significantly increased levels of expression in the Mecp2-null brain compared to the wildtype (Fig 6A). In contrast no differences in the expression levels of Actb, c-Myc or tyrosine hydroxylase mRNAs were observed (Fig 6A). On average, repetitive regions showed 1.6-fold overexpression in the MeCP2-null brain (4 biological replicates; KS-test p<0.0002), whereas the expression of these bona fide genes was unaffected (Fig 6B and Fig S4D). These results do not preclude the possibility that some genes show changes in the MeCP2-null brain. They argue, however, that one consequence of MeCP2-deficiency in neurons is an increase in transcriptional noise. This effect was not seen in embryonic brains from E18.5 mice (Fig S4C), suggesting that increased transcriptional noise is confined to the mature brain where MeCP2 is abundant enough to impact the whole genome.
Fig 6.
MeCP2 suppresses transcription from repetitive elements distributed throughout the genome. Nuclei were isolated from wildtype and Mecp2-null brains. RNA was extracted and cDNA was prepared, with and without reverse transcriptase. (A) Quantitative PCR was used to determine the expression levels of repetitive regions and genic regions. The data was normalised to GAPDH and shown as a ratio between Mecp2-null and wildtype nuclei. The horizontal line represents no change between wildtype and Mecp2-null mice. (B) Shows the pooled data, grouping repetitive regions and genic regions. Error bars indicate SEM +/− mean. The KS test was used to determine statistical significance. See Fig S4.
DISCUSSION
Mouse models and human neurological disorders suggest that appropriate levels of MeCP2 are essential for its proper function, as both deficiency and mild overexpression result in a neurological phenotype (Collins et al., 2004; Guy et al., 2001). Our data establish MeCP2 as one of the most abundant nuclear proteins in mature neurons. With an average density of one molecule every two nucleosomes, there is enough to bind approximately half of all methyl-CpG sites in the diploid neuronal genome. ChIP analysis of candidate regions and high-throughput sequencing confirms that MeCP2 is globally bound and tracks the density of methyl-CpG in the genome. As for all ChIP analyses, it is possible that there are transient interactions that are not captured using formaldehyde crosslinking (Schmiedeberg et al., 2009) and we may therefore be selectively visualising stable MeCP2-DNA interactions.
Our evidence supports earlier contentions that the large number of potential binding sites fits with a global regulatory role (Nan et al., 1997). Although provisional identification of MeCP2 “target genes” has been supported by MeCP2-ChIP near specific promoters, it is now apparent that most regions of the genome, including repetitive sequences and intergenic DNA, are associated with MeCP2 in neurons. The notion that MeCP2 resembles a transcription factor by being targeted to a specific subset of genes appears to be incorrect.
In line with its high abundance, MeCP2 impacts the chromatin state globally. Histone H1 is atypically low in neurons, only being sufficient to bind half of all neuronal nucleosomes, whereas in other cell types the stoichiometry is almost one-to-one (Allan et al., 1984; Pearson et al., 1984). We found that when MeCP2 is absent, the amount of neuronal H1 doubles, bringing it to the level typical of most cell types. There was no comparable effect in glia, where the density of chromosome-bound MeCP2 is much lower. MeCP2 can compete with H1 for binding to chromatin containing methylated DNA in vitro (Nan et al., 1997). Further in vitro studies have suggested that MeCP2 may function in a manner analogous to a linker histone as it can bind to entry-exit sites on nucleosomes and promote the formation of higher order structures (Ishibashi et al., 2008; Nikitina et al., 2006). It is tempting to propose that competition for chromatin binding sites between neuronal MeCP2 and H1 displaces the linker histone from half of its potential sites, leading to a reduction in the requirement for H1. In the Mecp2-null brain, where this competition does not occur, H1 is apparently restored to its conventional nucleosomal stoichiometry of 1:1 and may functionally compensate in part for MeCP2-deficiency. Further work is needed to clarify the role of MeCP2 as a putative alternative linker histone.
Elevated histone acetylation in MeCP2-deficient whole brain samples has been reported previously (Shahbazian et al., 2002). By sorting neuronal from glial nuclei, we show that this difference is entirely due to a large change in neurons, with no corresponding alteration detectable in glia. Using ChIP we show this increase is distributed throughout the genome, rather than at distinct sites. Early evidence suggested that MeCP2 can recruit the HDAC-containing complexes (Nan et al., 1998). A plausible model is therefore that MeCP2 attracts corepressors throughout the genome, which depress histone acteylation according to the local density of meCpG. The finding that the ratio of mutant to wildtype H3 acetylation matches the density profile of MeCP2 (Fig 5B) provides support for this contention. We propose that these global effects of MeCP2-deficiency on both H1 abundance and histone acetylation are seen exclusively in neurons because only in these cells is MeCP2 sufficiently abundant to coat the genome. In other cell types, the low density of MeCP2 occupancy is insufficient to impact bulk chromatin.
Studies with in vitro reconstituted chromatin have suggested that MeCP2 might play a structural role in chromatin regardless of DNA methylation (Georgel et al., 2003). Against this hypothesis, other studies have showed MeCP2 preferentially binds methylated DNA (Gregory et al., 2001; Nan et al., 1996). We establish that even in neurons, where MeCP2 abundance is equivalent to that of histone H1, DNA methylation dependence still holds. These in vivo data reinforce the view that MeCP2 binding depends upon the methylation status of chromosomal DNA. This dependence may account for the remarkable reversibility of severe neurological symptoms in mice when MeCP2 is restored following development under MeCP2-deficient conditions (Guy et al., 2007). If, as seems likely, DNA methylation patterns are laid down normally in the absence of MeCP2, then the restored protein is expected to distribute correctly according to these genomic cues and take up its usual function.
The global distribution of MeCP2 casts new light on gene expression studies, which initially detected few significant changes between Mecp2-null and wildtype brain (Jordan et al., 2007), but more recently have reported large numbers of subtle alterations comprising both increases and decreases in gene expression (Chahrour et al., 2008). Where a transcriptional regulator affects a restricted number of genes, coherent expression changes at those genes are expected when the protein is deficient. If, however, a large proportion of all genes are affected, it is more difficult to predict how deficiency will impact expression. The histone deacetylase inhibitor trichostatin A, for example, causes dramatic hyperacetylation of histones, but, despite the clear association between histone hyperacetylation and transcriptional activity, the drug usually causes equal numbers of genes to be up- and down-regulated (Peart et al., 2005). A potential explanation is that the nucleus contains limiting supplies of transcriptional machinery, so that increased expression at some loci must be matched by decreases at others, even if all genes are potentially activated by hyper-acetylation. By analogy, the high abundance of MeCP2 may mean that many genes are potentially affected, but analysis of stable gene expression patterns following long term absence of MeCP2 may not accurately reflect these effects.
Our hypothesis that high levels of MeCP2 affect global chromatin structure is supported by elevated transcriptional noise from repetitive elements in nuclei of the Mecp2-null brain. This effect is absent in embryonic brains, suggesting that the abundance of MeCP2 is key. Expression of bona fide genes, on the other hand, was not elevated, which may either mean that most transcription rates are little changed or that post-transcriptional controls are able to compensate. We suggest that globally bound MeCP2 dampens transcription in a genome-wide manner by recruiting HDAC activity and acting as a linker histone. Why might neurons have evolved a requirement for MeCP2 as a genome-wide transcriptional dampener, whereas other somatic tissues have not? One possibility is that the neurons are particularly sensitive to the levels of transcriptional noise from the bulk genome and therefore have a special need for MeCP2 to quench this. Another potential explanation is that neuronal plasticity and homeostasis depend upon the specific ability of MeCP2 to respond dynamically to activity. Neuronal firing leads to a burst of synaptic protein synthesis and nuclear transcription, which is required for development of synaptic systems (Bading, 2000). MeCP2 undergoes site-specific phosphorylation following synaptic firing, which is reported to alter its affinity for DNA and its nuclear distribution (Chen et al., 2003; Zhou et al., 2006). We speculate that phosphorylation may transiently relieve the genome-wide repressive chromatin structure maintained by MeCP2 and thereby facilitate appropriate gene expression. Further reports of post-translational modifications of MeCP2 (Tao et al., 2009), raise the possibility that MeCP2 is a signalling hub within neurons and as such may play a crucial role in neuronal maintenance. Recognition of MeCP2 as a global chromatin regulator that is responsive to neuronal activity promises to shed light on the molecular basis of Rett Syndrome.
EXPERIMENTAL PROCEDURES
Mouse Models
C57BL6 Mecp2tm1.1Bird mice were used with wildtype littermates as controls. Unless otherwise stated all mice were 6-8 weeks of age. Mice were euthanized by CO2 asphyxiation, tissues dissected and either frozen in nitrogen for ChIP or used directly for nuclear preparation.
Nuclei Preparation
Mouse brain nuclei were isolated as previously described (Klose and Bird, 2004). Recovered nuclei were resuspended in 1 × phosphate buffered saline supplemented with 20% glycerol, 10 mM sodium butyrate and complete protease inhibitors (Roche), frozen in liquid nitrogen and stored at −80 °C. Serial dilutions of nuclei were counted using a haemocytometer to determine concentration. Nuclei were lysed in Laemmli buffer and used directly for gel electrophoresis.
Fluorescence activated cell sorting
Detailed protocols are available in the Supplemental Data.
Gel Electrophoresis and Western Blotting
Proteins were resolved by SDS-PAGE and transferred to PVDF membrane. Visualization of reactive proteins was performed by enhanced chemiluminescence and quantitative infra-red imaging (Licor Odyssey). Intensity of proteins bands was quantified using NIH ImageJ and Licor software. Details of recombinant proteins are in the Supplementary Data.
Chromatin Immunoprecipitation and Solexa Analysis
Detailed protocols are available in the Supplemental Data. GEO Accession #GSE19786
Bisulfite Sequencing
Bisulfite treatment of DNA and sequencing was carried out as previously described (Illingworth et al., 2008).
Expression analysis
For whole-cell expression analysis, RNA was extracted using TriReagent (Sigma-Aldrich) from whole brain disrupted using a polytron (Janke and Kunkel). For nuclear expression analysis, equal numbers of nuclei were lysed in extraction buffer (30 mM Tris-HCl pH7.5, 300 mM NaCl, 10 mM EDTA, 1% SDS, 0.5 mg/ml proteinase K, 0.2 U/μl RNAsin) after which RNA was extracted using acid phenol, ethanol precipitated. After DNase1 Treatment (Ambion DNA-free), cDNA was transcribed using Moloney murine leukemia virus reverse transcriptase (Promega). Real time PCR was carried out using Quantace Sensimix Plus (primer sequences available on request).
Supplementary Material
HIGHLIGHTS.
Neuronal nuclei have about one molecule of MeCP2 for every two nucleosomes
MeCP2 in neurons binds chromosome-wide, tracking the density of DNA methylation
MeCP2-deficiency doubles both histone acetylation and histone H1 only in neurons
MeCP2 dampens transcriptional “noise” from repetitive elements and satellite DNA
ACKNOWLEDGEMENTS
We are grateful to Elizabeth Sheridan for testing the DNA sequencing protocol and Stuart Mclaren, Katherine Auger and Julian Parkhill (all Wellcome Trust Sanger Institute) for coordinating the DNA sequencing. We also thank Jim Selfridge, Jacky Guy, and Aimée Deaton for critical comments on the manuscript. This work was supported by the Wellcome Trust and by the “Epigenome” European Union Network of Excellence.
REFERENCES
- Allan J, Rau DC, Harborne N, Gould H. Higher order structure in a short repeat length chromatin. J Cell Biol. 1984;98:1320–1327. doi: 10.1083/jcb.98.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Armstrong DD. Neuropathology of Rett syndrome. Ment Retard Dev Disabil Res Rev. 2002;8:72–76. doi: 10.1002/mrdd.10027. [DOI] [PubMed] [Google Scholar]
- Bading H. Transcription-dependent neuronal plasticity the nuclear calcium hypothesis. Eur J Biochem. 2000;267:5280–5283. doi: 10.1046/j.1432-1327.2000.01565.x. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer D, Goldstine J, Rao YM, LaSalle JM. Elevated methyl-CpG-binding protein 2 expression is acquired during postnatal human brain development and is correlated with alternative polyadenylation. J Mol Med. 2003;81:61–68. doi: 10.1007/s00109-002-0396-5. [DOI] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Randall TE, Johnson CA, Khosla S, Hatada I, O’Neill LP, Turner BM, Feil R. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol Cell Biol. 2001;21:5426–5436. doi: 10.1128/MCB.21.16.5426-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hendrich BD, Brown CJ, Willard HF. Evolutionary conservation of possible functional domains of the human and murine XIST genes. Hum Mol Genet. 1993;2:663–672. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Thambirajah AA, Ausio J. MeCP2 preferentially binds to methylated linker DNA in the absence of the terminal tail of histone H3 and independently of histone acetylation. FEBS Lett. 2008;582:1157–1162. doi: 10.1016/j.febslet.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Jordan C, Li HH, Kwan HC, Francke U. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med Genet. 2007;8:36. doi: 10.1186/1471-2350-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J Biol Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- LaSalle JM, Goldstine J, Balmer D, Greco CM. Quantitative localization of heterogeneous methyl-CpG-binding protein 2 (MeCP2) expression phenotypes in normal and Rett syndrome brain by laser scanning cytometry. Hum Mol Genet. 2001;10:1729–1740. doi: 10.1093/hmg/10.17.1729. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Lubs H, Abidi F, Bier JA, Abuelo D, Ouzts L, Voeller K, Fennell E, Stevenson RE, Schwartz CE, Arena F. XLMR syndrome characterized by multiple respiratory infections, hypertelorism, severe CNS deterioration and early death localizes to distal Xq28. Am J Med Genet. 1999;85:243–248. doi: 10.1002/(sici)1096-8628(19990730)85:3<243::aid-ajmg11>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci U S A. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2006 doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, Bird A. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- Pearson EC, Bates DL, Prospero TD, Thomas JO. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur J Biochem. 1984;144:353–360. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, Holloway AJ, Johnstone RW. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L, Skene P, Deaton A, Bird A. A temporal threshold for formaldehyde crosslinking and fixation. PLoS One. 2009;4:e4636. doi: 10.1371/journal.pone.0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.