ABSTRACT
Rabbit hemorrhagic disease virus (RHDV) is a member of the Caliciviridae family (Lagovirus genus). RHDV is highly contagious and attaches to epithelial cells in the digestive or respiratory tract, leading to massive lesions with high mortality rates. A new variant of RHDV (termed RHDVb) recently has emerged, and previously vaccinated rabbits appear to have little protection against this new strain. Similar to human norovirus (Caliciviridae, Norovirus genus), RHDV binds histo-blood group antigens (HBGAs), and this is thought to be important for infection. Here, we report the HBGA binding site on the RHDVb capsid-protruding domain (P domain) using X-ray crystallography. The HBGA binding pocket was located in a negatively charged patch on the side of the P domain and at a dimeric interface. Residues from both monomers contributed to the HBGA binding and involved a network of direct hydrogen bonds and water-mediated interactions. An amino acid sequence alignment of different RHDV strains indicated that the residues directly interacting with the ABH-fucose of the HBGAs (Asp472, Asn474, and Ser479) were highly conserved. This result suggested that different RHDV strains also could bind HBGAs at the equivalent pocket. Moreover, several HBGA binding characteristics between RHDVb and human genogroup II norovirus were similar, which indicated a possible convergent evolution of HBGA binding interactions. Further structural studies with other RHDV strains are needed in order to better understand the HBGA binding mechanisms among the diverse RHDV strains.
IMPORTANCE We identified, for the first time, the HBGA binding site on an RHDVb P domain using X-ray crystallography. Our results showed that RHDVb and human genogroup II noroviruses had similar HBGA binding interactions. Recently, it was discovered that synthetic HBGAs or HBGA-expressing enteric bacteria could enhance human genogroup II norovirus infection in B cells. Considering that RHDVb and genogroup II norovirus similarly interacted with HBGAs, it may be possible that a comparable cell culture system also could work with RHDVb. Taken together, these new findings will extend our understanding of calicivirus HBGA interactions and may help to elucidate the specific roles of HBGAs in calicivirus infections.
INTRODUCTION
Rabbit hemorrhagic disease (RHD) was first reported in China in 1984 (1), and the etiological agent, rabbit hemorrhagic disease virus (RHDV; Caliciviridae family, Lagovirus genus), was identified subsequently (2–4). RHDV is highly contagious and is endemic in several countries with wild rabbit populations. The virus attaches to epithelial cells in the digestive or respiratory tract and can lead to massive lesions in the liver, trachea, and lungs of adult rabbits. High mortality rates in the range of 90% can lead to a drastic decimation of rabbit populations (reviewed in reference 5), which is an ecological threat and economic burden. A vaccine for RHDV was introduced in the 1990s (6), and this provided good coverage for the strains circulating at that time and up until recently (reviewed in reference 5). However, atypical RHD outbreaks in vaccinated rabbits and with high mortalities in young rabbits appear to be changing the epidemiological panorama of this disease, which coincided with the emergence and spread of a new variant RHDV (7–9).
RHDV is a single-stranded, positive-sense RNA virus with two open reading frames (ORFs). The first open reading frame encodes the nonstructural proteins and the capsid protein, whereas ORF2 encodes a minor structural protein (10). RHDV cannot be grown in cell culture, although the expression of the capsid protein in insect cells results in the formation of virus-like particles (VLPs) that are morphologically and antigenically similar to the native virion (11, 12). The capsid protein is divided into a shell (S) and protruding (P) domain, where the S domain forms a scaffold protecting the RNA from the environment and the P domain, which is more exposed, likely contains determinants for cell attachment and antigenic diversity.
RHDV strains usually are classified on the complete capsid amino acid sequences (13), and there appears to be two genetically distinct groups of RHDV strains, tentatively termed RHDV and RHDVb (8). Unlike human noroviruses (Caliciviridae family, Norovirus genus) that have a unified classification scheme (14), the RHDV strains have multiple nomenclatures. The subclusters in the distinct RHDV groups have been termed genogroups or groups 1 to 6 (G1 to G6 [15–18]), groups/clades 1 to 4 (15, 19), and clades A to D (13). The G6 cluster additionally was termed RHDVa (20). The RHDVb branch also is known as RHDV2 (8, 15, 21). This multiple naming of subclusters has caused confusion in the field, and for this study we used the nomenclature of RHDVa, RHDVb, RHDV G1 to G5, and RHDV (for all strains).
RHDV and human norovirus bind to histo-blood group antigens (HBGAs), and this is thought to be important for infection (22, 23). Recently, a genogroup II (GII) human norovirus was found to infect B cells, and the infection was enhanced by synthetic HBGAs or HBGA-expressing enteric bacteria (24). HBGAs are polymorphic carbohydrates that are synthesized by a stepwise addition of monosaccharides to different precursor structures, aided through the action of specific glycosyltransferases. The fecal-oral route is the major route of RHDV transmission. Rabbits were shown to express HBGAs (A, B, and H type 2 [Lewis Y {Ley}]) on the duodenum surface, although other areas, such as the trachea and biliary ducts, also weakly expressed several HBGA types (16). RHDV G1 to G5 and RHDVa were found to bind to HBGAs in a strain-dependent manner and with variable magnitudes (16). All strains bound B and H type 2, A type was not recognized by G2 or G3, and Lewis Y was recognized only by G1 and RHDVa. Little is known about RHDVb interactions with HBGAs, although hemagglutination assays using human O (H type), A, B, and AB erythrocytes indicated that RHDV G1 and RHDVb had variable interactions with the different blood types (8).
Recently, a pseudoatomic model of an RHDVa VLP was determined (12). Based on sequence alignment of RHDV representatives, seven regions of sequence variation on the RHDVa P domain were identified that could give rise to different HBGA binding specificities and antigenicity. As a result, three putative HBGA binding cavities were predicted on the outer surface of the RHDVa P domain (12). Here, we show the X-ray crystal structures of the new variant RHDVb P domain (also termed N11 strain [8]) in complex with Lewis Y and H type 2 HBGAs. We showed that HBGAs bound at the dimeric interface on the side of the RHDVb P dimer, which was distinctly different from the three predicted sites on the RHDVa P domain (12). Residues from both RHDVb monomers contributed to the HBGA binding and involved a network of hydrogen bonds. Sequence analysis showed that the HBGA binding residues were preserved among RHDVs, suggesting that other RHDV strains bind HBGA at the same pocket.
MATERIALS AND METHODS
Expression and purification of RHDVb P domain.
The RHDVb P domain (N11 strain; GenBank accession number JX133161) expression construct was designed from an alignment of a human norovirus P domain construct, which expressed well and diffracted to high resolution (25). Based on this alignment, amino acids 231 to 569 of the RHDVb capsid gene were codon optimized for Escherichia coli expression (Geneart), cloned into a modified pMal-c2X expression vector (New England BioLabs), and expressed as previously described, with slight modifications (25). Briefly, the P domain construct was transformed into BL21 cells, induced with 0.75 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.6, and then grown for 18 h at 20°C. Cells were harvested by centrifugation at 6,500 rpm for 15 min at 4°C and subsequently disrupted by sonication on ice. A His-tagged MBP-P domain fusion protein was purified over a Ni column (Qiagen) and digested with HRV-3C protease (Novagen) overnight at 4°C. The cleaved P domain was separated on the Ni column and then dialyzed against gel filtration buffer (50 mM Tris [pH 7.6] and 250 mM NaCl) overnight at 4°C. The P domain was further purified as a dimer by size-exclusion chromatography with a Superdex 200 column, concentrated to 9 mg/ml, and stored in gel filtration buffer at 4°C.
Crystallization of RHDVb P domain.
The RHDVb P domain was crystallized using the hanging-drop method. Briefly, a 1:1 ratio of P domain and mother solution containing 1 M lithium chloride, 24% polyethylene glycol 6000 (PEG 6000), and 0.1 M citric acid (pH 4) were mixed and incubated at 18°C. For cocrystallization experiments, we added an equal volume of P domain, mother solution, and a 30 to 60 molar excess of HBGA (Dextra, United Kingdom). Prior to flash freezing, crystals were transferred to a cryoprotectant containing mother liquor, a 10 to 30 molar excess of HBGA (for complexes), and 30% ethylene glycol.
Sequence analysis.
Amino acid sequences of the complete/partial capsid gene of RHDV strains representing different strains were aligned in Clustal X. The representative sequences were N11 (JX133161; RHDVb), CHA/97 (DQ205345; RHDVa), G1 (Z49271), G2 (AF402614), G3/G4 (FR823354), and G5 (AM085133).
Data collection, structure solution, and refinement.
X-ray diffraction data were collected at the European Synchrotron Radiation Facility, France, at beamlines BM30A and ID23-1, and they were processed with XDS (26). Structures were solved by molecular replacement in PHASER (27). Structures were refined in multiple rounds of manual model building in COOT (28) and further refined with PHENIX (29). The HBGAs were added at the final stages of structural refinement in order to exclude bias during refinement. Structures were validated with Procheck (30) and Molprobity (31). Glycosidic bonds for the oligosaccharides were defined in PHENIX during refinement. HBGA interactions were analyzed using Accelrys Discovery Studio (version 4.1), with hydrogen-bonding interaction distances between 2.4 and 3.5Å. Figures and protein contact potentials were generated using PyMOL (version 1.12r3pre).
Protein structure accession numbers.
The atomic coordinates of RHDVb unliganded, RHDVb Lewis Y tetrasaccharide (Ley-tetra), and RHDVb H type 2 trisaccharide (H2-tri) were deposited in the Protein Data Bank (PDB) under entries 4X1W, 4X1X, and 4X1Z, respectively.
RESULTS
Unbound structure of the variant RHDVb P domain.
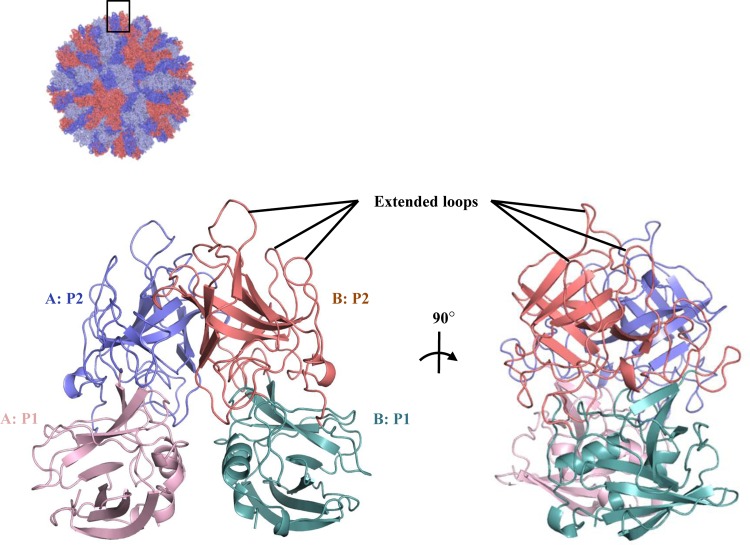
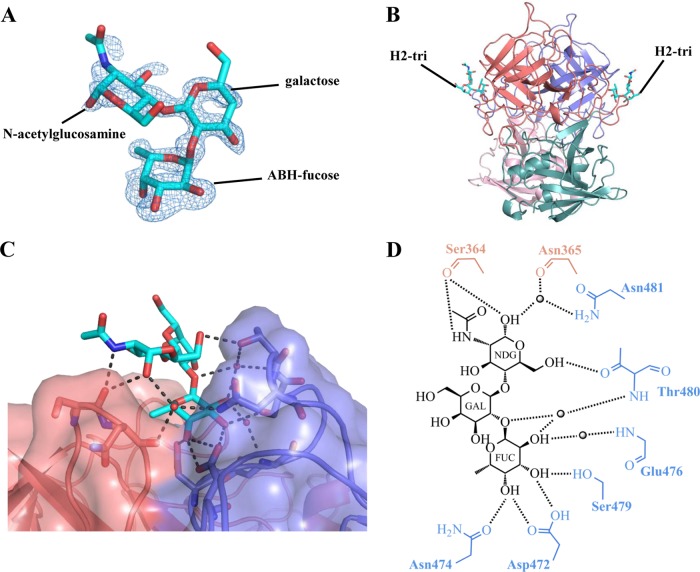
The RHDVb P domain coding sequence was expressed in E. coli, and the purified P dimer was crystallized. The unliganded RHDVb P domain was solved using molecular replacement with the previously determined RHDVa P domain (4EGT) as a search model. The structure was refined to a resolution of 1.95 Å, with two P dimers in the asymmetric unit (Table 1). The electron density was well defined for most of the RHDVb P dimers, having an overall B value of 25 Å2. Some regions showed weaker electron density. In particular, residues 477 to 482 on one chain had higher than average B values (between 50 and 110 Å2). The RHDVb P domain could be subdivided into P1 and P2 subdomains, as proposed for RHDVa (Fig. 1) (12). The RHDVb P1 subdomain contained an α-helix and several β-sheets, whereas the P2 subdomain contained six antiparallel β-sheets that formed a barrel-like structure. Three extended loops were located on the RHDVb P2 subdomain between residues 304 and 312, 343 and 377, and 382 and 390 (Fig. 1).
TABLE 1.
Data collection and refinement statistics of unliganded and complexed RHDVb P domain structures
| Parameter | Value(s) fora: |
||
|---|---|---|---|
| RHDVb unliganded (4X1W) | RHDVb Ley-tetra (4X1X) | RHDVb H2-tri (4X1Z) | |
| Data collection statistics | |||
| Space group | P1211 | P1211 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 59.19, 83.13, 118.29 | 59.44, 84.15, 62.66 | 76.17, 76.21, 107.68 |
| α, β, γ (°) | 90.00, 93.21, 90.00 | 90.00, 110.11, 90.00 | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 19.80–1.95 (2.02–1.95)* | 48.22–1.55 (1.61–1.55)* | 20.00–1.30 (1.35–1.30)* |
| Rmerge (%) | 16.61 (74.89)* | 6.87 (54.95)* | 6.57 (43.74)* |
| I/σ (I) | 7.82 (1.40)* | 10.85 (1.85)* | 22.20 (1.95)* |
| Completeness (%) | 99.67 (98.50)* | 98.02 (95.50)* | 98.30 (86.57)* |
| Redundancy | 4.6 (2.9)* | 3.0 (3.0)* | 13.5 (3.0)* |
| Refinement statistics | |||
| Resolution range (Å) | 19.80–1.95 | 48.22–1.60 | 20.00–1.36 |
| No. of unique reflections | 83191 | 82109 | 151410 |
| Rwork/Rfree (%) | 17.98/21.24 | 15.45/18.45 | 10.78/13.24 |
| No. of atoms | 10,593 | 5,583 | 5,773 |
| Protein | 9,665 | 4,869 | 4,856 |
| Ligand/ion | 27 | 51 | 86 |
| Water | 901 | 662 | 828 |
| Avg B factors (Å2) | |||
| Protein | 25.40 | 17.10 | 13.50 |
| Ligand/ion | 29.70 | 23.70 | 46.30 |
| Water | 30.50 | 26.60 | 26.00 |
| RMSD | |||
| Bond length (Å) | 0.005 | 0.005 | 0.007 |
| Bond angle (°) | 0.98 | 1.08 | 1.25 |
For RHDVb unliganded, two datasets were merged from two crystals. For RHDVb Ley-tetra, the data were collected from a single crystal. For RHDVb H2-tri, two datasets were merged from two different locations on a single crystal. Values in parentheses are for the highest-resolution shell.
FIG 1.
X-ray crystal structure of unliganded RHDVb P domain. The RHDVa pseudoatomic VLP model (T=3) was modeled with different monomer interactions (12), where each A, B, and C monomer was colored blue, salmon, and light blue, respectively. The boxed region shows the location of the P dimer. The unliganded RHDVb P domain dimer was colored according to monomers (chain A and B) and P1 and P2 subdomains, i.e., chain A, P1, light pink; chain A, P2, slate; chain B, P1, light teal; and chain B, P2, deep salmon. Three extended loops were located on the outer region of the P domain.
Comparison of RHDVa and RHDVb P domains.
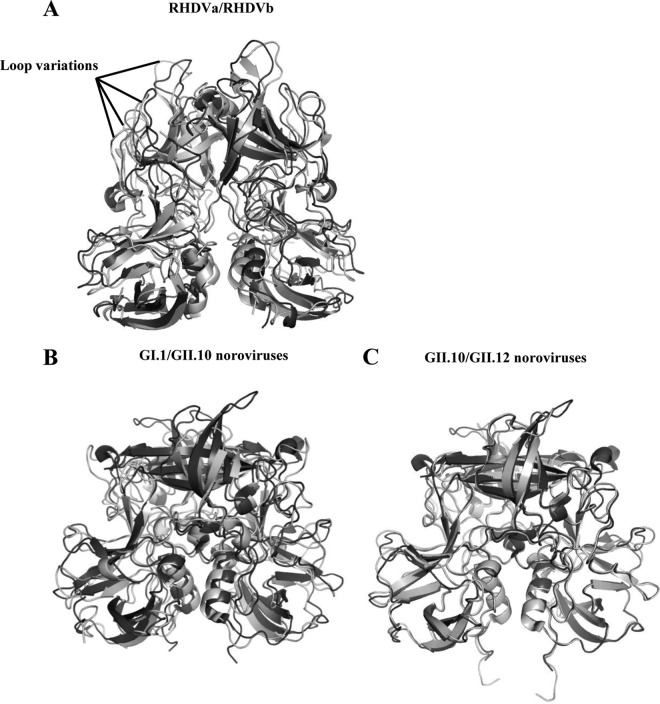
The sequence identity between RHDVa and RHDVb was 84%. Superposition of RHDVa and RHDVb showed that their overall structures were similar; however, the P1 subdomain helices were slightly shifted, and a number of P2 subdomain loops were oriented differently (Fig. 2A). The root mean square deviation (RMSD) for Cα atoms of RHDVa:RHDVb was calculated to be 2.0 Å, which indicated that the structures were moderately different. The high RMSD value was comparable to the RMSD of two different human norovirus genogroups (GI.1:GII.10 P domains; RMSD = 1.92 Å), which had 43% amino acid identity (Fig. 2B). On the other hand, the RHDVa:RHDVb RMSD was five times higher than that for two different norovirus genotypes (GII.10:GII.12 P domains; RMSD = 0.4 Å), which had 70% amino acid identity (Fig. 2C). These results implied that RHDVa and RHDVb P domain structures resembled distinct genogroups, although they had high (84%) amino acid identity.
FIG 2.
Structural comparisons of RHDV and norovirus P domains. (A) Superposition of RHDVa (light gray) and RHDVb (dark gray) P dimers. RHDVa and RHDVb P domains had an amino acid identity of 84% and an RMSD of 2.0 Å. The high RMSD likely related to the different loop variations. (B) Superposition of GI.1 (light gray; 2ZL5) and GII.10 (dark gray) norovirus P dimers. The GI.1 and GII.10 P domains had an amino acid identity of 43% and an RMSD of 1.92 Å and likely were antigenically distinct (40). (C) Superposition of GII.10 (dark gray) and GII.12 (light gray; 3R6J) norovirus P dimers. The GII.10 and GII.12 P domains had an amino acid identity of 70% and an RMSD of 0.4 Å and were antigenically distinct (41).
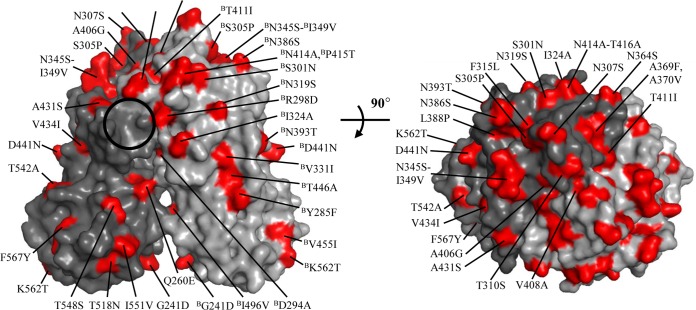
In order to better understand the structural variations between RHDVa and RHDVb P domains, amino acid differences were mapped on the RHDVb P dimer surface (Fig. 3). Amino acid changes were randomly distributed, although they were mainly concentrated on the outer surface of the P2 subdomain. Many of the amino acid changes were located on the three extended loop regions.
FIG 3.
Amino acid variations between RHDVa and RHDVb P dimers. Amino acid changes (red) were highlighted on the RHDVb P dimer (side and top views). The changes were numbered according to a change from RHDVa to RHDVb (chain B [denoted by a superscript “B”] is indicated for ease of viewing). The black circle indicates the position of the RHDVb HBGA binding pocket.
Complex structures of RHDVb P domain and HBGAs.
RHDV was the first calicivirus shown to interact with HBGAs (22), followed by noroviruses (23, 32). HBGAs can be subdivided into ABH and Lewis antigens (33), of which at least four were found to interact with RHDV (16). We determined the structures of the RHDVb P domain Ley-tetra and H2-tri complexes using X-ray crystallography (Table 1). These two HBGAs were chosen for this study because RHDV G1 and RHDVb interactions with H2 and Ley appeared variable (8). Electron densities were well defined for most of the RHDVb P dimer HBGA complexes and reminiscent of the unliganded RHDVb structure.
Structure of RHDVb P domain Ley-tetra complex.
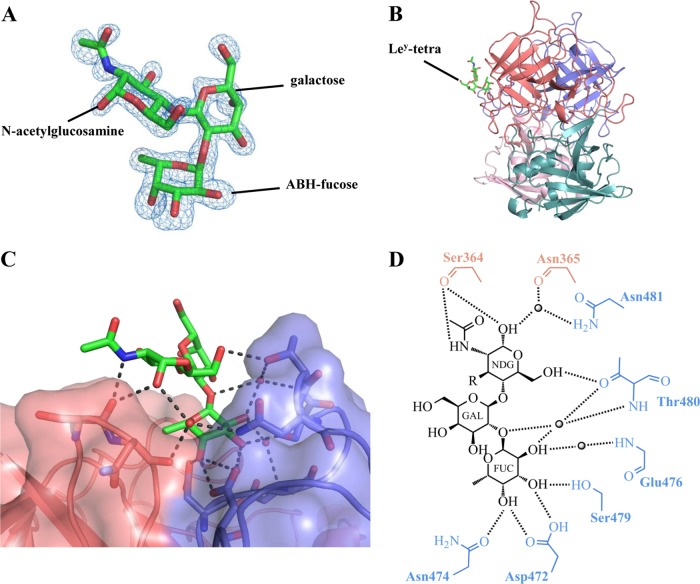
The electron density was easily discernible for the ABH-fucose, galactose, and N-acetylglucosamine of Ley-tetra, and further refinement led to a clearly defined orientation of these saccharides (Fig. 4A). The electron density for the peripheral Lewis-fucose of Ley-tetra was disordered and not modeled into the structure. The Ley-tetra bound at the RHDVb P2 subdomain and on the side of the dimer (Fig. 4B). Residues from both monomers contributed to Ley-tetra binding through a network of hydrogen bonds (Fig. 4C and D). The ABH-fucose was held with four direct hydrogen bonds from one monomer, two from the side chain of Asp472, one from the side chain of Asn474, and one from the side chain of Ser479. The galactose was positioned further away from the protein and showed no interaction with any residues. The N-acetylglucosamine was held with three direct hydrogen bonds from both monomers, two from the main chain of Ser364 and one from the side chain of Thr480. A number of main- and side-chain water-mediated interactions with ABH-fucose and N-acetylglucosamine also were observed (Fig. 4D). The reason why the terminal Lewis-fucose could not be modeled into the electron density could be the lack of binding interactions with the P dimer, since, based on the N-acetylglucosamine position, the Lewis-fucose likely was raised off the protein (Fig. 4C).
FIG 4.
RHDVb P domain and Ley-tetra interactions. The Ley-tetra is an α-l-fucose-(1-2)-β-d-galactose-(1-4)-N-acetyl-β-d-glucosamine-(3-1)-α-l-fucose. The reducing-end hydroxyl group was fixed in the α position in the Ley-tetra (underlined). (A) The Fo-Fc simulated annealing difference omit map (blue) was contoured at 3.0 σ. The ABH-fucose, galactose, and N-acetylglucosamine (green sticks) of Ley-tetra easily fit into the electron density. The Lewis-fucose was disordered and was not modeled into the structure. (B) The Ley-tetra bound to the dimeric interface on the side of the P dimer (colored as described in the legend to Fig. 1). (C) Closeup surface and ribbon representation of the RHDVb P dimer showing the bound Ley-tetra. (D) The Ley-tetra binding interaction with RHDVb P dimer, where the black dotted lines represent the hydrogen bonds and the sphere represents water molecules (fucose, FUC; galactose, GAL; N-acetylglucosamine, NDG). The Lewis-fucose was disordered and was not modeled into the structure (-R). For simplicity, only the backbone is shown for residues that were backbone mediated. Hydrogen bond distances were less than 3.2 Å, although the majority were ∼2.8 Å.
Superposition of the RHDVb P domain Ley-tetra complex and unliganded structures indicated that no major conformational changes occurred during HBGA binding. Only one Ley-tetra bound to the RHDVb P dimer, and the unoccupied pocket showed a possible steric clash with the neighboring subunit, which likely blocked the second Ley-tetra from binding (data not shown).
Structure of RHDVb P domain H2-tri complex.
The H2-tri electron density was clearly visible for the ABH-fucose and less defined for the β-galactose and N-acetylglucosamine (Fig. 5A). The H2-tri bound at the HBGA binding pocket on both sides of the dimer (Fig. 5B) and was positioned through a network of hydrogen bonds (Fig. 5C and D). The ABH-fucose was held with four direct hydrogen bonds from one monomer, two from the side chain of Asp472, one from the side chain of Asn474, and one from the side chain of Ser479. The galactose was not interacting with any residues. The N-acetylglucosamine was held with three direct hydrogen bonds, two from the main chain of Ser364 and one from the side chain of Thr480. Similar to the RHDVb Ley-tetra complex, water-mediated interactions with the ABH-fucose and N-acetylglucosamine also were observed (Fig. 5D).
FIG 5.
RHDVb P domain and H2-tri interactions. H2-tri is an α-l-fucose-(1-2)-β-d-galactose-(1-4)-N-acetyl-β-d-glucosamine. The reducing-end hydroxyl group was fixed in α position in H2-tri (underlined). (A) The Fo-Fc simulated annealing difference omit map (blue) was contoured at 3.0 σ. The electron density for H2-tri (cyan sticks) was clearly visible for ABH-fucose and less defined for galactose and N-acetylglucosamine. (B) H2-tri bound to the dimeric interface on the side of the P dimer. (C) Closeup surface-and-ribbon representation of the RHDVb P dimer showing the bound H2-tri. (D) The H2-tri binding interaction with RHDVb P dimer, where the black dotted lines represent the hydrogen bonds and the sphere represents water molecules (fucose, FUC; galactose, GAL; N-acetylglucosamine, NDG). For simplicity, only the backbone is shown for residues that were backbone mediated. Hydrogen bond distances were less than 3.2 Å, although the majority were ∼2.8 Å.
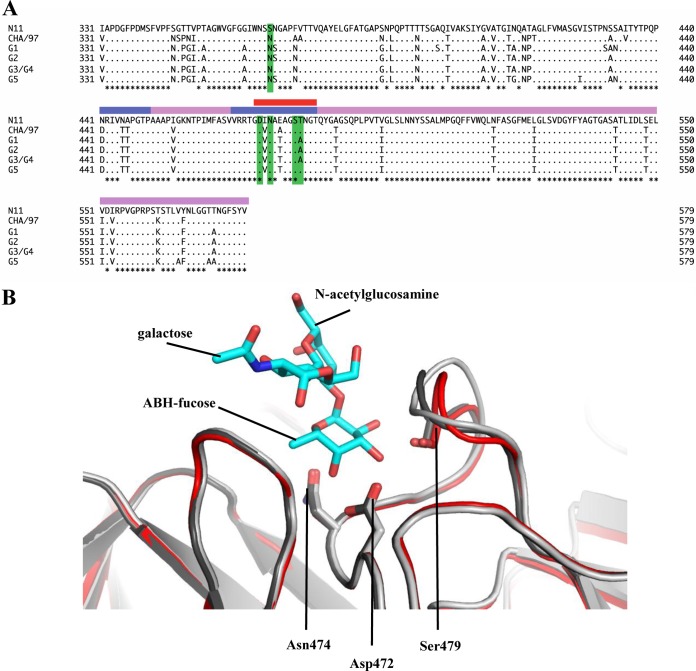
Sequence and structural analysis of the RHDVb HBGA binding pocket.
Comparisons of Ley-tetra and H2-tri structures indicated that the ABH-fucose was tightly held with three P domain residues (Asp472, Asn474, and Ser479). These three residues were highly conserved among different RHDV P domains (Fig. 6A). This result suggested that the different RHDV strains also could bind HBGAs at this pocket, although amino acid variations surrounding the pocket may influence the interactions (Fig. 3). Structural analysis of the RHDVb HBGA binding pocket showed that the three conserved residues were located on a single P2 subdomain loop. Superposition of the unbound RHDVb and RHDVb H2-tri revealed that the loop had slightly different conformations, especially between residues 477 and 482 (Fig. 6B). However, the loop on the alternative RHDVb H2-tri binding pocket did not show such conformational changes (data not shown). Moreover, in the unbound RHDVa structure (residues 472 to 483), this loop was partially unmodeled, since the electron density was disordered. Taken together, these results suggested that the loop was flexible. Further studies are needed to define how the flexible loop may have affected the HBGA binding interactions.
FIG 6.
Amino acid sequence alignment of the partial RHDV P domains and structural analysis of the loop that binds the ABH-fucose. (A) The P domains of different RHDV strains, N11 (JX133161; RHDVb), CHA/97 G6 (DQ205345; RHDVa), G1 (Z49271), G2 (AF402614), G3/G4 (FR823354), and G5 (AM085133), were aligned using Clustal X (partial sequences are shown). The P domain was divided into P1 (pink) and P2 (slate) subdomains as previously described (12). The residues having a direct hydrogen bond with HBGAs were highlighted (green). The flexible loop region was between residues 472 and 483 (red bar). The asterisks represent conserved amino acids residues. (B) Superposition and closeup view of the flexible loop (residues 472 to 483) of two unbound RHDVb dimers (light and dark gray) and one RHDVb H2-tri dimer (red), showing bound H2-tri (cyan sticks). The flexible loop, shown in various conformations, contained three residues (side chains of Asp472, Asn474, and Ser479) that were involved in binding of the ABH-fucose of H2-tri. The side chain of Ser479, which interacted with N-acetylglucosamine, was not modeled into the loop of one unbound RHDVb dimer due to poor electron density. However, on the corresponding loop of the second monomer, the electron density of the Ser479 side chain was clearly defined (data not shown).
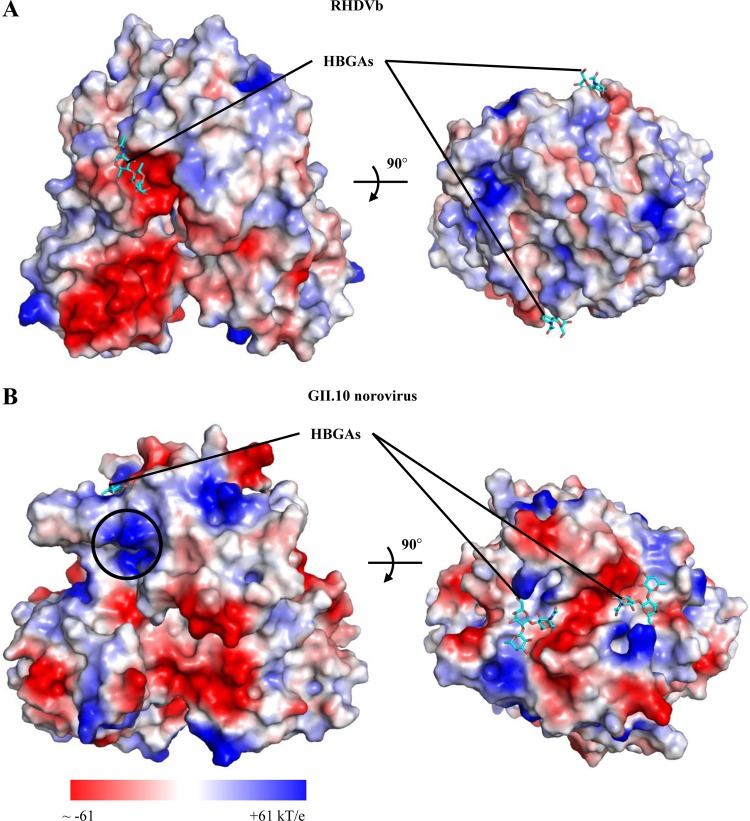
Protein contact potential of RHDVb P dimer.
The protein contact potential was calculated on the RHDVb P dimer using PyMOL (version 1.12r3pre) in order to better understand the variations in surface charge that could alter antigenicity and HBGA binding interactions. Four large patches of negative charge were found on the P dimer (Fig. 7A). These patches were located on the side of the P dimer and on the P1 and P2 subdomains. Other regions on the P dimer displayed a random distribution of charge. The HBGA binding pocket was situated in the negatively charged P2 subdomain patch and was distinctly different from a representative GII norovirus that showed patches of both negative and positive charge around the HBGA pocket (Fig. 7B). Interestingly, the equivalent RHDV HBGA binding pocket on the GII norovirus P dimer was mainly positively charged. It appeared that the HBGA binding pockets for RHDVb and GII norovirus were either negative regions or negative/positive patches, but they were never in exclusively positive regions.
FIG 7.
Protein contact potential of RHDVb and GII norovirus P dimers. Surface representation of protein contact potential of RHDVb and GII.10 norovirus P dimers with bound H2-tri (a second H2-tri was modeled into the GII.10 P domain to show both HBGA binding sites). RHDVb and GII norovirus bound HBGAs at different sites on the P domains. RHDVb bound HBGAs on the side of the P domain, whereas GII norovirus bound HBGAs on the outer surface of the P domain. (A) RHDVb P dimer protein contact potential (−53 to +53 kT/e). The region surrounding the ABH-fucose of H2-tri was mainly negatively charged. (B) The GII.10 norovirus (3Q3A) dimer protein contact potential (−61 to +61 kT/e). The region surrounding the HBGA pocket was both negatively and positively charged. The black circle indicates the position on the GII dimer that would correspond to the RHDVb HBGA binding pocket.
DISCUSSION
Despite an extensive vaccination and outbreak control strategy, RHDV is still a major economic and ecological burden in many parts of the world. Until recently, only one serotype was circulating (20); however, it seems possible that a new serotype (termed RHDVb or RHDV2) has emerged (7, 9, 15, 21). Significantly, the original RHDV vaccine has been reported to have low or no efficacy against RHDVb (7, 8). RHDVb has spread quickly and appears to be replacing the older RHDV strains on the Iberian Peninsula (8, 9) and in France, which is considered an important distribution source for RHDV (7, 16, 19). The drastic spread of RHDVb, combined with a changed virulence profile, lead to the assumption of an altered receptor specificity and viral capsid modifications (8).
The amino acid sequence identity between RHDVa and RHDVb was 84%. Structural comparisons of the RHDVa and RHDVb P domains showed that P1 subdomain helices and P2 subdomain extended loops were slightly shifted (Fig. 2A). These extended loops also contained amino acid variations, although there was a random distribution of amino acid changes on the P dimer surface (Fig. 3). The antigenicity among the earlier and variant RHDV strains still is poorly understood, but based on RMSD values, RHDVa and RHDVb P domains appeared distinctly different and likely represented different genogroups (Fig. 2B and C). Interestingly, the two different human norovirus genogroups (GI and GII) interact differently with HBGAs on the P domain, where GI binds HBGAs on a single monomer and GII binds HBGAs at a dimeric interface (25, 34, 35). Therefore, it is tempting to speculate that RHDVa and RHDVb also have different HBGA binding mechanisms, especially since RHDVa and RHDVb were found to have variable interactions with human blood types (8, 16). However, further structural studies of other RHDV strains are needed in order to determine the HBGA binding differences and similarities.
A previous study using nuclear magnetic resonance (NMR) spectroscopy showed that the fucose moiety of HBGAs is required as a minimal structural recognition motif for RHDV (36), which correlated well with our structural data. We showed that fucose was firmly held with three residues (Asp472, Asn474, and Ser479). These three amino acids were highly conserved among diverse RHDV strains (Fig. 6A). Superposition of unbound RHDVa and RHDVb H2-tri structures indicated that the RHDVa HBGA binding pocket was similar to that of RHDVb (data not shown). However, the loop interacting with the ABH-fucose appeared flexible in RHDVa and RHDVb (Fig. 6B). The reasons for the distinct loop flexibility were not known. However, flexibility in the HBGA binding pocket may provide a defense mechanism for RHDV by protecting the HBGA pocket and making it less vulnerable to the immune system. Moreover, the flexible loop could provide a selection mechanism for HBGA types and promote HBGA binding diversity among different RHDV strains (8). On the other hand, the RHDV flexible loop was the reverse of that of GII noroviruses, which usually have stable loops in the analogous fucose-binding site and flexible modulating loops at the peripheral saccharide site (data not shown).
In both RHDV and GII noroviruses, an aspartic acid residue coordinated the binding of fucose. However, the interacting hydroxyl groups of fucose were shifted from C2OH and C3OH in GII norovirus to C3OH and C4OH in RHDVb. Despite this difference, there were similar HBGA binding features. RHDVb and GII norovirus bound HBGAs at a dimeric interface, residues from both monomers interacted with HBGAs, the ABH-fucose was considered the minimal structural binding motif, and the binding pocket was mainly negatively charged (Fig. 7). These results implied a convergent evolution of HBGA binding interactions among caliciviruses, which was previously suggested (37, 38). Moreover, a question arises: if HBGAs served as a possible receptor or attachment factor for RHDV and noroviruses, what is preventing zoonosis events, especially since noroviruses are well known for genetic recombination (39)? Currently, the precise roles of HBGAs in infections are unknown for both RHDV and norovirus. However, it is speculated that other receptors/cofactors for virus infection of host cells are required, which could account for species specificity. Recently, it was shown that HBGAs or HBGA-expressing enteric bacteria enhanced human GII norovirus infection in B cells (24). Considering that RHDVb and GII norovirus interact similarly with HBGAs, it is possible that a comparable cell culture system also could work with RHDVb.
ACKNOWLEDGMENTS
The funding for this study was provided by the CHS foundation. K.P.D. was supported by grant AGL2013-48550-C2-1-R from the Spanish Ministerio de Economía y Competitividad, cofinanced by FEDER.
We acknowledge the European Synchrotron Radiation Facility (ID23-1 and BM30A) for the provision of synchrotron radiation facilities. We also thank Bishal Singh for assistance with structural refinements and Francisco Parra for helpful suggestions and phylogenetic analysis.
G.S.H. and M.M.L. designed the research; K.P.D. investigated and provided the RHDVb P domain sequence information; M.M.L. expressed and purified the RHDVb P domain; G.S.H. and M.M.L. finalized and analyzed the structures; and G.S.H. and M.M.L. wrote the manuscript, on which all authors commented.
REFERENCES
- 1.Liu SJ, Xue HP, Pu BQ, Qian NH. 1984. A new viral disease in rabbits. Anim Husbandry Vet Med 16:253–255. [Google Scholar]
- 2.Ohlinger VF, Haas B, Meyers G, Weiland F, Thiel HJ. 1990. Identification and characterization of the virus causing rabbit hemorrhagic disease. J Virol 64:3331–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parra F, Prieto M. 1990. Purification and characterization of a calicivirus as the causative agent of a lethal hemorrhagic disease in rabbits. J Virol 64:4013–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyers G, Wirblich C, Thiel HJ. 1991. Rabbit hemorrhagic disease virus–molecular cloning and nucleotide sequencing of a calicivirus genome. Virology 184:664–676. doi: 10.1016/0042-6822(91)90436-F. [DOI] [PubMed] [Google Scholar]
- 5.Abrantes J, van der Loo W, Le Pendu J, Esteves PJ. 2012. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res 43:12. doi: 10.1186/1297-9716-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arguello Villares JL. 1991. Viral haemorrhagic disease of rabbits: vaccination and immune response. Rev Sci Tech 10:459–480. [PubMed] [Google Scholar]
- 7.Le Gall-Recule G, Zwingelstein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, Decors A, Bertagnoli S, Guerin JL, Marchandeau S. 2011. Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet Rec 168:137–138. doi: 10.1136/vr.d697. [DOI] [PubMed] [Google Scholar]
- 8.Dalton KP, Nicieza I, Balseiro A, Muguerza MA, Rosell JM, Casais R, Alvarez AL, Parra F. 2012. Variant rabbit hemorrhagic disease virus in young rabbits, Spain. Emerg Infect Dis 18:2009–2012. doi: 10.3201/eid1812.120341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton KP, Nicieza I, Abrantes J, Esteves PJ, Parra F. 2014. Spread of new variant RHDV in domestic rabbits on the Iberian Peninsula. Vet Microbiol 169:67–73. doi: 10.1016/j.vetmic.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Meyers G, Wirblich C, Thiel HJ. 1991. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology 184:677–686. doi: 10.1016/0042-6822(91)90437-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibilia M, Boniotti MB, Angoscini P, Capucci L, Rossi C. 1995. Two independent pathways of expression lead to self-assembly of the rabbit hemorrhagic disease virus capsid protein. J Virol 69:5812–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Xu F, Liu J, Gao B, Liu Y, Zhai Y, Ma J, Zhang K, Baker TS, Schulten K, Zheng D, Pang H, Sun F. 2013. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog 9:e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinnear M, Linde CC. 2010. Capsid gene divergence in rabbit hemorrhagic disease virus. J Gen Virol 91:174–181. doi: 10.1099/vir.0.014076-0. [DOI] [PubMed] [Google Scholar]
- 14.Kroneman A, Vega E, Vennema H, Vinje J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. 2013. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol 158:2059–2068. doi: 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Gall-Recule G, Lavazza A, Marchandeau S, Bertagnoli S, Zwingelstein F, Cavadini P, Martinelli N, Lombardi G, Guerin JL, Lemaitre E, Decors A, Boucher S, Le Normand B, Capucci L. 2013. Emergence of a new lagovirus related to rabbit haemorrhagic disease virus. Vet Res 44:81. doi: 10.1186/1297-9716-44-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nystrom K, Le Gall-Recule G, Grassi P, Abrantes J, Ruvoen-Clouet N, Le Moullac-Vaidye B, Lopes AM, Esteves PJ, Strive T, Marchandeau S, Dell A, Haslam SM, Le Pendu J. 2011. Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog 7:e1002188. doi: 10.1371/journal.ppat.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Gall G, Arnauld C, Boilletot E, Morisse JP, Rasschaert D. 1998. Molecular epidemiology of rabbit haemorrhagic disease virus outbreaks in France during 1988 to 1995. J Gen Virol 79(Part 1):11–16. [DOI] [PubMed] [Google Scholar]
- 18.Le Gall-Recule G, Zwingelstein F, Laurent S, de Boisseson C, Portejoie Y, Rasschaert D. 2003. Phylogenetic analysis of rabbit haemorrhagic disease virus in France between 1993 and 2000, and the characterisation of RHDV antigenic variants. Arch Virol 148:65–81. doi: 10.1007/s00705-002-0908-1. [DOI] [PubMed] [Google Scholar]
- 19.Kerr PJ, Kitchen A, Holmes EC. 2009. Origin and phylodynamics of rabbit hemorrhagic disease virus. J Virol 83:12129–12138. doi: 10.1128/JVI.01523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capucci L, Fallacara F, Grazioli S, Lavazza A, Pacciarini ML, Brocchi E. 1998. A further step in the evolution of rabbit hemorrhagic disease virus: the appearance of the first consistent antigenic variant. Virus Res 58:115–126. doi: 10.1016/S0168-1702(98)00106-3. [DOI] [PubMed] [Google Scholar]
- 21.Puggioni G, Cavadini P, Maestrale C, Scivoli R, Botti G, Ligios C, Le Gall-Recule G, Lavazza A, Capucci L. 2013. The new French 2010 rabbit hemorrhagic disease virus causes an RHD-like disease in the Sardinian Cape hare (Lepus capensis mediterraneus). Vet Res 44:96. doi: 10.1186/1297-9716-44-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruvoen-Clouet N, Ganiere JP, Andre-Fontaine G, Blanchard D, Le Pendu J. 2000. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol 74:11950–11954. doi: 10.1128/JVI.74.24.11950-11954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutson AM, Atmar RL, Graham DY, Estes MK. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis 185:1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- 24.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansman GS, Biertumpfel C, Georgiev I, McLellan JS, Chen L, Zhou T, Katayama K, Kwong PD. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J Virol 85:6687–6701. doi: 10.1128/JVI.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26:795–800. doi: 10.1107/S0021889893005588. [DOI] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 31.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayukekbong JA, Fobisong C, Tah F, Lindh M, Nkuo-Akenji T, Bergstrom T. 2014. Pattern of circulation of norovirus GII strains during natural infection. J Clin Microbiol 52:4253–4259. doi: 10.1128/JCM.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bu W, Mamedova A, Tan M, Xia M, Jiang X, Hegde RS. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J Virol 82:5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rademacher C, Krishna NR, Palcic M, Parra F, Peters T. 2008. NMR experiments reveal the molecular basis of receptor recognition by a calicivirus. J Am Chem Soc 130:3669–3675. doi: 10.1021/ja710854r. [DOI] [PubMed] [Google Scholar]
- 37.Tan M, Xia M, Chen Y, Bu W, Hegde RS, Meller J, Li X, Jiang X. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. doi: 10.1371/journal.pone.0005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda N, Shimotake T, Ohama K. 2013. Duodenal perforation associated with norovirus and rotavirus gastroenteritis. Clin Case Rep 1:47–49. doi: 10.1002/ccr3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis 11:1079–1085. doi: 10.3201/eid1107.041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansman GS, Natori K, Shirato-Horikoshi H, Ogawa S, Oka T, Katayama K, Tanaka T, Miyoshi T, Sakae K, Kobayashi S, Shinohara M, Uchida K, Sakurai N, Shinozaki K, Okada M, Seto Y, Kamata K, Nagata N, Tanaka K, Miyamura T, Takeda N. 2006. Genetic and antigenic diversity among noroviruses. J Gen Virol 87:909–919. doi: 10.1099/vir.0.81532-0. [DOI] [PubMed] [Google Scholar]
- 41.Hansman GS, Doan LT, Kguyen TA, Okitsu S, Katayama K, Ogawa S, Natori K, Takeda N, Kato Y, Nishio O, Noda M, Ushijima H. 2004. Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch Virol 149:1673–1688. [DOI] [PubMed] [Google Scholar]