Abstract
The molecular mechanism of any tumor marker expression may shed a light on the mechanism of the particular tumorigenesis. This idea in mind, we have been pursuing the mechanism of specific induction of the placental type glutathione transferase (GST-P) gene during hepatocarcinogenesis of the rat. Making use of advanced technologies of molecular biology including proteomic analysis, gene cloning and production of specific transgenic rats etc., we were able to identify the enhancer and the activator proteins responsible for this tumor marker expression. Negative regulatory regions and modulatory proteins were also found. The overview of this long range study and the future outlook of the problem will be discussed.
Keywords: Hepatocarcinogenesis, glutathione transferase P (GST-P), tumor marker, precancerous lesion, transgenic rat
Proteomic analysis identifies the GST-P as a prominent tumor marker for hepatocarcinogenesis of the rat
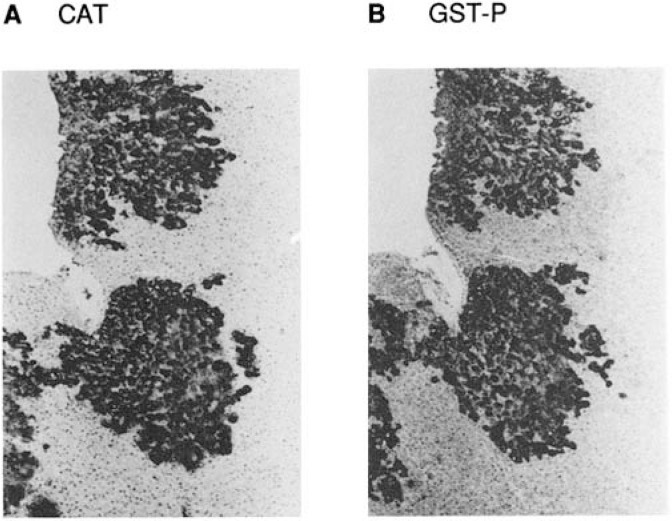
Tumor markers are known to be expressed rather specifically concomitant with the growth of the tumor. A number of tumor markers such as α-fetoprotein, carcinoembryonic antigen (CEA) and prostate specific antigen (PSA) are shown to have a diagnostic and prognostic value for different tumors. Hepatocarcinogenesis of the rat has long been employed as a model of chemical carcinogenesis due to its reproducible course of malignant transformation through the hyperplastic nodule, a precancerous stage.1) The experiment starts with a shot of 200 mg/kg diethylnitrosamine (DEN) followed by 2 weeks feeding with basal diet and the partial hepatectomy at the beginning of the third week. The experimental group is fed on basal diet containing 0.01% acetylaminofluorence (AAF) from the third week usually for 8 weeks, when numerous GST-P positive foci appear all over the liver (Fig. 1 A, B). In early 1980s, we were interested in the change of proteomics during this course using two-dimensional gel electrophoresis.2),3) Because further microanalyses with MALDI-TOF-MAS like technologies were not yet available at that time, we had to concentrate on a few prominent alterations of the polypeptide pattern among the normal liver, hyperplastic nodules and hepatocellular carcinomas.3) Among the several differences found, an array of dramatically increased spots were identified to be that of the placental type of glutathione transferase (GSTP) by immunological staining (Fig. 2 C, D). This tumor marker activation was independently found by enzymological studies and reported nearly one year ahead by K. Sato’s group.4) Our proteomic analysis clearly showed that GST-P dramatically increased at the early steps of hepatocarcinogenesis (at the stage of hyperplastic nodules) irrespective of the carcinogen used, e.g., diethylnitrosoamine or dimethylaminoazobenzene with the rare exception of nongenotoxic carcinogen such as clofibrate,5), 6) the reason of which has been elucidated later.6) The GSTP is not expressed in fetal liver and not induced in regenerating rat liver indicating that this marker is not a mere oncofetal antigen nor is related to cellular proliferation. GST-P positive foci of early hyperplastic nodules are shown in Fig. 1 B. Although there were a few more interesting spots that showed distinct changes (Fig. 2),3) we proceeded to analyze the molecular mechanisms of specific activation of GST-P gene during these processes.
Fig. 1.
- Experimental protocol for rat chemical hepatocarcinogenesis (Solt-Farber procedure). W, week; DEN, diethylnitrosamine; PH, partial hepatectomy; AAF, acetylaminofluorence; BD, basal diet.
- Appearance of the rat liver about 3 months after the Solt-Farber protocol stained with GST-P antibody. (Supplied by Dr. Tomoyuki Kitagawa at Cancer Institute)
Fig. 2.
- Control liver.
- Hyperplastic nodule.
- Hepatocellular carcinoma. Arrows show a newly appeared protein, whereas arrowheads show significantly increased proteins. Three arrowheads with approx. M. W. 26,000 coincide with the GST-P antibody as shown in D.
- Immunoblotting analysis of the 2-dimensional gel with anti-glutathione S-transferase P antibody. After 2-dimensional gel electrophoresis with 120 μg of total cellular protein from AAF-induced hepatocellular carcinoma, the polypeptides were transferred to nitrocellulose paper and incubated with rabbit anti-glutathione S-transferase P. The paper was further incubated with peroxidase-conjugated goat anti-rabbit IgG and developed with H2O2 and o-phynylenediamine. The colored spots were marked and then stained with Amido black. The marked spots coincide precisely with p26-6.9, p26-6.6, and p26-6.4, respectively. Positive spots with p26-6.9, p26-6.6 and p26-6.4 appear differentially phosphorylated form of GST-P.3)
Structural analysis of the GST-P gene demonstrates the presence of a strong enhancer element, GPE1, at about 2.5 Kb upstream of the gene
First, we cloned the GSTP gene from the rat7) and analyzed the transcriptional regulatory regions by so-called CAT analysis in dRLh84 cells with different partial deletion and substitution constructs.8)–10) The results are summarized in Fig. 3. The GST-P gene has the promoter with a typical TATA box at −27 upstream of the transcription start site. This promoter is accompanied by an ARE/TRE (antioxidant response element/TPA (phorbol 12-0-tetradecanoate 13-acetate)-response element) like sequence at −61, which appears important for the efficient promoter activity. A strong enhancer designated GPE1 was found at −2.5 Kb upstream from the promoter, which was found to be the major activator during the course of hepatocarcinogenesis (Fig. 3 A).9), 10) The structural requirements of the GPE1 were determined with CAT assay by changing characteristic nucleotide sequences with the results that GPE1 consisted of a pair of TRE (TPA-response element)-like sequences arranged in a palindromic manner with three bases in between (Fig. 3 B). The integrity of this structure was somewhat essential for the highest activity of the GPE1 as an enhancer in various cell lines such as dRLh cells derived from rat hepatoma, F9 cells derived from mouse embryonal carcinoma and HeLa cells derived from human uterus carcinoma.9),10) Although we found another enhancer element a little downstream of GPE1 and named GPE2, this was found to be minor having little activity as compared with GPE1. As shown in Fig. 3 A, we also found four negatively controlling regions in between −140 and −400 which were named GPS (GST-P silencers) 1 to 4 and found distinct proteins interacting with them.11),12) Osada, Imagawa and their collegues13)–15) identified SF-A as nuclear factor 1 (NF-1), SF-B as C/EBP and SF-C as zinc finger proteins (BTEB2, LKLF, TIEG1, MZFP and TFIIIA). These silencers may well be acting in normal liver to shut off the expression of GST-P under physiological conditions and contribute to the drastic expression of GST-P during hepatocarcinogenesis by some inhibition of their expression through some unknown mechanisms. This possibility certainly remains to be solved in future studies.16) Furthermore, Sakai and his collegues have noted a suppression of GST-P expression by glucocorticoid,17) modulation of GPE1 by Jun/Fos related products,17) and suppression of GST-P expression by peroxisome proliferators via interaction between Jun and PPAR α.6) These pheomena also remain to be further clarified.
Fig. 3.

- The upstream regulatory region of the rat GST-P gene. Enhancers GPE1, GPE2 (−2.5 kb), Silencers GPS-1, 2, 3 (−400 to −140) and the promoter with ARE/TRE (−61), GC-box (−47), and TATA-box (−27) are shown.
-
The structure of the GPE1 enhancer.The palindromic-like sequences designated the “core” are circumscribed in boxes.
Transgenic analysis unequivocally demonstrates the trans-activation of GST-P gene during hepatocarcinogenesis of the rat
If the GST-P gene activation occurs concomitantly with malignant transformation of hepatic cells, there must be a necessary molecular mechanism for this phenomenon. Two conceivable mechanisms are shown in Fig. 4. One is the cis-mechanism in which the GST-P gene and some yet-unidentified hepato-oncogene are co-activated by some local activation of chromatin. In this case, some responsible hepato-oncogene may be located near the GST-P gene and some abnormal activation of this region of chromatin may explain the malignant transformation and GST-P gene expression in the early stage of hepatocarcinogenesis (A). On the other hand, another mechanism which may be called trans-mechanism would suggest the mutation or activation of a gene which regulates both GST-P and malignant transformation by a common transactivator (B).
Fig. 4.
- A cis-mechanism, in which local activation of chromatin simultaneously activates the hepato-oncogene and the GST-P gene.
- A trans mechanism, in which the GST-P gene and the hepato-oncogene are not linked but share a trans-activator (regulator) and are coactivated by some mutation affecting its function or expression. More complex models for trans-activation are possible; only the simplest example is shown (see text).
In order to answer this question, we have made several independent lines of transgenic rats having the major upstream region (from the cap site up to −2.9 Kb) of the GST-P gene connected to the CAT expression plasmid (ECAT, Fig. 5 A).18), 19) The rats were subjected to the so-called Solt-Farber protocol of hepatocarcinogenesis (Fig. 1 A), and the activation of GPE1 in different organs was measured by CAT activity. The results shown in Fig. 5 B indicate that the CAT activity is dramatically increased in liver of rats under experimental diet but not in the control liver.18) The significant activity in the kidney of Line 4 may reflect some activation of GST-P in this organ under certain conditions. That the expression of ECAT and GST-P was occurring in the same cell was demonstrated by the immunohistochemical procedure as shown in Fig. 5 C.18)
Fig. 5A.
DNA constructs used in the experiments.19) Structure of GPE1 and normal/mutant GPE1 core are shown below. GPE1 has a core sequence consisting of the two AP-1 binding site-like sequences palindromically arranged at 3-bp spacing (arrows). Normal GPE1 core has only a short stretch of the core sequences. Each of the AP-1 binding site-like sequences (arrows) is 1 base different from the consensus AP-1 binding site sequence (TGAC/GTCA). Mutant GPE1 core has one point mutation, T to G, at the first letter of the downstream half-site (*), which is known to abolish the enhancer activity in a transfection system with cell culture.
Fig. 5B.
Liver tumor-specific expression of GST-P-CAT (ECAT) in transgenic rats.18)
Experimental. Tissues were taken from a rat that was subjected to the Solt-Farber protocol for 8 weeks. Control. Tissues were from an untreated rat (see Fig. 1). Lines 1, 4, and 5 are independent transgenic rat lines. Typical results from two or three experiments that showed similar data for each are shown.
Fig. 5C.
Immunohistochemical demonstration of the coincidental expression of ECAT and GST-P in focal lesions of altered hepatocytes.18) Serial sections of liver from a transgenic rat that was subjected to the Solt-Farber protocol for 8 weeks were immunostained with either rabbit anti-CAT antibody (A) or rabbit anti-GST-P antibody (B) with the immunoperoxidase staining method. (× 30).
Next, the structural requirement of this activation was examined by changing the transgene constructs (Fig. 5 A). As shown in Fig. 6, significant CAT activity was detected only in the livers of the rats transgenic with constructs having GPE1 (ECAT and Δ-56 CAT GPE1).19) Noteworthy was the fact that without GPE1 there was little activation anywhere and that abnormal positioning of the normal GPE1 core sequence (nCAT) without other upstream sequences decreases the enhancing activity drastically. The reason that only line 4 was positive may be due to the integration site effect. Fatal mutation of only one nucleotide in GPE1 (T to G) also abolishes the whole GPE1 activity (mCAT). This is consistent with the results obtained by the previous experiments with in vitro transfection10) analysis. Although the palindromic half-site is somewhat similar to the TRE, it is noteworthy that GPE1 is not activated by AP-1 or c-jun alone.
Fig. 6.
- CAT assay of experimental and control livers derived from ECAT or 1CAT (Fig. 5 A) transgenic rats.19) Solt-Farber +, samples obtained from whole liver containing foci at 8 weeks of Solt-Farber protocol; Solt-Farber −, samples obtained from control liver. Note that CAT was highly expressed in all the experimental whole livers of ECAT transgenic rats, while it was virtually not expressed in the whole experimental livers of 1CAT transgenic rats. The results were confirmed by two to three independent experiments. The degree of acetylation of the tumor samples was standardized by that of the control samples and the values indicated as CAT activity fold stimulation. Production of endogenous GST-P in the experimental group is also confirmed by Western blot, shown below the CAT assay, indicating that a carcinogenic process was under way normally.
- CAT assay of carcinogen-treated (Solt-Farber experiment) liver foci and control rat liver from Δ-56CAT orΔ-56CAT GPE1 (Fig. 5 A) transgenic rats. Solt-Farber +, samples obtained from whole liver obtained at 3 weeks or enucleated liver foci obtained at 16 weeks of Solt-Farber experiment; Solt-Farber −, samples obtained from control liver.
- CAT assay of carcinogen-treated (Solt-Farber experiment) liver foci and control rat liver from nCAT and mCAT (Fig. 5 A) transgenic rats. Liver samples were obtained from the control liver and enucleated Solt-Farber foci at 16 weeks.
The above set of transgenic rat experiments unequivocally demonstrated that the enhancer GPE1 is the major player for the GST-P gene activation during the hepatocarcinogenesis of the rat and is activated in trans by some activator (s). The next obligatory question is what the activators are?
Identification of the Nrf2/MafK as an activator of GPE-1
Because the GPE-1 consists of two TRE-like sequences with palindromic orientation and this sequence resembles those of ARE (antioxidant responsive element, -GTGACTTGGCA-)20), 21) and MARE (Maf recognition element, -TGCTGACTCAGCT-),22) interaction of transcription factors such as Jun, Fos, Nrf2, Maf and their family members was suspected, but only Nrf2 was found to be correlated well with GST-P expression. Nrf2 (NF-E2 related factor 2), a member of CNC (cap’n’ collar) family of transcription factors,23) dimerizes with small Maf proteins like MafK, MafG and MafF, and regulates the genes having ARE or MARE, including those of phase II detoxifying enzymes such as NQO1 and GST-Ya gene.24)–27) By using electrophoretic mobility shift assay (EMSA) and DNase I footprinting with recombinant Nrf2 and MafK proteins, Ikeda et al.28) were able to show their specific interaction to the GPE1 sequence, especially to the 3′-half of this palindrome (5′ TCAGTCA cta TGATTCA 3′) (Fig. 7 A and B). The importance of the 5′-half palindrome for full activity of GPE1 was also shown by the reporter transfection analysis (Fig. 7 C). In these experiments, F9 embryonal carcinoma cells that are devoid of c-Jun/c-Fos proteins were used for transfection to avoid possible noises with AP-1 on these sequences. Interestingly, TRE itself was found to bind with MafB strongly but not with Nrf2/MafK at all. Thus, these experiments with the GPE1 mutants corroborated the previous findings that the downstream TRE-like sequence was more important than the upstream one and the first T of this TRE-like sequence (5′ TCAGTCA cta TGATTCA-3′) was crucial to the GPE1 activity.9), 10) Not only that but also these experiments strongly suggested that at least one of the significant transactivators for the GPE1 was the Nrf2/MafK heterodimer.
Fig. 7.
- EMSA was performed with Nrf2/MafK and MafB proteins and GPE1, mGPE1, and mmGPE1. The nucleotide sequences of the probes are shown. Mutated positions are indicated by underlines.
- DNase I footprinting analysis of Nrf2/MafK with GPE1 probe. (+) and (−) indicate the probe with or without Nrf2/MafK proteins. Guanine and adenine residues of the same probe were cleaved by Maxam and Gilbert methods (M). Arrows indicate TRE-like sequences.
- Reporter transfection analysis of wild- and mutated-GPE1 in F9 cells. Indicated reporter plasmid or promoter-less luciferase plasmid (Vector, pGVB2) was cotransfected with expression plasmid of Nrf2 (black), c-Jun (gray), or without expression vector (open). Because of the abundance of MafK in F9 cells, MafK expressing plasmid was not used here, to avoid sequelching.
Nrf2 and MafKbind toGPE1in hyper-plastic nodules and hepatoma cells in vivo
To ascertain the binding of Nrf2/MafK to the GPE1 in vivo, chromatin immunoprecipitation (ChIP) assay was performed with anti-Nrf2 and anti-MafK antibodies on normal rat liver, livers with hyperplastic nodules (HN) and H4IIE and dRLh84 liver tumor cell lines. Figure 8 A clearly shows that anti-Nrf2 antibody precipitated the GPE1 sequence from the chromatin of cells from liver with hyperplastic nodules and of the two hepatomas examined, but not from the chromatin of normal liver cells.28) This is consistent with the notion that the Nrf2 protein was bound to the GPE1 only in the cells expressing GSTP but not in the cells not expressing this gene. The anti-Nrf2 antibody did not precipitate the proximal ARE/TRE-like sequence of the GST-P gene. Anti-Nrf2 antibody precipitated the NQO1 ARE from the chromatin of all the samples studied showing that the NQO1, a phase II detoxification enzyme, was expressed in the normal liver and also in hyperplastic nodule-bearing liver as well as in the hepatoma cell lines. When anti-MafK antibody was used, the results were exactly the same as seen with anti-Nrf2 antibody, demonstrating that MafK acted in the same manner as Nrf2, suggesting strongly that they bound to GPE1 as a heterodimer.
Fig. 8.
Nrf2 and MafK bind to GPE1 in hyperplastic nodules and hepatoma cells in vivo.28) ChIP analyses with anti-Nrf2 (A) and anti-MafK (B) antibodies on normal rat liver (Normal Liver), livers bearing hyperplastic nodules (HN), H4IIE cells, and dRLh84 cells. DNAs extracted from total sonicated nuclei (Input), those from Protein A-bound chromatin without antibody (−), with the indicated antibodies (Nrf2 or MafK) and with rabbit pre-immune serum (Preimmune) were analyzed. Specific enhancer and promoter regions were amplified by PCR (33 cycles) with specific primers of the ARE region of the NQO1 gene (NQO1 ARE, 5′-AGACCCAAGCGTGTACACCC-3′ and 5′-GTCCTTGGTCAGATGTGGGA-3′), GPE1 region of GST-P gene (GPE1, 5′-TGATTCTGCCATCTTTCTGC-3′ and 5′-CCAGCTTCTCTGGACAAACC-3′), and proximal ARE/TRE region of GST-P gene (ARE/TRE, 5′-CAGACTCCGGTCCAGCTGCT-3′ and 5′-CGCGAACTTACTAGCTGCTG-3′), respectively. The amplified regions of GST-P and NQO1 genes were schematically indicated in (C).
Thus, all the data from EMSA, reporter transfection and ChIP analyses point to the fact that Nrf2/MafK is the activator responsible for GST-P expression during hepatocarcinogenesis.
Negative regulation of GST-P gene by CCAAT enhancer binding protein (C/EBP) in rat liver
Some down regulation of GST-P gene by FosB protein through GPE1 enhancer17) and suppression by PPAR α through TRE (−61) site near the promoter which is activated by Jun family proteins6) have been reported, though their physiological significance is yet to be proven.
Recently, another breakthrough was opened when Ikeda et al.29) investigated the effect of the C/EBP on the transcription of GST-P gene under different conditions. First, they found that the expression of GST-P in a rat hepatoma derived cells was dramatically repressed by transfecting C/EBP α expressing vector (Fig. 9 A). This occurs with a GST-P reporter gene lacking GPS (silencer) region. However, when the main enhancer GPE1 was removed from the construct, the C/EBP α expression showed a rather stimulating effect on the GST-P gene, though the net effect was ten times lower. The results in Fig. 9 B also show that Nrf2 is required for GPE1 activity which is suppressed by C/EBP α strongly. In the F9 embryonal carcinoma cells that have neither AP-1 activity nor any GPE1 stimulating activity in them, a triplet of the C/EBP response element was also found to stimulate transcription extraordinarily by expressing C/EBP α (Fig. 9 C). Importantly, C/EBP α was found to bind also to GPE1 sequence as shown in Fig. 10.
Fig. 9.
- The indicated GST-P/luciferase or C/EBP-RE/luciferase constructs were transfected into H4IIE cells, with or without co-transfection with the C/EBP α or C/EBP β expression vectors (1 μg). C/EBP-RE/Δ50 Luc contains three tandem C/EBP binding elements from the mouse transthyretin gene joined to the Δ50 Luc vector.
- A 27-bp GPE1 core sequence was joined to the Δ50 Lucvector (GPE1/Δ50 Luc) and transfected into H4IIE cells, with or without co-transfection with the Nrf2, C/EBP α, or C/EBP β expression vectors, as indicated.
- The GPE1/Δ50 Luc construct was transfected into F9 cells, with or without co-transfection with the Nrf2 (1 μg) or C/EBP α (1 or 2 μg) expression vectors, as indicated.
Fig. 10.
- EMSA of C/EBP α and GPE1. Approximately 50 ng of the C/EBP α-MBP fusion protein was incubated with labeled probe (2 × 104 cpm). The probes were the multiple cloning site of pBluescirpt II vector (MCS), single or double C/EBP binding consensus sequences of the transthyretin gene (C/EBP-RE 1X, 2X), or the GPE1 core sequence (GPE1). In the competition analysis, increasing amounts (10X, 20X, and 50X for lanes 11–13, respectively) of unlabeled GPE1 core sequence were added to the binding mixtures.
- DNase I footprinting analysis of Nrf2/MafK and C/EBP α with the GPE1 probe. The indicated proteins (bovine serm albumin, Nrf2/MafK or C/EBP α fusion proteins) were incubated with the GPE1 probe and treated with DNase I. Guanine and adenine residues were modified and digested by the Maxam-Gilbert method (M). The vertical lines and dotted lines indicate the region protected by Nrf2/MafK and C/EBP α, respectively.
- he binding of Nrf2/MafK and C/EBP α to GPE1 is mutually exclusive. EMSAs were carried out with a fixed amount of C/EBP α (10 ng) and increasing amounts of Nrf2/MafK (0–50 ng, lanes 1–5) and a fixed amount of Nrf2/MafK (50 ng) and increasing amounts of C/EBP α (0–100 ng, lanes 6–10). GPE1 was used as the probe. The arrows indicate the Nrf2/MafK and C/EBP α-GPE1 complexes.
DNase I foot printing analysis shows that C/EBP α binds only 3′ half of the GPE1 enhancer core palindrome sequence, whereas Nrf2/MafK covers almost the whole GPE1 core sequence (Fig. 10 B). This is reasonable because the 3′-half of the GPE1 core does contain C/EBP-binding consensus like sequence.
The binding of Nrf2/MafK and C/EBP α is mutually exclusive as expected by the competitive nature of interaction (Fig. 10 C).
The fact that C/EBP α is found to GST-P gene chromatin in normal liver but is replaced by Nrf2 and MafK in the hepatoma H4IIE has also been shown clearly by the ChIP assay (not shown).29)
Conclusions and future overview
We have here looked back rather historically interesting aspects of the expression of a tumor marker GST-P, which is closely coincided with the malignant transformation of rat liver cells. Why has this enzyme to be expressed during the course, especially from the early stage, of hepatocarcinogenesis? Several groups including ours have been working to solve this problem and recently have reached the point where we can explain the molecular mechanisms by which the degree of GST-P expression is controlled in the normal liver, pre-cancerous lesions or hepatoma cells.
As described already, the structure of the GSTP gene is now fairly well analyzed including promoter, silencers and a strong enhancer, named GPE1. The expression level of GST-P in normal liver, hyperplastic nodules and hepatocellular carcinoma appears mostly, if not entirely, regulated by this enhancer.
The major reason for the extraordinary expression of the GST-P gene during hepatocarcinogenesis of the rat may be explained by the synergistic effects of positive and negative regulators on GPE1. Increase in activator complex Nrf2/MafK accompanied by decrease in the strong repressor, C/EBP α, may probably cause a flood of downstream product, GST-P. How these phenomena occur and are related to tumorigenity of the liver cells remain to be elucidated, although it is easy to understand that the down regulation of C/EBP α would change the cell environment to the promotion of cell cycle. The molecular mechanisms of irreversible downregulation of C/EBP α and upregulation of Nrf2 would be the next target in this direction.
In addition, the search for new genes differentially expressed during hepatocarcinogenesis by newly introduced microarray system would open a new field in cancer research.
References
- 1).Solt, D., and Farber, E. (1976) A new principle for the analysis of chemical carcinogenesis. Nature 263, 701–703. [Google Scholar]
- 2).O’Farrell, P. (1975) High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250, 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 3).Sugioka, Y., Fujii-Kuriyama, Y., Kitagawa, T., and Muramatsu, M. (1985) Changes in polypep-tide pattern of rat liver cells during chemical hepatocarcinogenesis. Cancer Res. 45, 365–378. [PubMed] [Google Scholar]
- 4).Sato, K., Kitahara, A., Satoh, K., Ishihara, T., Tatematsu, M., and Ito, N. (1984) The placental form of glutathione S-transferase as a new marker protein for preneoplasia in rat chemical hepatocarcinogenesis. Gann 75, 199–202. [PubMed] [Google Scholar]
- 5).Rao, M. S., Tatematsu, M., Subbarao, V., Ito, N., and Reddy, J. K. (1986) Analysis of peroxisome proliferators-induced preneoplastic and neoplastic lesions of rat liver for placental form of glutathione S-transferase and gamma-glutamyltrans-peptidase. Cancer Res. 46, 5287–5290. [PubMed] [Google Scholar]
- 6).Sakai, M., Matsushima-Hibiya, Y., Nishizawa, M., and Nishi, S. (1995) Suppression of rat glutathione transferase P expression by peroxisome proliferators: Interaction between Jun and peroxisome proliferators-activated receptor α.Cancer Res. 55, 5370–5376. [PubMed] [Google Scholar]
- 7).Okuda, A., Sakai, M., and Muramatsu, M. (1987) The structure of the rat glutathione S-transferase P gene and related pseudogenes. J. Biol. Chem. 262, 3858–3863. [PubMed] [Google Scholar]
- 8).Sakai, M., Okuda, A., and Muramatsu, M. (1988) Multiple regulatory elements and phorbol 12-O-tetradecanoate 13-acetate responsiveness of the rat placental glutathione transferase gene. Proc. Natl. Acad. Sci. USA 85, 9456–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Okuda, A., Imagawa, M., Maeda, Y., Sakai, M., and Muramatsu, M. (1989) Structural and functional analysis of an enhancer GPE1 having a phorbol 12-O-tetradecanoate 13-acetate responsive element-like sequence found in the rat glutathione transferase P gene. J. Biol. Chem. 264, 16919–16926. [PubMed] [Google Scholar]
- 10).Okuda, A., Imagawa, M., Sakai, M., and Muramatsu, M. (1990) Functional cooperativity between two TPA-responsive elements in undifferentiated F9 embryonic stem cells. EMBO J. 9, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Imagawa, M., Osada, S., Okuda, A., and Muramatsu, M. (1991) Silencer binding proteins function on multiple cis-elements in the glutathione transferase P gene. Nucleic Acids Res. 19, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Imagawa, M., Osada, S., Koyama, Y., Suzuki, T., Hirom, P. C., Diccianni, M. B., Morimura, S., and Muramatsu, M. (1991) SF-B that binds to a negative element in glutathione transferase P gene is similar or identical to trans-activator LAP/IL6-DBP. Biochem. Biophys. Res. Commun. 179, 293–300. [DOI] [PubMed] [Google Scholar]
- 13).Osada, S., Takano, K., Nishihara, T., Suzuki, T., Muramatsu, M., and Imagawa, M. (1995) CCAAT/enhancer-binding proteins alpha and beta interact with the silencer element in the promoter of glutathione S-transferase P gene during hepatocarcinogenesis. J. Biol. Chem. 270, 31288–31293. [DOI] [PubMed] [Google Scholar]
- 14).Osada, S., Daimon, S., Ikeda, T., Nishihara, T., Yano, K., Yamasaki, M., and Imagawa, M. (1997) Nuclear factor 1 family proteins bind to the silencer element in rat glutathione transferase P gene. J. Biochemistry 121, 355–363. [DOI] [PubMed] [Google Scholar]
- 15).Tanabe, A., Kurita, M., Oshima, K., Osada, S., Nishihara, T., and Imagawa, M. (2002) Functional analysis of zinc finger proteins that bind to the silencer element in the glutathione transferase P gene. Biol. Pharm. Bull. 25, 970–974. [DOI] [PubMed] [Google Scholar]
- 16).Sakai, M., and Muramatsu, M. (2005) Regulation of GST-P gene expression during hepatocarcino-genesis. Methods Enzymol. 401, 42–61. [DOI] [PubMed] [Google Scholar]
- 17).Oridate, N., Nishi, S., Inuyama, Y., and Sakai, M. (1994) Jun and Fos related gene products bind to and modulate the GPE-I, a strong enhancer element of the rat glutathione transferase P gene. Biochem. Biophys. Acta 1219, 499–504. [DOI] [PubMed] [Google Scholar]
- 18).Morimura, S., Suzuki, T., Hochi, S., Yuki, A., Nomura, K., Kitagawa, T., Nagatsu, I., Imagawa, M., and Muramatsu, M. (1993) Trans-activation of glutathione transferase P gene during chemical hepatocarcinogenesis of the rat. Proc. Natl. Acad. Sci. USA 90, 2065–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Suzuki, T., Imagawa, M., Hirabayashi, M., Yuki, A., Hisatake, K., Nomura, K., Kitagawa, T., and Muramatsu, M. (1995) Identification of an enhancer responsible for tumor marker gene expression by means of transgenic rat. Cancer Res. 55, 2651–2655. [PubMed] [Google Scholar]
- 20).Favreau, L. V., and Pickett, C. B. (1991) Transcriptional regulation of the rat NAD(P)H: quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J. Biol. Chem. 266, 4556–4561. [PubMed] [Google Scholar]
- 21).Jaiswal, A. K. (1994) Antioxidant response element. Biochem. Pharmacol. 48, 439–444. [DOI] [PubMed] [Google Scholar]
- 22).Kataoka, K., Noda, M., and Nishizawa, M. (1994) Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 14, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Moi, P., Chan, K., Asunis, I., Cao, A., and Kan, Y. W. (1994) Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus controlregion. Proc. Natl. Acad.Sci.USA 91, 9926–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Andrews, N. C., Kotkow, K. J., Ney, P. A., Erdjument-Bromage, H., Tempst, P., and Orkin, S. H. (1993) The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the vmaf oncogene. Proc. Natl. Acad. Sci. USA 90, 11488–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Igarashi, K., Kataoka, K., Itoh, K., Hayashi, N., Nishizawa, M., and Yamamoto, M. (1994) Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367, 568–572. [DOI] [PubMed] [Google Scholar]
- 26).Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I.et al. (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322. [DOI] [PubMed] [Google Scholar]
- 27).Jaiswal, A. K. (2000) Regulation of genes encoding NAD (P) H: quinone oxidoreductases. Free Radic. Biol. Med. 29, 254–262. [DOI] [PubMed] [Google Scholar]
- 28).Ikeda, H., Nishi, S., and Sakai, M. (2004) Transcription factor Nrf2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. Biochem. J. 380, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Ikeda, H., Omoteyama, K., Yoshida, K., Nishi, S., and Sakai, M. (2006) CCAAT enhancer-binding protein α suppresses the rat placental glutathione S-transferase gene in normal liver. J. Biol. Chem. 281, 6734–6741. [DOI] [PubMed] [Google Scholar]