ABSTRACT
Neutralizing antibodies (NAbs) targeting glycoprotein E2 are important for the control of hepatitis C virus (HCV) infection. One conserved antigenic site (amino acids 412 to 423) is disordered in the reported E2 structure, but a synthetic peptide mimicking this site forms a β-hairpin in complex with three independent NAbs. Our structure of the same peptide in complex with NAb 3/11 demonstrates a strikingly different extended conformation. We also show that residues 412 to 423 are essential for virus entry but not for E2 folding. Together with the neutralizing capacity of the 3/11 Fab fragment, this indicates an unexpected structural flexibility within this epitope. NAbs 3/11 and AP33 (recognizing the extended and β-hairpin conformations, respectively) display similar neutralizing activities despite converse binding kinetics. Our results suggest that HCV utilizes conformational flexibility as an immune evasion strategy, contributing to the limited immunogenicity of this epitope in patients, similar to the conformational flexibility described for other enveloped and nonenveloped viruses.
IMPORTANCE Approximately 180 million people worldwide are infected with hepatitis C virus (HCV), and neutralizing antibodies play an important role in controlling the replication of this major human pathogen. We show here that one of the most conserved antigenic sites within the major glycoprotein E2 (amino acids 412 to 423), which is disordered in the recently reported crystal structure of an E2 core fragment, can adopt different conformations in the context of the infectious virus particle. Recombinant Fab fragments recognizing different conformations of this antigenic site have similar neutralization activities in spite of converse kinetic binding parameters. Of note, an antibody response targeting this antigenic region is less frequent than those targeting other more immunogenic regions in E2. Our results suggest that the observed conformational flexibility in this conserved antigenic region contributes to the evasion of the humoral host immune response, facilitating chronicity and the viral spread of HCV within an infected individual.
INTRODUCTION
An estimated 180 million people worldwide are infected by hepatitis C virus (HCV), and the majority of infected patients (70 to 80%) develop chronic infection that leads to progressive liver disease (1). Major advances in HCV therapy during the last decade resulted in combination therapies consisting of direct-acting antivirals (DAAs) with sustained virological response rates of >90% (reviewed in reference 2). Nevertheless, the lack of availability of this HCV therapy in developing countries illustrates the urgent need to design a safe and efficient HCV vaccine, a process that is hampered by our limited understanding of the key epitopes inducing a protective neutralizing immune response.
The majority of neutralizing antibodies (NAbs) identified to date target the major envelope glycoprotein E2 (reviewed in reference 3), which binds the cellular receptors CD81 and scavenger receptor BI (SR-BI) (4, 5). The glycoprotein contains hypervariable regions (HVRs), termed HVR1, HVR2, and igVR (intergenotypic variable region) (6, 7), the deletion of which does not affect the overall glycoprotein conformation. The structurally flexible HVR1 located at the N terminus of E2 (8) is dispensable for virus infectivity in chimpanzees (9). Recent structural studies have shown that HCV E2 has a core fragment with an Ig superfamily fold flanked by a front layer and a back layer containing β-sheets, random coils, and short α-helices (10, 11). Of note, both structures were obtained by using an E2 fragment lacking HVR1; thus, an interaction of HVR1 with the E2 core cannot be excluded.
The neutralizing antibody AR3C binds to a large part of the front layer (amino acids [aa] 426 to 446) and residues within the CD81 binding loop (aa 528 to 531). Further insights into the recognition of E2 neutralizing epitopes came from structural studies that cocrystallized Fab fragments derived from anti-E2 NAbs recognizing two regions comprising residues 430 to 446 and 412 to 423, respectively, in complex with their respective epitope peptides (12–18). In these complexes, the peptide comprising aa 430 to 446 adopts a short α-helical conformation, with extended segments in either direction (12, 16), and was proposed to adopt two discrete conformations in the context of the viral particle based on these peptide structures (11, 13). The segment comprising aa 412 to 423 adopts a β-hairpin in complex with three independent broadly neutralizing antibodies (HCV1, AP33, and Hu5B3.v3), suggesting a flexible flap-like structure (14, 15, 17, 18). The antigenic site spanning aa 412 to 423 contains highly conserved epitopes targeted by monoclonal antibodies (MAbs) neutralizing HCV strains of all major genotypes (19–23) and is positioned downstream of HVR1. All described epitopes include a tryptophan residue at position 420 that plays a critical role in CD81 recognition (24); nonetheless, a surprisingly weak immune response against this antigenic site was reported for infected patients (22, 25).
Here we report the crystal structure of the epitope comprising aa 412 to 423 in complex with the neutralizing anti-HCV E2 antibody 3/11, unexpectedly revealing an extended peptide conformation that is strikingly different from the previously reported β-hairpin. We demonstrate that the segment spanning aa 412 to 423 is not required for the native overall fold of a soluble E2 (sE2) ectodomain but is essential for the infectivity of HCV pseudoparticles (HCVpp) and cell culture-derived HCV (HCVcc). A comparative functional analysis of Fab fragments derived from NAbs 3/11 and AP33, exemplary for NAbs recognizing the β-hairpin, reveals similar neutralization activities for both NAbs and Fab fragments in spite of strikingly different binding kinetics. In combination with the neutralizing activity of MAb 3/11, our results illustrate the structural flexibility of this region within HCV E2 and provide novel insights into the recognition of conserved neutralizing epitopes, with implications for vaccine design.
MATERIALS AND METHODS
Production and purification of recombinant Fabs and glycoproteins.
Synthetic genes of the Fab coding regions of MAbs 3/11 and AP33 were cloned into a Drosophila melanogaster S2 Fab expression vector described previously (26). The HCV E2 full-length ectodomain (sE2), the ectodomain lacking hypervariable region 1 (sE2 ΔHVR1) (amino acids 412 to 715 of the HCV polyprotein), and the ectodomain lacking the N-terminal 42 residues (sE2426–717) from strain UKN2B2.8 (GenBank accession number AY734983) were expressed in Drosophila S2 cells as previously described (27, 28). Briefly, Drosophila S2 cells were transfected as reported previously (63), amplified, and induced with 4 μM CdCl2 at a density of ∼7 × 106 cells/ml for 8 days for large-scale production. Proteins were purified from the supernatant by affinity chromatography using a StrepTactin Superflow column followed by size exclusion chromatography (SEC) using a Superdex200 column. Pure monomeric proteins were concentrated to ∼20 mg/ml.
Neutralizing capacity of recombinant Fab fragments and infectivity of HCVcc and HCVpp Δaa384–425 deletion mutants.
HCVcc of H77/JFH-1 and J6/JFH-1 were produced as previously described (29). A mutant HCV J6/JFH1 clone lacking HVR1 (aa 384 to 410) was generated by using plasmid pFL-J6/JFH1 as the template (29) and the Q5 site-directed mutagenesis kit (NEB) (primers are available upon request). Huh7.5 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–10% fetal calf serum (FCS) and harvested at 24, 48, and 72 h following transfections. NS5A expression, as an indirect indicator for replication/translation, was assessed by immunostaining of electroporated cells using MAb 9E10. Supernatants were then used to infect naive cells, and infectivity was assessed by NS5A immunostaining after 48 h.
Neutralization assays were performed by mixing 100 focus-forming units (FFU) of HCVcc in the absence or presence of the indicated concentrations of antibodies for 1 h before addition to Huh7.5 cells (a kind gift from C. Rice, Rockefeller University) and incubation for 72 h at 37°C. Cells were then fixed and stained for NS5A, and the number of infected foci was determined by counting immunofluorescence (IF)-positive cells. The number of infected cells was compared to untreated samples, or to samples in the presence of control IgG, to calculate the percentage of inhibition.
RNA transcripts were produced and electroporated into Huh7.5 cells as previously described (29). A plasmid encoding the E1/E2 primary isolate UKN1A20.8 (GenBank accession number EU155192) was used as the template for the production of HCV pseudoparticles containing wild-type (wt) or ΔHVR1 E2, as previously described (30). Infectivity in Huh7 cells was determined after 48 h by reading the luciferase activity in lysed cells (Promega).
Peptides and complex formation.
A synthetic peptide comprising residues 412 to 423 (QLINTNGSWHVN) of genotype 1a strain Glasgow (GenBank accession number AY885238), which differs from the H77 reference sequence by a V422I substitution (31), was synthesized by the American Peptide Company (>98% purity) and dissolved in water plus 5% dimethyl sulfoxide (DMSO) at 10 mg/ml. A complex containing 1.5 mg/ml peptide plus 9 mg/ml Fab was formed overnight at 293 K.
Crystallization, data collection, structure determination, and refinement.
Complex crystals were grown at 293 K by using the hanging-drop vapor diffusion method with drops containing 1 μl complex solution (10.6 mg/ml in 10 mM Tris [pH 8.0], 100 mM NaCl) mixed with 1 μl reservoir solution. This reservoir solution contained 100 mM Tris (pH 7.5), 27% polyethylene glycol 4000 (PEG 4000), and 100 mM Na-acetate or 100 mM Tris (pH 8.5) and 66% 2-methyl-2,4-pentanediol (MPD) for crystals in space group P1 or P21, respectively. Diffraction quality crystals appeared after 1 week and were flash-frozen in mother liquor with (space group P1) or without (space group P21) 22% glycerol. Space groups and cell dimensions of the crystals, the number of complexes per asymmetric unit, resolution limits, data collection details, and refinement statistics are summarized in Table 2.
TABLE 2.
Data collection and refinement statistics
| Parameter | Value for complexa |
|
|---|---|---|
| Fab 3/11 + peptide reported under PDB accession no. 4WHY | Fab 3/11 + peptide reported under PDB accession no. 4WHT | |
| Data collection statistics | ||
| Space group | P21 | P1 |
| No. of complexes per AUb | 4 | 12 |
| Cell dimensions | ||
| a, b, c (Å) | 64.76, 205.51, 69.02 | 64.79, 128.23, 163.63 |
| α, β, γ (°) | 90, 103.18, 90 | 88.79, 94.36, 96.15 |
| Resolution range (Å) | 50.00–2.62 (2.76–2.62) | 48.74–2.22 (2.34–2.22) |
| Rmerge | 0.068 (0.444) | 0.072 (0.383) |
| I/σI | 9.9 (1.6) | 9.0 (1.8) |
| Completeness (%) | 97.0 (87.5) | 95.4 (81.4) |
| Redundancy | 3.2 (2.1) | 2.4 (1.8) |
| Refinement statistics | ||
| Resolution range (Å) | 47.97–2.62 | 32.50–2.22 |
| No. of reflections | 50,781 | 246,026 |
| Rwork/Rfree | 0.204/0.258 | 0.209/0.243 |
| No. of atoms | ||
| Protein | 13,022 | 39,256 |
| Ligand | ||
| Water | 156 | 850 |
| No. of residues per AU | 1,710 | 5,169 |
| Mean temp factors (B factors) | ||
| Protein | 59.61 | 41.72 |
| Ramachandran statistics (%) | ||
| Favored | 96.4 | 97.2 |
| Allowed | 3.0 | 2.5 |
| Outliers | 0.6 | 0.3 |
| RMSD | ||
| Bond length (Å) | 0.01 | 0.01 |
| Bond angles (°) | 1.25 | 1.23 |
Values in parentheses correspond to the highest-resolution shell.
AU, asymmetric unit.
Data were collected on the beamline Proxima-1 at Synchrotron Soleil, processed, scaled, and reduced by using XDS (32), Pointless (33), and programs from the CCP4 suite (34). The crystal structures of the Fab complexes were determined by the molecular replacement method using Phaser (35). We used separate variable and constant regions of a hypothetical Fab fragment assembled from the best sequence match in the Protein Data Bank (PDB), the light chain (LC) reported under PDB accession number 1NLD and the heavy chain (HC) reported under PDB accession number 3EOT, as a search model for the P21 crystals and the refined P21 structure as a search model for the P1 crystal form. Model building was performed by using Coot (36), and refinement was done by using AutoBuster (37).
Crystal structure analysis.
The two peptides derived from the different crystal forms were superposed by using an iterative alignment process pruning long atom pairs until no pair exceeded 0.5 Å, implemented in the MatchMaker algorithm of Chimera (38) and the MultiProt server (39). Root mean square deviations (RMSDs) between the peptides derived from different crystal forms of the 3/11 complex were calculated either over all atoms per residue or by taking into account only the main-chain atoms (N, CA, C, and O), using Chimera.
Buried solvent-accessible surface areas for the interfaces as well as for individual residues within the peptides were calculated by using the PISA server (40). Shape complementarity was calculated by using programs of the CCP4 suite (34). Interactions were determined by using the Protein Interactions Calculator (PIC) (41). Figures were prepared with Pymol (http://www.pymol.org/).
Surface plasmon resonance analysis.
Real-time surface plasmon resonance (SPR) assays were performed by using a Biacore 2000 instrument (GE Healthcare) equilibrated at 25°C in phosphate-buffered saline (PBS) supplemented with 0.1 mg/ml bovine serum albumin (BSA). To determine the affinity of the peptide for Fabs 3/11 and AP33, these Fab fragments (in 10 mM acetate [pH 4.5]) were covalently coupled to the carboxy methyl moieties of CM5 sensorchips by using a Biacore 2000 instrument and the Amine Coupling kit (GE Healthcare), achieving immobilization density (Rimmo) values of 3,000 to 5,000 resonance units (RU) (1 RU equals 1 pg mm−2). We used Fab e137 (40) as a negative control, because it recognizes an unrelated conformational epitope within HCV E2. A triplicate series of 10 concentrations of peptides (1 to 2,000 nM) was injected over the Fab surfaces and an empty reference flow cell for 5 min at a flow rate of 50 μl · min−1. After monitoring dissociation for 10 min, the surfaces were regenerated by a 30-s wash with 0.1% SDS. To determine the affinity of native and denatured glycoproteins for the Fab fragments, monoclonal antibody C23.21 directed against Strep-Tag (in 10 mM acetate [pH 5.5]) was covalently coupled to all 4 flow cells of CM5 sensorchips, achieving an Rimmo of 7,500 to 9,000 RU. Subsequently, full-length or truncated (native or denatured) HCV sE2 of genotype 1a strain H77 was captured via its Strep-Tag to a density of 300 to 400 RU. A series of 10 concentrations of Fabs 3/11 and AP33 (1.5 to 1,500 nM) was injected over the HCV sE2 surfaces and an unliganded C23.21 reference flow cell for 8 min at a flow rate of 30 μl · min−1. After monitoring dissociation for 5 min, the surfaces were regenerated by two 1-min washes with 10 mM glycine-HCl (pH 2.0) and one wash with 0.1% SDS.
The real-time interaction profiles were double referenced by using Scrubber 2.0 software (BioLogic Software), i.e., the signals from both reference surfaces and blank experiments using PBS-BSA instead of peptide or Fab. The association rate (kon), dissociation rate (koff), and the equilibrium dissociation constant (Kd) were determined by using BIAevaluation 4.1 software (GE Healthcare), by globally fitting the processed experimental curves to a 1:1 Langmuir model. To facilitate direct comparisons of the kinetics of binding to different Fab fragments, the binding response was normalized and expressed as the occupancy of binding sites. The latter was determined by dividing the experimental SPR signals measured on each surface by the corresponding maximal binding capacity (Rmax).
Pulldown assays.
To analyze the conformation of sE2 in complex with Fab 3/11, UKN2b2.8 sE2 ΔHVR1 was bound to a StrepTactin Superflow minicolumn, followed by an initial washing step. Subsequently, a 3-fold molar excess of Fab 3/11 was added, followed by a second washing step and, afterwards, the addition of a 3-fold molar excess of CBH-4D antibody (collection of human material, Institut Pasteur no. DC-2010-1197/7). After extensive washing, the complex was eluted and analyzed by SDS-PAGE and Coomassie blue staining.
To determine the interaction of sE2426–717 with conformation-sensitive antibodies and its receptor CD81, sE2426–717 was bound to the column, followed by an extensive wash step. Subsequently, a 3-fold molar excess of the large extracellular loop of human CD81 (CD81-LEL) (produced as described previously [5]), a Fab fragment (e137) (42), or MAb CBH-4D (43) directed against HCV E2 was added. After extensive washing, the complex was eluted and analyzed by SDS-PAGE followed by Coomassie blue staining.
Protein structure accession numbers.
The atomic coordinates and structure factors for two crystal structures were deposited in the Protein Data Bank (http://www.pdb.org/) under accession numbers 4WHT and 4WHY.
RESULTS AND DISCUSSION
Crystallization of the Fab-peptide complex and structure determination.
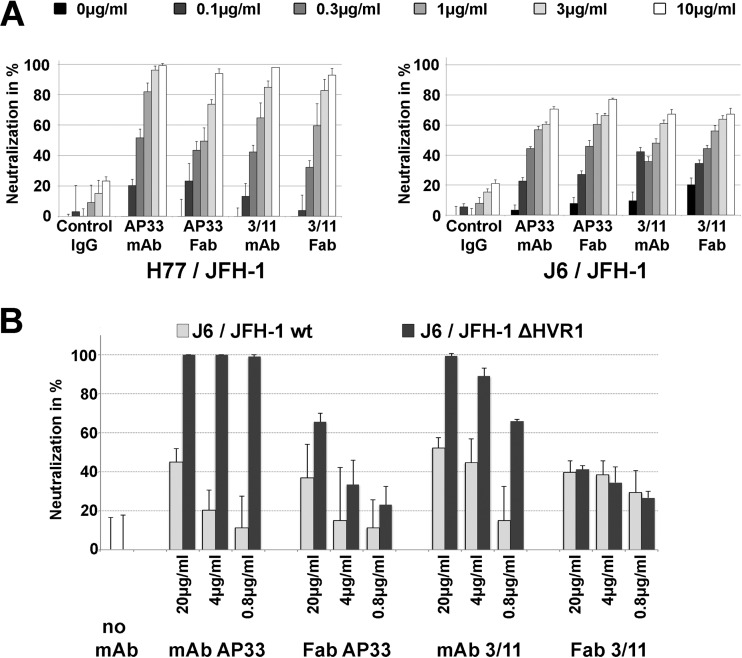
We expressed the Fab fragments derived from MAbs 3/11 and AP33 (the latter one was used as the control throughout this study) in Drosophila S2 cells. Both Fab fragments were able to neutralize cell culture-derived HCV (HCVcc) harboring HCV envelope glycoproteins of genotypes 1a (strain H77) and 2a (strain J6) in a dose-dependent manner albeit slightly less efficiently than the parental antibodies, reflecting the reduced avidity of the Fab fragments (Fig. 1 and Table 1). We performed cocrystallization trials for a complex containing Fab 3/11 and a peptide comprising residues 412 to 423 (QLINTNGSWHVN) of genotype 1a strain Glasgow and obtained diffraction quality crystals in two different space groups (P1 and P21, respectively) (Table 2). The peptide sequence used in this study was chosen because it was previously used for cocrystallization with Fab AP33 (18). The structure of the Fab 3/11-peptide complex was determined by the molecular replacement method (Fig. 2A), using the variable and constant regions of unrelated Fab fragments as separate search models (see Materials and Methods) in the P21 crystal form. Difference maps calculated after refinement of the recombinant Fab molecules revealed well-defined electron density for the peptide, which allowed manual building of an atomic model for the peptide (Fig. 3A). The resulting structure of the Fab 3/11-peptide complex is displayed in Fig. 2A, with Fab 3/11 being almost planar, with an elbow angle of 171.7°. The peptide conformation is identical in all complexes in the asymmetric unit (4 and 12 complexes in space groups P21 and P1, respectively). For further analysis, the peptide comprising all residues with the lowest mean B value after crystallographic refinement, indicating the highest degree of order in this part of the structure, from the higher-resolution structure (space group P1) was selected.
FIG 1.
Neutralization activity of MAbs and Fabs 3/11 and AP33. HCVcc (genotype 1a [H77/JFH] and genotype 2a [J6/JFH {A} or J6/JFH lacking hypervariable region 1 {J6/JFH ΔHVR1} {B}]) was preincubated with increasing antibody concentrations for 1 h and then used to infect Huh7.5 cells. Cultures were incubated for 72 h and then methanol fixed, and infection was enumerated by counting NS5A-positive foci by immunofluorescence. Results are presented as means and standard deviations of data from duplicate (A) or triplicate (B) experiments.
TABLE 1.
IC50 values for Fabs and MAbs 3/11 and AP33a
| Antibody | IC50 (μg/ml) (IC50 [nM]) for strain |
|
|---|---|---|
| H77 | J6 | |
| Fab 3/11 | 0.43 (8.48) | 2.57 (50.65) |
| MAb 3/11 | 0.39 (2.6) | 1.89 (12.6) |
| Fab AP33 | 0.57 (10.98) | 0.83 (15.98) |
| MAb AP33 | 0.23 (1.53) | 1.05 (7) |
| Control IgG | 61.76 (411.73) | 107.3 (715.33) |
IC50, 50% inhibitory concentration.
FIG 2.
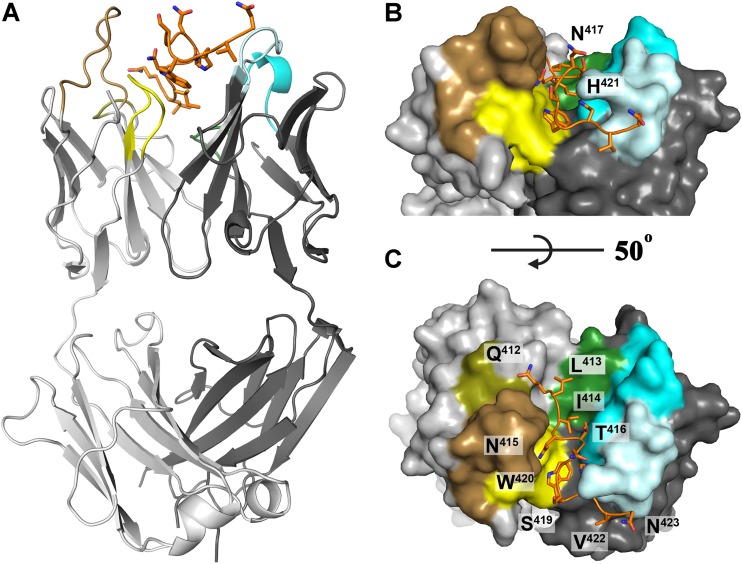
Crystal structure of Fab 3/11 in complex with its peptide epitope. (A) The crystal structure of the Fab 3/11-peptide complex was determined and refined to a 2.2-Å resolution (shown as a cartoon). The peptide (orange) adopts an extended conformation and interacts with the Fab mainly in the cleft between the heavy (dark gray) and light (light gray) chains. Complementarity-determining regions 1, 2, and 3 are shown in cyan, light cyan, and dark green, respectively, for the heavy chain and in sand, olive, and yellow, respectively, for the light chain. The peptide N terminus interacts mostly with the light chain, and its C-terminal part makes a right-angle turn around complementarity-determining region 2 of the heavy chain (CDR-H2). C-terminal residues H421, V422, and N423 interact exclusively with the heavy chain. (B and C) View on the paratope of the Fab 3/11-peptide complex from two angles illustrating how deeply the peptide immerses into the cleft between the heavy and light chains. The molecular surface, the peptide, and the complementarity-determining regions of the Fab fragment are colored as described above for panel A.
FIG 3.
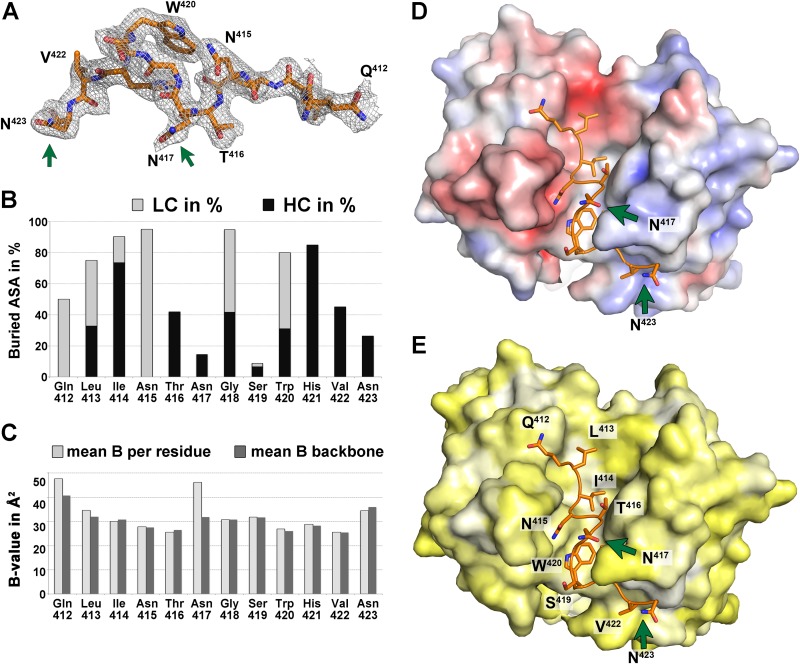
Analysis of the peptide comprising aa 412 to 423. (A) Electron density of a composite omit map of the Fab 3/11-peptide complex contoured at 1 σ, allowing for manual building of the peptide model. The peptide is shown as sticks and is colored by atom type (orange, red, and blue for carbon, oxygen, and nitrogen, respectively). Green arrows indicate asparagine residues N417 and N423 that carry N-linked glycans in the infectious virus particle. (B) Percentages of accessible surface area (ASA) buried in the complex, calculated by using PISA (40), represented per residue as stacked columns for heavy (dark gray) and light (light gray) chains of Fab 3/11. (C) Average temperature factor values after crystallographic refinement of the peptide plotted per residue (light gray) and taking into account only backbone atoms (dark gray) illustrate that only the terminal peptide residues are less ordered, suggesting that they contribute less to antigen binding. In addition, the side chain of N417, the only really exposed residue, displays a higher temperature factor value. (D and E) The paratope of Fab 3/11 is shown as a molecular surface, and its peptide epitope is shown as sticks, colored as described above for panel A. The molecular surface of the paratope is colored according to its electrostatic potential (−5 kT/e [red] to 5 kT/e [blue]) across the molecular surface of the paratope, calculated by using the adaptive Poisson-Boltzmann solver (D) or according to a normalized hydrophobicity scale from white (hydrophobic) to bright yellow (hydrophilic) (E).
Molecular determinants of the Fab 3/11 interaction with its peptide epitope.
The peptide corresponding to residues 412 to 423 binds to Fab 3/11 in an extended conformation that is deeply immersed within the cleft between the heavy and light chains (Fig. 2B and C). Clear electron density was observed for all peptide residues, indicating a highly ordered interface, with a surface area buried by antigen binding of 871.3 Å2 on the 3/11 paratope (492.3 and 379.0 Å2 on the heavy chain [HC] and light chain [LC], respectively) and a shape complementarity index of 0.78. The N-terminal part of the peptide interacts mostly with the light chain, and its C-terminal part makes a right-angle turn around complementarity-determining region 2 of the heavy chain (CDR-H2). The central part of the peptide comprising residues N415 to S419 bulges out, resulting in the side chains of N417 and S419 being exposed (Fig. 2C). Because the side chain of N423 is also exposed, this overall peptide conformation is in line with the fact that N417 and N423 represent N-linked glycosylation sites (44), which, in the context of the infectious virus particle, need to accommodate the two N-linked glycans and therefore may not be involved in the antibody binding face of the epitope (Fig. 4A).
FIG 4.
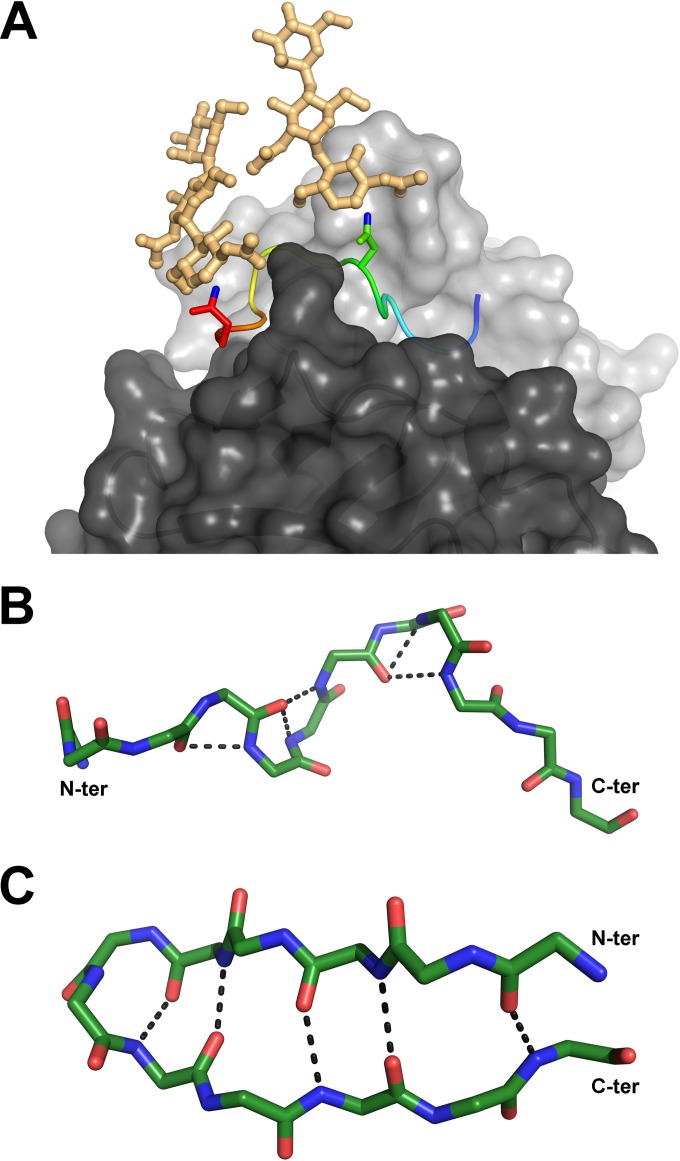
Relevant conformations of the antigenic site spanning aa 412 to 423. (A) Compatibility of the Fab 3/11-peptide complex structure with the position of N-linked glycans attached to N417 and N423 of the native glycoprotein. Shown is a cartoon representation of the peptide, ramp colored from blue to red through yellow, from the N to the C termini, with the two asparagine side chains shown as sticks and the ND2 atoms, to which the sugar chains are linked, shown in blue. Hypothetical glycan chains containing two N-acetylglucosamine moieties and one mannose moiety (light orange) are modeled to visualize the extended conformation of the native glycoprotein required for 3/11 binding. (B and C) Comparison of both peptide conformations observed for aa 412 to 423. The backbone atoms of the peptides bound to Fab 3/11 (B) and Fab HCV1 (C) (15), as an example of the β-hairpin conformation of aa 412 to 423, are shown as sticks and colored by atom type (green, red, and blue for carbon, oxygen, and nitrogen, respectively) to illustrate the differences in the backbone conformations observed for the two structures. The five hydrogen bonds stabilizing the backbone conformation in both structures are indicated as dotted lines.
Analysis of the surface area per residue that is buried by Fab binding confirmed that Q412, L413, W420, and, in particular, N415 are mostly in contact with the light chain, whereas I414, T416, H421, and V422 interact predominantly with the heavy chain (Fig. 3B). Analysis of the mean temperature factors (B-factors) per residue (calculated over the main chain and over all atoms) suggested a higher degree of disorder at both ends of the peptide (Fig. 3C). The high B-factor value calculated over all atoms of N417 compared to its main-chain B-factor value suggests a greater flexibility of the side chain, in agreement with its role as an attachment site for an N-linked glycan.
A number of cell culture-adaptive mutations or antibody escape mutants have been identified within this antigenic site, including the N415Y (selected with MAb AP33), N415D, T416A, N417S, G418D, and I422L neutralization-resistant escape mutants. Of these mutations, N415D and N417S severely impaired 3/11 binding (45). For the N415D mutation, this is likely due to an electrostatic repulsion between the negatively charged side chains of the aspartic acid and E31 within the CDR-L1 (EL31). Given that 95% of the solvent-accessible surface of N415 is buried, the interface between 3/11 and E2 cannot accommodate bulky amino acids larger than asparagine at this position, explaining why the N415Y mutation abolishes virus neutralization by 3/11 (46). The N417S mutation shifts the N-linked glycosylation site from N417 to N415 (17), where the glycan chain cannot be accommodated within the interface. These results indicate an antibody-antigen complex interface that is dominated by residues N415, W420, and H421, in line with data from our previously reported alanine scanning mutagenesis analysis (20), supporting the notion that our structure reflects the native antibody-antigen interaction.
Conformation of the E2 peptide comprising aa 412 to 423.
In spite of the extended conformation, the bulge at the peptide center results in the same overall number of backbone hydrogen bonds found in the β-hairpin; however, no similarities between hydrogen bonding networks or the corresponding peptide conformations were observed (Fig. 3). In solution, the peptide is likely to adopt several different conformations that are in equilibrium, and different antibodies bind to it according to the principle of conformational selection. All four antibodies for which the peptide structure has been reported (HCV1, AP33, Hu5B3.v3, and 3/11) neutralize HCV infection (i.e., they bind to their respective epitopes at the surface of infectious virions), suggesting that the peptide spanning aa 412 to 423 adopts different conformations. Our structure therefore illustrates that the previously reported structural flexibility of HVR1 (8) extends to the downstream neutralizing antigenic site (aa 412 to 423), in line with the recently reported increased deuterium exchange rate in the context of a soluble E2 ectodomain (10). The fact that all four antibodies neutralize HCV infection by inhibiting the E2-CD81 interaction (19, 21, 47) suggests that both conformations can be accessible at the virus surface, likely in a dynamic equilibrium that can be shifted into either direction by antibody binding. The dose-dependent neutralization of both MAbs 3/11 and AP33 (>98% of HCVcc H77) (Fig. 1A) supports this hypothesis. Our results suggest that any of the four antibodies binding to aa 412 to 423 at the virus surface will contribute to neutralization by shifting the equilibrium into the respective direction.
One possible explanation for the observed difference in peptide conformations is that the epitope structure depends on the polypeptide sequence up- and/or downstream of the antigenic site, implying that different strains/isolates present the epitope in different conformations. Evidence for such a strain-specific modulation of neutralization profiles was reported for a group of broadly neutralizing human MAbs targeting aa 412 to 423 (HC33 antibodies) in spite of an identical amino acid sequence within this site (22). It should be noted, however, that NAbs targeting this antigenic site neutralize a broad range of HCV genotypes independent of the recognized conformation (19, 20), suggesting that the isolate-specific amino acid sequences are unlikely to determine the conformation of the antigenic site spanning aa 412 to 423 but rather modulate the neutralization efficiency by minor changes in epitope presentation.
Another possible explanation could be a conformational change that E2, in particular the antigenic site spanning aa 412 to 423, undergoes during virus entry, suggesting that the two conformations may represent snapshots of the same region at different functional stages of the entry process. All four antibodies (HCV1, AP33, Hu5B3.v3, and 3/11) neutralize HCV infection by interfering with CD81 binding (19, 21, 47), suggesting that they all neutralize HCV upstream of the E2-CD81 interaction. However, a more detailed interpretation will require further experiments to analyze MAb interactions with infectious virus particles in a quantitative and time-resolved manner (e.g., by precipitation).
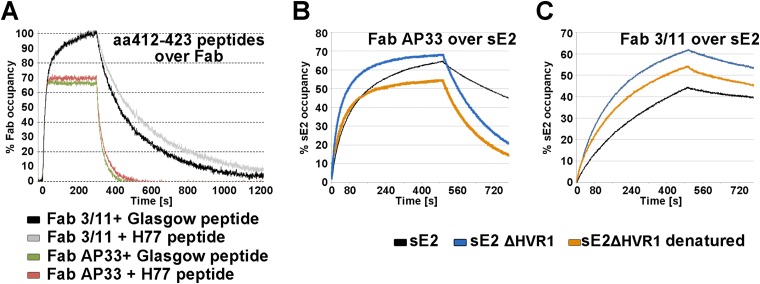
Affinity of Fabs 3/11 and AP33 for their epitope peptide and native soluble E2.
To further investigate the role of the distinct epitope conformations in antigen binding, we determined the equilibrium dissociation constants (Kd) as well as association (kon) and dissociation (koff) rates for the binding of Fabs 3/11 and AP33 to the monomeric full-length E2 ectodomain (wt sE2) and epitope peptides by surface plasmon resonance (SPR). SPR analysis was performed by using the crystallized peptide as well as the corresponding peptide derived from the H77 reference strain to allow direct comparison of peptide and sE2 binding kinetics and revealed no major differences in binding kinetics between the two peptides, in line with the facts that the V422I substitution is the only difference between the two peptides and that I422 is not a major determinant of the Fab binding interface. The affinity of Fab 3/11 for the H77 peptide was ∼10-fold higher than that for sE2 of strain H77 (Kd values of 6.5 nM and 65 nM for the peptide and sE2, respectively) (Table 3 and Fig. 5), suggesting that the extended conformation observed in our structure can be adopted more easily by the peptide than by sE2, likely due to the presence of the surrounding N- and C-terminal polypeptide chains. Similar values for glycoprotein-Fab affinity were observed with inversed SPR settings with the Fab fragment covalently coupled to the surface (data not shown), confirming the obtained kinetic parameters. The relatively slow association and high stability of the complex are in line with the peptide being deeply immersed into the cleft between the heavy and light chains. Fab AP33 bound the peptide with a lower affinity than did Fab 3/11 (Kd, 50.9 nM) but bound wt sE2 with an ∼2-fold-higher affinity (Kd, 38 nM). The binding was in both cases characterized by faster association and dissociation than for Fab 3/11 (Table 3). A closer look at the kinetic parameters revealed that Fab 3/11 dissociates more slowly from its antigen than does Fab AP33, whereas Fab AP33 associates with its antigen faster than does Fab 3/11 (Fig. 5B and C). One possible explanation for these differences in kinetic parameters could be a high prevalence of the β-hairpin, compared to the low prevalence of the extended conformation, although quantitative binding experiments using Fab and infectious virus particles are necessary to support this hypothesis. These differences in binding kinetics may explain the apparent contradiction in previous affinity comparisons of the two antibodies obtained by an enzyme-linked immunosorbent assay (ELISA)-based method using IgG (20), which likely detected a higher kon of AP33 than of 3/11 but failed to account for koff values. Alternatively, the bivalency of the antibody could compensate for the lower stability of the AP33 complex, whereas it does not affect the lower association rate of 3/11. In summary, we observed similar overall affinities for the binding of both Fabs to wt sE2, but kinetic binding analysis revealed marked differences in association and dissociation rates.
TABLE 3.
Kinetic parameters of Fab 3/11 and AP33 binding to peptide and sE2
| Affinity | kon (M−1 · s−1) ± SE | koff (s−1) ± SE | Kd (nM) (±SE) |
|---|---|---|---|
| “Glasgow” peptide over: | |||
| Fab AP33a,b | 5.9 × 105 ± 0.5 × 105 | 3.1 × 10−2 ± 0.2 × 10−2 | 55.3 (±3.6) |
| Fab 3/11a,b | 4.6 × 105 ± 0.4 × 105 | 3.6 × 10−3 ± 0.3 × 10−3 | 7.9 (±0.7) |
| “H77” peptide over: | |||
| Fab AP33a,b | 5.1 × 105 ± 0.3 × 105 | 2.5 × 10−2 ± 0.1 × 10−2 | 50.9 (±3.3) |
| Fab 3/11a,b | 3.9 × 105 ± 0.2 × 105 | 2.6 × 10−3 ± 0.2 × 10−3 | 6.5 (±0.4) |
| Fab 3/11 over: | |||
| sE2b | 4.1 × 103 | 2.7 × 10−4 | 65 |
| sE2 ΔHVR1b | 15 × 103 | 4.1 × 10−4 | 26 |
| Denatured sE2 ΔHVR1b | 11 × 103 | 5.0 × 10−4 | 44 |
| Fab AP33 over: | |||
| sE2b | 35 × 103 | 13 × 10−4 | 38 |
| sE2 ΔHVR1b | 45 × 103 | 43 × 10−4 | 97 |
| Denatured sE2 ΔHVR1b | 29 × 103 | 57 × 10−4 | 196 |
Fab affinities for the peptide were determined in three independent experiments and are shown as mean values with standard errors.
koff values were determined by globally fitting dissociation profiles alone, while kon and Kd values were determined by globally fitting association and dissociation profiles simultaneously.
FIG 5.
SPR analysis of Fab binding to aa 412 to 423. To facilitate direct comparisons of the kinetics of binding to different Fab fragments, the binding response was normalized in each case, as described in Materials and Methods, and expressed as the occupancy of binding sites. (A) Real-time SPR analysis of the interaction of the epitope peptides derived from genotype 1a strain Glasgow and H77 with immobilized Fab fragments. The respective epitope peptide was injected over a surface of covalently immobilized Fab at a flow rate of 50 μl/ml, and the binding response in resonance units (RU) was recorded as a function of time. To compensate for the small signal (due to the low molecular mass of the peptide ligand of ∼2 kDa), three independent experiments were performed, and the mean values with standard errors are presented in Table 3. The representative association/dissociation time course profile shown corresponds to an injection of 125 nM peptide over each of the Fabs, illustrating the different kinetic properties of the two Fab-peptide complexes. (B and C) Real-time SPR analysis of the binding of Fabs AP33 (B) and 3/11 (C) to the immobilized HCV E2 ectodomain. Fabs were injected over HCV sE2 immobilized by using an anti-Strep-Tag antibody at a flow rate of 30 μl/ml, and the binding response in RU was recorded as a function of time. Representative association/dissociation time course profiles correspond to the injection of Fabs (355 nM) over full-length immobilized HCV sE2 or native and denatured HCV sE2 ΔHVR1.
The kinetic binding parameters together with the ∼2-fold-higher neutralization potency of MAb AP33 (Fig. 1A and Table 1) suggest that the latter is determined mainly by the higher on-rate. A similar relationship between kinetic binding parameters and biological activity was reported for affinity-maturated Fab molecules against envelope protein F of respiratory syncytial virus (RSV), where kon was the crucial kinetic parameter that determined the biological activity of the IgG molecules, whereas slow dissociation increased the overall affinity but did not improve the neutralizing capacity (48, 49). For other antibodies, a direct link between a lower koff rate and improved biological activity has been established (50–52), but the correlation is not necessarily linear (50), and in some cases, only specificity determines the biological activity, in spite of a low binding affinity (53).
An important role in shielding neutralizing epitopes, in particular of the CD81 binding site, has been attributed to HVR1 (54, 55). To analyze the contribution of this region to Fab binding and neutralization, we first determined the kinetic parameters for the interaction between Fabs 3/11 and AP33 and an E2 ectodomain lacking HVR1 (encoding H77 aa 412 to 717) (sE2 ΔHVR1). Fab 3/11 bound to the mutant with an affinity similar to that for the peptide (Kd, 26 nM) and with a 4-fold-higher association rate than that for wt sE2, suggesting that the presence of HVR1 restricts Fab 3/11 binding to wt sE2. In contrast, Fab AP33 bound sE2 ΔHVR1 with a 2.5-fold-lower affinity than that for the wt glycoprotein due to a higher dissociation rate (Table 3 and Fig. 5B and C). These results suggest that the presence of HVR1, while hampering Fab 3/11 binding to wt sE2, contributed to the stability of the AP33-glycoprotein complex. This could be due to additional contact residues or a direct stabilizing effect of HVR1 on the β-hairpin.
MAbs and patient sera targeting the CD81 binding site within E2 neutralize ΔHVR1 HCVcc more efficiently than they neutralize wt virus (54). Surprisingly, the neutralizing activities of Fab 3/11, but not that of MAb 3/11, were almost identical for both wt and ΔHVR1 viruses (Fig. 1B), despite the faster association with sE2 ΔHVR1. Similarly, while both Fab AP33 and MAb AP33 neutralized ΔHVR1 virus more efficiently than they neutralized wt virus (Fig. 1B), this difference was more pronounced for MAb AP33 (∼9-fold) than for the Fab fragment (∼2-fold). These results suggest that, independent of the kinetic binding parameters, bivalent IgG molecules neutralize ΔHVR1 virus more efficiently than do the corresponding monovalent Fab molecules, whereas for wt virus, this appears to depend on the particular NAb. Of note, corresponding IgG and Fab molecules do not necessarily utilize identical neutralization mechanisms (56). In summary, our results support a model in which the removal of HVR1 results in increased exposure of the CD81 binding site, an effect that is likely to be more pronounced for larger MAbs (molecular mass of ∼150 kDa) than for smaller Fab molecules (molecular mass of ∼50 kDa).
Requirement of aa 412 to 423 for protein folding and infectivity.
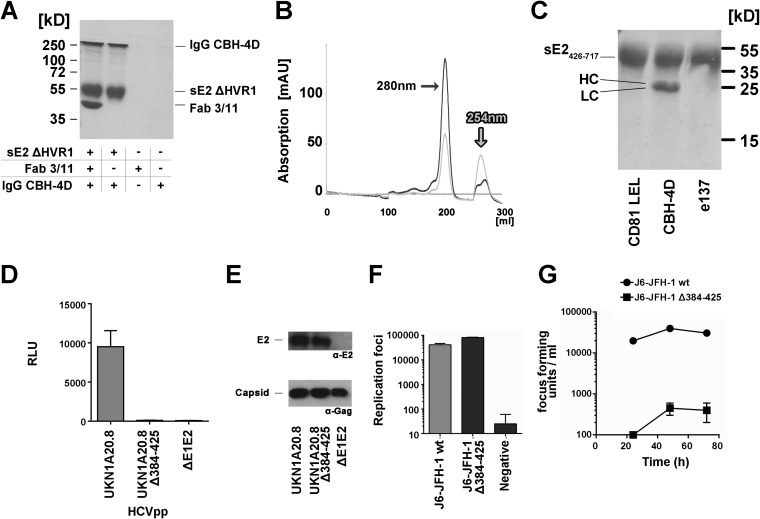
The very low RMSD of 0.8 Å between Cα atoms of the two E2 core fragments, one containing and one lacking the 44 residues downstream of HVR1 (aa 412 to 456) (10, 11), indicates that this region is not required for E2 folding and therefore that the binding of MAb 3/11 is unlikely to affect the overall fold of E2. To test this hypothesis, we determined the binding of the conformation-sensitive, nonneutralizing human antibody CBH-4D (43) to a Fab 3/11-sE2 ΔHVR1 complex by a coprecipitation assay. SDS-PAGE analysis of the eluted fractions demonstrated that sE2 ΔHVR1 in complex with Fab 3/11 efficiently bound the CBH-4D antibody (Fig. 6A), indicating that 3/11 binding does not affect the overall fold of the glycoprotein.
FIG 6.
Effect of deletion of aa 412 to 425 on protein folding and infectivity. (A) Pulldown assay of the conformation-dependent nonneutralizing human antibody CBH-4D (43) by sE2 ΔHVR1 and the Fab 3/11-sE2 ΔHVR1 complex indicating that Fab 3/11 binding does not affect the overall glycoprotein fold. (B) Elution profile of sE2426–717 (UKN2B2.8) from a Superdex200 size exclusion column. AU, absorption units. (C) Pulldown assay of CD81-LEL, Fab CBH-4D, and Fab e137 by sE2426–717 reveals an intact overall glycoprotein fold but severely impaired binding to the CD81 binding site. (D to G) Infectivity of deletion mutants in HCVcc and HCVpp models of infection. (D) The E1/E2 genes of the highly infectious primary isolate UKN1A20.8 were used as the template for the deletion of amino acids 384 to 425. HCVpp possessing wt and mutant glycoproteins were used to infect Huh7 cells. Infectivity measurements were performed with a luciferase reporter gene. Only target cells inoculated with wild-type HCVpp exhibited luciferase activity. RLU, relative light units. (E, top) Western blot analysis of HCVpp using the linear anti-E2 MAb ALP98 reveals similar E2 incorporation levels for wt and mutant HCVpp (lanes 1 and 2, respectively). (Bottom) The amount of particles was verified by Western blotting using an anti-Gag antibody. (F and G) A plasmid encoding a chimeric J6/JFH-1 virus was used as the template for the deletion of amino acids 384 to 425. RNA transcripts of wt and mutant viruses were electroporated into Huh7.5 cells. Results are presented as means and standard deviations from duplicate experiments. (F) RNA replication was assessed by intracellular staining for the presence of HCV NS5a in electroporated cells. Both constructs replicated similarly, with the deletion mutant having slightly enhanced replication. (G) Cell supernatants harvested 24, 48, and 72 h after transfection were assessed for the presence of infectious virus by infecting naive Huh7.5 cells. Infectious wild-type virus was produced, while no infectivity was observed for the mutant virus.
Deletion of the N-terminal 72 residues of E2 abolishes binding to its cellular receptor CD81 (10), whereas deletion of HVR1 does not affect CD81 binding (7). To analyze the role of aa 412 to 423, we expressed recombinant soluble E2 lacking the N-terminal 42 residues (encoding aa 426 to 717) (sE2426–717). As anticipated, size exclusion chromatography (SEC) analysis revealed a majority of the purified protein eluting at a volume corresponding to a monomer, confirming that the deletion of aa 412 to 425 does not lead to multimer formation via nonproper interchain disulfides and therefore likely does not impair protein folding (Fig. 6B). To analyze the adopted conformation of the deletion mutant, we used pulldown assays to analyze the interaction of sE2426–717 with the soluble portion of CD81 (CD81-LEL) and Fab fragments derived from nonoverlapping conformation-sensitive human MAbs (CBH-4D and e137), one derived from a broadly neutralizing antibody overlapping the CD81 binding site (e137) (42) and one derived from a nonneutralizing antibody (CBH-4D). As expected, Fab CBH-4D bound sE2426–717, but no interaction between sE2426–717 and Fab e137 or CD81-LEL was observed (Fig. 6C), suggesting that sE2426–717 adopts a native overall fold but that the binding capacity of its CD81 binding site is impaired.
The role of aa 412 to 425 in the viral cycle was further investigated by using retroviral particles pseudotyped with envelope glycoproteins (HCVpp) derived from primary HCV isolate UKN1A20.8 lacking the region encompassing aa 384 to 425. HCVpp carrying glycoproteins from this isolate were shown to be highly infectious (20). HCVpp infectivity was ablated by the E2Δaa384–425 mutant (Fig. 6D), in spite of the incorporation of E2 into purified HCVpp (Fig. 6E), suggesting that the deletion of aa 384 to 425 affected the entry of retroviral pseudoparticles, which is in line with impaired CD81 binding. Since HCVpp have been shown to be more sensitive to glycoprotein mutations than HCVcc (45), we introduced the same deletion into an infectious HCVcc chimera (J6/JFH1). Despite clear evidence of intracellular replication of the virus genome (Fig. 6F), no infectious particles were released from cells transfected with the mutant genome (Fig. 6G). These results confirm the essential role of the antigenic site comprising aa 412 to 423 during HCV entry.
Concluding remarks.
In summary, the striking structural flexibility displayed by the antigenic site at aa 412 to 423 confirms that this region of E2 is not part of a structured domain. As described above, the glycan chain attached to N417 requires the peptide chain to adopt an extended conformation to allow binding of MAb 3/11. The neutralizing activity of MAb 3/11 indicates that this extended conformation is present at the surface of infectious particles during virus entry and that the N-terminal 40 residues within E2 are structurally flexible. This is in agreement with the recently observed elevated deuterium exchange rates for the N-terminal 72 residues within E2 (10) and with the finding that the reported conformation of the E2 ectodomain core front layer (corresponding to aa 412 to 491) is likely to be partially induced by MAb AR3C binding (11).
The structural flexibility of this antigenic site is likely to contribute to its reduced immunogenicity in patients (22, 25). Human immunodeficiency virus (HIV) has been reported to utilize conformational flexibility in the CD4 binding site within the envelope protein gp120 as an immune evasion mechanism (reviewed in reference 57). For NAbs targeting the protruding (P) domain of the capsid of murine norovirus type 1 (MNV-1), escape mutants were shown to switch conformations in the two structurally flexible P domain loops, resulting in a deterioration of the stereochemical fit of the NAb paratope to the P domain (58). It is tempting to speculate that HCV also employs structural flexibility as an immune evasion mechanism to prevent NAbs targeting this conserved receptor binding domain from being elicited. While our structural analysis suggests that the antigenic site spanning aa 412 to 423 can adopt at least two different conformations, the majority of monoclonal antibodies characterized so far recognize the β-hairpin. In addition, our SPR analysis highlights differences in the kinetic behaviors of MAbs 3/11 and AP33 binding wt sE2 and sE2 ΔHVR1, respectively, suggesting that the β-hairpin might be the predominant conformation that could be stabilized by interactions within E2 and/or E1 at the virus surface. Our data have implications for vaccine design, because the flexibility in this antigenic site could prevent this epitope from being an ideal candidate for an efficient vaccine. In view of a number of potent broadly neutralizing antibodies directed against different regions within E2 (23, 42, 59–62), other antigenic sites could potentially be more advantageous for vaccine design. Our results provide insights into the recognition of E2 by broadly neutralizing antibodies and are an important step on the way to developing an efficient vaccine against HCV.
ACKNOWLEDGMENTS
This work was funded by the CNRS, an ANRS grant and recurrent funding from the Institut Pasteur to F.A.R., and an ANRS grant to T.K.
We thank Ahmed Haouz and Patrick Weber from the crystallization platform for help in crystallization, Richard Urbanowicz for help with initial experiments and valuable discussions, and staff of the synchrotron beamline Proxima-1 at Synchrotron Soleil for help during data collection.
REFERENCES
- 1.Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Schinazi R, Halfon P, Marcellin P, Asselah T. 2014. HCV direct-acting antiviral agents: the best interferon-free combinations. Liver Int 34(Suppl 1):S69–S78. doi: 10.1111/liv.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball JK, Tarr AW, McKeating JA. 2014. The past, present and future of neutralizing antibodies for hepatitis C virus. Antiviral Res 105:100–111. doi: 10.1016/j.antiviral.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 5.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Sekiya H, Hijikata M, Shimotohno K. 1992. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem Biophys Res Commun 189:119–127. doi: 10.1016/0006-291X(92)91533-V. [DOI] [PubMed] [Google Scholar]
- 7.McCaffrey K, Boo I, Poumbourios P, Drummer HE. 2007. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J Virol 81:9584–9590. doi: 10.1128/JVI.02782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore EE, Peterson DA, Mishiro S. 1993. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology 195:297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 9.Forns X, Thimme R, Govindarajan S, Emerson SU, Purcell RH, Chisari FV, Bukh J. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc Natl Acad Sci U S A 97:13318–13323. doi: 10.1073/pnas.230453597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng L, Zhong L, Struble E, Duan H, Ma L, Harman C, Yan H, Virata-Theimer ML, Zhao Z, Feinstone S, Alter H, Zhang P. 2013. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci U S A 110:7418–7422. doi: 10.1073/pnas.1305306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng L, Ma L, Virata-Theimer ML, Zhong L, Yan H, Zhao Z, Struble E, Feinstone S, Alter H, Zhang P. 2014. Discrete conformations of epitope II on the hepatitis C virus E2 protein for antibody-mediated neutralization and nonneutralization. Proc Natl Acad Sci U S A 111:10690–10695. doi: 10.1073/pnas.1411317111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong L, Giang E, Nieusma T, Robbins JB, Deller MC, Stanfield RL, Wilson IA, Law M. 2012. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J Virol 86:13085–13088. doi: 10.1128/JVI.01939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, Giang E, Robbins JB, Stanfield RL, Burton DR, Wilson IA, Law M. 2012. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci U S A 109:9499–9504. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krey T, Meola A, Keck Z-Y, Damier-Piolle L, Foung SKH, Rey FA. 2013. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog 9:e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantua H, Diao J, Ultsch M, Hazen M, Mathieu M, McCutcheon K, Takeda K, Date S, Cheung TK, Phung Q, Hass P, Arnott D, Hongo J-A, Matthews DJ, Brown A, Patel AH, Kelley RF, Eigenbrot C, Kapadia SB. 2013. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol 425:1899–1914. doi: 10.1016/j.jmb.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck Z-Y, Lau P, Foung SKH, Taylor GL, Patel AH. 2012. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol 86:12923–12932. doi: 10.1128/JVI.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. 2009. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol 83:12473–12482. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJP, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601. doi: 10.1002/hep.21088. [DOI] [PubMed] [Google Scholar]
- 21.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol 73:6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, Xia J, Patel AH, Bukh J, Foung SK. 2013. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol 87:37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabo MC, Luca VC, Prentoe JC, Hopcraft SE, Blight KJ, Yi M, Lemon SM, Ball JK, Bukh J, Evans MJ, Fremont DH, Diamond MS. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol 85:7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol 80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarr AW, Owsianka AM, Jayaraj D, Brown RJP, Hickling TP, Irving WL, Patel AH, Ball JK. 2007. Determination of the human antibody response to the epitope defined by the hepatitis C virus-neutralizing monoclonal antibody AP33. J Gen Virol 88:2991–3001. doi: 10.1099/vir.0.83065-0. [DOI] [PubMed] [Google Scholar]
- 26.Backovic M, Johansson DX, Klupp BG, Mettenleiter TC, Persson MAA, Rey FA. 2010. Efficient method for production of high yields of Fab fragments in Drosophila S2 cells. Protein Eng Des Sel 23:169–174. doi: 10.1093/protein/gzp088. [DOI] [PubMed] [Google Scholar]
- 27.Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, England P, Persson MA, Martin A, Rey FA. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog 6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarr AW, Lafaye P, Meredith L, Damier-Piolle L, Urbanowicz RA, Meola A, Jestin J-L, Brown RJP, McKeating JA, Rey FA, Ball JK, Krey T. 2013. An alpaca nanobody inhibits hepatitis C virus entry and cell-to-cell transmission. Hepatology 58:932–939. doi: 10.1002/hep.26430. [DOI] [PubMed] [Google Scholar]
- 29.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 30.Tarr AW, Urbanowicz RA, Hamed MR, Albecka A, McClure CP, Brown RJ, Irving WL, Dubuisson J, Ball JK. 2011. Hepatitis C patient-derived glycoproteins exhibit marked differences in susceptibility to serum neutralizing antibodies: genetic subtype defines antigenic but not neutralization serotype. J Virol 85:4246–4257. doi: 10.1128/JVI.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel AH, Wood J, Penin F, Dubuisson J, McKeating JA. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J Gen Virol 81:2873–2883. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. 1988. Automatic indexing of rotation diffraction patterns. J Appl Crystallogr 21:67–72. [Google Scholar]
- 33.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.McCoy LE, Quigley AF, Strokappe NM, Bulmer-Thomas B, Seaman MS, Mortier D, Rutten L, Chander N, Edwards CJ, Ketteler R, Davis D, Verrips T, Weiss RA. 2012. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J Exp Med 209:1091–1103. doi: 10.1084/jem.20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek P, Roversi P, Sharff A, Smart O, Vonrhein C, Womack T. 2010. BUSTER version 2.9. Global Phasing Ltd, Cambridge, United Kingdom. [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Shatsky M, Nussinov R, Wolfson HJ. 2004. A method for simultaneous alignment of multiple protein structures. Proteins 56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Tina KG, Bhadra R, Srinivasan N. 2007. PIC: Protein Interactions Calculator. Nucleic Acids Res 35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perotti M, Mancini N, Diotti R, Tarr AW, Ball JK, Owsianka A, Adair R, Patel AH, Clementi M, Burioni R. 2008. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J Virol 82:1047. doi: 10.1128/JVI.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hadlock KG, Lanford RE, Perkins S, Rowe J, Yang Q, Levy S, Pileri P, Abrignani S, Foung SK. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol 74:10407–10416. doi: 10.1128/JVI.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goffard A, Callens N, Bartosch B, Wychowski C, Cosset F-L, Montpellier C, Dubuisson J. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol 79:8400–8409. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhillon S, Witteveldt J, Gatherer D, Owsianka AM, Zeisel MB, Zahid MN, Rychłowska M, Foung SKH, Baumert TF, Angus AGN, Patel AH. 2010. Mutations within a conserved region of the hepatitis C virus E2 glycoprotein that influence virus-receptor interactions and sensitivity to neutralizing antibodies. J Virol 84:5494–5507. doi: 10.1128/JVI.02153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gal-Tanamy M, Keck Z-Y, Yi M, McKeating JA, Patel AH, Foung SKH, Lemon SM. 2008. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc Natl Acad Sci U S A 105:19450–19455. doi: 10.1073/pnas.0809879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owsianka AM, Clayton RF, Loomis-Price LD, McKeating JA, Patel AH. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J Gen Virol 82:1877–1883. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, Pfarr DS, Tang Y, An L-L, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. 2005. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol 350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 49.Bates JT, Keefer CJ, Utley TJ, Correia BE, Schief WR, Crowe JE. 2013. Reversion of somatic mutations of the respiratory syncytial virus-specific human monoclonal antibody Fab19 reveal a direct relationship between association rate and neutralizing potency. J Immunol 190:3732–3739. doi: 10.4049/jimmunol.1202964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Keck ZY, Saha A, Xia J, Conrad F, Lou J, Eckart M, Marks JD, Foung SK. 2011. Affinity maturation to improve human monoclonal antibody neutralization potency and breadth against hepatitis C virus. J Biol Chem 286:44218–44233. doi: 10.1074/jbc.M111.290783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanCott TC, Bethke FR, Polonis VR, Gorny MK, Zolla-Pazner S, Redfield RR, Birx DL. 1994. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. J Immunol 153:449–459. [PubMed] [Google Scholar]
- 52.Rani M, Bolles M, Donaldson EF, Van Blarcom T, Baric R, Iverson B, Georgiou G. 2012. Increased antibody affinity confers broad in vitro protection against escape mutants of severe acute respiratory syndrome coronavirus. J Virol 86:9113–9121. doi: 10.1128/JVI.00233-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck Z-Y, Foung SKH, Pécheur E-I, Pietschmann T. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol 84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prentoe J, Jensen TB, Meuleman P, Serre SBN, Scheel TKH, Leroux-Roels G, Gottwein JM, Bukh J. 2011. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol 85:2224–2234. doi: 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klasse PJ, Sattentau QJ. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J Gen Virol 83:2091–2108. [DOI] [PubMed] [Google Scholar]
- 57.Pantophlet R, Burton DR. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol 24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 58.Kolawole AO, Li M, Xia C, Fischer AE, Giacobbi NS, Rippinger CM, Proescher JB, Wu SK, Bessling SL, Gamez M, Yu C, Zhang R, Mehoke TS, Pipas JM, Wolfe JT, Lin JS, Feldman AB, Smith TJ, Wobus CE. 2014. Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization. J Virol 88:4543–4557. doi: 10.1128/JVI.03685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MAA. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci U S A 104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keck Z, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen TH, Li AY-J, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SK. 2012. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog 8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med 14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 62.Keck Z-Y, Olson O, Gal-Tanamy M, Xia J, Patel AH, Dreux M, Cosset F-L, Lemon SM, Foung SKH. 2008. A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. J Virol 82:6067–6072. doi: 10.1128/JVI.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson DX, Krey T, Andersson O. 2012. Production of recombinant antibodies in Drosophila melanogaster S2 cells. Methods Mol Biol 907:359–370. doi: 10.1007/978-1-61779-974-7_21. [DOI] [PubMed] [Google Scholar]