ABSTRACT
The herpesviral nuclear egress complex (NEC), consisting of pUL31 and pUL34 homologs, mediates efficient translocation of newly synthesized capsids from the nucleus to the cytosol. The tail-anchored membrane protein pUL34 is autonomously targeted to the nuclear envelope, while pUL31 is recruited to the inner nuclear membrane (INM) by interaction with pUL34. A nuclear localization signal (NLS) in several pUL31 homologs suggests importin-mediated translocation of the protein. Here we demonstrate that deletion or mutation of the NLS in pseudorabies virus (PrV) pUL31 resulted in exclusively cytosolic localization, indicating active nuclear export. Deletion or mutation of a predicted nuclear export signal (NES) in mutant constructs lacking a functional NLS resulted in diffuse nuclear and cytosolic localization, indicating that both signals are functional. pUL31 molecules lacking the complete NLS or NES were not recruited to the INM by pUL34, while site-specifically mutated proteins formed the NEC and partially complemented the defect of the UL31 deletion mutant. Our data demonstrate that the N terminus of pUL31, encompassing the NLS, is required for efficient nuclear targeting but not for pUL34 interaction, while the C terminus, containing the NES but not necessarily the NES itself, is required for complex formation and efficient budding of viral capsids at the INM. Moreover, pUL31-ΔNLS displayed a dominant negative effect on wild-type PrV replication, probably by diverting pUL34 to cytoplasmic membranes.
IMPORTANCE The molecular details of nuclear egress of herpesvirus capsids are still enigmatic. Although the key players, homologs of herpes simplex virus pUL34 and pUL31, which interact and form the heterodimeric nuclear egress complex, are well known, the molecular basis of this interaction and the successive budding, vesicle formation, and scission from the INM, as well as capsid release into the cytoplasm, remain largely obscure. Here we show that classical cellular targeting signals for nuclear import and export are important for proper localization and function of the NEC, thus regulating herpesvirus nuclear egress.
INTRODUCTION
Herpesvirus assembly and maturation proceed in two different cellular compartments. After capsid formation and genome packaging in the nucleus, nucleocapsids are translocated to the cytoplasm, where final maturation and egress from the host cell occur. To exit the nucleus, nucleocapsids bud at the inner nuclear membrane (INM), resulting, after scission of the vesicles, in primary enveloped virions in the perinuclear space. The primary envelope subsequently fuses with the outer nuclear membrane (ONM), thereby releasing the capsids into the cytoplasm (reviewed in references 1 to 4). The nuclear egress complex (NEC), which is composed of proteins homologous to herpes simplex virus 1 (HSV-1) pUL31 and pUL34, is the key player in this process and is conserved among all members of the family Herpesviridae studied to date. The NEC recruits viral and cellular kinases to phosphorylate, and thereby soften, the nuclear lamina to allow access of nucleocapsids to the budding sites at the INM. It also mediates membrane bending and vesicle scission. Absence of either pUL31 or pUL34 significantly impairs nuclear egress, while coexpression of both proteins is sufficient for membrane budding and scission not only in eukaryotic cells (reviewed in references 1 to 4) but also in model membrane systems, e.g., giant unilamellar vesicles (5), demonstrating that no other viral or cellular proteins are required for these steps.
While the pUL34 homologs are tail-anchored membrane proteins residing in the nuclear membrane, pUL31 homologs constitute phosphoproteins localizing to the nucleus (6–11). In the absence of pUL34, pUL31 is found diffusely distributed in the nucleus but is recruited to the INM by its complex partner pUL34 (6, 11–17). However, the molecular details of this interaction are still not fully understood and no crystal structures of the NEC or of the single proteins are available yet. Deletion and substitution analyses for different pUL34 homologs showed that the N-terminal part, which is well conserved between the different homologs, is necessary for pUL31 binding while the C terminus comprising the transmembrane domain is required for correct positioning but can be functionally substituted by heterologous sequences as long as a membrane anchor is provided (12, 18–23).
Comparison of the various pUL31 homologs from different herpesvirus subfamilies resulted in the identification of four conserved regions (CR1 to CR4) (Fig. 1) (11). The N-terminal part, which is variable in length in the different homologs, shows low sequence conservation but comprises a classical mono- or bipartite nuclear localization signal (NLS) in many of the pUL31 homologs (6, 9, 11, 24). The NLS function was experimentally mapped to amino acids (aa) 18 to 27 in human cytomegalovirus (HCMV) pUL53 (24) and aa 1 to 106 in murine cytomegalovirus (MCMV) M53 (11). Since all pUL31 homologs are present in the nuclei of transfected or infected cells, it is reasonable to speculate that the protein is actively imported by the classical importin-mediated cellular pathway.
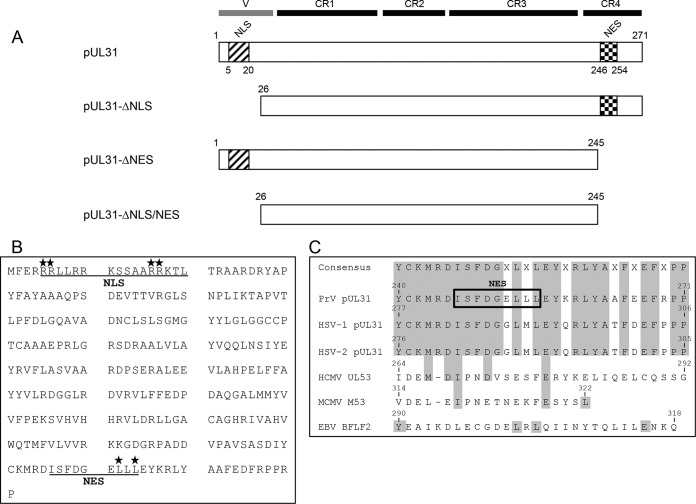
FIG 1.
pUL31 mutant constructs. (A) Schematic representation of PrV pUL31 with a variable (V) region and CR1 to CR4 (11). The pUL31 NLS and NES deletion mutant constructs used in this study are also shown. (B) Amino acid sequence of PrV pUL31. Underlined are the nuclear trafficking signals (NLS and NES) predicted by NLS (37) and NES (40) prediction software. Amino acids that were mutated to alanine are indicated by asterisks. (C) Alignment of CR4 of selected pUL31 homologs in three alphaherpesviruses (PrV, HSV-1, HSV-2), two betaherpesviruses (HCMV, MCMV), and a gammaherpesvirus (EBV). The predicted NES in PrV pUL31 is boxed, and conserved amino acids are highlighted.
The pUL34 interaction domain of pUL31 was localized to CR1 in HSV-1, pseudorabies virus (PrV), HCMV, MCMV, and Epstein-Barr virus (EBV) homologs by protein complementation assays (25). An amphipathic helix within this region predicted for HCMV pUL53 is supposed to be responsible for complex formation by a combination of hydrophobic and charge-charge interactions (26). Nevertheless, sequences in CR2 and CR3 in HSV-1 pUL31 and MCMV M53 also influence the interaction with the respective pUL34 homologs (27, 28). While CR3 of HSV-1 pUL31 was supposed to be involved in generating the curvature of the INM (27), mutagenesis within CR4 of MCMV M53 yielded dominant negative mutant forms with defects in nuclear capsid release (29). Yeast two-hybrid studies with PrV indicated that the N terminus (up to aa 41) comprising the predicted NLS is dispensable for pUL34 interaction in this artificial system (6). In addition to its well-known function in nuclear egress, pUL31 might also be involved in capsid assembly and/or the DNA cleavage and packaging process (28, 30), although this could not be corroborated for PrV (6).
In this study, we performed mutational analyses of the predicted NLS and nuclear export signal (NES) in PrV pUL31 to further elucidate their role in NEC formation and function during nuclear egress.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney (RK13) cells were cultivated in Dulbecco's modified Eagle's minimum essential medium supplemented with 10% fetal calf serum. Wild-type strain PrV Kaplan (PrV-Ka) (31) was grown on RK13 cells, while PrV-ΔUL31 and PrV-ΔUL31/US3 were propagated on RK13-UL31 cells (6). Mutant PrV-ΔUL31/US3 was generated after the cotransfection of PrV-ΔUL31 genomic DNA (6) and plasmid pΔUS3gfp (32) into RK13-UL31 cells. Green fluorescing plaques were purified to homogeneity and tested for correct deletion of UL31- and US3-specific sequences (data not shown).
Preparation of pUL31-NLS and pUL31-NES mutant constructs.
Truncated versions of pUL31 were generated by PCR with Pfx DNA polymerase (Invitrogen), the primers listed in Table 1, and pcDNA-UL31 (6) as the template. For cloning, PCR products were digested with EcoRI and ligated with EcoRI-cleaved mammalian expression vector pcDNA3 (Invitrogen). Point mutations were introduced by site-directed mutagenesis with the Agilent Technologies QuikChange II XL site-directed mutagenesis kit as described recently (20) with pcDNA-UL31 as the template. Correct amplification, cloning, and mutagenesis of all constructs were verified by sequencing.
TABLE 1.
Primers used for mutagenesis of PrV pUL31
| Name | Sequence (5′ to 3′)a | Location in PrV-Ka (nt)b |
|---|---|---|
| UL31for | CTGAATTCACACGCTCGGCAGCTATGTTTG | 29581–29573 |
| UL31rev | CTGAATTCTTCGCGGCGCTCACGG | 29771–29766 |
| UL31-ΔNLSfor | CACAGAATTCATGGATCGCTACGCGCCCTA | 29506–29490 |
| UL31-ΔNESrev | CTGAATTCCGCTCAGTCCCGCATCTTGCAATAAATG | 28868–28847 |
| UL31-NLSPM1-1 | GCTATGTTTGAGCGAGCGGCGCTCCTGCGGCGCAAG | 29584–29549 |
| UL31-NLSPM1-2 | CTTGCGCCGCAGGAGCGCCGCTCGCTCAAACATAGC | 29549–29584 |
| UL31-NLSPM2-1 | AAGTCGTCGGCCGCGGCGGCCAAGACGCTGACGCGC | 29551–29516 |
| UL31-NLSPM2-2 | GCGCGTCAGCGTCTTGGCCGCCGCGGCCGACGACTT | 29516–29551 |
| UL31-NESPM1-1 | GCTTCGACGGGGAGGCGCTGGCGGAGTACAAAAGATTG | 28842–28805 |
| UL31-NESPM2-2 | CAATCTTTTGTACTCCGCCAGCGCCTCCCCGTCGAAGC | 28805–28842 |
Restriction sites introduced for convenient cloning are underlined, start and stop codons are shown in bold, and mismatches for site-directed mutagenesis in italics.
Positions correspond to the sequence with GenBank accession number JQ809328. nt, nucleotides.
Generation of stably expressing cell lines.
For complementation studies, cell lines stably expressing the pUL31-NLS and pUL31-NES mutant constructs were generated. To this end, RK13 cells were transfected with the mutated pcDNA-UL31 plasmids by calcium phosphate precipitation (19, 33). Cells were selected in medium containing 0.5 mg/ml G418, and correct protein expression was verified by indirect immunofluorescence and Western blot analysis.
Western blot analysis.
For Western blot analysis, transgenic cell lines were harvested by scraping the cells into the medium. Cells were pelleted by centrifugation, washed twice with PBS, resuspended in sodium dodecyl sulfate (SDS) sample buffer, and disrupted by supersonic lysis. Cell lysates were separated by SDS–10% polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. After blocking with 3% skim milk, the membrane was incubated with a monospecific polyclonal rabbit serum directed against pUL31 (6). Bound antibody was visualized by enhanced chemiluminescence (Super Signal West Pico; Thermo Scientific) and subsequently recorded in an image analyzer (Bio-Rad).
Laser scanning confocal microscopy.
To analyze the localization of the mutated pUL31, RK13 cells seeded onto coverslips were transfected with the corresponding expression plasmids by calcium phosphate precipitation either singly or cotransfected with pcDNA-UL34 (34). For analysis of protein localization during infection, transgenic cell lines were infected with PrV-Ka or PrV-ΔUL31 under plaque assay conditions. Cells were fixed with 3% paraformaldehyde 2 days after transfection or infection, washed with 50 mM NH4Cl in PBS, and permeabilized with 0.5% Triton X-100 in PBS. Fixed cells were incubated with rabbit anti-pUL31 serum (6) and mouse anti-pUL34 polyclonal serum (35). Bound antibodies were detected by Alexa Fluor 488-conjugated goat anti-rabbit and Alexa Fluor 555-conjugated goat anti-mouse serum (Invitrogen). Fluorescent signals were recorded with a laser scanning confocal microscope (Leica SP5).
Complementation assays.
To investigate the functional complementation of pUL31 constructs during infection, cell lines stably expressing pUL31 or the corresponding mutant proteins were infected on ice with PrV-Ka or PrV-ΔUL31 at a multiplicity of infection (MOI) of 3. One hour after infection, the inoculum was replaced with prewarmed medium and samples were incubated at 37°C for an additional hour. Subsequently, extracellular virus was inactivated by low-pH treatment (36) and cells were further kept at 37°C. At 24 h after infection, cells and supernatants were harvested and progeny virus was titrated on RK13-UL31. Mean values of three independent experiments and the corresponding standard deviations are shown. The statistical significance of differences between PrV-Ka and PrV-ΔUL31 infections was determined by Student's t test.
Ultrastructural analysis.
For ultrastructural analysis, transgenic cell lines were infected at an MOI of 1 with PrV-Ka, PrV-ΔUL31, or PrV-ΔUL31/US3. Samples were fixed at 14 h p.i. and processed for transmission electron microscopy as described previously (34).
RESULTS
Generation of pUL31-NLS and pUL31-NES mutant constructs.
Computer analysis predicted a bipartite NLS between aa 5 and 20 in PrV pUL31 (6, 24, 37) (Fig. 1). To investigate its role in pUL31 localization and function, we either deleted the first N-terminal 25 aa (Fig. 1A) or changed four arginine residues within this motif to alanines by site-directed mutagenesis (Fig. 1B). The localization of the resulting protein product was determined by confocal microscopy after the transfection of RK13 cells with the corresponding expression plasmids (Fig. 2). In contrast to wild-type pUL31, which is located in the nucleus, N-terminally truncated, as well as point mutated, pUL31 was found exclusively in the cytoplasm, although its predicted molecular mass (<28 kDa) should actually permit passive diffusion through the nuclear pore (38, 39). This implied not only that the predicted NLS is functional but also that both NLS-deficient pUL31 constructs are actively excluded from the nucleus. To test for corresponding export motifs, the amino acid sequence of PrV pUL31 was analyzed by NES prediction software (NetNES 1.1 server, http://www.cbs.dtu.dk/services/NetNES/) (40), which disclosed a leucine-rich NES between aa 246 and 254 within CR4 (Fig. 1B). Although the sequences within CR4 are well conserved in HSV-1 and -2 pUL31, and also partly in murine and HCMV M53 and pUL53 (Fig. 1C), the software failed to detect a NES for these homologs.
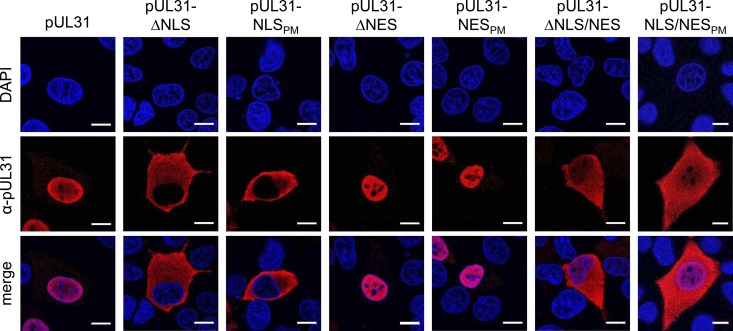
FIG 2.
Localization of pUL31-NLS and pUL31-NES mutant constructs. RK13 cells were transfected with expression plasmids for the different pUL31 mutant constructs indicated and then analyzed by confocal laser scanning microscopy 2 days posttransfection by incubation with monospecific rabbit anti-pUL31 serum and Alexa Fluor 555-conjugated secondary antibodies (red). 4′,6-Diamidino-2-phenylindole (DAPI; blue) was used for better visualization of the nucleus. Bars, 10 μm.
To test for the function of the predicted NES in PrV pUL31, the C terminus, including the predicted NES, was deleted (aa 246 to the end; Fig. 1A) or codons for two leucine residues within this motif were mutated to alanines, resulting in pUL31-ΔNES and pUL31-NESPM, respectively (Fig. 1B). The corresponding pUL31 expression vectors were transfected into RK13 cells and analyzed by confocal microscopy (Fig. 2). Neither the NES deletion nor the leucine-to-alanine change in pUL31 showed an impact on the nuclear localization of the respective proteins, which were, like wild-type pUL31, detectable predominantly in the nucleus. However, when the NLS and NES mutations were combined, resulting in pUL31-ΔNLS/NES and pUL31-NLS/NESPM (Fig. 1), the proteins were found diffusely distributed throughout the cell (Fig. 2), indicating that both the NLS and the NES are functional.
Interaction of pUL31-NLS and pUL31-NES mutant constructs with pUL34.
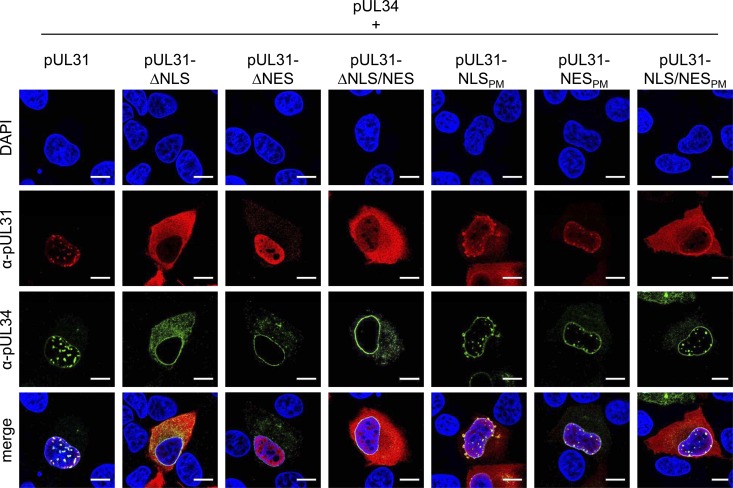
To analyze for interaction and vesicle formation, the pUL31-NLS and pUL31-NES expression plasmids were cotransfected with wild-type pcDNA-UL34 and analyzed by confocal microscopy (Fig. 3). Coexpression of wild-type pUL31 and pUL34 leads to recruitment of nucleoplasmic pUL31 to the nuclear rim and the development of pUL31- or pUL34-containing speckles indicative of the formation of vesicles within the perinuclear cleft (35). pUL31-ΔNLS failed to form speckles with pUL34, which was not surprising since the protein was excluded from the nucleus (Fig. 2). Speckle formation was also not observed after cotransfection of pcDNA-UL34 with pcDNA-UL31-ΔNES and pcDNA-UL31-ΔNLS/NES, although the corresponding proteins are in the nucleus. Interestingly, a larger proportion of cytoplasmic pUL34 was observed after coexpression with pUL31-ΔNLS than after coexpression with wild-type pUL31 or the other mutant constructs.
FIG 3.
Colocalization of transiently expressed pUL31 mutant constructs with wild-type pUL34. To investigate the interaction of pUL31 constructs with pUL34, RK13 cells were cotransfected with the corresponding expression plasmids and colocalization was analyzed by confocal laser scanning microscopy of indirect immunofluorescence reactions with DAPI for labeling of chromatin and monospecific rabbit anti-pUL31 (red) and murine anti-pUL34 (green) sera. Bars, 10 μm.
Speckle formation could be observed after cotransfection of pcDNA-UL34 with pcDNA-UL31-NLSPM. This was surprising since the corresponding protein was, like pUL31-ΔNLS, detected exclusively in the cytoplasm after single expression (Fig. 2) but found partially in the nucleus in the presence of pUL34 (Fig. 3). However, the speckles exhibited a different morphology, irregular in size and bulging toward the cytoplasmic side. After cotransfection of pcDNA-UL34 with pcDNA-UL31-NESPM or pcDNA-UL31-NLS/NESPM, speckles indistinguishable from those of wild-type pUL31- and pUL34-transfected cells were present, indicating that the point mutations in pUL31-NLS or pUL31-NES do not abrogate pUL31-pUL34 interaction and vesicle formation.
Impact of mutations in pUL31-NLS and pUL31-NES on PrV replication.
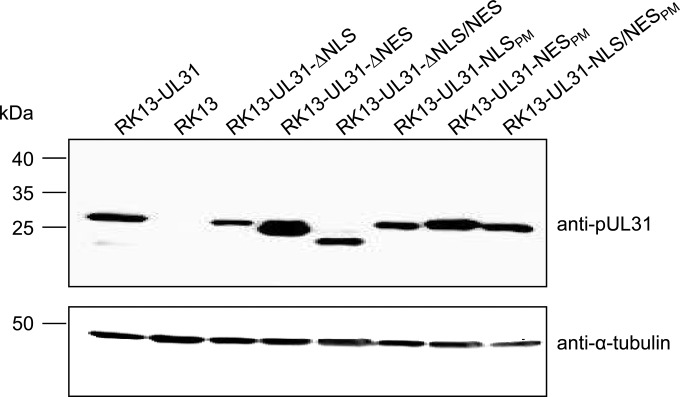
To analyze the importance of the NLS and NES regions in PrV pUL31 for virus replication, RK13 cell lines stably expressing the different truncated or mutated proteins were generated. Western blot analysis of cell lysates demonstrated protein expression at sizes expected from the mutations introduced. Expression levels were largely comparable to those of RK13-UL31, with the exception of RK13-UL31-ΔNES, which appeared to express a larger amount of pUL31 (Fig. 4).
FIG 4.
Western blot analysis of transgenic RK13 cells expressing pUL31 NLS and NES mutant proteins. RK13 cells stably expressing native pUL31 or pUL31 mutant proteins were lysed, and proteins were separated by SDS–10% polyacrylamide gel electrophoresis. After transfer onto a nitrocellulose membrane, the blot was incubated with polyclonal monospecific rabbit anti-pUL31 serum. A control blot was incubated with a murine anti-α-tubulin monoclonal antibody.
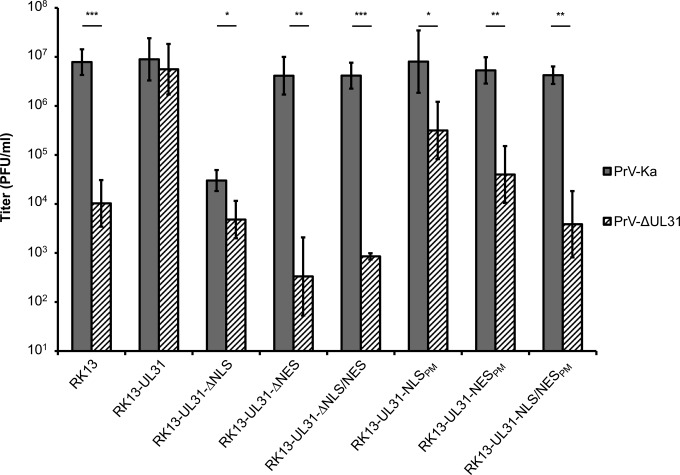
To test for complementation, the stably expressing cell lines were infected with PrV-Ka or PrV-ΔUL31 at an MOI of 3. Viral progeny titers were determined at 24 h p.i. on RK13-UL31 cells (Fig. 5). All of the truncated mutant constructs (pUL31-ΔNLS, pUL31-ΔNES, and pUL31-ΔNLS/NES) failed to complement the defect of PrV-ΔUL31, which was expected, since no pUL34 interaction was detectable (Fig. 3). Furthermore, pUL31-ΔNLS, which is found exclusively in the cytoplasm, exerted a significant dominant negative effect on PrV-Ka replication and virus titers were comparable to those of PrV-ΔUL31 on RK13 cells, indicating that pUL31-ΔNLS impairs the formation of a functional NEC and, consequently, nuclear egress.
FIG 5.
Functional complementation of PrV-ΔUL31 by pUL31-NLS and pUL31-NES mutant proteins. RK13 cells expressing pUL31 mutant proteins were infected with PrV-Ka or PrV-ΔUL31 at an MOI of 3, and viral progeny was harvested 24 h after infection. Titers were determined by plaque assays on RK13-UL31 cells. Shown are the means of three independent experiments with standard deviations. Statistically significant differences between PrV-Ka and PrV-ΔUL31 infections are indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Infectious progeny with only approximately 10-fold lower titers than those from RK13-UL31 cells could be found after infection of RK13-UL31-NLSPM with PrV-ΔUL31. Approximately 100-fold reduced titers were detectable after infection of RK13-UL31-NESPM cells, while titers from cells expressing pUL31-NLS/NESPM were comparable to those derived after infection of nontransgenic RK13 cells, pointing to an additive effect of the mutations in NLS and NES (Fig. 5).
Ultrastructural analysis.
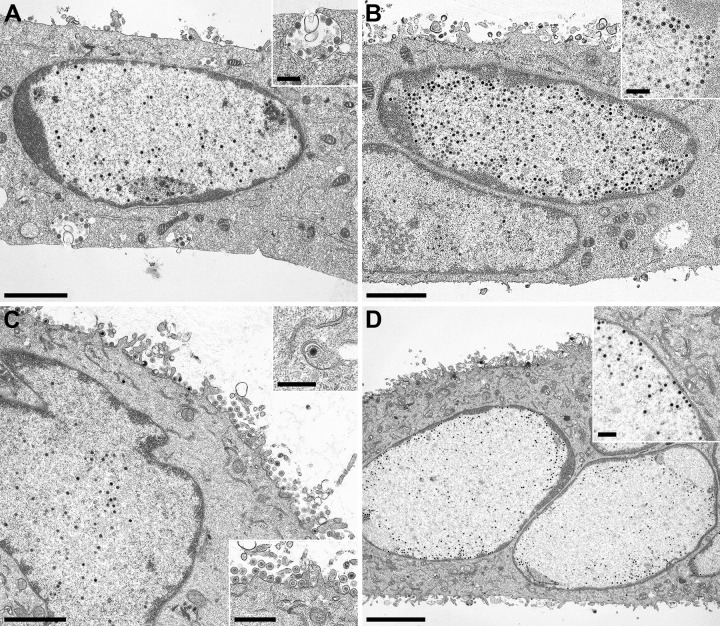
For in-depth analysis of the effect of the pUL31-NLS and pUL31-NES mutations, transgenic cell lines were infected with PrV-ΔUL31 or PrV-ΔUL31/US3 at an MOI of 1, fixed 14 h later, and processed for high-resolution transmission electron microscopy. In addition, RK13-UL31-ΔNLS cells were infected with PrV-Ka (Fig. 6B). On RK13 cells expressing pUL31-ΔNES (Fig. 6A), pUL31-ΔNLS, and pUL31-ΔNLS/NES (not shown), PrV-ΔUL31 showed the same phenotype as on nontransgenic RK13 cells (6). Capsids remained diffusely distributed in the nucleus, and neither budding at the INM nor primary enveloped virions in the perinuclear cleft nor final virus maturation stages in the cytoplasm were observed, despite the formation of capsidless L particles (Fig. 6A), indicating that nuclear egress does not ensue. The same phenotype was found after infection of RK13-UL31-ΔNLS with PrV-Ka (Fig. 6B), indicating that the cytoplasmic location of pUL31-ΔNLS precludes the formation of a functional NEC at the INM. All stages of herpesvirus morphogenesis, including primary envelopment at the INM, primary enveloped virions in the perinuclear space, secondary envelopment in the cytoplasm, and virions at the plasma membrane, were detectable after infection of RK13-UL31-NLSPM (Fig. 6C), demonstrating that disruption of the nuclear import signal by site-specific mutagenesis does not block nuclear egress.
FIG 6.
Ultrastructural analysis of transgenic cell lines. RK13-UL31-ΔNES (A), RK13-UL31-NLSPM (C), or RK13-UL31-NESPM (D) cells were infected with PrV-ΔUL31 and RK13-UL31-ΔNLS cells were infected with PrV-Ka (B) at an MOI of 1 for 14 h and processed for electron microscopic analysis. Bars, 2 μm in panels A to C; 4 μm in panel D, 500 nm in the insets in panels A, B, and D and the upper inset in panel C, and 1 μm in the lower inset in panel C.
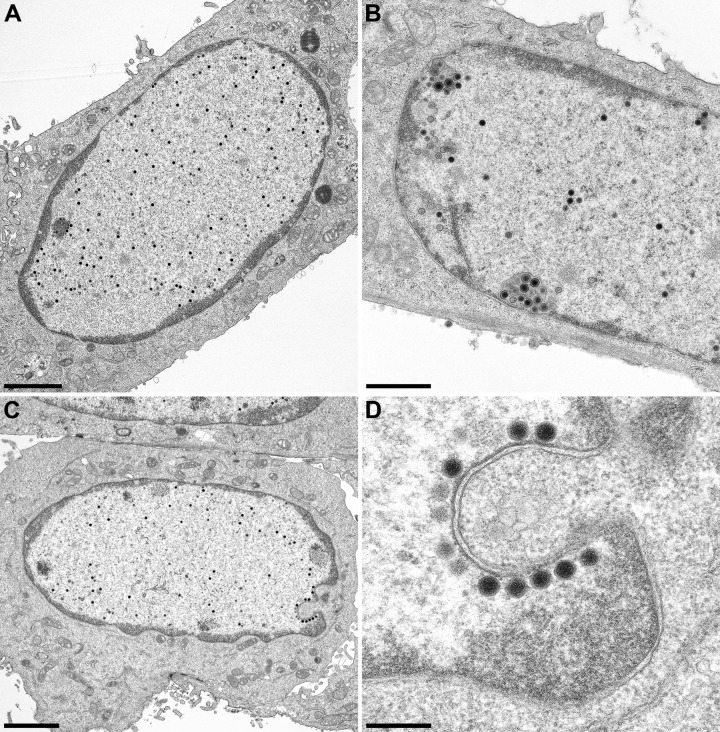
In contrast, nuclear egress was observed only rarely in RK13-UL31-NESPM cells infected with PrV-ΔUL31. However, a striking number of nucleocapsids were found closely attached to the INM, with only a few primary enveloped particles in the perinuclear cleft (Fig. 6D), indicating that the sequence containing the NES is dispensable for docking of nucleocapsids at the INM but required for primary envelopment. To investigate this in more detail, RK13-UL31-NESPM cells were infected with PrV-ΔUL31/US3 (Fig. 7). Deletion of US3 results in the accumulation of primary virions in the perinuclear cleft, which should enhance the observation of any effect of the mutations on these virus maturation intermediates (32). An accumulation of primary enveloped virions was evident after the infection of RK13-UL31 with PrV-ΔUL31/US3 (Fig. 7B). In PrV-ΔUL31/US3-infected RK13-UL31-NESPM cells, numerous nucleocapsids were detected closely attached to the INM, which was especially evident at invaginations of the nuclear membrane (Fig. 7C and D). These data confirm that mutation of the two leucine residues within the NES does not inhibit the docking of nucleocapsids at the INM but interferes with efficient budding.
FIG 7.
Ultrastructural analysis after infection with PrV-ΔUL31/US3. RK13 (A), RK13-UL31 (B), and RK13-UL31-NESPM (C and D) cells were infected at an MOI of 1, and samples for ultrastructural analysis were prepared at 14 h p.i. Bars, 2 μm in panels A and C, 1 μm in panel B, and 300 nm in panel D.
Localization of pUL34 and mutated pUL31 during infection.
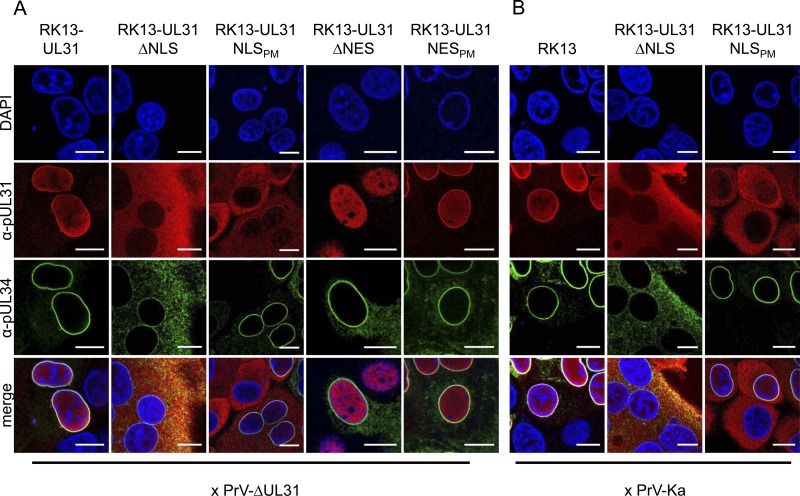
To explain the discrepancy between the truncated and site-specifically mutated pUL31 proteins, localization of the NEC components was analyzed during infection. To this end, cell lines stably expressing the different pUL31 mutant constructs were infected with PrV-ΔUL31 under plaque assay conditions and analyzed by confocal microscopy 2 days postinfection (p.i.) (Fig. 8A). During infection of wild-type pUL31-expressing cells, both proteins colocalize at the nuclear rim, as described earlier (6), with pUL34 only faintly detectable in cytoplasmic structures. In contrast, after infection of cells expressing the truncated molecules pUL31-ΔNLS and pUL31-ΔNES, pUL34 was detectable with a punctate pattern in the cytoplasm in addition to its normal nuclear-rim staining, while the complex partner pUL31 was not recruited to the nuclear envelope. Nuclear-rim staining was evident for pUL34, pUL31-NLSPM, and pUL31-NESPM, indicating that these proteins interact, thus confirming the data of the transient-expression studies (Fig. 3). The most drastic difference in pUL34 distribution is evident after infection of RK13-UL31-ΔNLS and pUL31-NLSPM-expressing cells, where pUL34 seems to be bound to the nuclear membrane in the presence of pUL31-NLSPM but is mislocalized in cells expressing pUL31-ΔNLS, indicating that the N-terminal 25 aa of pUL31 are important for correct and efficient nuclear-envelope targeting of pUL34.
FIG 8.
Localization of mutated pUL31 and wild-type pUL34 during PrV infection. To investigate the interaction of mutated pUL31 with pUL34 during virus infection, the corresponding cell lines were incubated with PrV-ΔUL31 (A) or PrV-Ka (B) under plaque assay conditions and fixed 2 days p.i. Localization of pUL31 and pUL34 was analyzed by confocal laser scanning microscopy after an indirect immunofluorescence assay with monospecific rabbit anti-pUL31 (red) and murine anti-pUL34 (green) sera. DAPI (blue) was included to visualize nuclei. Bars, 10 μm.
To test whether this mislocalization of pUL34 also occurs during wild-type PrV infection, RK13, RK13-UL31-ΔNLS, and RK13-UL31-NLSPM cells were infected with PrV-Ka under plaque assay conditions and analyzed by confocal microscopy 2 days p.i. (Fig. 8B). As observed after infection with PrV-ΔUL31, in PrV-Ka-infected RK13-UL31-ΔNLS cells, pUL34 was also found with a significant cytoplasmic localization pattern, suggesting that mislocalization of pUL34 leads to the dominant negative effect of pUL31-ΔNLS.
DISCUSSION
The NEC components homologous to HSV pUL31 and pUL34 are sufficient for membrane deformation and scission of membranous vesicles not only from the INM in eukaryotic cells but also in model membrane systems such as giant unilamellar vesicles, indicating that no other viral or cellular protein is necessary for this process (5, 35, 41). However, the spatiotemporal regulation of NEC formation and function and how nucleocapsids are recruited into these vesicles remain incompletely understood. Mutational studies of pUL34 have already shed some light on the mechanism of NEC formation and function (12, 18–23), but corresponding data on pUL31 homologs are less comprehensive.
Many of the pUL31 homologs of herpesviruses comprise a predicted or experimentally proven classical NLS in their N-terminal domain, suggesting importin-mediated nuclear translocation of the protein (6, 9, 11, 24). In the nucleus, pUL31 encounters and interacts with INM-targeted pUL34, forming the NEC (6, 13, 14, 17), which then drives primary envelopment as a prerequisite for nuclear egress of herpesvirus nucleocapsids. In order to further investigate the role of pUL31 during nuclear egress, we performed mutational analyses and tested mutated pUL31 for localization, interaction with pUL34, and function during nuclear egress.
Disabling of the predicted NLS in PrV pUL31 by either deletion of the N-terminal 25 aa or site-specific mutagenesis of four arginine residues within this motif resulted in exclusion from the nucleus and the exclusive presence of pUL31 in the cytoplasm, which demonstrates the significance of the nuclear import signal. Mutation of only two adjacent arginine residues, R5 and R6 or R16 and R17, did not impair the nuclear targeting or nuclear egress function (data not shown), indicating that one stretch of basic amino acids is probably sufficient to mediate efficient nuclear import. This is consistent with data from other pUL31 homologs, where functional NLSs were reported to reside in the variable N terminus (9, 11, 24). By in silico analysis, a leucine-rich NES could be uncovered for PrV pUL31 (aa 246 to 254) within C-terminal CR4 (Fig. 1). PrV pUL31 mutant constructs simultaneously lacking the predicted NLS and NES sequences, pUL31-ΔNLS/NES and pUL31-NLS/NESPM, were indeed found diffusely distributed throughout the cell, indicating that nuclear exclusion is not due to the size or overall shape of the protein but that active export is involved. However, despite high sequence homology, at least within the alphaherpesvirus homologs (Fig. 1C), a NES was predicted by the program NetNES only for PrV pUL31. Nevertheless, NLS mutations in HSV-2 pUL31, MCMV M53, and HCMV pUL53 resulted in cytoplasmic localization patterns similar to those of PrV pUL31, indicating that these proteins are also actively exported from the nucleus (9, 11, 24).
Although both PrV pUL31-NLS mutant constructs were detected exclusively in the cytoplasm when expressed singly, the presence of complex partner pUL34 resulted in speckle formation at the nuclear rim with pUL31-NLSPM, indicating that the protein reaches the nucleus, where it encounters and is retained by interaction with pUL34, resulting in reorganization of the INM with speckle formation. However, the INM-derived speckles detected by confocal microscopy after cotransfection of pcDNA-UL34 and pcDNA-UL31-NLSPM exhibited a phenotype different from those formed by coexpression of the wild-type proteins, with striking protuberances reaching into the cytoplasm. Despite the different localization in transfected cells, pUL31-NLSPM complemented the defect of PrV-ΔUL31 to a significant extent, resulting in titers approximately 100-fold higher than those in noncomplementing cells, indicating that, even without a functional NLS, sufficient pUL31 is present in the nucleus to support a certain level of nuclear egress.
Speckle formation from the nuclear membrane could not be observed after coexpression of pUL34 with pUL31-ΔNLS, lacking the complete N terminus up to aa 25. In contrast to the regular smooth nuclear rim staining (34, 35), pUL34 was found predominantly in the cytoplasm in a pattern reminiscent of the endoplasmic reticulum, indicating that pUL31-ΔNLS blocks pUL34 from reaching the INM or that the complete NEC is exported via the pUL31 NES function. It was therefore not surprising that pUL31-ΔNLS did not complement the defect of PrV-ΔUL31. Above all, the efficient depletion of pUL34 from the INM and relocation to cytoplasmic membranes, not only in transfected but also in infected cells (Fig. 8), most likely explain the dominant negative effect of pUL31-ΔNLS on PrV-Ka replication.
The different phenotypes of pUL31-ΔNLS and pUL31-NLSPM indicate that not only does the functional NLS play a role in nuclear egress but sequences beyond the mutated arginine residues also participate in this process. Roller et al. (27) suggested that the NEC exists in two different conformations involving different parts of the proteins, one necessary for correct targeting of the complex and one necessary for membrane bending during primary envelopment. It might be speculated that complex formation involving the N terminus of pUL31 is required to prevent premature pUL34 escape from the INM as a prerequisite to allow multimerization, eventually resulting in membrane bending. For HSV-1 pUL31, six phosphorylation sites were identified in the N-terminal domain (42). Analysis of the corresponding region deleted in PrV pUL31-ΔNLS revealed the presence of two serine residues (S12, S13) that are found in a context resembling the consensus sequences for phosphorylation by pUS3 (43). We are currently investigating whether these sites are indeed targeted by phosphorylation and whether this modification contributes to the different phenotypes of pUL31-ΔNLS and pUL31-NLSPM.
In contrast to the N terminus containing the NLS, the C-terminal 26 aa of PrV pUL31 are required for pUL34 interaction. Despite wild-type-like nuclear localization, pUL31-ΔNES is not recruited to the INM by coexpressed pUL34. This is in line with results of yeast two-hybrid studies where only pUL31 fragments comprising the complete C terminus were fished with pUL34 as bait (6). Mutation of two leucine residues within the predicted NES impaired efficient nuclear export, indicating that the signal is functional but might usually be masked or overcome by the NLS. Interestingly, the NES point mutations did not abrogate pUL34 interaction and vesicle formation either in the presence of the functional NLS (pUL31-NESPM) or in its absence (pUL31-NLS/NESPM). However, despite complex formation, the titers of PrV-ΔUL31 derived from RK13-UL31-NESPM were approximately 100-fold lower than those of PrV-Ka and only background levels are produced in RK13-UL31-NLS/NESPM, pointing to an additive effect of the two mutations.
In ultrastructural analyses, budding at the INM and presence of primary virions are only rarely observed in RK13-UL31-NESPM infected with PrV-ΔUL31, although numerous nucleocapsids were found closely attached to the INM, indicating that docking of nucleocapsids at the INM takes place but budding is impaired. Infection of RK13-UL31-NESPM with PrV-ΔUL31/US3 resulted in a similar pattern, but in addition, inward bulges of both nuclear membranes, studded by nucleocapsids, are evident. The INM at these structures is lined with electron-dense material most likely representing the NEC as part of the primary tegument, but budding and scission obviously do not occur.
Interestingly, nucleocapsids closely attached to the INM were also observed after infection with PrV-ΔUL25 (44) or HSV-1-ΔUL25 (45) lacking one component of the capsid vertex-specific complex (CVSC). It was shown recently for HSV-1 that pUL25 interacts with pUL31, thereby selecting capsids for primary envelopment (30), but pUL31 binding to capsids obviously also occurs in the absence of the protein (46), consistent with data for PrV (47). Thus, it might be speculated that pUL31-pUL25 interaction is important for the envelopment of mature capsids and that mutation of the NES interferes with this interaction.
A charge cluster mutation in the amino terminus of HSV-1 pUL34 (D35A, E37A) also resulted in an impairment of primary envelopment of nucleocapsids (27). The authors speculated that docking of capsids to the NEC at the INM triggers a change in NEC structure allowing interaction between the N terminus of pUL34 and the C terminus of pUL31. This structural change might be a prerequisite for multimerization of complexes, which then drives curvature around the capsids probably involving the CVSC (27). Consistent with this model, capsid docking at the INM was not impaired by deletion of pUL25, mutations in the N terminus of pUL34, or mutation of the leucine residues in the C terminus of pUL31 (this study; 27, 44). This might also explain the discrepancies between the pUL31-pUL34 interaction domain determined in vitro (48) and by in vivo mutational analyses (19, 20, 22, 23, 27). Additionally, the phosphorylation status of the pUL31 N terminus may regulate the budding step (5, 42).
Nevertheless, the presence of neither capsids nor pUL25 is essential for primary envelopment, since the NEC components alone are sufficient for the formation of primary envelopes from the INM (35, 41) or vesicle formation from artificial membrane systems (5).
In summary, we show here that PrV pUL31 contains not only a functional nuclear import signal but also a NES. Deletion of these motifs abrogates NEC formation and impairs viral replication while inactivation of the corresponding motifs still allows partial NEC function. The N-terminal 25 aa of pUL31, including the NLS, are required for efficient nuclear localization and proper positioning of the NEC but not for interaction with pUL34. An intact NLS is not required, but sequences within the N-terminal 25 aa are essential for vesicle formation, whereas the C-terminal 26 aa are necessary for interaction with pUL34. In addition, the leucine residues within the NES motif play a major role in the budding process. However, it remains unclear if the NES acting as a NES involves exportin binding or has an as-yet-unknown function in primary envelopment at the INM during herpesvirus nuclear egress. Since the mutated leucine residues are also part of a predicted SUMO binding site (Eukaryotic Linear Motif; http://elm.eu.org/) (49), the regulation of pUL31 and/or NEC function via corresponding protein modifications needs further investigation.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/12-1).
We thank Cindy Meinke, Maximilian Sell, and Petra Meyer for technical help and Mandy Jörn for photographic assistance.
REFERENCES
- 1.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Mettenleiter TC, Müller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 4.Mettenleiter TC. 2002. Herpesvirus assembly and egress. J Virol 76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J Virol 76:364–378. doi: 10.1128/JVI.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dal Monte P, Pignatelli S, Zini N, Maraldi NM, Perret E, Prevost MC, Landini MP. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J Gen Virol 83(Pt 5):1005–1012. [DOI] [PubMed] [Google Scholar]
- 8.Chang YE, Roizman B. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol 67:6348–6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu HY, Yamada H, Jiang YM, Yamada M, Nishiyama Y. 1999. Intracellular localization of the UL31 protein of herpes simplex virus type 2. Arch Virol 144:1923–1935. doi: 10.1007/s007050050715. [DOI] [PubMed] [Google Scholar]
- 10.Gonnella R, Farina A, Santarelli R, Raffa S, Feederle R, Bei R, Granato M, Modesti A, Frati L, Delecluse HJ, Torrisi MR, Angeloni A, Faggioni A. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J Virol 79:3713–3727. doi: 10.1128/JVI.79.6.3713-3727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lötzerich M, Ruzsics Z, Koszinowski UH. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J Virol 80:73–84. doi: 10.1128/JVI.80.1.73-84.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bubeck A, Wagner M, Ruzsics Z, Lotzerich M, Iglesias M, Singh IR, Koszinowski UH. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J Virol 78:8026–8035. doi: 10.1128/JVI.78.15.8026-8035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamauchi Y, Shiba C, Goshima F, Nawa A, Murata T, Nishiyama Y. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J Gen Virol 82(Pt 6):1423–1428. [DOI] [PubMed] [Google Scholar]
- 15.Camozzi D, Pignatelli S, Valvo C, Lattanzi G, Capanni C, Dal Monte P, Landini MP. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J Gen Virol 89:731–740. doi: 10.1099/vir.0.83377-0. [DOI] [PubMed] [Google Scholar]
- 16.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 17.Lake CM, Hutt-Fletcher LM. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99–106. doi: 10.1016/j.virol.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Schuster F, Klupp BG, Granzow H, Mettenleiter TC. 2012. Structural determinants for nuclear envelope localization and function of pseudorabies virus pUL34. J Virol 86:2079–2088. doi: 10.1128/JVI.05484-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paßvogel L, Trübe P, Schuster F, Klupp BG, Mettenleiter TC. 2013. Mapping of sequences in pseudorabies virus pUL34 that are required for formation and function of the nuclear egress complex. J Virol 87:4475–4485. doi: 10.1128/JVI.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paßvogel L, Janke U, Klupp BG, Granzow H, Mettenleiter TC. 2014. Identification of conserved amino acids in pUL34 which are critical for function of the pseudorabies virus nuclear egress complex. J Virol 88:6224–6231. doi: 10.1128/JVI.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott M, Tascher G, Hassdenteufel S, Zimmermann R, Haas J, Bailer SM. 2011. Functional characterization of the essential tail anchor of the herpes simplex virus type 1 nuclear egress protein pUL34. J Gen Virol 92:2734–2745. doi: 10.1099/vir.0.032730-0. [DOI] [PubMed] [Google Scholar]
- 22.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J Virol 77:7601–7610. doi: 10.1128/JVI.77.13.7601-7610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milbradt J, Auerochs S, Sevvana M, Muller YA, Sticht H, Marschall M. 2012. Specific residues of a conserved domain in the N terminus of the human cytomegalovirus pUL50 protein determine its intranuclear interaction with pUL53. J Biol Chem 287:24004–24016. doi: 10.1074/jbc.M111.331207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmeiser C, Borst E, Sticht H, Marschall M, Milbradt J. 2013. The cytomegalovirus egress proteins pUL50 and pUL53 are translocated to the nuclear envelope through two distinct modes of nuclear import. J Gen Virol 94:2056–2069. doi: 10.1099/vir.0.052571-0. [DOI] [PubMed] [Google Scholar]
- 25.Schnee M, Ruzsics Z, Bubeck A, Koszinowski UH. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J Virol 80:11658–11666. doi: 10.1128/JVI.01662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sam MD, Evans BT, Coen DM, Hogle JM. 2009. Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J Virol 83:2996–3006. doi: 10.1128/JVI.02441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roller RJ, Bjerke SL, Haugo AC, Hanson S. 2010. Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J Virol 84:3921–3934. doi: 10.1128/JVI.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogoda M, Bosse JB, Wagner FM, Schauflinger M, Walther P, Koszinowski UH, Ruzsics Z. 2012. Characterization of conserved region 2-deficient mutants of the cytomegalovirus egress protein pM53. J Virol 86:12512–12524. doi: 10.1128/JVI.00471-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popa M, Ruzsics Z, Lotzerich M, Dolken L, Buser C, Walther P, Koszinowski UH. 2010. Dominant negative mutants of the murine cytomegalovirus M53 gene block nuclear egress and inhibit capsid maturation. J Virol 84:9035–9046. doi: 10.1128/JVI.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan AS, Vatter AE. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 32.Klupp BG, Granzow H, Mettenleiter TC. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J Gen Virol 82(Pt 10):2363–2371. [DOI] [PubMed] [Google Scholar]
- 33.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 34.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol 74:10063–10073. doi: 10.1128/JVI.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A 104:7241–7246. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter TC. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. 2009. NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terry LJ, Wente SR. 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell 8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panté N, Kann M. 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 41.Desai PJ, Pryce EN, Henson BW, Luitweiler EM, Cothran J. 2012. Reconstitution of the Kaposi's sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J Virol 86:594–598. doi: 10.1128/JVI.05988-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mou F, Wills E, Baines JD. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J Virol 83:5181–5191. doi: 10.1128/JVI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leader DP, Deana AD, Marchiori F, Purves FC, Pinna LA. 1991. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases A and C. Biochim Biophys Acta 1091:426–431. doi: 10.1016/0167-4889(91)90210-O. [DOI] [PubMed] [Google Scholar]
- 44.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol 80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhn J, Leege T, Granzow H, Fuchs W, Mettenleiter TC, Klupp BG. 2010. Analysis of pseudorabies and herpes simplex virus recombinants simultaneously lacking the pUL17 and pUL25 components of the C-capsid specific component. Virus Res 153:20–28. doi: 10.1016/j.virusres.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 46.Yang K, Wills E, Lim HY, Zhou ZH, Baines JD. 2014. Association of herpes simplex virus pUL31 with capsid vertices and components of the capsid vertex-specific complex. J Virol 88:3815–3825. doi: 10.1128/JVI.03175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leelawong M, Guo D, Smith GA. 2011. A physical link between the pseudorabies virus capsid and the nuclear egress complex. J Virol 85:11675–11684. doi: 10.1128/JVI.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang L, Baines JD. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J Virol 79:3797–3806. doi: 10.1128/JVI.79.6.3797-3806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, Jodicke L, Dammert MA, Schroeter C, Hammer M, Schmidt T, Jehl P, McGuigan C, Dymecka M, Chica C, Luck K, Via A, Chatr-Aryamontri A, Haslam N, Grebnev G, Edwards RJ, Steinmetz MO, Meiselbach H, Diella F, Gibson TJ. 2012. ELM—the database of eukaryotic linear motifs. Nucleic Acids Res 40:D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]