Abstract
ATP is synthesized by F-type proton-translocating ATPases (F-ATPases) coupled with an electrochemical proton gradient established by an electron transfer chain. This mechanism is ubiquitously found in mitochondria, chloroplasts and bacteria. Vacuolar-type ATPases (V-ATPases) are found in endomembrane organelles, including lysosomes, endosomes, synaptic vesicles, etc., of animal and plant cells. These two physiologically different proton pumps exhibit similarities in subunit assembly, catalysis and the coupling mechanism from chemistry to proton transport through subunit rotation. We mostly discuss our own studies on the two proton pumps over the last three decades, including ones on purification, kinetic analysis, rotational catalysis and the diverse roles of acidic luminal organelles. The diversity of organellar proton pumps and their stochastic fluctuation are the important concepts derived recently from our studies.
Keywords: ATP, F-type ATPase, V-type ATPase, endomembrane organelle, rotational catalysis, acidic lumen
Introduction
Most of the cellular energy currency, ATP, is synthesized through oxidative phosphorylation in mitochondria and bacterial membranes or photophosphorylation in chloroplasts.1)–3) In both mechanisms, an electron transfer chain establishes electrochemical proton gradient, which subsequently drives a proton-pumping ATPase for ATP synthesis. This ATPase was first recognized as factors (FoF1) required for ATP synthesis through mitochondrial oxidative phosphorylation: F1 stands for “factor one” or “coupling factor one” and Fo for a “factor giving oligomycin sensitivity” to F1 (Fig. 1).4) F-ATPase, a name based on “factors”, has been used widely (Fig. 1),5) although physiologically they are ATP synthase in most organisms. Other name such as F-ATP synthase, H+-translocating ATPase and H+ ATPase have also been used.
Fig. 1.
A schematic model of F-ATPase.
A model of F-ATPase is shown; the catalytic hexamer (α3β3) and membrane domain are connected by a stalk domain. The membrane extrinsic sector (F1) and transmembrane domain (Fo) are shown together with schematic energy coupling between ATP synthesis/hydrolysis and proton transport through subunit rotation (yellow arrow).
Up to the early 1970s, common materials used for studying ATP synthesis were bovine mitochondria and spinach chloroplasts, and researchers in the field did not pay much attention to Escherichia coli.3), 4) We became interested in bacterial F-ATPase, considering that the genetic approach combined with biochemistry would contribute greatly to a better understanding of the structure and mechanism of the complicated enzyme. Although E. coli F-ATPase was introduced later, its subunit assembly, amino acid sequence, catalytic residues and proton pathway were determined before those of most other organisms.6) These results together with the recent bovine crystal structure7) have established that F-ATPase is a unique membrane enzyme in coupling chemistry and proton transport through subunit rotation.1)–3)
Proton pumping vacuolar-type ATPases or V-ATPases,5) initially found in fungal vacuoles, are widely distributed in animals and plants,8)–10) and are also called VoV1 based on the similarities to the subunit structure and mechanism of F-ATPases (FoF1) (Fig. 1). Furthermore, V-ATPases have provided methods and ideas for studying the roles of the acidic luminal pH in diverse organelles, including lysosomes and endosomes. It should be noted that the two ATPases are different from the mammalian stomach proton pump H+/K+ ATPase that is P-ATPase5) forming acylphosphate intermediate. We have shown the similarity of the ATPase to Na+/K+ ATPase in gene organization and amino acid sequences,11)–13) and its specific expression in the parietal cells of the gastric lumen.12), 14), 15)
The proton pumping F-ATPases and V-ATPases have attracted interest in a wide range of research areas, and have been the most competitively studied enzymes. We mostly discuss our own studies on the two enzymes over the last three decades. Our initial interest was obviously to define F-ATPase biochemically, especially its subunits and their primary structures. This approach was extended to understand mechanism and catalysis using mutational analysis. We were interested in diverse endomembrane organelles with acidic lumen from early stage of our research.16) Since fungal V-ATPases were introduced into the field of proton transporting ATPases, we were interested in their similarities to F-ATPase and diverse roles corresponding to different unique organelles. Results not discussed in detail here can be found in related review articles1)–3), 8)–12) and those in the book edited by us.17)
1. F-ATPase: from genes to stochastic catalysis
Biochemistry of E. coli F-ATPase
Only limited information was available on E. coli F-ATPase when we started its purification and reconstitution. Our interest in the ATPase was originated from studies on active transport. Membrane vesicles obtained from bacterial plasma membranes were important for studying the transport in vitro.18) Everted or inside–out membrane vesicles could be obtained by disrupting the bacterial cells with a French press,18) whereas right-side out vesicles were obtained by osmotic lysis of spheroplasts.18)–20) The F-ATPase was localized in everted vesicles with its F1 sector exposed to the outer surface, transporting protons into the vesicles, upon hydrolysis of ATP added to the solution.21) The F1 sector having ATPase activity could be released by washing the everted vesicles with dilute buffer, leaving Fo bound to the membranes.21) The F-ATPase could be reconstituted by combining solubilized F1 and Fo in the washed membranes.
To define subunit organization, the F1 sectors with different subunit assemblies, 5-subunit (α, β, γ, δ, ε) and 4-subunit (α, β, γ, ε) F1, were purified from the soluble fraction using conventional procedures (Fig. 2A).21) Both F1 sectors had ATPase activity, but only the 5-subunit F1, the first purified active E. coli F1, could bind to washed membranes, and form F-ATPase capable of ATP synthesis and ATP-hydrolysis dependent proton transport. These results indicate that the δ subunit is essential for F1 binding to Fo. The purified F1 could be dissociated at 4 °C in the presence of chaotropic agent such as KNO3 or NaNO3, and all five subunits were purified to apparent homogeneity (Fig. 2B).22), 23) The α3β3γ complex having ATPase activity could be reconstituted from the three purified subunits in the presence of Mg2+ and ATP.22) Combination of the α3β3γ complex with the δ and ε subunits allowed reconstitution of functional F1 that could bind to Fo.23) The purified α and β subunits had nucleotide binding sites,23), 24) consistent with the results of studies on the binding of ATP analogues. Localization of the F1 sector on the cytoplasmic surface of the E. coli plasma membrane was shown using antibodies against 5-subunit F1.18)
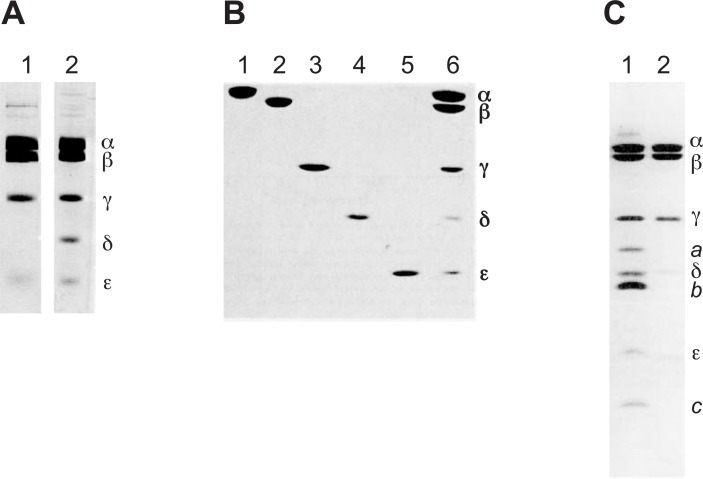
Fig. 2.
Preparations of E. coli F-ATPase and its subunits.
Polyacrylamide gel electrophoresis of the 4-subunit and 5-subunit F1 sectors (A), isolated F1 subunits (B), and FoF1 (C), are shown. The positions of subunits (α, β, γ, δ, ε, a, b and c) are indicated. The 5-subunit F1 (A, lane 2) could bind to Fo to reconstitute F-ATPase. The F1 sector could be reconstituted from isolated subunits (B, lanes 1, 2, 3, 4, and 5), and FoF1 (C, lane 1) could be reconstituted in liposomes capable of ATP synthesis and ATP-dependent proton transport. F1 sectors are also shown as controls (B, lane 6, C, lane 2). Modified from previous results.21)–23), 26)
Purification of the entire F-ATPase was difficult because of its complex subunit structure including transmembrane proteins. F-ATPase solubilized from membranes with a detergent had to be subjected to “state of art” procedures to obtain an active pure protein assembly.25) However, rapid one-step purification of the detergent-solubilized F-ATPase became possible26) from an overproducing strain (DK8/pBWU13), in which ∼ 30% of the inner membrane protein was F-ATPase.27) The purified F-ATPase (α, β, γ, δ, ε, a, b, and c subunits) reconstituted into liposomes could form a proton gradient or membrane potential upon ATP hydrolysis (Fig. 2C).
Genetics of F-ATPase
Using E. coli, F-ATPase could be studied utilizing genetic approaches that are difficult to use for those from mammalian mitochondria or plant chloroplasts. Mutants with defective F-ATPase are unable to grow on succinate because they can not synthesize ATP through oxidative phosphorylation. They can grow, however, on glucose, synthesizing ATP through glycolysis.3) This growth phenotype led to an easy protocol for isolating mutants. Gibson and coworkers introduced the term unc (uncoupled) genes coding subunits of F-ATPase, since they isolated unc mutants: cells with defective unc genes have a normal electron transfer chain (respiratory chain), but exhibit no ATP synthesis, thus being uncoupled in oxidative phosphorylation.28) Many unc mutations have been isolated by our group, and their gene cluster, the unc operon, was found to be localized around 82.5 min (between the asn and bgl genes) on the linkage map, where origin of the chromosome replication (OriC) was also mapped29) (Fig. 3). Interest in OriC resulted in the isolation of a series of transducing λ phages carrying a chromosome segment around the 82.5 min region.29)
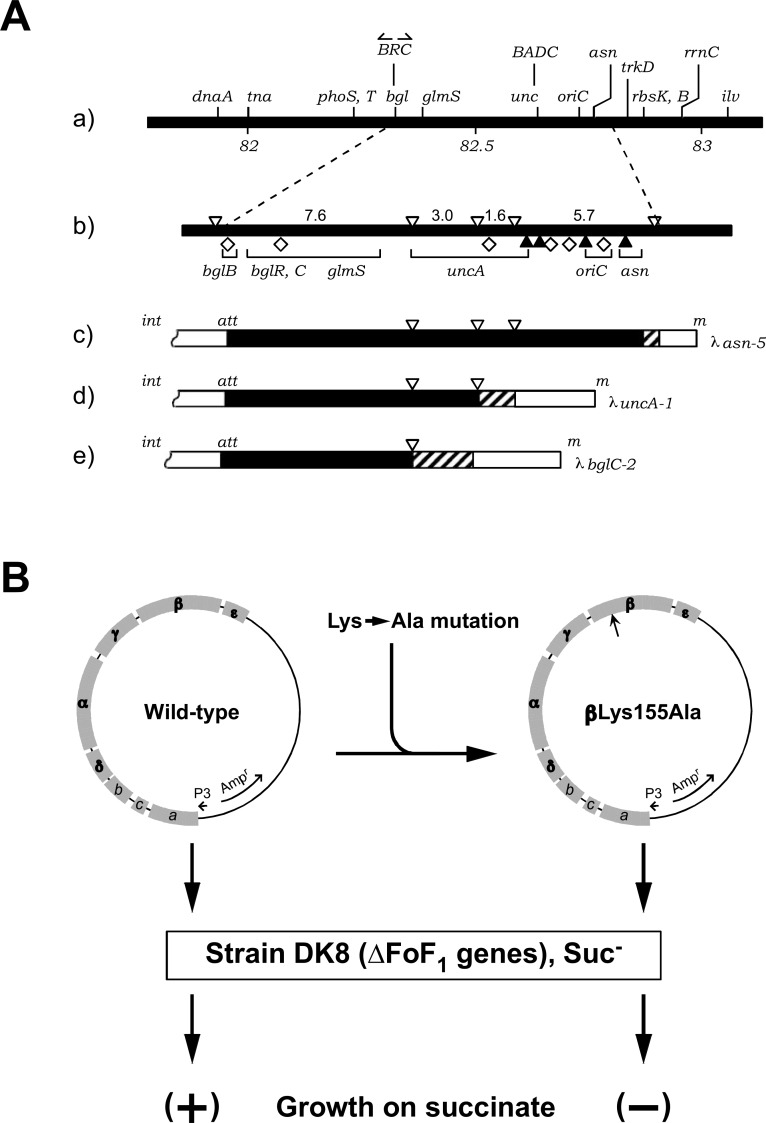
Fig. 3.
From the unc operon to transducing λ phages and recombinant plasmids.
(A) Transducing λ phages are shown with an E. coli linkage map (a, b). The transducing λ phages used to study F-ATPase are also shown: λasn-5 (c), carrying the F-ATPase gene (unc operon); λuncA1 (d), carrying part of the unc operon; λbglC, not carrying the unc operon. (B) pBWU13, a plasmid carrying the wild-type unc operon, is shown together with the positions of subunit genes (B, left). An example of a plasmid carrying a mutant β subunit gene is also shown (B, right). Plasmids were introduced into E. coli DK8 lacking unc operon (ΔFoF1 genes). Cells with the mutant gene could not grow on succinate. Modified from previous results.27), 30), 31)
We found that transducing phage λasn-5 could complement all unc mutants available, whereas other phages, such as λbgl-2 or λunc-1 could only complement limited numbers of the mutants (Fig. 3A).30) After induction of lysogenic λasn-5 phage, the F-ATPase activity, amount of Fo (as F1 binding site), and proton pathway were all increased,30) indicating that the phage carries the genes for an entire F-ATPase. All eight polypeptides in the purified F-ATPase were shown to be authentic subunits, since they were overproduced stoichiometrically upon induction.25) Complementation of available unc mutants was tested with a series of transducing phages and their DNA segments, and the unc operon was mapped to a ∼ 7.5 kb DNA segment.31)
Finally, DNA sequences of the eight genes encoding the amino acid sequences of the individual subunits were obtained.32)–35) The unc operon is formed from three genes for Fo (B, E, and F) and five for F1 (H, A, G, D, and C) organized in that order downstream of the promoter,6), 36) encoding the a, c, b, δ, α, γ, β, and ε subunits, respectively. Thus, subunits of F-ATPase were defined with their primary structures. It became easier, after sequencing of the entire operon, to map mutations to subunits, followed by identification of amino acid replacements. Before the gene sequence became available, methods for identifying the subunits with mutations were limited.37)
Although it was not easy to construct a recombinant plasmid carrying all the genes of the unc operon directly from the bacterial DNA, such a plasmid (pBWU13) was constructed by combining DNA fragments derived from λasn-5 (Fig. 3B).27) pBWU13 with or without mutation has been used widely for biochemical studies.
To detect a DNA fragment with point mutations, we have used the mobility shift on gel electrophoresis.38), 39) A similar method was applied later to identify human DNA polymorphisms.40)
Catalysis in the β subunit
As described above, purified α and β subunit had nucleotide binding sites, and could reconstitute α3β3γ complex having ATPase activity. Since the DNA sequence of F-ATPase genes was available, it became possible to study the catalytic site by F-ATPase with defined amino acid substitutions. Mutants of the α and β subunits often exhibit reduced synthesis and hydrolysis of ATP,41), 42) consistent with the notion that the catalytic site is present at the interface between the two subunits.2)
Although the steady state ATPase activity of the purified F1 sector had been assayed in the presence of a large excess of ATP, uni-site catalysis with a small amount of ATP just sufficient for a single catalytic site was introduced.43) Steady state catalysis is ∼106 fold faster than uni-site catalysis. Mutations in the α subunit often lowered the steady state rate more than 1000-fold, but uni-site catalysis was maintained.42), 44) Mixed reconstitution experiments suggested that the steady state ATPase activity was lost when one α or β subunit from the defective mutant was introduced into the α3β3γ complex.45) Similarly, the steady state ATPase was lost when one of the three catalytic subunits was chemically modified or affinity-labeled.46)–48) These results suggest that the three sites cooperatively synthesize or hydrolyze ATP.
The β subunit has a conserved sequence named the phosphate-binding P-loop (E. coli sequence, GlyGlyAlaGlyValGlyLysThr, Gly149 ∼ Thr156) (Fig. 4A). The P-loop (consensus, Gly-X-X-X-XGly-Lys-Thr/Ser), also called the glycine-rich sequence, is conserved among ATP- or GTP-binding proteins such as p21 Ras protein, adenylate kinase, V-ATPase subunit A, etc.49) One of the randomly isolated mutants with a low steady state ATPase was mapped to the P-loop (Gly149 → Ala).49) Furthermore, the β subunit sequence could be replaced by that of p21 Ras, indicating the similar structures and roles of the P-loop in the two proteins. However, the sequence with Gly between Lys and Thr, such as that of adenylate kinase, was not functional,50) possibly because the orientation of the Thr156 residue was altered by the insertion. The ATP analogue adenosine triphosphopyridoxal bound to Lys155, suggesting that this residue is close to the γ phosphate moiety of ATP.48) Furthermore, the GERXXE (Gly180–Glu185) sequence of the β subunit is conserved in F1 from different species and in the A subunit of V-ATPase. DCCD (dicyclohexylcarbodiimide) bound to Glu-181 in the sequence.46)
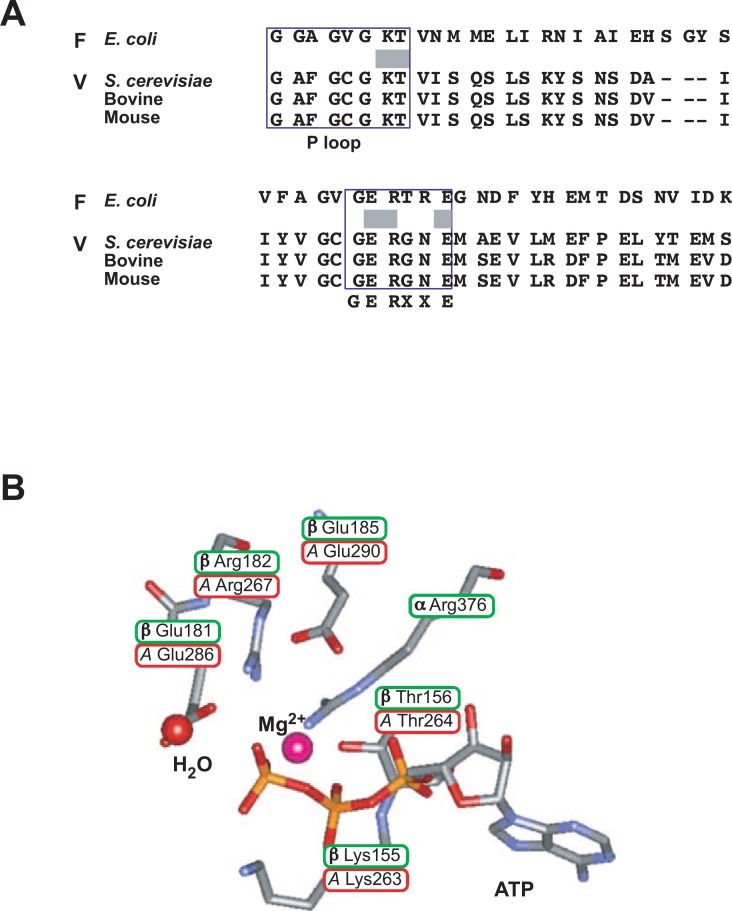
Fig. 4.
Catalytic sites of F-ATPases and V-ATPases.
(A) Conserved residues in the catalytic subunits of F- and V-ATPases. The boxed areas denote the P-loop and the GERXXE sequence containing catalytic residues Lys155, Thr156 and Gln181 of F-ATPase. (B) The catalytic residues of E. coli F-ATPase and the corresponding residues of yeast V-ATPase are shown. Their positions are cited according to the bovine crystal structure.7) Modified from reviews.1), 56)
Based on these results, we systematically replaced conserved residues in the β subunit one by one, especially between Gly149 and Glu185. Mutations were introduced into the gene carried by the recombinant plasmid (pBWU13)27) (Fig. 3B), and overproduced mutant F1 sectors were purified for biochemical analysis including of kinetics. The mutants (Lys155 → Ala, Ser or Thr; Thr156 → Ala, Cys, Asp) showed low steady state catalysis rates (≤ 10−5 of the wild-type level) and lower k1 rates (≤ 10−2) of ATP binding in single site kinetics, suggesting that Lys155 and Thr156 are catalytic residues.51)–53) Similarly, kinetic analysis of mutant F1 suggested that Glu181 and Arg182 are catalytic residues.53), 54)
All replacements of Glu185 except that with Asp caused loss of steady catalysis (< 0.2% of wild-type rate), whereas the enzyme with an Asp residue or a chemically introduced S-carboxymethyl cysteine at position 185 exhibited a substantial steady state.54) Gln185 or Cys185 mutant F1 retained similar single site catalysis to that of the wild-type, suggesting that a carboxyl moiety at position 185 is essential for catalytic cooperativity.54) Similarly, substitution of Arg373 of the α subunit resulted in 2 × 10−3-fold lower steady state activity than the wild-type, but still exhibited wild-type single site catalysis.55) The two residues are related to information transfer between the three catalytic sites, although they do not directly participate in hydrolysis/synthesis.
The crystal structure of bovine F1 was determined by Walker and coworkers during our mutational studies.7) Since the amino acid sequences of the β subunits of E. coli and beef are about 70% homologous, the bacterial residues could be located in the higher-ordered structure. As expected, the catalytic residues and others discussed above are located close to the bound ATP in the β subunit (Fig. 4B).7) It should be emphasized that the mutational approach applied to the conserved residues has been fruitful.
Role of the γ subunit in energy coupling
Mutational studies suggested that γ subunit (Fig. 5) participates energy coupling between catalysis and proton transport as well as regulation of ATPase activity.56) Such studies are briefly discussed below. The amino acid sequence of the γ subunit is less conserved among different species; early alignment of the known γ subunit sequence revealed 28 conserved residues. Eighteen of them were located within 50 residues of the carboxy terminus, and four of them are in the amino terminal region. The results of sequential deletion of residues suggested that the carboxyl terminal region functions in catalysis and is coupled with proton translocation.5), 57), 58)
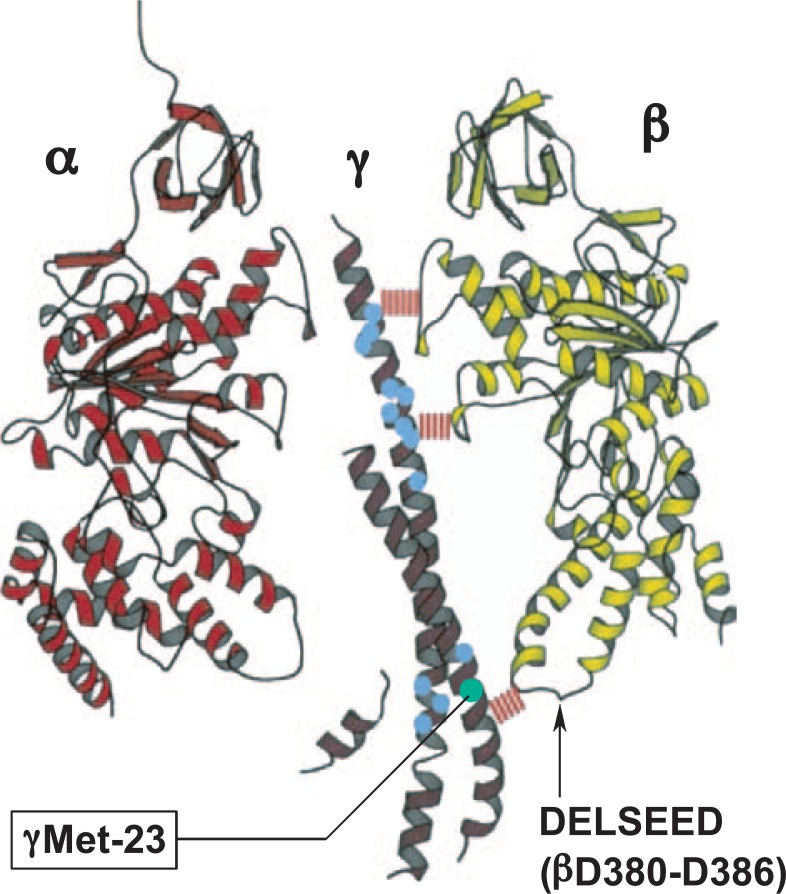
Fig. 5.
Mutational studies of the γ subunit.
The results of mutational studies of the γ subunit are summarized and shown on the bovine crystal structure.7) The Met-23Lys mutant of the γ subunit of F-ATPase exhibited wild-type ATPase activity, but reduced proton translocation. This defect was restored by one of the second mutations mapped distantly (blue circles). Cited from previous studies.60)–62)
The importance of the conserved amino terminal region was suggested by deletion of residues between Lys21 and Ala27, which resulted in the loss of F1 assembly and proton leakage through membrane via Fo.59) We systematically introduced mutations in the conserved amino terminal region.60) Most of the changes only slightly affected ATP synthesis and ATPase activity. Interesting exceptions were the Met23 → Arg and Met23 → Lys mutants (Fig. 5). They could not grow on succinate through oxidative phosphorylation, and their membranes showed very low proton pumping. However, they had similar ATPase activities to that of the wild-type, indicating that the mutants exhibited decreased efficiency in energy coupling. The defect caused by the Met23Lys mutation was reversed by the second mutation mapped to the γ subunit carboxyl terminus (residues at positions 242 and 269–280) (Fig. 5).61) Furthermore, defects with mutations in the carboxyl terminus (γGln269 → Glu and γThr273 → Val) were suppressed by single amino acid replacements at the amino terminus.61), 62)
These mutation results could be correlated to the structures of the β and γ subunits, once the X-ray structure became available.7) The residues of which substitution suppressed the initial mutation, Met23Lys, were located in the γ subunit region interacting with the β subunit. Typically, the γ subunit Met23 residue is located close to the β subunit Asp380–Asp386 loop (called the DELSEED loop, the sequence with comprising one letter symbols). The mutant Lys23 could form an ionized hydrogen bond with β subunit Glu381. On the other hand, the γ subunit carboxyl terminal residues, such as Glu269 or Thr293, do not directly interact with the first mutation. The occurrence of such suppression over a long distance suggested that the two α helices of the γ subunit undergo long-range conformational changes during catalysis. Consistent with this notion, the crystal structure has shown different conformations of the γ subunit relative to the ATP-bound, ADP-bound and empty β subunit (abbreviated as βT, βD and βE, respectively).7)
Rotation to proton pathway
After the sequences of the Fo subunits had been obtained, the possible secondary structures of the subunits were estimated, also using other information6), 36): c subunit, two hairpin-like transmembrane helices connected by a central polar sequence that is exposed to the cytoplasm; a subunit, five trans-membrane helices; b subunit, embedded in the membrane through an amino terminal helix and a long helical domain extending into the cytoplasm. These estimated structures were supported by extensive later studies.63) The mutations of the subunit a and the c subunit impaired proton transport and F1 binding,64), 65) and the entire carboxyl terminal region of the b subunit was shown to be necessary for assembly of the functional Fo.66) These results indicated the roles of Fo subunits in energy coupling and assembly of the proton pathway.
Mutations in the a or c subunit caused defective proton transport, consistent with the notion that a proton pathways was formed from the two subunits.1)–3) The defective proton transport of the c subunit mutant could be complemented with high copy number plasmids carrying the wild-type gene, but not with low copy number ones. This was one of the earliest suggestions that a functional unit of the c subunit is an oligomer.65) We proposed, based on an image obtained on atomic force microscopy (AFM), that the multiple c subunits form a ring structure.67) Consistent with these early suggestions, the structure of yeast F1 attached to the c subunit ring with ten monomers was solved by X-ray crystallography.68) It has been established that a proton pathway is formed by the Asp61 and Arg210 residues of the c and a subunits, respectively.69)
Our series of experiments together with those of other laboratories suggested that three β subunits sequentially participate in catalysis through conformation changes. Furthermore, the different orientation of the γ subunit as to the three β subunits in the crystal structure7) strongly supports γ subunit rotation.
The ATP hydrolysis-dependent continuous rotation of an actin filament connected to the γ subunit of Bacillus F1 was video-recorded recently.70) We confirmed the γ rotation in E. coli F1 using the same method,71) and subsequently studied the entire F-ATPase to determine the physiological significance of the rotation (Fig. 6).
Fig. 6.
Rotational catalysis of the F1 sector and F-ATPase.
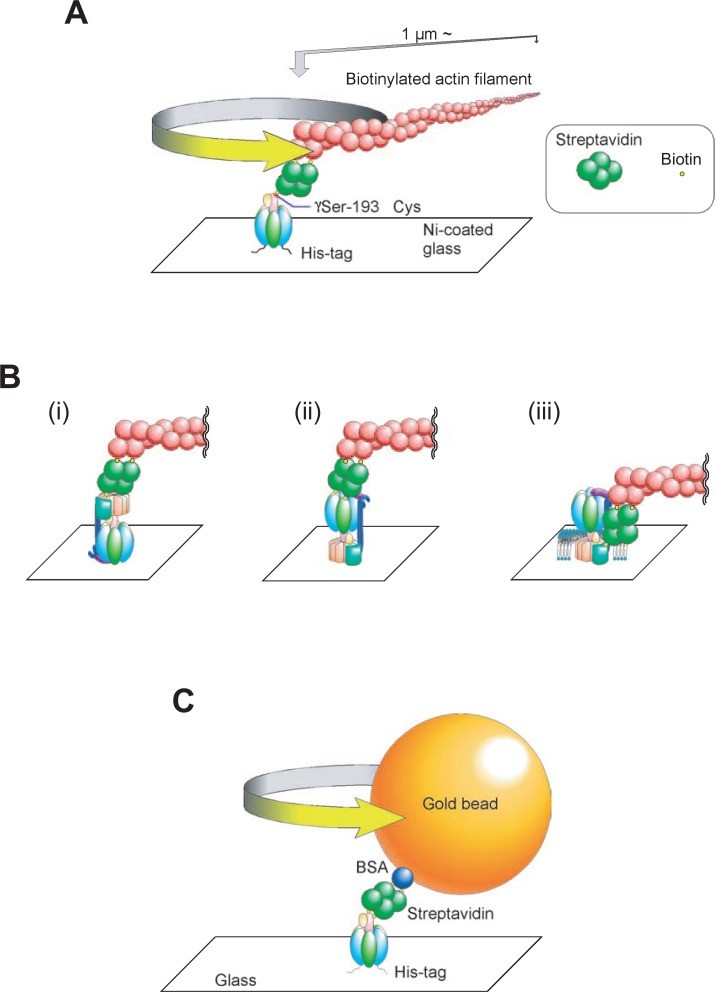
Experiments showing rotational catalysis are summarized: (A) Immobilized F1 sector with an actin filament attached; (B) F-ATPase holoenzyme immobilized through the α subunit (i) and the c ring (ii), and F-ATPase in a planar membrane immobilized through the c ring and an actin filament attached to subunit a (iii); (C) immobilized F1 with gold beads attached. The direction of ATP-dependent rotation observed is shown by arrows.
As a part of the F-ATPase mechanism, the γ rotation should be transmitted to the Fo sector to complete ATP hydrolysis-dependent proton pumping. Reversibly in ATP synthesis, proton transport through Fo should lead to rotation of the γ subunit. To examine the rotation of the Fo sector with the γ subunit, purified F-ATPase was immobilized on a glass surface through His residues introduced into the α subunit. An actin filament connected to the c ring continuously rotated in an anticlockwise direction, indicating that the c ring forms a rotor with the γ subunit (Fig. 6B).72) An actin filament connected to the β subunit also rotated in the F-ATPase immobilized through the c ring.73) Furthermore, an actin probe connected to the β subunit rotated in F-ATPase embedded in a planar membrane.74) The relative rotation of the c ring as to the a subunit was also shown in the membrane F-ATPase (Fig. 6B, iii). These studies established the rotational catalysis of F-ATPase, showing that γεc10 and α3β3δab2 were an interchangeable rotor and a stator, respectively.75), 76)
Toward rotational mechanism
The mechanism of rotation and generation of torque became of interest. The maximal rotational rate of an actin filament (∼ 1 μm long) was ∼ 10 sec−1, much slower than that expected from ATPase turnover, possibly due to the high viscous drag due to the filament.70)–74) It is widely accepted that the ε subunit is inhibitory as to F1 ATPase activity, and its inhibitory domain was mapped to the carboxyl terminus.77) However, we could not observe effects of the ε subunit on rotation using the actin filament.78) Similarly, the same probe was not useful for characterizing the mutant F1, which exhibited significantly lower ATPase activity than the wild-type.79) Although an actin probe has been useful for demonstrating rotary catalysis qualitatively, probes with lower viscous drag are required for further studies to establish the rotation mechanism.
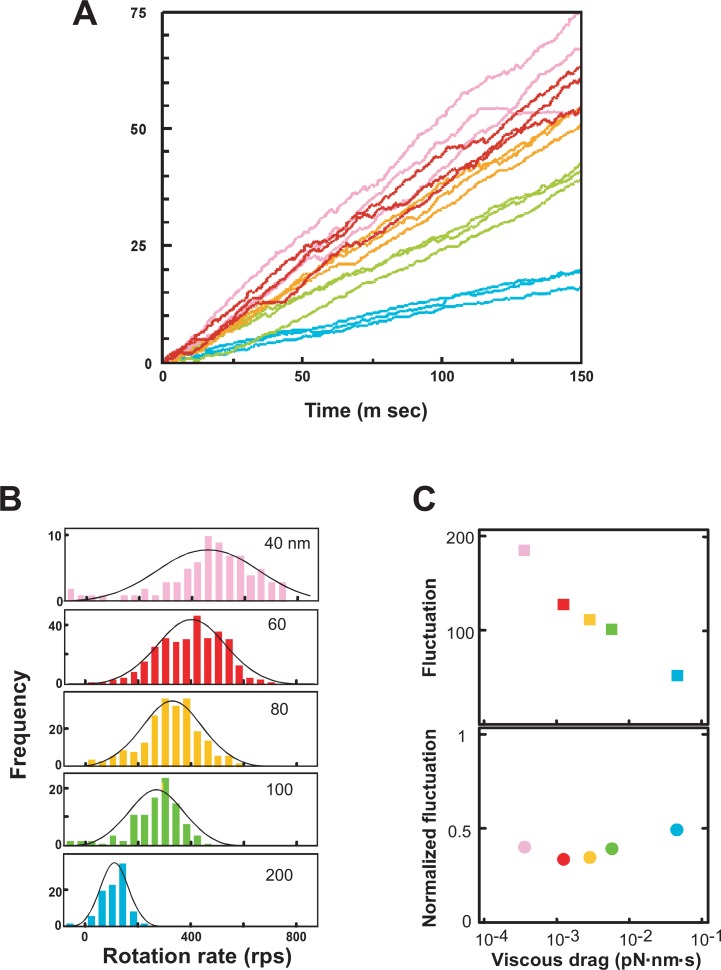
We used 40 and 60 nm gold beads for further studies (Fig. 6C). Since the diameter of the F1 sector is ∼ 10 nm, the beads were 4 ∼ 6 fold larger than that of F1, a much smaller probe than an actin filament. We observed that the rotation rates of 40 and 60 nm diameter gold beads were significantly higher than those of 100 and 200 nm ones (Fig. 7A).78), 80) Since the rates of the single beads were variable (Fig. 7A), we estimated the rates every 10 m sec. The resulting histograms showed a Gaussian distribution, and clearly indicated the stochastic fluctuation of the rates when the results for ∼20 beads were combined (Fig. 7B, C) or single beads were followed.81) The maximal and average rates for 40 ∼ 60 nm beads were ∼700 and 380 rps (revolutions/sec), respectively.78) The average rate was ∼ 10-fold higher than the value expected from that of the steady state ATPase, indicating that ∼ 10% of F1 is rotating in m sec time resolution.
Fig. 7.
Stochastic fluctuation of the rotation of gold beads attached to the F1 γ subunit.
(A) Time courses of gold bead rotation: pink, 40 nm; red, 60 nm; yellow, 80 nm; green, 100 nm; blue, 200 nm. The rates of rotation of beads were estimated every 10 m sec. (B) Histograms of the rotation speeds fitted to a Gaussian distribution are shown. (C) The stochastic fluctuation of bead rotation is shown (upper) together with normalized values (lower). Modified from previous studies.78), 80)
The mutants with the β subunit Ser174Phe and Ser174Leu substitutions are interesting ones isolated earlier.27), 41) They could not synthesize ATP and had ∼ 10% the ATPase activity of the wild-type. However, rotation of an actin probe attached to the mutant was not strikingly different from those in the case of the wild-type79): Ser174Leu and Ser174Phe generated similar and ∼ 1/2 torque compared to the wild-type, respectively. The mutant rotations were re-evaluated using 60 nm gold beads.81) The mutant rates were ∼ 10 rps due to the higher tendency of pausing at 120° steps than in the case of the wild-type. Since the rotation was assayed in the presence of a high concentration of ATP, the pausing occurred before release of the product. The pausing dwell times were stochastically variable. The Ser174Phe mutation was suppressed by the Gly149Ala substitution, and Ser174Phe/Gly149Ala became essentially similar to the wild-type.82) These results indicated the active role of the region between β-Sheet 4 and the P-loop, where Ser174 and Gly149 are located, respectively. Consistent with the surface location of Ser174 near the α subunit, the Ser174Phe mutation was also suppressed by a second mutation in the α subunit (Arg276Cys).83)
2. Vacuole type ATPases (V-ATPases) and acidic compartments
Similarities to F-ATPase
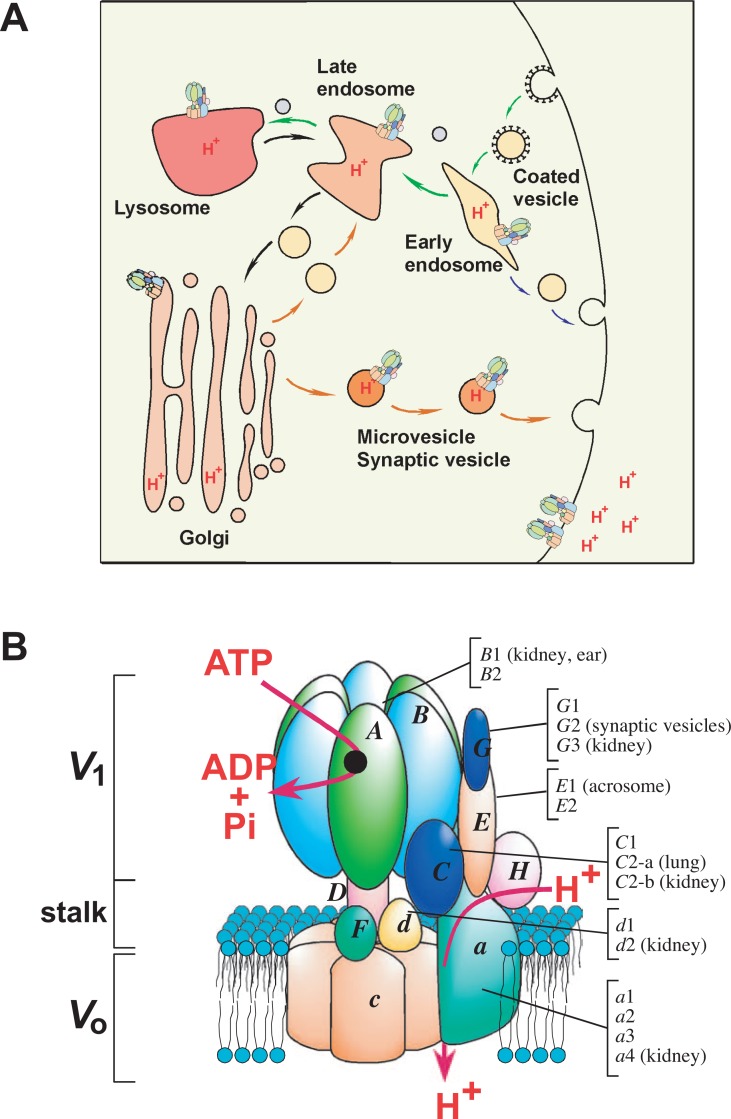
V-ATPases are localized in various endomembrane organelles including early endosomes, lysosomes, the Golgi apparatus, synaptic vesicles, and plasma membranes (Fig. 8). They are biochemically similar to F-ATPases, although their physiological roles are apparently different.8)–10) The V-ATPase A and B subunits are homologues of the F-ATPase β and α subunits, respectively, V-ATPase catalytic site of A subunit may be similar to that of the F-ATPase β subunit (Fig. 4). The subunit D corresponds to the γ as judged from the results of sequence and cross-linking experiments84) (Fig. 8B). Similar to in F-ATPases, three catalytic sites in the A3B3 hexamer showed cooperativity.85) A ring structure similar to the c ring may be formed from the c, c′ and c″ subunits in yeast V-ATPase.8) The mammalian ring may be slightly different, because c′ was not found in mice.86) Subunit c, c′ and c″ are duplicated forms of which the amino and carboxyl terminal halves are homologous to those of the F-ATPase c subunit.
Fig. 8.
Localization of diverse V-ATPases in endomembrane organelles and plasma membranes. (A) Organelles are shown schematically together with endocytic and exocytic vesicle trafficking. V-ATPase localizations in endomembrane organelle and plasma membranes are shown. Examples of plasma membrane localization in osteoclasts and kidney proximal tubules are shown. Multiple V-ATPases are present in organelles and plasma membranes, although only one model is shown in each compartment because of space limitation. The luminal pH is more acidic when the cargo reaches its destination. Denser colors indicate more acidic pH. (B) A schematic model of a V-ATPase is shown together with mouse isoforms, including subunit a isoform a1, a2, a3 and a4. Tissue- or cell-specific isoforms are indicated. The isoforms not specified are ubiquitously found in mouse tissues. See text for references.
In the ATPase family, only V-ATPases are sensitive to macrolide antibiotics Bafilomycin A1 and Concanamycin A.87) The initial finding of high affinity Bafilomycin binding to the Vo sector was made for chromaffin granule enzyme.85) These antibiotics became essential reagents for studying acidic organelles in mammalian cells, as described below.
The biochemical similarities of the two enzymes suggested that the V-ATPase catalysis may be reversible. As expected, yeast V-ATPase synthesized ATP from ADP and phosphate in vacuolar vesicles when an electrochemical proton gradient was established in engineered vacuoles.88) The gradient was generated upon hydrolysis of pyrophosphate by the Arabidopsis thaliana proton-transporting pyrophosphatase, which was expressed in the vacuoles. The ATP synthesis was sensitive to Bafilomycin A1, similar to ATP hydrolysis. It should be noted that V-ATPases do not synthesize ATP physiologically, since they are localized in the organelles that can not generate a sufficient electrochemical proton gradient.
Encouraged by rotation of the F-ATPase holoenzyme, and finding of its interchangeable rotor and stator,72)–74) we immobilized V-ATPase through the c subunit. An actin filament connected to subunit G rotated continuously, generating essentially the same torque as one connected to F-ATPase.89) It should be noted that subunit G exhibits some homology to subunit b, which forms a stator with the a subunit in F-ATPase. Thus, the rotation of G relative to c was reasonable to observe experimentally. Concanamycin A inhibited rotation, confirming the close coupling between trans-membrane proton transport and subunit rotation, since the antibiotic binds to the Vo sector. The details of the rotation mechanism must await further analysis with probes exhibiting low viscous drag.
V-ATPase, essential for animal development
Yeast mutant cells with deletion of one of the VMA (Vacuolar Membrane ATPase) genes encoding V-ATPase subunits could not grow at neutral pH, whereas they could grow at acidic pH, possibly forming acidic compartments through constitutive endocytosis.8) These results indicate that acidic compartments generated by V-ATPase are essential for yeast growth. The obvious and interesting question is whether V-ATPase is required for the development of higher organisms.
The C. elegans C subunit is encoded by a single gene90) and has no isoforms, whereas subunit a has four isoforms expressed in different cells.91) The worm became embryonic lethal upon injection of the double-stranded RNA corresponding to the C subunit. The RNAs corresponding to the three subunit a isoforms generated either lethal embryos (unc-32 gene) or caused death at a specific larval stage (vha-5 or vha-6), whereas the vha-7 RNA had no effect.91) These results indicated that V-ATPase is essential for worm embryogenesis.
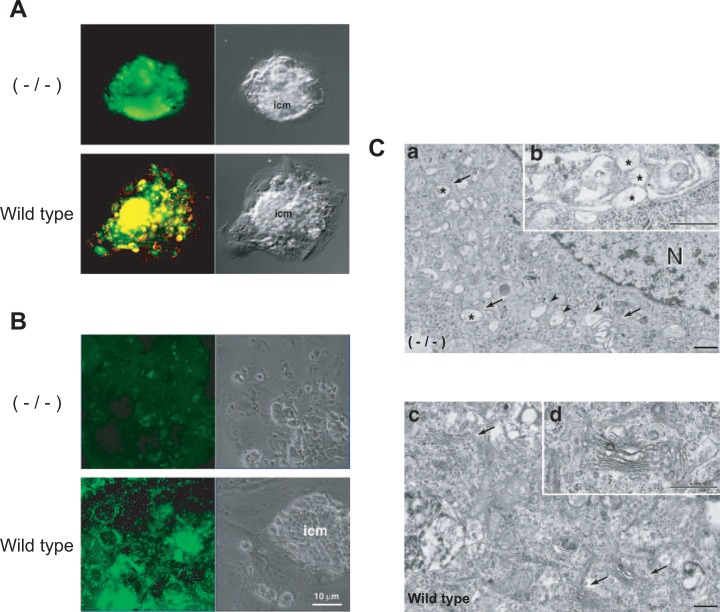
Mouse embryos have extensively developed acidic compartments in the cytoplasm from the single cell stage, which can be stained with a pH indicator dye.92) It is of interest as to whether these compartments are required for early embryonic development. Knock-out mice as to the c subunit could be used to answer this question, since the mammalian c subunit is encoded by a single gene and has no isoform.93) Mouse embryos lacking the c subunit genes (homozygote)94) could grow up to the blastocyst stage, but could not become implanted, leading to an embryonic lethal phenotype.92) The mutant blastocyst cells cultured in vitro had no acidic organelles inside, and thus could not take up dextran (Fig. 9A, B).92) The endomembrane organelles of the mutant cells were altered and extensively vacuolated (Fig. 9C). These results clearly indicate that the electrochemical proton gradient established by V-ATPase is essential for early mammalian development.
Fig. 9.
Acidic compartments in blastocysts from wild-type and c subunit null mutant mice. (A) Wild-type and c subunit null mutant (−/−) blastocysts were grown in vitro, and then stained with acridine orange. No acidic compartments were observed in the outgrowths from null blastocysts, whereas those of wild-type are stained. (B) Wild-type and c subunit null mutant (−/−) blastocysts were incubated with fluorescent dextran. No uptake of the dextran was obserrrved in c/c blastocysts, whereas it was taken up by wild-type. The respective Nomarski images are shown together (right). (C) Electron microscopy of null −/− (a, b) and wild-type (c, d) blastocyst cells is also shown. Vacuolated Golgi areas are observed in −/− mice (a, b). Modified from Sun-Wada et al.92)
Transport driven by V-ATPase
The proton gradient and membrane potential established by V-ATPase drive active transport into organelles such as synaptic vesicles, microvesicles and secretory granules.95)–99) Glutamate is accumulated in synaptic vesicles or norepinephrine in microvesicles coupled with the membrane potential,96) whereas serotonin or γ amino butyric acid is transported coupled with the proton gradient.95) As expected, this transport was abolished by ionophores, uncouplers, Bafilomycin, or Concanamycin A. The neurotransmitters accumulated in these vesicles are released into the synaptic cleft upon their fusion with the presynaptic plasma membrane.
The pH gradient also led to the accumulation of lipophilic amines in an acidic compartment.100) Their non-protonated forms pass through membranes freely because of the hydrophobicity. However, they become unable to pass through membranes, and thus, are accumulated in the organellar lumen once protonated at acidic pH. Lipophilic amines such as acridine orange or quinacrine had been widely used to assay the formation of an electrochemical proton gradient in membrane vesicles or organelles.30) Following this mechanism, local anesthetics and antineoplastic drugs were accumulated in acidic organelles, and lowered their luminal pH.100) The accumulation may be related to the drug action mechanism.
Bafilomycin and Concanamycin have been useful for studying the roles of acidic compartments established by V-ATPases. In the presence of Bafilomycin, epidermal growth factor (EGF) was transported to lysosomes, but its proteolysis in lysosomes was inhibited.101) Similarly, diphtheria toxin was transported to endosomes, but its processing was inhibited.102) The organellar enzymes became defective when the acidic pH was abolished, although the cargo (EGF or toxin) was transported to its destination in the absence of an acidic pH.
Diverse acidic compartments
Due to interest in the physiological roles of mammalian V-ATPases, we have cloned mouse cDNA homologous to yeast subunit genes. A number of isoforms has been found for the subunits of Vo sectors (a, d) and the stalk domain (C, E, G) connecting V1 and Vo (Fig. 8B), although only the B1 and B2 isoforms were known before our studies.103) The isoforms of stalk domain subunits (C, E, G) are as follows: C2-a, lamaller bodies of lung type II alveolar cells104); C2-b, plasma membrane of renal α and β intercalated cells104);E1, specific for acrosome and testis105), 106); G2, synaptic vesicles; and G3, kidney.107) The ubiquitously expressed isoforms are C1, E2 and G1, respectively.
The cDNAs of the stalk subunits could complement the yeast mutant lacking the corresponding genes. Thus, we could study the mouse subunit in a mouse/yeast hybrid V-ATPase. Interestingly, the hybrid with E1 could not transport protons at higher temperature, although that with E2 could.105) Since the enzyme with E1 and E2 had the same ATPase activity, the E1 hybrid was uncoupled at higher temperature. The V-ATPase with the C2 − a or C2 − b subunit showed a lower Km and proton transport than that with C1 or Vma5p (yeast C subunit).104) These results imply that the stalk region plays roles by regulating ATPase and in energy coupling.
Yeast Vo has three subunits, c, c′ and c″, whereas Fo only has c.8) C. elegans has two genes for the c subunit, although it was difficult to demonstrate correspondence to yeast c and c′ from their sequence similarities.108) Only c and c″ were found in mammalians.86) Four isoforms, a1, a2, a3 and a4, were found for mammalian subunit a.109), 110) Isoform a4 was found immunochemically in the apical and basolateral plasma membranes of cortical α- and β- intercalated cells in the kidney-collecting duct,110) whereas a1, a2, and a3, exhibiting 48–52% sequence identity, were expressed ubiquitously.109) Similarly, d2 is expressed in kidneys, whereas d1 is ubiquitous.107) Thus, unique V-ATPases are related to cell- or organelle-specific physiology. Some of the genes were identified as disease-related ones. Human genes for V-ATPase subunits have been summarized together with their aliases and synonyms.111) It is noteworthy that only isoforms of subunit a (Stv1 and Vph1) were found in yeast.112) Thus, the diverse V-ATPase isoforms in higher eukaryotes may be due to the evolution of cells and organelles.
Osteoclasts
The acidic lumen formed by V-ATPases is essential for many cell biological and physiological processes. V-ATPases play roles other than in the transport of protons? Their subunits, especially transmembrane subunits, may play roles to localize V-ATPase to the specific organellar membranes. In this regard, the results for osteoclasts are of interest to review.
Bone homeostasis in vertebrates is maintained through an equilibrium between bone formation and resorption. Resorption activity is provided by osteoclasts, fully differentiated multinuclear cells of monocyte-macrophage lineage.113) An acidic pH is required in the bone resorption lacuna, the compartment for dissolving hydroxyapatite and degradading the bone matrix. A V-ATPase, highly expressed in the osteoclast plasma membrane, pumps protons into the lacuna. It was of interest to determine which V-ATPase subtype is present in osteoclasts.
We found that the a3 isoform is specifically localized in the plasma membrane and its vicinity of osteoclasts derived from bone marrow cells (Fig. 10).109) On the other hand, a1 and a2 were found in cytoplasmic organelles of the same cells. The presence of a3 in osteoclasts was confirmed by immuno-histochemical staining of sections of tibiae and femora. Consistent with this localization, disruption of the ATP6 gene encoding for mouse a3 causes severe osteopetrosis.113), 114) These results suggested that a3 is a key subunit of a V-ATPase for its plasma membrane localization.
Fig. 10.
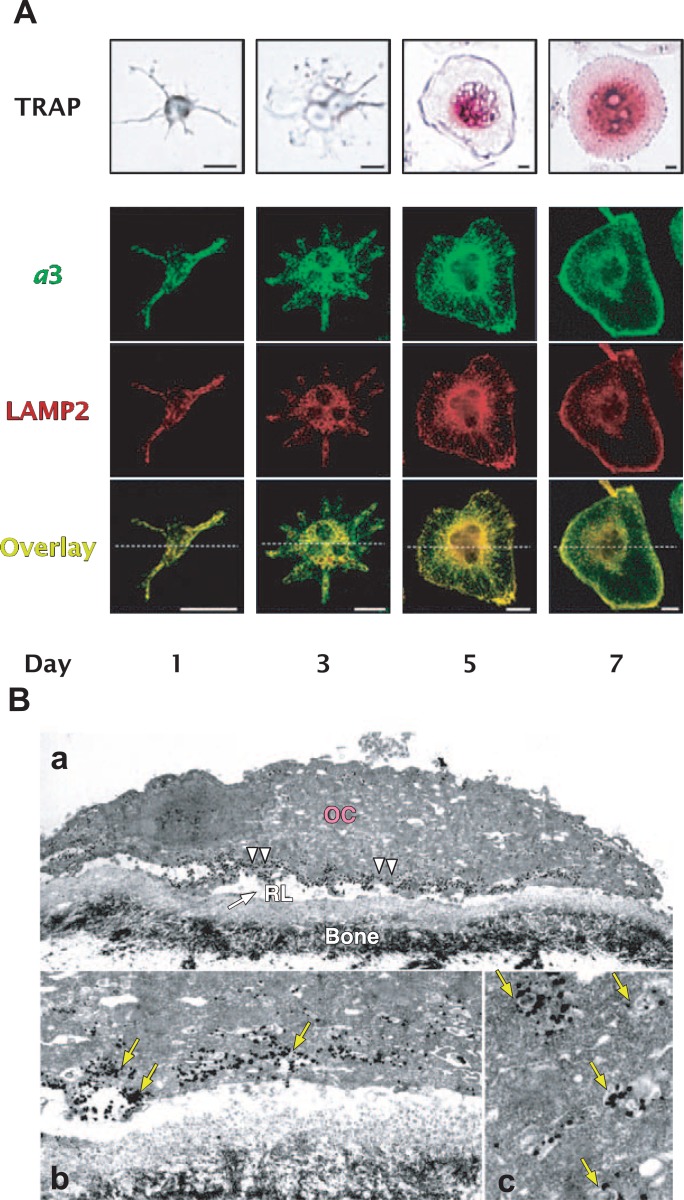
Immunochemical localization of a3 and lamp2 during differentiation of RAW264.7 cells into osteoclast-like cells. (A) RAW264.7 cells were cultured for seven days in medium containing sRANKL (solubilized RANKL) and M-CSF. Osteoclast-like cells exhibit staining for tartarate-resistant acid phosphatase (TRAP). The localization of a3 and lamp2 (LAMP2) was visualized by immunostaining. Both a3 and lamp2 were localized in the cytoplasm of the projenitor cells (Day 1), but found in the plasma membranes of the differentiated cells (Day 7). (B) Electron microscopic localization of the V-ATPase with an a3 isoform in RAW264.7 cells grown on a bone surface. Localization of the a3 isoform is shown immunochemically by electron dense silver-enhanced gold particles (for examples of gold particle, see arrow heads). The plasma membrane facing the bone (b) is densely labeled (arrows). A higher magnification for organellar localization is also shown (arrows) (c). Scale bars: a, 10 μm; b and c, 1 μm. RL, bone resorption lacuna; OC, osteoclast. Modified from previously shown.9), 106), 115)
However, the a3 isoform is not restricted to osteoclasts, being expressed in all tissues examined.109) V-ATPase with an a3 isoform was localized to late endosomes and lysosomes in non-osteoclast cells such as NIH3T3 cells and other cell lines.115) RAW264.7 cells, an established macrophage cell line, can form multinuclear cells, when cultured with sRANKL (the extra cellular domains of receptor activators of nuclear factor κB ligand). The differentiated cells express osteoclast markers including tartarate-resistant acid phosphatase (TRAP) and cathepsin K, indicating that RAW264.7 cells can be a good model for studying localization of V-ATPase during the differentiation. However, in contrast to in osteoclasts, the a3 isoform and lysosome marker proteins were localized in the same organelles before the addition of sRANKL and one day after addition (Fig. 10A, Day 1), indicating that the a3 isoform is localized to lysosomes and late endosomes of the osteoclast projenitor or an early stage after stimulation.
Upon stimulation, the a3 isoform and lamp2 (lysosome-associated membrane protein 2) were localized in the same organelles associated with a filamentous structure extending to the cell surface (Fig. 10A, Days 3, and 5).115) After 7 days’ incubation, the two markers were localized mostly on the plasma membranes of mature multi-nuclear osteoclast-like cells (Fig. 10A, Day 7). As shown on immunoelectron microscopy, the a3 signal was highly concentrated at the cell periphery and plasma membrane facing to bone matrix (Fig. 10B). These results suggest that V-ATPases of the a3 isoform localized in late endosomes/lysosomes were transported to the cell periphery, and finally assembled into the plasma membrane of osteoclasts. The a3 isoform may play a role in targeting lysosomes to plasma membranes.
Endocytosis and exocytosis
The exocytosis and endocytosis pathways allows the secretion and uptake of proteins, and small molecular ligands and transmitters. The kidney plays important roles in mammalian homeostasis, including in acid secretion, bicarbonate uptake, and protein reabsorption. Immunoprecipitation from a kidney extract revealed the presence of two type V-ATPases formed from kidney-specific isoforms B1, G3, d1, a4 and C2b and other subunits, and ones formed from ubiquitously expressed B2, C1, G1 and a1, a2 or a3 are present in the kidney (Fig. 8B).104) These results suggest that V-ATPases have unique subunit isoforms for renal ion homeostasis.
An acidic pH is required in the lumen of organelles along the endocytic or exocytic pathway (Fig. 8A). A pH-sensor that may couple the luminal pH to the formation of transport vesicles has been postulated.116) During endocytosis/exocytosis, carrier vesicles bud from early endosomes and transfer cargos to late endosomes (Fig. 8A). Both GTP-binding protein Arf6 and its cognate GDP/GTP exchange factor, ARNO (ADP-ribosylation factor nucleotide site opener), have been implicated in carrier vesicle coat formation. The recruitment of Arf6 and ARNO to the endosomal membrane is driven by an intra-endosomal acidic pH. This pH-sensitive mechanism involves interaction of Arf6 with the c subunit and ARNO with the a2 isoform.
V-ATPase with an a3 isoform is highly expressed in pancreatic islet β cells and is localized in insulin secretory vesicles.117) The oc/oc mutant mouse with deletion of a part of the a3 gene exhibited low blood insulin level together with severe osteopetrosis. The osteoporosis is due to the defective proton transport into bone resorption lacuna, as discussed above. The mutant Langerhans islets contained mature insulin, but its secretion in response to glucose or depolarization was impaired. These observations suggest that the oc/oc mouse is defective in the exocytosis of secretory vesicles. βTC9 cells, having the characteristics of β cells, could secrete insulin even after Bafilomycin treatment. These results indicate that the secretion does not require acidification, but is dependent on the presence of V-ATPase with an a3 isoform in the membranes of secretory vesicles. These results suggest that the subunits of V-ATPase play roles in regulating membrane trafficking in exocytosis or endocytosis.
We have discussed our studies over the three decades on proton pumping F-ATPases and V-ATPases. The important results include the stochastic properties of F-ATPases and diverse V-ATPases. Furthermore, the results obtained suggest that V-ATPase subunit isoforms play roles in targeting to organellar membranes. These findings emphasize a new concept of the enzyme.
Acknowledgments
I wish to thank the coworkers whose names appear in the references. I am especially grateful to Drs. H. Kanazawa, Masatomo Maeda, Yoh Wada, G.-H. Sun-Wada, A. Iwamoto-Kihara, Y. Sambongi, Y. Moriyama, and M. Nakanishi-Matsui. Their contributions to the results discussed in this article are most appreciated.
We are also greatly indebted to the work of other laboratories, most of which could not be cited because of the nature of this article and the limited space. I thank Dr. Ikuko Taira-Fuji for preparing the figures in this article. Research in our laboratories has been supported by Grants-in-Aid the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Japan Science and Technology Agency.
Profile
Masamitsu Futai was born in 1940, and started his research career in 1963 as a graduate student at the Faculty of Pharmaceutical Sciences, University of Tokyo, studying ribonucleases, phosphodiesterases, and related enzymes in E. coli and rat liver emphasizing their localizations. After obtaining PhD degree, he worked in University of Wisconsin and Cornell University as a postdoctoral fellow. He was promoted to Professor of Microbiology at the Faculty of Pharmaceutical Sciences, Okayama University in 1977. He moved, in 1985, to Department of Biological Sciences, Institute of Scientific and Industrial Research, Osaka University. Between 2000 and 2003, he was a Director of the Institute.

He has been studying proton pumping ATPases including F-ATPase (ATP synthase), mammalian V-ATPase (vacuolar type ATPase) and gastric proton pump (H+/K+ ATPase). He is a pioneer of the field of E. coli bioenergetics especially of F-ATPase, establishing its purification and identifying gene sequences, and contributed its mechanism of energy coupling, catalytic site and proton transport/rotational mechanism. He has shown functional and structural diversity of V-ATPase in mice and C. elegans, and contributed to the understanding the roles of acidic lumens of organelles. He also made important contribution on gene expression of the gastric proton pump.
He was awarded American Cancer Society Fellowship, Academic Award of Mochida Memorial Foundation, and Academic Prize of Japanese Pharmaceutical Society. He has served Editorial Board of J. Microbiology, J. Biochemistry, J. Bacteriology, and J. Biological Chemistry and Molecular Biology, and is an honorary member of the Japanese Biochemical Society. He is, a Special Research Scientist at Microbial Chemistry Research Center, Microbial Chemistry Research Foundation since 2003.
References
- 1).Futai, M., Sun-Wada, G.-H., and Wada, Y. (2003) Proton-translocating ATPases: Introducing unique enzymes coupling catalysis and proton translocation through mechanical rotation. InHandbook of ATPases: Biochemistry, Cell Biology, Pathophysiology (eds. Futai M., Wada Y., and Kaplan J.), Wiley-VCH Verlag GmbH & Co, KgaA, Weinheim, Germany, pp. 237–260. [Google Scholar]
- 2).Futai, M., Omote, H., Sambongi, Y., and Wada, Y. (2000) ATP synthase (H+ ATPase): coupling between catalysis, mechanical work, and proton translocation. Biochim. Biophys. Acta 1458, 276–288. [DOI] [PubMed] [Google Scholar]
- 3).Futai, M., Noumi, T., and Maeda, M. (1989) ATP synthase (H+-ATPase): Results by combined biochemical and molecular biological approaches. Ann. Rev. Biochem. 58, 111–136. [DOI] [PubMed] [Google Scholar]
- 4).Racker, E. (1976) A New Look at Mechanisms of Bioenergetics. Academic Press, New York-San Francisco-London. [Google Scholar]
- 5).Pedersen, P. L., and Carafoli, E. (1987) Ion motive ATPase. I. Ubiquity, properties, and significance to cell function. Trends Biochem. Sci. 12, 146–150. [Google Scholar]
- 6).Futai, M., and Kanazawa, H. (1983) Structure and function of proton-translocating ATPase (FoF1): biochemical and molecular biological approaches. Microbiol. Rev. 47, 285–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Abrahams, J. P., Leslie, A. G. W., Lutter, R., and Walker, J. (1994) Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628. [DOI] [PubMed] [Google Scholar]
- 8).Nelson, N., and Harvey, W. R. (1999) Vacuolar and plasma membrane proton-adenosinetriphosphatases. Physiol. Rev. 79, 361–385. [DOI] [PubMed] [Google Scholar]
- 9).Sun-Wada, G.-H., Wada, Y., and Futai, M. (2003) Vacuolar-type proton pump ATPase: subunit isoforms and tissue-specific function. InHandbook of ATPases: Biochemistry, Cell Biology, Pathophysiology (eds. Futai M., Wada Y., and Kaplan J.), Wiley-VCH Verlag GmbH & Co, KgaA, Weinheim, Germany, pp. 379–394. [Google Scholar]
- 10).Futai, M., Oka, T., Sun-Wada, G.-H., Moriyama, Y., Kanazawa, H., and Yada, Y. (2000) Luminal acidification of diverse organelles by V-ATPase in animal cells. J. Exp. Biol. 203, 107–116. [DOI] [PubMed] [Google Scholar]
- 11).Maeda, M., Oshiman, K., Tamura, S., and Futai, M. (1990) Human gastric (H+ + K+)-ATPase gene: Similarity to (Na+ + K+)-ATPase genes in exon/intron organization but difference in control region. J. Biol. Chem. 265, 9027–9032. [PubMed] [Google Scholar]
- 12).Maeda, M., Oshiman, K., Tamura, S., Kaya, S., Mahmood, M., Reuben, M., Sachs, G., and Futai, M. (1991) The rat H+/K+-ATPase β subunit gene and recognition of its control region by gastric DNA-binding protein. J. Biol. Chem. 266, 21584–21588. [PubMed] [Google Scholar]
- 13).Maeda, M., Ishizaki, J., and Futai, M. (1988) cDNA cloning and sequence determination of pig gastric (H+ + K+)-ATPase. Biochem. Biophys. Res. Commun. 157, 203–209. [DOI] [PubMed] [Google Scholar]
- 14).Tamura, S., Wang, X., Maeda, M., and Futai, M. (1993) Gastric DNA-binding proteins recognize upstream sequence motifs of parietal cell-specific genes. Proc. Natl. Acad. Sci. USA 90, 10876–10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Tamura, S., Oshiman, K., Nishi, T., Mori, M., Maeda, M., and Futai, M. (1992) Sequence motif in control regions of the H+/K+-ATPase α and β subunit genes recognized by gastric specific nuclear protein. FEBS Lett. 298, 137–141. [DOI] [PubMed] [Google Scholar]
- 16).Futai, M., Tsung, P.-K., and Mizuno, D. (1972) Possible heterogeneity of the distribution of lysosomal marker enzymes among “lysosomal particles” of rat liver. Biochim. Biophys. Acta 261, 508–516. [DOI] [PubMed] [Google Scholar]
- 17).Futai, M., Wada, Y., and Kaplan, J. (2003) Handbook of ATPases; Biochemistry, Cell Biology, Pathophysiology. Wiley-VCH Verlag, Gmbh & Co., Kga A, Weinheim, Germany. [Google Scholar]
- 18).Futai, M. (1974) Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J. Membrane Biol. 15, 15–28. [DOI] [PubMed] [Google Scholar]
- 19).Futai, M. (1974) Reconstitution of transport dependent on D-lactate or glycerol 3-phosphate in membrane vesicles of Escherichia coli deficient in the corresponding dehydrogenases. Biochemistry (Washington) 13, 2327–2333. [DOI] [PubMed] [Google Scholar]
- 20).Futai, M. (1974) Stimulation of transport into Escherichia coli membrane vesicles by internally generated reduced nicotinamide adenine dinucleotide. J. Bacteriol. 120, 861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Futai, M., Sternweis, P. C., and Heppel, L. A. (1974) Purification and properties of reconstitutively active and inactive adenosinetriphosphatase from Escherichia coli. Proc. Natl. Acad. Sci. USA 71, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Futai, M. (1977) Reconstitution of ATPase activity from the isolated α, β, and γ subunits of the coupling factor, F1, of Escherichia coli. Biochem. Biophys. Res. Commun. 79, 1231–1237. [DOI] [PubMed] [Google Scholar]
- 23).Dunn, S. D., and Futai, M. (1980) Reconstitution of a functional coupling factor from the isolated subunits of Escherichia coli F1 ATPase. J. Biol. Chem. 255, 113–118. [PubMed] [Google Scholar]
- 24).Hirano, M., Takeda, K., Kanazawa, H., and Futai, M. (1984) ATP-dependent conformational change of the β subunit of Escherichia coli H+-ATPase. Biochemistry (Washington) 23, 1652–1656. [DOI] [PubMed] [Google Scholar]
- 25).Foster, D. L., Mosher, M. E., Futai, M., and Fillingame, R. H. (1980) Subunits of the H+-ATPase of Escherichia coli: Overproduction of an eight-subunit F1Fo-ATPase following induction of a λ transducing phage carrying the unc operon. J. Biol. Chem. 255, 12037–12041. [PubMed] [Google Scholar]
- 26).Moriyama, Y., Iwamoto, A., Hanada, H., Maeda, M., and Futai, M. (1991) One-step purification of Escherichia coli H+-ATPase (FoF1) and its reconstitution into liposomes with neurotransmitter transporters. J. Biol. Chem. 266, 22141–22146. [PubMed] [Google Scholar]
- 27).Iwamoto, A., Omote, H., Hanada, H., Tomioka, N., Itai, A., Maeda, M., and Futai, M. (1991) Mutation in Ser-174 and the glycine-rich sequence (Gly-149, Gly-150 and Thr-156) in the β subunit of Escherichia coli H+-ATPase. J. Biol. Chem. 266, 16350–16355. [PubMed] [Google Scholar]
- 28).Downie, J. A., Gibson, F., and Cox, G. B. (1979) Membrane adenosine triphosphatases of prokaryotic cells. Ann. Rev. Biochem. 48, 103–131. [DOI] [PubMed] [Google Scholar]
- 29).Miki, T., Hiraga, S., Nagata, T., and Yura, T. (1978) Bacteriophage λ carrying the Escherichia coil chromosomal region of the replication origin. Proc. Natl. Acad. Sci. USA 75, 5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Kanazawa, H., Miki, T., Tamura, F., Yura, T., and Futai, M. (1979) Specialized transducing phage λ carrying the genes for couplng factor of oxidative phosphorylation of Escherichia coli: Increased synthesis of coupling factor on induction of prophage λasn. Proc. Natl. Acad. Sci. USA 76, 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Kanazawa, H., Tamura, F., Mabuchi, K., Miki, T., and Futai, M. (1980) Organization of unc gene cluster of Escherichia coli coding for the proton-translocating ATPase of oxidative phosphorylation. Proc. Natl. Acad. Sci. USA 77, 7005–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Kanazawa, H., Mabuchi, K., Kayano, T., Tamura, F., and Futai, M. (1981) Nucleotide sequence of genes coding for dicyclohexylcarbodiimide-binding protein and the α subunit of proton-translocating ATPase of Escherichia coli. Biochem. Biophys. Res. Commun. 100, 219–225. [DOI] [PubMed] [Google Scholar]
- 33).Kanazawa, H., Kayano, T., Mabuchi, K., and Futai, M. (1981) Nucleotide sequence of the genes coding for α, β and γ subunits of proton-translocating ATPase of Escherichia coli. Biochem. Biophys. Res. Commun. 103, 604–612. [DOI] [PubMed] [Google Scholar]
- 34).Kanazawa, H., Mabuchi, K., Kayano, T., Noumi, T., Sekiya, T., and Futai, M. (1981) Nucleotide sequence of the genes for Fo components of the proton-translocating ATPase from Escherichia coli: Prediction of the primary structure of Fo subunits. Biochem. Biophys. Res. Commun. 103, 613–620. [DOI] [PubMed] [Google Scholar]
- 35).Kanazawa, H., Kayano, T., Kiyasu, T., and Futai, M. (1982) Nucleotide sequence of the genes for β and ε subunits of proton-translocating ATPase from Escherichia coli. Biochem. Biophys. Res. Commun. 105, 1257–1264. [DOI] [PubMed] [Google Scholar]
- 36).Kanazawa, H., and Futai, M. (1982) Structure and function of H+ATPase: What we have learned from Escherichia coli H+-ATPase. Ann. N. Y. Acad. Sci. 402, 45–64. [DOI] [PubMed] [Google Scholar]
- 37).Kanazawa, H., Saito., S., and Futai, M. (1978) Coupling factor ATPase from Escherichia coli: An uncA mutant (uncA401) with defective α subunit. J. Biochem. 84, 1513–1517. [DOI] [PubMed] [Google Scholar]
- 38).Kanazawa, H., Noumi, T., and Futai, M. (1986) Analysis of Escherichia coli mutants of the proton-translocating ATPase F1Fo: Identification of altered domains and residues. Methods in Enzymol. 126, 595–603. [DOI] [PubMed] [Google Scholar]
- 39).Noumi, T., Futai, M., and Kanazawa, H. (1984) Replacement of serine-373 by phenylalanine in the α subunit of Escherichia coli F1-ATPase results in loss of steady state catalysis by the enzyme. J. Biol. Chem. 259, 10076–10079. [PubMed] [Google Scholar]
- 40).Orita, M., Iwanaga, H., Kanazawa, H., Hayashi, K., and Sekiya, T. (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. USA 86, 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Noumi, T., Mosher, M. E., Natori, S., Futai, M., and Kanazawa, H. (1984) A phenylalanine for serine substitution in the β subunit of Escherichia coli F1-ATPase affects dependency of its activity on divalent cations. J. Biol. Chem. 259, 10071–10075. [PubMed] [Google Scholar]
- 42).Soga, S., Noumi, T., Takeyama, M., Maeda, M., and Futai, M. (1989) Mutational replacements of conserved amino acid residues in the α subunit change the catalytic properties of Escherichia coli F1-ATPase. Arch. Biochem. Biophys. 268, 643–648. [DOI] [PubMed] [Google Scholar]
- 43).Penefsky, H. S., and Cross, R. L. (1991) Structure and mechanism of FoF1-type ATP synthases and ATPases. Enzymol. Related Area Mol. Biol. 64, 173–214. [DOI] [PubMed] [Google Scholar]
- 44).Noumi, T., Futai, M., and Kanazawa, H. (1984) Replacement of Ser373 by phenylalanine in the α subunit of Escherichia coli F-ATPase results in loss of steady-state catalysis by the enzyme. J. Biol. Chem. 259, 10076–10079. [PubMed] [Google Scholar]
- 45).Noumi, T., Taniai, M., Kanazawa, H., and Futai, M. (1986) Replacement of arginine 246 by histidine in the β subunit of Escherichia coli H+-ATPase resulted in loss of multi-site ATPase activity. J. Biol. Chem. 261, 9196–9201. [PubMed] [Google Scholar]
- 46).Yoshida, M., Allison, W. S., Esch, F. S., and Futai, M. (1982) The specificity of carboxyl group modification during the inactivation of E. coli F1 ATPase with dicyclohexyl 14C carbodiimide. J. Biol. Chem. 257, 10033–10037. [PubMed] [Google Scholar]
- 47).Noumi, T., Tagaya, M., Miki-Takeda, K., Maeda, M., Fukui, T., and Futai, M. (1987) Loss of unisite and multisite catalyses by Escherichia coli F1 through modification with adenosine trior tetraphosphopyridoxal. J. Biol. Chem. 262, 7686–7692. [PubMed] [Google Scholar]
- 48).Ida, K., Noumi, T., Maeda, M., Fukui, T., and Futai, M. (1991) Catalytic site of F1-ATPase of Escherichia coli: Lys-155 and Lys-201 of the β subunit are located near the γ phosphate group of ATP in the presence of Mg2+. J. Biol. Chem. 266, 5424–5429. [PubMed] [Google Scholar]
- 49).Hsu, S.-Y., Noumi, T., Takeyama, M., Maeda, M., Ishibashi, S., and Futai, M. (1987) β subunit of Escherichia coli F1-ATPase: An amino acid replacement within a conserved sequence (G-X-XX-X-G-K-T/S) of nucleotide-binding proteins. FEBS Lett. 218, 222–226. [DOI] [PubMed] [Google Scholar]
- 50).Takeyama, M., Ihara, K., Moriyama, Y., Noumi, T., Ida, K., Tomioka, N., Itai, A., Maeda, M., and Futai, M. (1990) The glycine-rich sequence of the β subunit of Escherichia coli H+-ATPase is important for activity. J. Biol. Chem. 265, 21279–21284. [PubMed] [Google Scholar]
- 51).Omote, H., Maeda, M., and Futai, M. (1992) Effects of mutations of conserved Lys-155 and Thr-156 residues in the phosphate-binding glycine-rich sequence of the F1-ATPase β subunit of Escherichia coli. J. Biol. Chem. 267, 20571–20576. [PubMed] [Google Scholar]
- 52).Iwamoto, A., Park, M.-Y., Maeda, M., and Futai, M. (1993) Domains near ATP phosphate in the catalytic site of H+-ATPase: Model proposed from mutagenesis and inhibitor studies. J. Biol. Chem. 268, 3156–3160. [PubMed] [Google Scholar]
- 53).Park, M.-Y., Omote, H., Maeda, M., and Futai, M. (1994) Conserved Glu-181 and Arg-182 residues of Escherichia coli H+ -ATPase (ATP synthase) β subunit are essential for catalysis: Properties of 33 mutants between β Glu-161 and β Lys-201 residues J. Biochem. 116, 1139–1145. [DOI] [PubMed] [Google Scholar]
- 54).Omote, H., Le, N. P., Park, M.-Y., Maeda, M., and Futai, M. (1995) β subunit Glu-185 of Escherichia coli H+-ATPase (ATP synthase) is an essential residue for cooperative catalysis. J. Biol. Chem. 270, 25656–25660. [DOI] [PubMed] [Google Scholar]
- 55).Le, N. P., Omote, H., Wada, Y., Al-Shawi, M. K., Nakamoto, R., and Futai, M. (2000) Escherichia coli ATP synthase α subunit Arg-376: the catalytic site arginine does not participate in the hydrolysis/synthesis reaction, but is required for promotion to steady state. Biochemistry (Washington) 39, 2778–2783. [DOI] [PubMed] [Google Scholar]
- 56).Nakamoto, R. K., Shin, K., Iwamoto, A., Omote, H., Maeda, M., and Futai, M. (1992) Escherichia coli FoF1-ATPase: residues involved in catalysis and coupling. Ann. N. Y. Acad. Sci. 671, 335–344. [DOI] [PubMed] [Google Scholar]
- 57).Iwamoto, A., Miki, J., Maeda, M., and Futai, M. (1990) H+-ATPase γ subunit from Escherichia coli: Role of the conserved carboxyl terminal region. J. Biol. Chem. 265, 5043–5048. [PubMed] [Google Scholar]
- 58).Miki, J., Takeyama, M., Noumi, T., Kanazawa, H., Maeda, M., and Futai, M. (1986) Escherichia coli H+-ATPase: Loss of the carboxyl terminal region of the γ subunit causes defective assembly of the F1 portion. Arch. Biochem. Biophys. 251, 458–464. [DOI] [PubMed] [Google Scholar]
- 59).Kanazawa, H., Hama, H., Rosen, B. P., and Futai, M. (1985) Deletion of seven amino acid residues from the γ subunit of Escherichia coli H+-ATPase causes total loss of F1 assembly on membranes. Arch. Biochem. Biophys. 241, 364–370. [DOI] [PubMed] [Google Scholar]
- 60).Shin, K., Nakamoto, R. K., Maeda, M., and Futai, M. (1992) FoF1-ATPase γ subunit mutations perturb the coupling between catalysis and transport. J. Biol. Chem. 267, 20835–20839. [PubMed] [Google Scholar]
- 61).Nakamoto, R. K., Maeda, M., and Futai, M. (1993) The γ subunit of the Escherichia coli ATP synthase: mutations in the carboxyl-terminal region restore energy coupling to the amino-terminal mutant γMet-23 → Lys. J. Biol. Chem. 268, 867–872. [PubMed] [Google Scholar]
- 62).Nakamoto, R. K., Al-Shawi, M., and Futai, M. (1995) The ATP synthase γ subunit: suppressor mutagenesis reveals three helical regions involved in energy coupling. J. Biol. Chem. 270, 14042–14046. [DOI] [PubMed] [Google Scholar]
- 63).Stock, D., Gibbones, C., Arechaga, I., Leslie, A. G. W., and Walker, J. E. (2000) The rotary mechanism of ATP synthase. Curr. Opinion Structure. Biol. 10, 672–679. [DOI] [PubMed] [Google Scholar]
- 64).Eya, S., Noumi, T., Maeda, M., and Futai, M. (1988) Intrinsic membrane sector (Fo) of H+-ATPase (FoF1) from Escherichia coli: Mutations in the α subunit give Fo with impaired proton translocation and F1 binding. J. Biol. Chem. 263, 10056–10062. [PubMed] [Google Scholar]
- 65).Tamura, F., Kanazawa, H., Tsuchiya, T., and Futai, M. (1981) Structural gene coding for the dicyclohexylcarbodiimide-binding protein in the proton-translocating ATPase of Escherichia coli: Locus of the gene in the F1Fo gene cluster. FEBS Lett. 127, 48–52. [DOI] [PubMed] [Google Scholar]
- 66).Takeyama, M., Noumi, T., Maeda, M., and Futai, M. (1988) Fo portion of Escherichia coli H+-ATPase: Carboxyl terminal region of the b subunit is essential for assembly of functional Fo. J. Biol. Chem. 263, 16106–16112. [PubMed] [Google Scholar]
- 67).Takeyasu, K., Omote, H., Nettikadan, S., Tokumasu, F., Iwamoto, A., and Futai, M. (1996) Molecular imaging of Escherichia coli FoF1-ATPase in reconstituted membranes using atomic force microscopy. FEBS Lett. 392, 110–113. [DOI] [PubMed] [Google Scholar]
- 68).Stock, D., Leslie, A. G. W., and Walker, J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705. [DOI] [PubMed] [Google Scholar]
- 69).Fillingame, R. H., Angevien, C. M., and Dmitriev, O. Y. (2003) Mechanics of coupling proton movement to c-ring rotation in ATP synthase. FEBS Lett. 555, 29–34. [DOI] [PubMed] [Google Scholar]
- 70).Noji, H., Yasuda, R., Yoshida, N., and Kinoshita, K.Jr. (1997) Direct observation of the rotation of F1-ATPase. Nature 386, 299–302. [DOI] [PubMed] [Google Scholar]
- 71).Omote, H., Sambonmatsu, N., Saito, K., Sambongi, Y., Iwamoto-Kihara, A., Yanagida, T., Wada, Y., and Futai, M. (1999) The γ subunit rotation and torque generation in F1-ATPase from wild-type or uncoupled mutant Escherichia coli. Proc. Natl. Acad. Sci. USA 96, 7780–7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Sambongi, Y., Iko, Y., Tanabe, M., Omote, H., Iwamoto-Kihara, A., Ueda, I., Yanagida, T., Wada, Y., and Futai, M. (1999) Mechanical rotation of the c subunit oligomer in ATP synthase (FoF1): direct observation. Science 286, 1722–1724. [DOI] [PubMed] [Google Scholar]
- 73).Tanabe, M., Nishio, K., Iko, Y., Sambongi, Y., Iwamoto-Kihara, A., Omote, H., Ueda, I., Wada, Y., and Futai, M. (2001) Rotation of a complex of the γ subunit and c ring of Escherichia coli ATP synthase: the rotor and stator are interchangeable. J. Biol. Chem. 276, 15269–15274. [DOI] [PubMed] [Google Scholar]
- 74).Nishio, K., Iwamoto-Kihara, A., Yamamoto, A., Wada, Y., and Futai, M. (2002) Subunit rotation of ATP synthase embedded in membranes: α or β subunit rotation relative to the c subunit ring. Proc. Natl. Acad. Sci. USA 99, 13448–13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Sambongi, Y., Ueda, I., Wada, Y., and Futai, M. (2000) A biological molecular motor, proton-translocating ATP synthase: Multidiciplinary approach for a unique membrane enzyme. J. Bioenerg. Biomemb. 32, 441–448. [DOI] [PubMed] [Google Scholar]
- 76).Wada, Y., Sambongi, Y., and Futai, M. (2000) Biological nano motor, ATP synthase FoF1: From catalysis to γεc10–12 subunit assembly rotation. Biochim. Biophys. Acta 1459, 499–505. [DOI] [PubMed] [Google Scholar]
- 77).Kuki, M., Noumi, T., Maeda, M., Amemura, A., and Futai, M. (1988) Functional domains of ε subunit of Escherichia coli H+-ATPase (FoF1). J. Biol. Chem. 263, 17437–17442. [PubMed] [Google Scholar]
- 78).Nakanishi-Matsui, M., Kashiwagi, S., Hosokawa, H., Cipriano, D. J., and Futai, M. (2006) Stochastic high-speed rotation of the Escherichia coli ATP synthase F1 sector: The ε subunit sensitive rotation. J. Biol. Chem. 281, 4126–4131. [DOI] [PubMed] [Google Scholar]
- 79).Iko, Y., Sambongi, Y., Tanabe, M., Iwamoto-Kihara, A., Saito, K., Ueda, I., Wada, Y., and Futai, M. (2001) ATP synthase F1 sector rotation; defective torque generation in the β subunit Ser-174 to Phe mutant and its suppression by second mutations. J. Biol. Chem. 276, 47508–47511. [DOI] [PubMed] [Google Scholar]
- 80).Nakanishi-Matsui, M., and Futai, M. (2006) Stochastic proton pumping ATPase: from single molecules to diverse physiological roles. IUBMB Life 58, 318–322. [DOI] [PubMed] [Google Scholar]
- 81).Nakanishi-Matsui, M., Kashiwagi, S., Ubukata, T., Iwamoto-Kihara, A., Wada, Y., and Futai, M. (2006) Rotational catalysis of Escherichia coli ATP synthase F1 sector: A key domain of β subunit (in preparation). [DOI] [PubMed]
- 82).Iwamoto, A., Omote, H., Hanada, H., Tomioka, N., Itai, A., Maeda, M., and Futai, M. (1991) Mutations in Ser174 and glycine-rich sequence (Gly149, Gly150 and Thr156) in the β subunit of Escherichia coli H+ ATPase. J. Biol. Chem. 266, 16350–16355. [PubMed] [Google Scholar]
- 83).Omote, H., Park, M.-Y., Maeda, M., and Futai, M. (1994) The α/β subunit interaction in H+-ATPase (ATP synthase): an Escherichia coil α subunit mutation (αArg-296 → Cys) restored coupling efficiency to the deleterious β subunit mutant (βSer-174 → Phe). J. Biol. Chem. 269, 10265–10269. [PubMed] [Google Scholar]
- 84).Arata, Y., Baleja, J. D., and Forgac, M. (2002) Localization of subunits D, E, and G in the yeast V-ATPase complex using cystine-mediated cross-linking to subunit B. Biochemistry (Washington) 41, 11301–11307. [DOI] [PubMed] [Google Scholar]
- 85).Hanada, H., Moriyama, Y., Maeda, M., and Futai, M. (1990) Kinetic studies of chromaffin granule H+-ATPase and effects of bafilomycin A1. Biochem. Biophys. Res. Commun. 170, 873–878. [DOI] [PubMed] [Google Scholar]
- 86).Sun-Wada, G.-H., Murakami, H., Nakai, H., Wada, Y., and Futai, M. (2001) Mouse Atp6f, the gene encoding the 23-kDa proteolipid of vacuolar proton-translocating ATPase. Gene 274, 93–99. [DOI] [PubMed] [Google Scholar]
- 87).Bowman, E. M., Siebers, A., and Altendorf, K. (1998) Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 85, 7972–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Hirata, T., Nakamura, N., Omote, H., Wada, Y., and Futai, M. (2000) Regulation and reversibility of vacuolar H+-ATPase. J. Biol. Chem. 275, 386–389. [DOI] [PubMed] [Google Scholar]
- 89).Hirata, T., Iwamoto-Kihara, A., Okajima, T., Wada, Y., and Futai, M. (2003) Subunit rotation in vacuolar-type proton pumping ATPase: relative rotation of G as to the c subunit. J. Biol. Chem. 278, 23714–23719. [DOI] [PubMed] [Google Scholar]
- 90).Oka, T., and Futai, M. (2000) Requirement of V-ATPase for ovulation and embryogenesis in Caenorhabditis elegans. J. Biol. Chem. 275, 29556–29561. [DOI] [PubMed] [Google Scholar]
- 91).Oka, T., Toyomura, T., Honjo, K., Wada, Y., and Futai, M. (2001) Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase: Cell-specific expression during development. J. Biol. Chem. 276, 33079–33085. [DOI] [PubMed] [Google Scholar]
- 92).Sun-Wada, G.-H., Murata, Y., Yamamoto, A., Kanazawa, H., Wada, Y., and Futai, M. (2000) Acidic endomembrane organelles are required for mouse post-implantation development. Dev. Biol. 228, 315–325. [DOI] [PubMed] [Google Scholar]
- 93).Hanada, H., Hasebe, M., Moriyama, Y., Maeda, M., and Futai, M. (1991) Molecular cloning of cDNA encoding the 16 kDa subunit of vacuolar H+-ATPase from mouse cerebellum. Biochem. Biophys. Res. Commun. 176, 1062–1067. [DOI] [PubMed] [Google Scholar]
- 94).Inoue, H., Noumi, T., Nagata, M., Murakami, M., and Kanazawa, H. (1999) Targeted disruptions of the gene encoding the proteolipid subunit of mouse vacuolar H+-ATPase leads to early embryonic lethality. Biochim. Biophys. Acta 1413, 130–138. [DOI] [PubMed] [Google Scholar]
- 95).Moriyama, Y., and Futai, M. (1990) Presence of 5′-hydroxytryptamine (serotonine) transport coupled with vacuolar type H+-ATPase in neurosecretory granules from bovine posterior pituitary. J. Biol. Chem. 265, 9165–9169. [PubMed] [Google Scholar]
- 96).Moriyama, Y., Maeda, M., and Futai, M. (1990) Energy coupling of L-glutamate transport and vacuolar H+-ATPase in brain synaptic vesicles. J. Biochem. 108, 689–693. [DOI] [PubMed] [Google Scholar]
- 97).Moriyama, Y., and Futai, M. (1990) H+-ATPase, a primary pump for accumulation of neurotransmitters, is a major constituent of brain synaptic vesicles. Biochem. Biophys. Res. Commun. 173, 443–448. [DOI] [PubMed] [Google Scholar]
- 98).Moriyama, Y., Yamamoto, A., Yamada, H., Tashiro, Y., Tomochika, K., Takahashi, M., Maeda, M., and Futai, M. (1995) Microvesicles isolated from bovine posterior pituitary accumulate norepinephrine. J. Biol. Chem. 270, 11424–11429. [DOI] [PubMed] [Google Scholar]
- 99).Moriyama, Y., Amakatsu, K., and Futai, M. (1993) Uptake of the neurotoxin, 4-methyphenylpyridinium, into chromaffin granules and synaptic vesicles: A proton gradient drives its uptake through monoamine transporter. Arch. Biochem. Biophys. 305, 271–277. [DOI] [PubMed] [Google Scholar]
- 100).Moriyama, Y., Manabe, T., Yoshimori, T., Tashiro, Y., and Futai, M. (1994) ATP-dependent uptake of anti-neoplastic agents by acidic organelles. J. Biochem. 115, 213–218. [DOI] [PubMed] [Google Scholar]
- 101).Yoshimori, T., Yamamoto, A., Moriyama, Y., Futai, M., and Tashiro, Y. (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712. [PubMed] [Google Scholar]
- 102).Umata, T., Moriyama, Y., Futai, M., and Mekada, E. (1990) The cytotoxic action of diphtheria toxin and its degradation in intact Vero cells are inhibited by bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase. J. Biol. Chem. 265, 21940–21945. [PubMed] [Google Scholar]
- 103).Nelson, R. D., Guo, X.-L., Masood, K., Brown, D., Kalkbrenner, M., and Gluck, S. (1992) Selectively amplified expression of an isoform of the vacuolar H+-ATPase 56-kilodalton subunit in renal intercalated cells. Proc. Natl. Acad. Sci. USA 89, 3541–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104).Sun-Wada, G.-H., Murata, Y., Namba, M., Yamamoto, A., Wada, Y., and Futai, M. (2004) Mouse proton pump ATPase C subunit isoforms (C2-a and C2-b) specifically expressed in kidney and lung. J. Biol. Chem. 278, 44843–44851. [DOI] [PubMed] [Google Scholar]
- 105).Sun-Wada, G.-H., Imai-Senga, Y., Yamamoto, A., Murata, Y., Hirata, T., Wada, Y., and Futai, M. (2002) A proton pump ATPase with a testis-specific E1 subunit isoform required for acrosome acidification. J. Biol. Chem. 277, 18098–18105. [DOI] [PubMed] [Google Scholar]
- 106).Imai-Senga, Y., Sun-Wada, G.-H., Wada, Y., and Futai, M. (2002) A human gene, ATP6E1, encoding a testis-specific isoform of H+ ATPase subunit E. Gene 289, 7–12. [DOI] [PubMed] [Google Scholar]
- 107).Sun-Wada, G.-H., Yoshimizu, T., Imai-Senga, Y., Wada, Y., and Futai, M. (2003) Diversity of mouse proton-translocating ATPase: Presence of multiple isoforms of the C, d and G subunits. Gene 302, 147–153. [DOI] [PubMed] [Google Scholar]
- 108).Oka, T., Yamamoto, R., and Futai, M. (1998) Multiple genes for vacuolar-type ATPase proteolipids in Caenorhabditis elegans: a new gene, vha-3, has a distinct cell-specific distribution. J. Biol. Chem. 273, 22570–22576. [DOI] [PubMed] [Google Scholar]
- 109).Toyomura, T., Oka, T., Yamaguchi, C., Wada, Y., and Futai, M. (2000) Three subunit a isoforms of mouse vacuolar H+-ATPase: Preferential expression of the a3 isoform during osteoclast differentiation. J. Biol. Chem. 275, 8760–8765. [DOI] [PubMed] [Google Scholar]
- 110).Oka, T., Murata, Y., Namba, M., Yoshimizu, T., Toyomura, T., Yamamoto, A., Sun-Wada, G.-H., Hamasaki, N., Wada, Y., and Futai, M. (2001) a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J. Biol. Chem. 276, 40050–40054. [DOI] [PubMed] [Google Scholar]
- 111).Smith, A. N., Lovering, R., Futai, M., Takeda, J., Brown, D., and Karet, F. E. (2003) The vacuolar-type H+-ATPase: a revised nomenclature. Mol. Cell 12, 801–803. [DOI] [PubMed] [Google Scholar]
- 112).Manolson, M. F., Proteau, D., Preston, R. A., Stenbit, A., Roberts, B. T., Hoyt, M. A., Preus, D., Mulholland, J., Botstein, D., and Jones, E. W. (1992) The VPHl gene encodes a 95-kDa integral membrane polypeptide required for in vitro assembly of the yeast vacuolar H+-ATPase. J. Biol. Chem. 267, 14294–14303. [PubMed] [Google Scholar]
- 113).Väänänen, H. K., Zhao, H., Mulon, M., and Halleen, J. M. (2000) Commentary; The cell biology of osteoclast function. J. Cell Sci. 113, 377–381. [DOI] [PubMed] [Google Scholar]
- 114).Sobacchi, C., Franttini, A., Orchard, P., Porras, O., and Tezcan, I. (2001) The mutational spectrum of human malignant autosomal recessive osteopetrosis. Hum. Mol. Genet. 10, 1767–1773. [DOI] [PubMed] [Google Scholar]
- 115).Toyomura, T., Murata, Y., Yamamoto, A., Oka, T., Sun-Wada, G.-H., Wada, Y., and Futai, M. (2003) From lysosomes to plasma membrane: Localization of vacuolar type H+-ATPase with the a3 isoform during osteoclast differentiation. J. Biol. Chem. 278, 22023–22030. [DOI] [PubMed] [Google Scholar]
- 116).Hurtado-Lorenzo, A., Skinner, M., Anna, J. E., Futai, M., Sun-Wada, G.-H., Bourgoin, S., Casanova, J., Wildman, A., Bechaova, S., Ausiello, D. A.et al. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degrative pathway. Nature Cell Biol. 8, 124–136. [DOI] [PubMed] [Google Scholar]
- 117).Sun-Wada, G.-H., Toyomura, T., Murata, Y., Yamanoto, A., Futai, M., and Wada, Y. Vacuolar-type proton pump with a 3 isoforms is involved in insulin secretion from pancreatic β cells. J. Cell Sci. (in press). [DOI] [PubMed] [Google Scholar]