ABSTRACT
The development of a panel of mucosally transmissible simian-human immunodeficiency virus (SHIV) challenge stocks from multiple virus clades would facilitate preclinical evaluation of candidate HIV-1 vaccines and therapeutics. The majority of SHIV stocks that have been generated to date have been derived from clade B HIV-1 env sequences from viruses isolated during chronic infection and typically required serial animal-to-animal adaptation for establishing mucosal transmissibility and pathogenicity. To capture essential features of mucosal transmission of clade C viruses, we produced a series of SHIVs with early clade C HIV-1 env sequences from acutely HIV-1-infected individuals from South Africa. SHIV-327c and SHIV-327cRM expressed env sequences that were 99.7 to 100% identical to the original HIV-1 isolate and did not require in vivo passaging for mucosal infectivity. These challenge stocks infected rhesus monkeys efficiently by both intrarectal and intravaginal routes, replicated to high levels during acute infection, and established chronic setpoint viremia in 13 of 17 (76%) infected animals. The SHIV-327cRM challenge stock was also titrated for both single, high-dose intrarectal challenges and repetitive, low-dose intrarectal challenges in rhesus monkeys. These SHIV challenge stocks should facilitate the preclinical evaluation of vaccines and other interventions aimed at preventing clade C HIV-1 infection.
IMPORTANCE We describe the development of two related clade C SHIV challenge stocks. These challenge stocks should prove useful for preclinical testing of vaccines and other interventions aimed at preventing clade C HIV-1 infection.
INTRODUCTION
HIV-1 does not typically infect nonhuman primates, and thus chimeric SHIVs have been constructed consisting of the simian immunodeficiency virus (SIV) backbone with HIV-1 env, tat, rev, and vpu (1–3). Such SHIVs are widely used for preclinical testing of candidate vaccines and therapeutics, but the majority of currently available SHIVs have been constructed with chronic clade B env genes (4–7). Transmission of HIV-1 across an intact mucosal barrier is relatively inefficient, and in most cases a single or a small number of transmitted/founder viruses are responsible for establishing infection (8–10). SHIVs containing early HIV-1 env's might therefore be mucosally transmissible and potentially useful for testing interventions that aim to block mucosal infection.
Clade C is the dominant HIV-1 subtype in the world accounting for nearly half of all global infections (11). To date, only a limited number of clade C SHIV stocks have been reported (12–16) and typically have required in vivo passaging for mucosal transmissibility and pathogenicity (1, 17–19). As expected, multiple amino acid changes in env accumulated in these viruses that diverged from the original env sequence (14, 16, 19). Without in vivo adaptation, most SHIVs replicate poorly in rhesus monkey cells (17, 20, 21), possibly as a result of reduced ability to bind rhesus CD4 (21). However, some natural HIV-1 env's have been shown to interact with both rhesus and human CD4.
To generate mucosally transmissible clade C SHIV challenge stocks with minimal env mutations, we produced multiple candidate virus stocks without in vivo passaging and evaluated their in vitro and in vivo characteristics. Two related early clade C SHIV challenge stocks, SHIV-327c and SHIV-327cRM, reliably established mucosal infection in rhesus monkeys and led to high peak acute viral loads, consistent CD4+ T cell declines during acute infection, and chronic setpoint viremia in the majority of animals.
MATERIALS AND METHODS
Animals.
Rhesus monkeys (Macaca mulatta) of Indian origin were used in the present study. Twelve Mamu-A*01-negative adult male and female animals were housed at Bioqual (Rockville, MD), and six Mamu-A*01-positive adult female animals were housed at the New England Primate Research Center (Southborough, MA). The animals were maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animals with appropriate approvals from the relevant Institutional Animal Care and Use Committees.
Cells.
Human and rhesus monkey peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque gradient purification, followed by stimulation with concanavalin A (ConA; 6.25 μg/ml) and 20 U/ml human interleukin-2 (IL-2; AIDS Research and Reference Reagent Program) overnight. The cells were cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 U/ml of IL-2. TZM-bl cells (also called JC53-bl clone 13 cells) are derived from a HeLa cell line (JC.53) that stably expressed CD4 and HIV coreceptors, as well as luciferase and β-galactosidase under the control of the HIV-1 long terminal repeat. GHOST cell lines expressed CD4 alone or with different chemokine receptors and were obtained from the AIDS Research and Reference Reagent Program.
Construction of SHIV molecular clones.
Fifteen early clade C env sequences (072c, 140c, 327c, 349c, 405c, 426c, 431c, 459c, 590c, 706c, 756c, 823c, 823D6c, 885c, and 939c) derived from acutely HIV-1-infected participants in the HVTN503 clinical trial in South Africa (22) were used to generate the SHIV molecular clones. A KB9-AC backbone containing a ClaI restriction site immediately upstream of the env ATG start codons and an AgeI restriction site upstream of the 3′ HIV-SIV recombination junction was provided by Norman Letvin, Beth Israel Deaconess Medical Center, Boston, MA. In order to facilitate the cloning using the unique ClaI and AgeI sites, KB9-AC sequences from the ClaI site to the env ATG start codons were introduced upstream of the early clade C env sequences with the AgeI site designed at the corresponding 3′ end. The clade C env sequences were synthesized by GeneArt (GeneArt, Invitrogen, Darmstadt, Germany) and were digested and subcloned into the corresponding regions of the KB9-AC backbone using ClaI and AgeI (Fig. 1). Plasmids containing the early SHIV infectious molecular clones were transfected into 293T cells using LipoD293 (SignaGen Laboratories, Rockville, MD). Cell culture supernatants were collected after 48 h and clarified through a 0.45-μm-pore-size filter.
FIG 1.
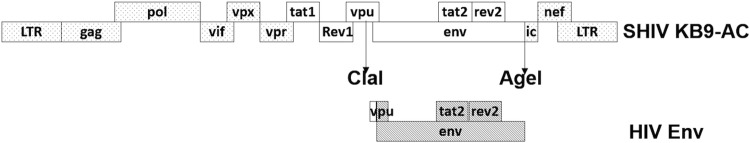
Cloning strategy for the generation of SHIVs expressing early HIV-1 env sequences. Two unique restriction sites, ClaI and AgeI, in the SHIV-KB9 infectious clone were utilized. Fragments containing KB9 sequence from the ClaI site to the env ATG start codons and early clade C env sequences with AgeI site designed at the corresponding 3′ end were synthesized and replaced the corresponding regions in the SHIV-KB9-AC.
Generation of large-scale SHIV stocks.
Five infectious molecular clones (327c, 405c, 459c, 590c, and 823cD6) that replicated to the highest levels in human PBMC were selected for production of large-scale challenge stocks. For the generation of each virus stock, PBMC were isolated from 120 ml of human or rhesus monkey blood. Cell culture supernatants harvested from transiently transfected 293T cells were used to inoculate to ConA-stimulated PBMC in the presence of 20 U/ml human IL-2 (AIDS Research and Reference Reagent Program). SHIV-327cRM was prepared by propagation of the human stock in rhesus monkey PBMC. The medium was replaced and collected every 3 days. Virus was quantified by SIV p27 enzyme-linked immunosorbent assay (ELISA; Zeptometrix), and the 50% tissue culture infectious dose (TCID50) was determined in TZM-bl cells.
TCID50 titer in TZM-bl cells.
Virus stocks were assessed for infectivity by inoculation in TZM-bl cells at serial 1:4 dilutions in the presence of 40 μg of DEAE-dextran hydrochloride (Sigma, St. Louis, MO)/ml. Virus infectivity was determined 48 h postinfection by measuring the level of luciferase activity expressed in infected cells. Each experiment was performed in triplicate. The TCID50 was calculated as the dilution point at which 50% of the cultures were infected.
Coreceptor studies.
The inhibitors TAK-779 and AMD-3100 were obtained from the AIDS Research and Reference Reagent Program and were diluted per manufacturer instructions. TZM-bl cells were plated at 3 × 104 cells/well in 96-well flat-bottom plates. TZM-bl cells were incubated for 1 h in either 10-fold dilutions of TAK-779 ranging from 5 to 500,000 ng/ml or 10-fold dilutions of AMD-3100 ranging from 5 to 50,000 ng/ml. After 1 h, 100 TCID50 of SHIV stocks were as added to the wells, and 48 h later the cells were lysed and analyzed by a Steady-Glo (Promega) luciferase assay with luminescence measured on a Victor 3 luminometer (Perkin-Elmer). GHOST cells expressing coreceptors CXCR4, CCR5, and CCR5/CXCR4 and the parental strain were obtained from the AIDS Research and Reference Reagent Program. These were maintained in Dulbecco modified Eagle medium with 10% fetal calf serum and penicillin-streptomycin and supplemented with 500 μg/ml G418, 100 μg/ml hygromycin, and 1 μg/ml puromycin. Puromycin was excluded for culture of the parental cell line. Cells were plated at 104 cells per ml in 96-well round-bottom wells in medium supplemented with 20 μg/ml Polybrene. One hundred TCID50 of each virus stock was added to each well, followed by incubation overnight. After being washed with phosphate-buffered saline (PBS) and replacement with fresh culture medium, cell culture supernatants were harvested 4 days after incubation and assessed for SIV p27 levels by ELISA (Zeptometrix).
Neutralization assay.
Neutralization assays were performed in triplicate in TZM-bl cells, as previously described (23). Monoclonal antibodies (MAbs) 4E10, 2G12, 2F5, b12, PG9, PG16, and VRC01 (Polymun Scientific) and soluble human CD4 protein (Progenics) were obtained commercially. 3BNC117, 8ANC195, 10-1074, 3BC176, and 45-46W were provided by Michel Nussenzweig (Rockefeller University, New York, NY). PGT121, PGT126, and PGT128 were provided by Dennis Burton (The Scripps Research Institute, La Jolla, CA). Purified immunoglobulin preparations from clade C HIV-1-positive plasma samples were obtained from the South Africa National Blood Services (Johannesburg, South Africa) and were provided by Lynn Morris.
i.r. inoculation of clade C SHIVs.
Animals received a single intrarectal (i.r.) inoculation with 1 ml of undiluted SHIV-327c, SHIV-327cRM, SHIV-405c, or SHIV-459c stocks. For the SHIV-327cRM in vivo titration, one or multiple i.r. challenges with 1 ml of 1:10- and 1:100-diluted virus stock were performed. All animals were monitored for viral loads and CD4+ T cell counts.
i.vag. inoculation of SHIV-327c and SHIV-327cRM.
Four weeks before intravaginal (i.vag.) challenge with SHIV-327c and SHIV-327cRM, a single dose of Depo-Provera (30 mg/kg) was administered by intramuscular injection to six rhesus monkeys. Each animal then received a single i.vag. inoculation with 1 ml of undiluted SHIV-327c or SHIV-327cRM stock. All animals were monitored for viral loads and CD4+ T cell counts.
CD4+ T cell counts.
EDTA-anticoagulated whole blood was stained using anti-CD3-APC, anti-CD4-PE, and anti-CD8-FITC MAbs and analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Cell counts were determined using the BD TrueCount tubes according to the manufacturer's instructions (BD Biosciences).
Lymphocyte immunophenotyping.
PBMC isolated from Indian-origin rhesus monkeys at week 0, 4 and 12 were stained for flow cytometric analysis using combinations of the following fluorochrome-conjugated MAbs: anti-CD3-APC, anti-CD8-APC CY7, anti-CD4-AMCYAN, anti-CD95-PerCP-Cy5.5, anti-CD28-PE-Cy7, anti-CCR7-FITC, and anti-CCR5-PE. The samples were analyzed by four-color flow cytometry (FACSCalibur; BD Biosciences Immunocytometry Systems). Data analysis was performed by using FlowJo (Tree Star, San Carlos, CA). CD3+ CD4+ T cells were gated based on CD28 and CD95 expression to define memory CD4+ T cell subpopulations: naive (CD28+ CD95−, central/transitional memory (CD28+ CD95+), and effector memory (CD28+ CD95+) cells. The central/transitional memory cells were further gated based on the expression of CCR7 and CCR5 to distinguish transitional memory (CD28+ CD95+ CCR7− CCR5+) and central memory (CD28+ CD95+ CCR7+) CD4 T cells subsets. To detect the specific CD8+ T responses in Mamu-A*01-positive animals after SHIV infection, whole blood specimens were stained with Mamu-A*01-p11C-PE for 15 min at room temperature. The cells were then stained with a mixture of APC-anti-CD3 and FITC-anti-CD8α MAbs for 15 min. Whole blood specimens (100 μl) were lysed using a Q-prep workstation (Beckman-Coulter) before being fixed in 1.5% formaldehyde. Data were acquired using a FACSCalibur flow cytometer and analyzed using CellQuest software (BD Biosciences).
Measurement of plasma viral RNA levels.
Viral RNA was isolated from cell-free plasma using the QIAamp viral RNA isolation kit (Qiagen) and quantitated essentially as previously described (24). Quantitative reverse transcription-PCR was conducted in a two-step process. First, RNA was reverse transcribed in parallel with an SIV gag RNA standard using the gene-specific primer sGag-R 5′-CACTAGGTGTCTCTGCACTATCTGTTTTG-3′. All samples were then treated with RNase H (Stratagene) for 20 min at 37°C. Amplification primers, including the forward primer SGAG21 (5′-GTCTGCGTCATPTGGTGCATTC-3′), reverse primer SGAG22 (5′-CACTAGKTGTCTCTGCACTATPTGTTTTG-3′), 100-nm probe pSGAG23 [5′-(FAM)CTTCPTCAGTKTGTTTCACTTTCTCTTCTGCG-(BHQ1)-3′], and 50-nm passive reference dye [5′-(FAM)TTTTTTTTTT(ROX)(C3-blocked)-3′] were used. All reactions were carried out on a 7300 ABI real-time PCR system with 1.25 U of TaqGold polymerase (Applied Biosystems) in triplicate according to the manufacturer's protocols. Ultrasensitive viral load assays were performed essentially as described previously (25).
Single-genome amplification (SGA) and sequencing.
Viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen). RNA was eluted and immediately subjected to cDNA synthesis. Reverse transcription of RNA to single-stranded cDNA was performed using SuperScript III reverse transcriptase according to the manufacturer's recommendations (Invitrogen). In brief, each cDNA reaction mixture included 1× RT buffer, a 0.5 mM concentration of each deoxynucleoside triphosphate, 5 mM dithiothreitol, 2 U/ml RNaseOUT (RNase inhibitor), 10 U/ml SuperScript III reverse transcriptase, and 0.25 mM antisense primer. The mixture was incubated at 50°C for 60 min, followed by an increase in temperature to 55°C for an additional 60 min. The reaction mix was then heat inactivated at 70°C for 15 min and treated with 2 U of RNase H at 37°C for 20 min. The newly synthesized cDNA was used immediately or frozen at −80°C. cDNA was serially diluted and distributed among wells of replicate 96-well plates to identify a dilution where PCR-positive wells constituted <30% of the total number of reaction wells, as previously described (8, 26). At this dilution, most positive wells would contain amplicons derived from a single cDNA molecule. This was confirmed for every positive well by direct sequencing of the amplicon and inspection of the sequence for mixed bases, which would be indicative of priming from more than one original template or the introduction of PCR error in early cycles. Any sequence with mixed bases was excluded from further analysis. PCR amplification was performed in the presence of 1× high-fidelity polymerase in a 20-μl reaction mix (Invitrogen). First-round PCR primers included the sense primer and the antisense primer. PCR was performed in MicroAmp 96-well reaction plates (Applied Biosystems) with the following PCR parameters: 1 cycle of 94°C for 2 min; 35 cycles of a denaturing step of 94°C for 15 s, an annealing step of 54°C for 30 s, and an extension step of 68°C for 4 min; and a final extension step of 68°C for 10 min. Next, 2 μl of the first-round PCR product was added to a second-round PCR mix. The second-round PCR was performed under the same conditions used for first-round PCR, but with a total of 45 cycles. env amplicons were inspected on precast 1% agarose 96-well E-gels (Invitrogen). Both DNA strands of env amplicons were directly sequenced using partially overlapping fragments.
Nucleotide sequence accession numbers.
The env sequences of the amplicons were deposited in GenBank under accession numbers KP126997 to KP127067.
RESULTS
Generation of clade C SHIV stocks.
Eleven of fifteen clade C early HIV-1 env sequences from acutely HIV-1-infected individuals from South Africa were successfully cloned into the SHIV-KB9-AC backbone (Fig. 1). Transfection of 293T cells yielded high SIV p27 levels (data not shown) and virus infectivity titers of 2.5 × 103 to 1.56 × 107 TCID50/ml (Table 1). These cultures were then used to assess virus growth in human and rhesus PBMC. As expected, these viruses replicated well in human PBMC but replicated poorly in rhesus PBMC (Table 1).
TABLE 1.
Cloning and replication of 293T-derived SHIVs in human and rhesus PBMCa
| HIV-1 Env | Cloning in SHIV-KB9 | Infectivity titer (TCID50/ml) |
||
|---|---|---|---|---|
| 293T cells | Human PBMC | Rhesus PBMC | ||
| 072c | Yes | 1.25 × 105 | 1.25 × 104 | 0 |
| 140c | Yes | 5.00 × 105 | 1.25 × 104 | 0 |
| 327c | Yes | 3.125 × 106 | 5.25 × 107 | 0 |
| 349c | Yes | 1.56 × 107 | 1.10 × 105 | 0 |
| 405c | Yes | 1.25 × 105 | 2.15 × 107 | 0 |
| 426c | No | ND | ND | ND |
| 431c | Yes | 1.25 × 105 | 0 | 0 |
| 459c | Yes | 6.25 × 105 | 3.25 × 106 | 0 |
| 590c | Yes | 1.25 × 105 | 1.25 × 108 | 0 |
| 706c | Yes | 2.50 × 103 | 3.10 × 102 | 0 |
| 756c | No | ND | ND | ND |
| 823c | Yes | 2.50 × 104 | 1.50 × 103 | 0 |
| 823cD6 | Yes | 1.50 × 107 | 1.50 × 105 | 0 |
| 885c | No | ND | ND | ND |
| 939c | No | ND | ND | ND |
Supernatants were collected 2 days after plasmid transfection in 293T cells or at 9 days postinoculation of the stocks into human or rhesus PBMC. ND, not done.
We selected five SHIVs (327c, 405c, 459c, 590c, and 823D6c) for generating large-scale challenge stocks by inoculation of the 293T transfection-derived supernatants into primary human PBMC. The large-scale challenge stocks ranged in infectivity from 1 × 106 to 2 × 109 TCID50/ml, and viral loads ranged from 2 × 108 to 5 × 109 RNA copies/ml (Table 2). Interestingly, three of these stocks also replicated at low levels in rhesus PBMC, particularly SHIV-327c (Table 2). We then prepared a large-scale challenge stock of this virus in primary rhesus PBMC, resulting in SHIV-327cRM. Although the virus titer of the rhesus PBMC-derived SHIV-327cRM stock was 16-fold lower than the human PBMC-derived SHIV327c stock, SHIV-327cRM replicated 24-fold more efficiently than the SHIV-327c stock in rhesus PBMC (Table 2).
TABLE 2.
Summary of large-scale SHIV stocksa
| SHIV stock | PBMC utilized | Infectivity titer (TCID50/ml) | Human p27 PBMC (ng/ml) | Viral load (RNA copies/ml) | Rhesus p27 PBMC (ng/ml) | Coreceptor usage |
|---|---|---|---|---|---|---|
| 327c | Human | 5 × 107 | 176.4 | 2.1 × 109 | 3.13 | CCR5 |
| 405c | Human | 2 × 109 | 312.4 | 4.9 × 109 | 0.10 | CCR5 |
| 459c | Human | 1 × 106 | 120.8 | 2.2 × 108 | 0.15 | CCR5 |
| 590c | Human | 1 × 106 | 108.17 | 2.1 × 108 | 0 | CCR5 |
| 823cD6 | Human | 5 × 106 | 87.75 | 2.1 × 109 | 0 | CCR5 |
| 327cRM | Rhesus monkeys | 3 × 106 | 75.53 | 2.2 × 109 | 75.53 | CCR5 |
PBMC utilized for stock generation, infectivity in TZM-bl cells (TCID50/ml), SIV p27 levels in human PBMC, viral load (RNA copies/ml), and SIV p27 levels after inoculation of rhesus monkey PBMC and coreceptor usage are shown.
We utilized SGA to analyze the env sequences of the SHIV-327c and SHIV-327cRM challenge stocks compared to the original HIV-1 env patient sequence, which was obtained on day 71 of acute HIV-1 infection from an individual in South Africa (Fig. 2). Among 36 SGA-derived sequences obtained from the human PBMC-derived SHIV-327c stock, five amplicons showed one to three sporadic nucleotide differences from the original human env early consensus sequence. The rest of the amplicons from the SHIV-327c stock were 100% identical to the original sequence. For the rhesus PBMC-derived SHIV-327cRM stock, there were two consistent mutations in the majority of the 35 SGA-derived sequences, including an L124F mutation in the CD4-binding domain (observed in 25/35 amplicons) and a G408R mutation in the V4 loop (observed in 29/35 amplicons), as well as several additional sporadic nucleotide differences (Fig. 2). These data show that the env sequences in the SHIV-327c stock were very similar to the original early human env sequence, whereas the SHIV-327cRM stock differed primarily by two amino acids.
FIG 2.
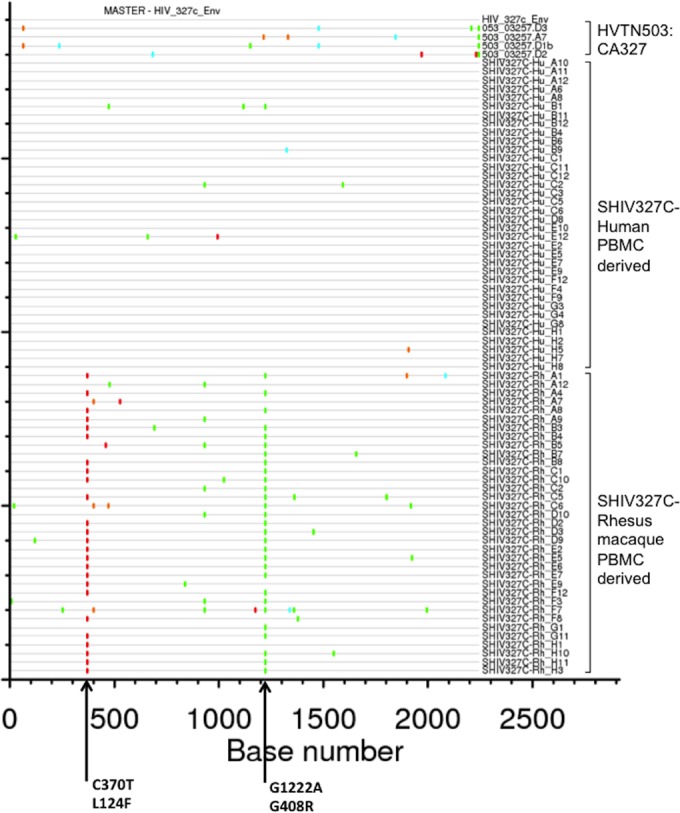
Highlighter amino acid sequence alignment of env derived from the SHIV-327c and SHIV-327cRM stocks and the parental CA327 HIV-1 env. Amino acid substitutions that differed from the parental HIV-1 CA327 env sequence (GenBank accession number JN681220) are indicated in color. Dashes indicate amino acid sequences identical to the parental sequence. The two relevant mutations in SHIV-327cRM (L124F, G408R) are indicated. A total of 36 sequences from SHIV-327c and 35 sequences from SHIV-327cRM were generated by SGA.
Phenotypic properties of the clade C SHIV stocks.
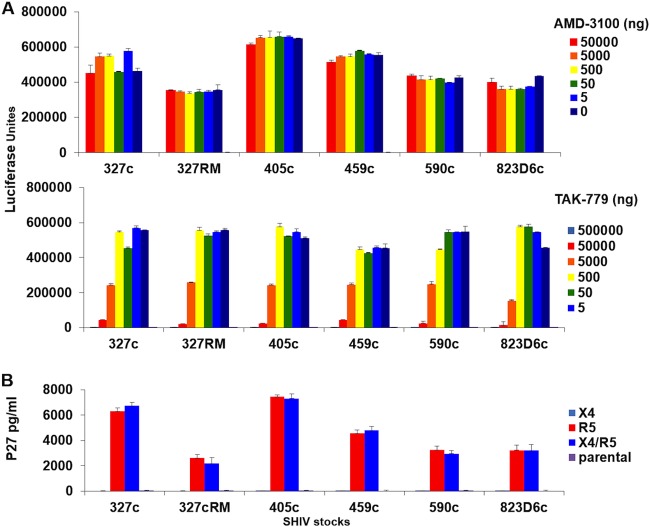
The TZM-bl reporter cell line and pharmacologic inhibitors of HIV-1 coreceptors were utilized to evaluate coreceptor usage of the early clade C SHIV stocks. All of these SHIVs were inhibited by the CCR5 inhibitor TAK-779 but not the CXCR4 inhibitor AMD-3100 in TZM-bl infectivity assays (Fig. 3A). Using GHOST cells expressing alternate coreceptors, we also demonstrated that these SHIV stocks only replicated in R5 or dual X4/R5 expressing cells and thus did not appreciably utilize X4 alone for entry (Fig. 3B). The results demonstrate that all of these SHIV stocks utilized CCR5 as their coreceptor.
FIG 3.
Coreceptor tropism of early SHIV stocks. (A) TZM-bl cells were incubated for 1 h with different concentrations of the CXCR4 inhibitor AMD-3100 or the CCR5 inhibitor TAK-779 and subsequently were infected with 100 TCID50 of the indicated SHIV stocks. The luciferase activity was quantified after 48 h. (B) GHOST cell lines expressing CXCR4 (X4) and/or CCR5 (R5) coreceptors were used, and inoculated with 100-TCID50 SHIV stocks. Cell culture supernatant was collected for SIV p27 determination after 4 days of infection. All assays were done in triplicate. The means with the standard deviations are shown.
The antigenic properties of these SHIV stocks were characterized in neutralizing antibody assays in TZM-bl cells utilizing a panel of broadly reactive MAbs and purified immunoglobulin (Ig) samples from clade C HIV-1-infected individuals (Table 3). The human PBMC-derived SHIV stocks exhibited variable sensitivity to MAbs targeting the major neutralization epitopes but overall appeared relatively neutralization resistant. The rhesus PBMC-derived SHIV-327cRM stock also appeared relatively neutralization resistant, although it showed greater sensitivity than SHIV-327c to soluble human CD4 but reduced sensitivity to certain MAbs such as PG9 and PG16.
TABLE 3.
Neutralization properties of SHIV stocks in TZM-bl assaysa
| Specificity | Antibody | SHIV stock (neutralization sensitivity) |
|||||
|---|---|---|---|---|---|---|---|
| 327c | 327cRM | 405c | 459c | 590c | 823cD6 | ||
| CD4 | sCD4 | >50 | 0.07 | >50 | 48.66 | >50 | >50 |
| CD4bs | b12 | >50 | >50 | >50 | >50 | >50 | >50 |
| 3BNC117 | 1.82 | 2.49 | 4.94 | 5.97 | 0.99 | 5.37 | |
| VRC01 | 2.23 | 1 | 13.31 | 11.95 | 2.96 | 1.99 | |
| 45-46W | 0.22 | 0.085 | 1.83 | 3.12 | 0.358 | 0.751 | |
| V3/CD4i | 3BNC176 | >50 | >50 | >50 | >50 | >50 | >50 |
| gp120 (V3/V4) | 2G12 | >50 | >50 | >50 | >50 | >50 | >50 |
| V1/V2 glycan | PG9 | 30.2 | >50 | 17.41 | >50 | 0.39 | 2.12 |
| PG16 | 15.38 | >50 | 2.05 | 6.7 | <0.023 | 0.089 | |
| V3 glycan | PGT128 | >50 | >50 | 45.94 | 0.65 | 0.18 | 0.12 |
| PGT121 | 1.38 | 0.292 | <0.023 | 0.173 | <0.023 | 0.05 | |
| PGT126 | >50 | >50 | >50 | 42.9 | 0.153 | 0.528 | |
| 10-1074 | 0.056 | <0.023 | <0.023 | 0.427 | 0.032 | 0.398 | |
| gp41/CD4bs | 8ANC195 | >50 | >50 | 18.13 | >50 | 44.05 | 0.461 |
| gp41 (MPER) | 2F5 | >50 | >50 | >50 | >50 | >50 | >50 |
| 4E10 | >50 | 24.01 | 18.25 | >50 | >50 | >50 | |
| 10E8 | 23.66 | 3.61 | 0.983 | 16.03 | >50 | >50 | |
| Polyclonal | HIVIG-B | >2,500 | 603.01 | >2,500 | >2,500 | >2,500 | >2,500 |
| HIVIG-C | 593.8 | 85.66 | 361.51 | 354.42 | 140.42 | 361.64 | |
| HIVIG-C2 | 2,147.37 | 97.07 | 392.16 | >2,500 | 848.05 | >2,500 | |
| HIVIG-C8 | 1,246.4 | 99.68 | 971.81 | 1,301.66 | 459.49 | 1,949.13 | |
| HIVIG-C44 | 893.5 | 175.36 | >2,500 | 1,427.02 | 565.25 | 1,490.73 | |
| HIVIG-C62 | 1,232.82 | 368.21 | 1,101.63 | >2,500 | 1,486.69 | >2,500 | |
| HIVIG-C72 | >2,500 | 127.36 | >2,500 | >2,500 | >2,500 | >2,500 | |
| HIVIG-C74 | >2,500 | 84.79 | 2,385.27 | >2,500 | 855.51 | >2,500 | |
CD4bs, CD4 binding site; CD4i, CD4 induced site; HIVIG, purified immunoglobulin obtained from clade C HIV+ plasma samples.
i.r. infectivity of four SHIV stocks in rhesus monkeys.
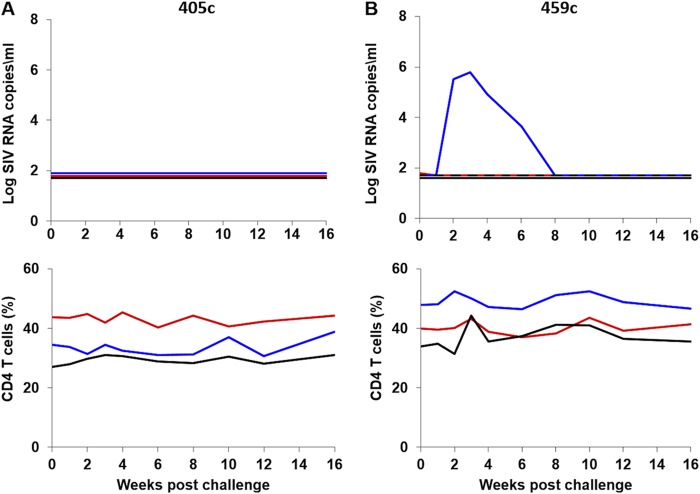
We next evaluated the infectivity in rhesus monkeys of the four early clade C SHIV stocks that exhibited detectable replication in rhesus PBMC (405c, 459c, 327c, and 327cRM). 12 adult rhesus monkeys were inoculated with a single i.r. inoculation of 1 ml of each of these virus stocks (n = 3/group). None of the animals inoculated with SHIV-405c (Fig. 4A), and only one of three animals inoculated with SHIV-459C, exhibited detectable viremia (Fig. 4B). The one animal that was productively infected with SHIV-459c exhibited transient viremia with a peak viral load of 5.8 log RNA copies/ml at week 3 postchallenge and undetectable viral loads by week 10. Thus, SHIV-405c and SHIV-459c proved poorly infectious in vivo.
FIG 4.
i.r. challenge with the clade C SHIV-405c and SHIV-459c stocks in rhesus monkeys. Six animals were challenged once with 1 ml of undiluted SHIV-405c (n = 3) (A) or SHIV-459c (n = 3) (B) stocks by the i.r. route. The upper panel shows plasma viral loads, and the lower panel shows the percentage of CD4+ T cells in peripheral blood. The dotted line reflects the limit of detection of the assay (50 RNA copies/ml).
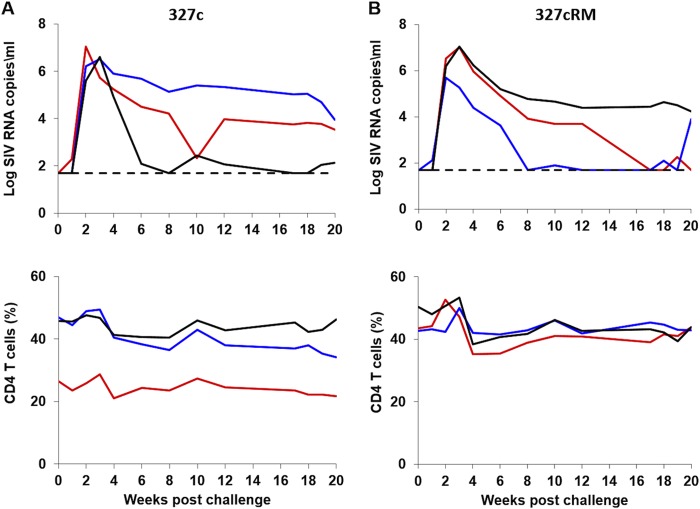
In contrast, all three animals inoculated with the SHIV-327c stock and all three animals inoculated with the SHIV-327cRM stock were productively infected (Fig. 5). These animals exhibited robust peak viral loads ranging from 5.6 to 7.0 log SIV RNA copies/ml at 2 to 3 weeks after inoculation. These viral loads are comparable to those observed with the well-studied clade B SHIV-SF162P3 and SHIV-AD8 stocks (1, 18). Viral loads declined and reached chronic setpoint levels by approximately week 10. Setpoint viral loads varied widely among animals (1.9 to 5.4 log SIV RNA copies/ml), but 5 of 6 infected animals exhibited chronic viremia at week 20, and 3 of 6 animals demonstrated consistent moderate- or high-setpoint viral loads > 3 log RNA copies/ml. During acute infection, CD4+ T cell numbers declined by 5 to 16% in all SHIV-327c and SHIV-327cRM inoculated animals but did not subsequently exhibit further decline. The CD4+ T cell decline was primarily driven by declines in CCR5+ effector and transitional memory CD4+ T cells (data not shown).
FIG 5.
i.r. challenge with the clade C SHIV-327c and SHIV-327cRM stocks in rhesus monkeys. Six animals were challenged once with 1 ml of undiluted SHIV-327c (n = 3) (A) or SHIV-327cRM (n = 3) (B) stocks by the i.r. route. The upper panel shows plasma viral loads, and the lower panel shows the percentage of CD4+ T cells in peripheral blood. The dotted line reflects the limit of detection of the assay (50 RNA copies/ml).
i.r. titration of the SHIV-327cRM stock in rhesus monkeys.
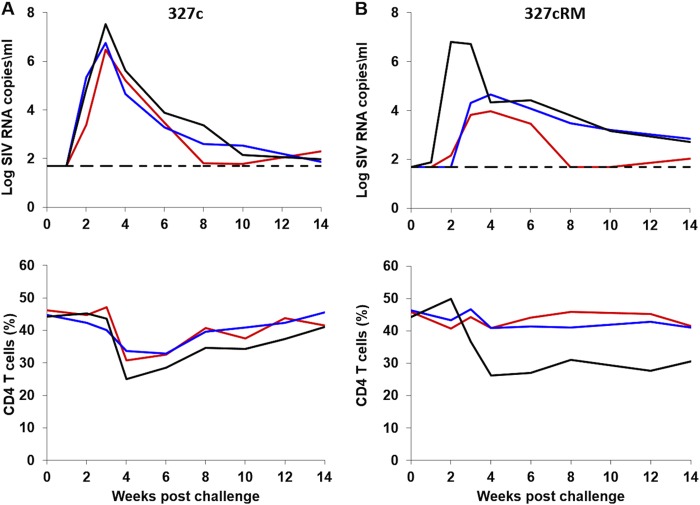
We next performed an in vivo titration to evaluate whether lower doses of the SHIV-327cRM stock would also prove infectious by the i.r. route. Five adult rhesus monkeys received a single i.r. inoculation of 1 ml of a 1:10 (n = 3) or a 1:100 (n = 2) dilution of the SHIV-327cRM stock. All three animals that received a single i.r. challenge with the 1:10 dilution were productively infected with peak viral loads ranging from 6.5 to 7.5 log RNA copies/ml, CD4+ T cell declines of 5.9 to 12.6% during acute infection, and chronic setpoint viremia in all animals (Fig. 6). Neither of the animals inoculated i.r. with the 1:100 dilution was infected after the first challenge. We therefore continued to perform a total of six repetitive i.r. challenges in these two animals using the 1:100 dilution of the challenge stock. These animals became infected after three and six challenges with peak viral loads of 6.3 and 6.8 log RNA copies/ml, respectively, and CD4+ T cell declines of 12% during acute infection (Fig. 7). These data show that both undiluted and a 1:10 dilution of the SHIV-327cRM stock reliably infected rhesus monkeys by the i.r. route and that a 1:100 dilution may prove optimal for repetitive i.r. challenge protocols.
FIG 6.
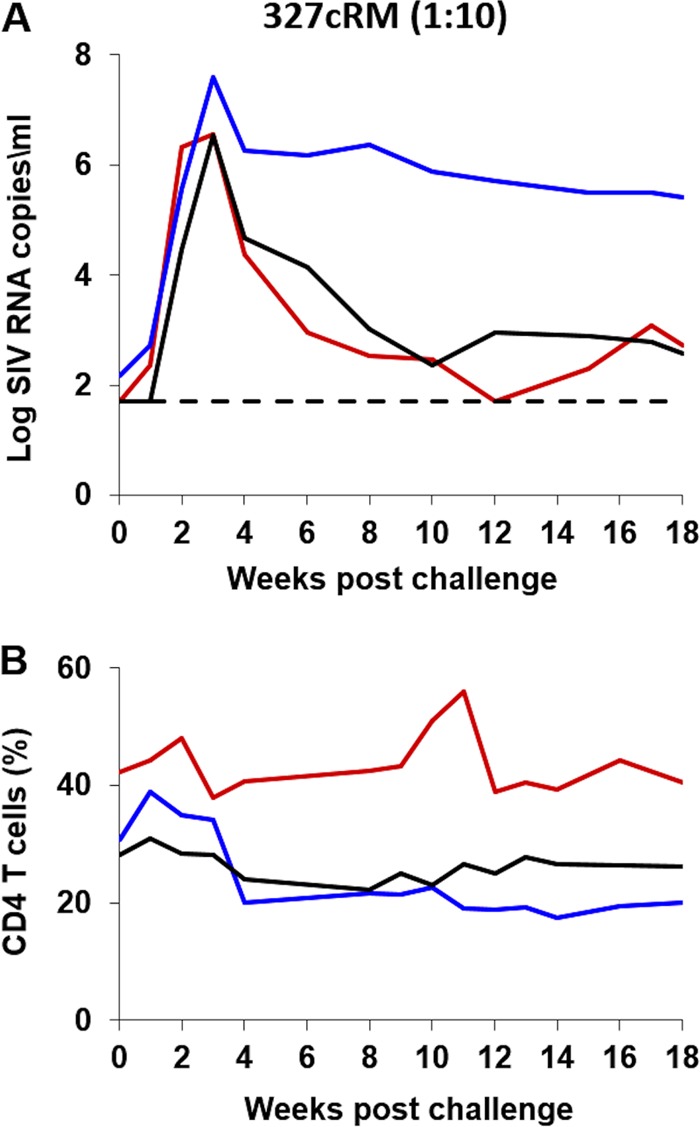
i.r. titration of the SHIV-327cRM stock in rhesus monkeys (1:10 dilution). Three animals were challenged once with 1 ml of 1:10-diluted SHIV-327cRM stock by the i.r. route. The upper panel shows plasma viral loads, and the lower panel shows the percentage of CD4+ T cells in peripheral blood. The dotted line reflects the limit of detection of the assay (50 RNA copies/ml).
FIG 7.
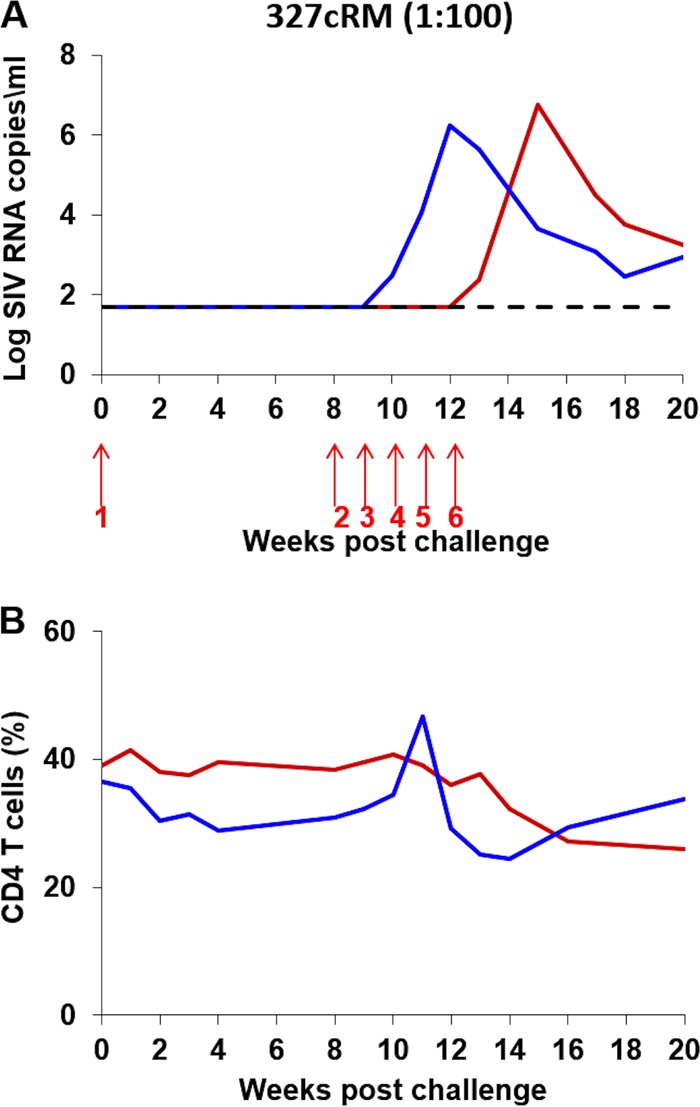
i.r. titration of the SHIV-327cRM stock in rhesus monkeys (1:100 dilution). Two animals were challenged six times (red arrows) with 1 ml of 1:100-diluted SHIV-327cRM stock by the i.r. route. The upper panel shows plasma viral loads, and the lower panel shows the percentage of CD4+ T cells in peripheral blood. The dotted line reflects the limit of detection of the assay (50 RNA copies/ml).
i.vag. infectivity of the SHIV-327c or SHIV-327cRM stocks in rhesus monkeys.
We next assessed the infectivity of the SHIV-327c and SHIV-327cRM stocks by the i.vag. route. 6 female monkeys were pretreated with Depo-Provera 1 month prior to challenge and were then infected with a single i.vag. inoculation of 1-ml undiluted SHIV-327c (n = 3) or SHIV-327cRM (n = 3) stocks. All monkeys were infected by the i.vag. route and exhibited high peak viral loads of 6.5 to 7.5 log RNA copies/ml in four animals and lower peak viral loads of 4.0 to 4.6 log RNA copies/ml in two animals (Fig. 8). During the acute phase of infection, CD4+ T cells declined by 5 to 18%. These data show that the SHIV-327c and SHIV-327cRM stocks reliably infected rhesus monkeys by both the i.r. and the i.vag. routes.
FIG 8.
i.vag. challenge with the clade C SHIV-327c and SHIV-327cRM stocks in rhesus monkeys. Six animals were pretreated 4 weeks prior to challenge with Depo-Provera and were then challenged once with 1 ml of undiluted (A) SHIV-327c (n = 3) or (B) SHIV-327cRM (n = 3) stocks by the i.vag. route. The upper panel shows plasma viral loads, and the lower panel shows the percentage of CD4+ T cells in peripheral blood. The dotted line reflects the limit of detection of the assay (50 RNA copies/ml).
DISCUSSION
In this study, we report the generation of mucosally transmissible clade C SHIV challenge stocks expressing early HIV-1 env sequences derived from acute HIV-1 infection. These SHIV-327c and SHIV-327cRM challenge stocks proved highly infectious in rhesus monkeys by both i.r. and i.vag. routes (Table 4) and resulted in high acute viral loads, moderate CD4+ T cell declines during primary infection, and chronic setpoint viremia in 13 of 17 (76%) of animals. These clade C challenge stocks were generated without in vivo passaging, and the SHIV-327c and SHIV-327cRM stocks encoded env sequences that matched the original human env sequence by 99.7 to 100%. These novel clade C SHIVs may prove useful in the preclinical evaluation of clade C HIV-1 vaccines, MAbs, and other prophylactic or therapeutic interventions.
TABLE 4.
Summary of in vivo infectivity of SHIV-327c and SHIV-327cRM stocks in rhesus monkeys
| Route | Dilution | Infectivity rate (%) |
|
|---|---|---|---|
| SHIV-327c | SHIV-327cRM | ||
| i.r. | 1:1 | 3/3 (100) | 3/3 (100) |
| 1:10 | ND | 3/3 (100) | |
| 1:100 | ND | 0/2 (0), 1/2 (50), 2/2 (100)a | |
| i.vag. | 1:1 | 3/3 (100) | 3/3 (100) |
The results for challenges 1, 3, and 6, respectively, are indicated.
Both SHIV-327c and SHIV-327cRM challenge stocks proved 100% infectious by i.r. and i.vag. routes when inoculated at a high dose and led to robust acute viral replication in nearly all animals. Infection also resulted in consistent declines of CD4+ T cell counts during acute infection and led to the establishment of setpoint viral loads in the majority of animals, but spontaneous virologic control was observed in a subset of animals. Serial in vivo passaging (1, 16, 27) could be performed to increase the pathogenicity of these challenge stocks, but in vivo adaptation would also likely lead to additional env mutations that would result in increased divergence from the original HIV-1 env sequence.
Two early clade C SHIVs, 1157i and 109F.PB4, have previously been reported (14, 19). The SHIV-1157i stock was generated using a rapid animal-to-animal adaptation strategy by transferring virus at peak viremia from one animal to the next, and mucosal transmissibility and pathogenicity was demonstrated after five serial passages with SHIV-1157ip (28) and with the late-stage virus SHIV-1157ipd3N4. Compared to the original infectious SHIV-1157i clone, mutations in the passaged viruses were present in variable loops resulting in decreased 2G12 sensitivity, and a 33-amino-acid insertion was observed in gp41 (29). Similarly, although the SHIV-C109F.PB4 clone was infectious, the mucosal transmissibility was demonstrated with the in vivo passaged viruses SHIV-C109P3, SHIV-C109P4, and SHIV-C109P3N, which contained amino acid changes in the variable loops and potential N-linked glycosylation sites (14). Our data extend these prior studies with the development of two additional early clade C SHIV challenge stocks, SHIV-327c and SHIV-327cRM, which express an early HIV-1 env from South Africa and did not require in vivo passaging.
In summary, we have generated clade C SHIV-327c and SHIV-327cRM challenge stocks that contain env sequences that were 99.7% to 100% identical to the original env sequence from acute HIV-1 infection. These stocks proved mucosally transmissible by both i.r. and i.vag. routes, CCR5 dependent, and relatively neutralization resistant and were titrated for use in single, high-dose and repetitive, low-dose mucosal challenge studies in rhesus monkeys. These clade C SHIV stocks should prove useful for the preclinical evaluation of vaccines, MAbs, and other interventions aimed at blocking acquisition of clade C HIV-1 infection.
ACKNOWLEDGMENTS
We thank J. Liu, K. E. Stephenson, K. M. Smith, E. N. Borducchi, C. Cabral, S. Blackmore, J. R. Perry, M. Beck, J. Kramer, and M. G. Lewis for advice, assistance, and reagents. The Human IL-2 and Ghost cells were obtained from the NIH AIDS Research and Reference Reagent Program.
We acknowledge support from the Bill and Melinda Gates Foundation (OPP1083689), the National Institutes of Health (AI096040, AI095985, AI084794, and AI078526), the HVTN Laboratory Program, and the Ragon Institute of MGH, MIT, and Harvard.
REFERENCES
- 1.Harouse JM, Gettie A, Eshetu T, Tan RC, Bohm R, Blanchard J, Baskin G, Cheng-Mayer C. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J Virol 75:1990–1995. doi: 10.1128/JVI.75.4.1990-1995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y, Salvato MS, Pauza CD, Li J, Sodroski J, Manson K, Wyand M, Letvin N, Jenkins S, Touzjian N, Chutkowski C, Kushner N, LeFaile M, Payne LG, Roberts B. 1996. Utility of SHIV for testing HIV-1 vaccine candidates in macaques. J Acquir Immune Defic Syndr Hum Retrovirol 12:99–106. [DOI] [PubMed] [Google Scholar]
- 3.Siddappa NB, Watkins JD, Wassermann KJ, Song R, Wang W, Kramer VG, Lakhashe S, Santosuosso M, Poznansky MC, Novembre FJ, Villinger F, Else JG, Montefiori DC, Rasmussen RA, Ruprecht RM. 2010. R5 clade C SHIV strains with tier 1 or 2 neutralization sensitivity: tools to dissect env evolution and to develop AIDS vaccines in primate models. PLoS One 5:e11689. doi: 10.1371/journal.pone.0011689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aidoo M, Otten RA, Rodriguez V, Sariol CA, Martinez M, Kraiselburd E, Robinson H, Folks T, Butera S, Ellenberger D. 2007. Absence of SHIV infection in gut and lymph node tissues in rhesus monkeys after repeated rectal challenges following HIV-1 DNA/MVA immunizations. Vaccine 25:6474–6481. doi: 10.1016/j.vaccine.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Shen Z, Li X, Bai J, Zeng L, Tian M, Song YJ, Du S, Ren D, Liu C, Zhu N, Sun D, Li Y, Jin N. 2012. Protection against SHIV-KB9 infection by combining rDNA and rFPV vaccines based on HIV multiepitope and p24 protein in Chinese rhesus macaques. Clin Dev Immunol 2012:958404. doi: 10.1155/2012/958404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsibris AM, Pal U, Schure AL, Veazey RS, Kunstman KJ, Henrich TJ, Klasse PJ, Wolinsky SM, Kuritzkes DR, Moore JP. 2011. SHIV-162P3 infection of rhesus macaques given maraviroc gel vaginally does not involve resistant viruses. PLoS One 6:e28047. doi: 10.1371/journal.pone.0028047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver EA, Nehete PN, Nehete BP, Buchl SJ, Palmer D, Montefiori DC, Ng P, Sastry KJ, Barry MA. 2009. Protection against mucosal SHIV challenge by peptide and helper-dependent adenovirus vaccines. Viruses 1:920. doi: 10.3390/v1030920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog 8:e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemelaar J, Gouws E, Ghys PD, Osmanov S, WHO-UNAIDS Network for HIV Isolation and Characterisation . 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia A, Siddappa NB, Li Q, Haase AT, Paul K, Stroud F, Zhang X, Fountain JA, Villinger F, Novembre FJ, Else JG, Evan Secor W, Ruprecht RM. 2010. AIDS and optic neuritis in a rhesus monkey infected with the R5 clade C SHIV-1157ipd3N4. J Med Primatol 39:356–360. doi: 10.1111/j.1600-0684.2010.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, Li Y, Yang G, Dai J, Ruprecht RM, Shao Y. 2011. Molecularly cloned SHIV-CN97001: a replication-competent, R5 simian/human immunodeficiency virus containing env of a primary Chinese HIV-1 clade C isolate. J medical primatology 40:427–436. doi: 10.1111/j.1600-0684.2011.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren W, Mumbauer A, Gettie A, Seaman MS, Russell-Lodrigue K, Blanchard J, Westmoreland S, Cheng-Mayer C. 2013. Generation of lineage-related, mucosally transmissible subtype C R5 simian-human immunodeficiency viruses capable of AIDS development, induction of neurological disease, and coreceptor switching in rhesus macaques. J Virol 87:6137–6149. doi: 10.1128/JVI.00178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siddappa NB, Song R, Kramer VG, Chenine AL, Velu V, Ong H, Rasmussen RA, Grisson RD, Wood C, Zhang H, Kankasa C, Amara RR, Else JG, Novembre FJ, Montefiori DC, Ruprecht RM. 2009. Neutralization-sensitive R5-tropic simian-human immunodeficiency virus SHIV-2873Nip, which carries env isolated from an infant with a recent HIV clade C infection. J Virol 83:1422–1432. doi: 10.1128/JVI.02066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol 80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol 74:4433–4440. doi: 10.1128/JVI.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautam R, Nishimura Y, Lee WR, Donau O, Buckler-White A, Shingai M, Sadjadpour R, Schmidt SD, LaBranche CC, Keele BF, Montefiori D, Mascola JR, Martin MA. 2012. Pathogenicity and mucosal transmissibility of the R5-tropic simian/human immunodeficiency virus SHIV(AD8) in rhesus macaques: implications for use in vaccine studies. J Virol 86:8516–8526. doi: 10.1128/JVI.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humbert M, Rasmussen RA, Song R, Ong H, Sharma P, Chenine AL, Kramer VG, Siddappa NB, Xu W, Else JG, Novembre FJ, Strobert E, O'Neil SP, Ruprecht RM. 2008. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology 5:94. doi: 10.1186/1742-4690-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Huang Y, Zhao X, Skulsky E, Lin D, Ip J, Gettie A, Ho DD. 2000. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina). J Virol 74:6501–6510. doi: 10.1128/JVI.74.14.6501-6510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humes D, Emery S, Laws E, Overbaugh J. 2012. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol 86:12472–12483. doi: 10.1128/JVI.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray GE, Allen M, Moodie Z, Churchyard G, Bekker LG, Nchabeleng M, Mlisana K, Metch B, de Bruyn G, Latka MH, Roux S, Mathebula M, Naicker N, Ducar C, Carter DK, Puren A, Eaton N, McElrath MJ, Robertson M, Corey L, Kublin JG, HPS Team . 2011. Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect Dis 11:507–515. doi: 10.1016/S1473-3099(11)70098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh WW, Rahman I, Hraber P, Coffey RT, Nevidomskyte D, Giri A, Asmal M, Miljkovic S, Daniels M, Whitney JB, Keele BF, Hahn BH, Korber BT, Shaw GM, Seaman MS, Letvin NL. 2010. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J Virol 84:6018–6032. doi: 10.1128/JVI.02741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DI, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. 2014. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitney JB, Hraber PT, Luedemann C, Giorgi EE, Daniels MG, Bhattacharya T, Rao SS, Mascola JR, Nabel GJ, Korber BT, Letvin NL. 2011. Genital tract sequestration of SIV following acute infection. PLoS Pathog 7:e1001293. doi: 10.1371/journal.ppat.1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reimann KA, Parker RA, Seaman MS, Beaudry K, Beddall M, Peterson L, Williams KC, Veazey RS, Montefiori DC, Mascola JR, Nabel GJ, Letvin NL. 2005. Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J Virol 79:8878–8885. doi: 10.1128/JVI.79.14.8878-8885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen RA, Ong H, Song R, Chenine AL, Ayash-Rashkovsky M, Hu SL, Polacino P, Else JG, Novembre FJ, Ruprecht RM, Clade CPP. 2007. Efficacy of a multigenic protein vaccine containing multimeric HIV gp160 against heterologous SHIV clade C challenges. AIDS 21:1841–1848. doi: 10.1097/QAD.0b013e32828684ea. [DOI] [PubMed] [Google Scholar]
- 29.Chenine AL, Siddappa NB, Kramer VG, Sciaranghella G, Rasmussen RA, Lee SJ, Santosuosso M, Poznansky MC, Velu V, Amara RR, Souder C, Anderson DC, Villinger F, Else JG, Novembre FJ, Strobert E, O'Neil SP, Secor WE, Ruprecht RM. 2010. Relative transmissibility of an R5 clade C simian-human immunodeficiency virus across different mucosae in macaques parallels the relative risks of sexual HIV-1 transmission in humans via different routes. J Infect Dis 201:1155–1163. doi: 10.1086/651274. [DOI] [PMC free article] [PubMed] [Google Scholar]