ABSTRACT
Tripartite motif-containing protein 5α (TRIM5α) is considered to be a potential target for cell-based gene modification therapy against human immunodeficiency virus type 1 (HIV-1) infection. In the present study, we used a relevant rhesus macaque model of infection with simian immunodeficiency virus from sooty mangabey (SIVsm) to evaluate the effect of TRIM5α restriction on clinical outcome. For macaques expressing a restrictive TRIM5 genotype, the disease outcomes of those infected with the wild-type TRIM-sensitive SIVsm strain and those infected with a virus with escape mutations in the capsid were compared. We found that TRIM5α restriction significantly delayed disease progression and improved the survival rate of SIV-infected macaques, supporting the feasibility of exploiting TRIM5α as a target for gene therapy against HIV-1. Furthermore, we also found that preservation of memory CD4 T cells was associated with protection by TRIM5α restriction, suggesting memory CD4 T cells or their progenitor cells as an ideal target for gene modification. Despite the significant effect of TRIM5α restriction on survival, SIV escape from TRIM5α restriction was also observed; therefore, this may not be an effective stand-alone strategy and may require combination with other targets.
IMPORTANCE Recent studies suggest that it may be feasible not only to suppress viral replication with antiviral drugs but also potentially to eliminate or “cure” human immunodeficiency virus (HIV) infection. One approach being explored is the use of gene therapy to introduce genes that can restrict HIV replication, including a restrictive version of the host factor TRIM5α. TRIM5 was identified as a factor that restricts HIV replication in macaque cells. The rhesus gene is polymorphic, and some alleles are restrictive for primary SIVsm isolates, although escape mutations arise late in infection. Introduction of these escape mutations into the parental virus conferred resistance to TRIM5 on macaques. The present study evaluated these animals for long-term outcomes and found that TRIM5α restriction significantly delayed disease progression and improved the survival rate of SIV-infected macaques, suggesting that this could be a valid gene therapy approach that could be adapted for HIV.
INTRODUCTION
Although the implementation of combination antiretroviral therapy (CART) has converted human immunodeficiency virus type 1 (HIV-1) infection from a fatal disease to a chronic disorder, the antiretroviral drugs still have negative consequences (1). In addition, despite effective suppression of viremia, patients receiving CART still experience immune activation and clinical effects due to ongoing low-level viral replication (2, 3). The case of the “Berlin patient” who was functionally cured of HIV-1-infection by receiving a hematopoietic stem cell transplant from a donor with a defective HIV CCR5 coreceptor (4) suggested gene therapy as an attractive method of curing HIV infection. Gene therapy involves genetic modification of CD4 T cells or hematopoietic progenitor cells to create HIV-resistant cells. Transplantation of these cells into HIV-infected patients has the potential to eradicate HIV infection (5). In addition to CCR5, host restriction proteins that inhibit HIV replication are considered candidate targets for gene modification. Among the host restriction proteins identified, including apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G (APOBEC3G), tetherin/bone marrow stromal cell antigen 2 (BST2), tripartite motif-containing protein 5α (TRIM5α), SAM domain and HD domain-containing protein 1 (SAMHD1), and Mx2, TRIM5α is the most promising target protein for gene therapy. TRIM5 was first identified as a restriction factor by the effect of the rhesus gene in blocking HIV-1 infection in primate cells (6, 7). Although human TRIM5α does not block HIV infection, restriction can be acquired by modifying the human SPRY/B30.2 domain through mutations or deletions, substitutions of critical rhesus TRIM5α sequence, or fusion with human cyclophilin A (CypA) to form a chimeric TRIM-CypA protein. Several studies have reported that resistance to HIV infection was achieved by the expression of modified TRIM5α in both cell lines and primary CD4 T cells (8–12). In the humanized mouse model, human CD34+ hematopoietic stem cells expressing chimeric rhesus/human TRIM5α were also successfully engrafted and gave rise to multilineage progenitors that were resistant to HIV infection (13).
Prior to the pursuit of TRIM5 as a gene therapy target, it is critical to assess the potential effect of a restrictive gene on disease progression. For example, before the use of the CCR5-delta32 mutant for gene therapy, retrospective clinical studies demonstrated a correlation between the expression of this allele and a reduction in HIV transmission risk and delay of disease progression in human populations (14–16). In contrast, the effect of restrictive TRIM5α alleles on clinical outcome has not been evaluated in vivo, since none of the known human TRIM5 variants are particularly restrictive for HIV-1. Although two genetic polymorphisms of human TRIM5α, H43Y and R136Q, have been observed in human populations (17, 18), they are associated with only a slight increase in the anti-HIV activity of human TRIM5α in vitro and had no effect or only a modest effect on HIV disease progression (17–22). However, rhesus macaques express polymorphic TRIM5 genes that are associated with various degrees of restriction of primary SIVsm (simian immunodeficiency virus from sooty mangabey) strains, and some alleles show a degree of restriction for SIVsm similar to that for HIV-1 (23). Hence, studies in the SIV/macaque model as a surrogate are feasible for evaluation of the relation between TRIM5α restriction and disease progression. In our previous studies, we found that an insertion/deletion polymorphism at amino acid positions 339 to 341 of the rhesus TRIM5α B30.2/SPRY domain, resulting in TFP/Q amino acid polymorphism, affected SIVsm replication in rhesus macaques. Viral loads were significantly lower in macaques with restrictive TRIM5TFP and TRIM5CypA alleles than in macaques with permissive TRIM5αQ alleles (23). This study did not allow us to evaluate the effect of expression of a restrictive gene on disease progression, due to the lack of clinical endpoints for many of the animals studied. However, we observed that restrictive alleles selected for escape mutants with common amino acid substitutions in the capsid late in infection (23, 24). The introduction of these amino acid substitutions into SIVsm resulted in enhanced virus acquisition and replication in rhesus macaques with restrictive alleles (24). In the present study, we evaluated whether such TRIM5α restriction also affects clinical outcome and disease progression after SIV infection by comparing the disease outcomes of macaques expressing a restrictive TRIM genotype infected with the wild-type SIVsm strain or with a virus with escape mutations in the capsid.
MATERIALS AND METHODS
Animals.
Colony-bred rhesus macaques of Indian origin (Macaca mulatta) were housed in a biosafety level 2 (BSL2) facility using BSL3 practices. This study was carried out in accordance with the recommendations of the Office of Laboratory Animal Welfare, National Institutes of Health, and the U.S. Department of Agriculture described in the Guide for the Care and Use of Laboratory Animals (25). All animal work was approved by the NIAID Division of Intramural Research IACUC (Institutional Animal Care and Use Committees) in Bethesda, MD (protocol LMM-6). The animal facility is accredited by the American Association for Accreditation of Laboratory Animal Care. All procedures were carried out under ketamine anesthesia by trained personnel under the supervision of veterinary staff, and all efforts were made to promote the animals' welfare and to minimize their suffering. Early-endpoint criteria, as specified by the IACUC-approved parameters, were used to determine when animals should be humanely euthanized. Macaques were euthanized if they lost >20% of their body weight or developed intractable diarrhea that was unresponsive to supportive or antibiotic treatment, respiratory signs with radiographic evidence of pneumonia, persistent anorexia and lethargy, or neurologic signs. Representative samples were taken for formalin fixation. All sections were stained with hematoxylin and eosin for routine histopathology.
The TRIM5 and major histocompatibility complex class I (MHC I) genotypes of rhesus macaques were determined as described previously (24). Twelve simian T-cell leukemia virus (STLV)-, simian retrovirus (SRV)-, and SIV-seronegative rhesus macaques with a TRIM5TFP/TFP genotype were divided into two groups and were inoculated intrarectally with a 1:50 dilution of SIVsmE543-3 or SIVsmE543-3 S37S98 virus stocks in peripheral blood mononuclear cells (PBMC) (1,000 50% tissue culture infective doses [TCID50]; 5 × 105 RNA copies). Four weeks later, animals that had not become infected were challenged weekly with the same dose of virus until viral RNA was detected in plasma samples. Blood and plasma were collected sequentially, and the viral RNA levels in plasma were determined by quantitative reverse transcriptase PCR (RT-PCR) as described previously (42).
Flow cytometric analysis.
Cryopreserved PBMC samples were thawed, washed in RPMI 1640 medium plus 10% fetal bovine serum (FBS), and incubated at 37°C for 2 h. After an extensive wash with phosphate-buffered saline (PBS), the PBMC were stained with the following combination of fluorochrome-conjugated monoclonal antibodies as described previously (42): Alexa Fluor 700-conjugated anti-human CD3, allophycocyanin (APC)-Cy7-conjugated anti-human CD20, APC-conjugated anti-human CD4, Pacific Blue-conjugated anti-human CD8, phycoerythrin (PE)-Cy5-conjugated anti-human CD95, electron-coupled dye (ECD)-conjugated anti-human CD28, and PE-conjugated anti-CCR5. Live and dead cells were distinguished by the absence of staining with an amine-reactive dye (Live/Dead Aqua). Labeled cells were fixed with 0.5% paraformaldehyde and were analyzed with an LSRFortessa fluorescence-activated cell sorting (FACS) system (BD Biosciences). Data were analyzed with FlowJo.
RT-PCR and gag sequence analysis.
Virus RNA was isolated from plasma samples of infected macaques with the QiaAmp viral RNA kit (Qiagen, Germany). Viral RNA was reverse transcribed to single-stranded cDNA using the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA) with primer Gag-R (5′-GCT GAT GAT TCA ATT GTA ACA GG-3′) according to the manufacturer's instructions. PCR products (1.8 kb) covering the full gag region were amplified by PCR using Platinum Taq high-fidelity polymerase (Invitrogen). PCR was performed using 1 μl of bulk cDNA with primers Nar-F (5′-GGT TGG CGC CCG AAC AGG GAC TT-3′) and Gag-R (5′-GCT GAT GAT TCA ATT GTA ACA GG-3′) under the following cycling conditions: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 2 min, with a final extension of 68°C for 5 min. The PCR products were purified on agarose gels and were sequenced. Capsid sequences were aligned using ClustalX2 and were compared with those of the parental SIVsmE543-3 clone (GenBank accession number U72748).
Statistical analyses.
All statistical analyses and graphic analyses were performed using GraphPad Prism, version 5 (GraphPad Software, La Jolla, CA). The cumulative survival rates of macaques infected with SIVsmE543-3 or the TRIM5α-resistant mutant SIVsmE543-3 S37S98 were plotted as Kaplan-Meier curves and were compared by a log rank test. The CD4+ T cell kinetics in these two groups were compared by two-way analysis of variance (ANOVA).
Nucleotide sequence accession numbers.
All sequences determined in this study have been deposited in GenBank under accession numbers KP162150 to KP162163.
RESULTS
TRIM5α restriction delayed disease progression and improved the survival rate of SIV-infected macaques.
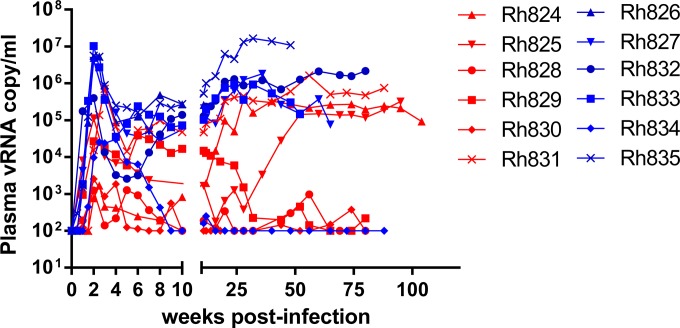
To investigate the impact of TRIM5α restriction on disease progression and clinical outcome, we inoculated 12 rhesus macaques with SIVsmE543-3 or its mutant SIVsmE543-3 S37S98. All inoculated macaques expressed homozygous TRIM5αTFP alleles. SIVsmE543-3, originally isolated from a sooty mangabey, has been through 2 passages in rhesus macaques (26) but is nevertheless sensitive to TRIM5αTFP restriction (23, 24). Two amino acid substitutions in the capsid region of Gag (P37S and R98S) seen in viruses (cloned from animals) that had escaped TRIM5αTFP restriction were introduced into the wild-type virus to generate SIVsmE543-3 S37S98. These two substitutions conferred resistance to TRIM5αTFP restriction in vitro (24). Thus, the viruses used in this study were isogenic except for these two amino acid substitutions. Our previous report followed the acute phase of infection of these macaques. The macaques infected with the TRIM5α-resistant mutant SIVsmE543-3 S37S98 showed a higher frequency of acquisition of infection and significantly higher plasma viremia during acute infection than those infected with the wild-type, TRIM5α-sensitive virus SIVsmE543-3 (24). In the present study, we followed these macaques clinically and virologically for 120 weeks postinfection (p.i.). Plasma samples were collected, and viremia was monitored sequentially (Fig. 1A). Among macaques infected with the TRIM5α-sensitive virus SIVsmE543-3, three (Rh828, Rh829, and Rh830) controlled virus replication after the acute phase of infection. Plasma viremia was suppressed below 103 RNA copies per ml in Rh828 and Rh830 after 16 weeks p.i. and in Rh829 after 32 weeks p.i. The suppression of virus replication could not be explained solely by MHC restriction, since only macaque Rh828 expressed restrictive MHC alleles (Mamu-A01 and Mamu-B17) (Table 1). Two macaques (Rh824 and Rh825) maintained plasma viremia above 105 RNA copies per ml after transient viremia suppression below 103 RNA copies per ml for 10 to 12 weeks. Macaque Rh831 maintained stable viremia above 104 to 105 RNA copies per ml after the acute phase of infection. In contrast, plasma viremia was considerably more robust and consistent in macaques infected with the TRIM5α-resistant variant SIVsmE543-3 S37S98. Only one macaque (Rh834) controlled virus replication and suppressed plasma viremia to 103 RNA copies per ml after the acute phase of infection. This macaque also expressed restrictive MHC allele Mamu-B17, suggesting a possible role of MHC restriction in virus suppression (Table 1). The other five macaques in this group maintained stable viremia above 105 RNA copies per ml.
FIG 1.
Replication of SIVsmE543-3 and SIVsmE543-3 S37S98 in macaques with the TRIM5TFP/TFP genotype. Viral loads were quantified and are shown as RNA copies in rhesus macaques infected with SIVsmE543-3 (red) or SIVsmE543-3 S37S98 (blue). vRNA, viral RNA.
TABLE 1.
Summary of clinical and pathological findings
| Inoculated virus | Macaque | MHC genotypea | Time to death (weeks p.i.) | Clinical and pathological findings | Opportunistic agent |
|---|---|---|---|---|---|
| SIVsmE543-3 | Rh824 | A08, B01 | 105 | Seizures, interstitial pneumonia, splenomegaly, PML | SV40 |
| Rh825 | A02, B01 | Surviving | N/Ab | N/A | |
| Rh828 | A01, A08, B17 | Surviving | N/A | N/A | |
| Rh829 | Negative | Surviving | N/A | N/A | |
| Rh830 | A08 | Surviving | N/A | N/A | |
| Rh831 | B17 | 95 | Lymphoproliferative disorder (kidney, lymph nodes, spleen), enteritis | N/Dc | |
| SIVsmE543-3 S37S98 | Rh826 | B01 | 11 | Significant weight loss, diarrhea, severe enterocolitis | Brachyspira |
| Rh827 | A02, A08 | 63 | Severe diarrhea, severe enterocolitis | Cytomegalovirus | |
| Rh832 | A01, B01 | 93 | Diarrhea, lymphoproliferative disorder (kidney, lymph nodes, spleen), enteritis | N/D | |
| Rh833 | A08, B01, B08 | 54 | Diarrhea, PML, esophagitis, enterocolitis | Candida, SV40 | |
| Rh834 | B17 | Surviving | N/A | N/A | |
| Rh835 | Negative | 41 | Severe diarrhea, enterocolitis, focal pyogranulomatous lymphadenitis | N/D |
Restrictive alleles Mamu-A01, -B08, and -B17 are shown in boldface.
N/A, not available.
N/D, not detected.
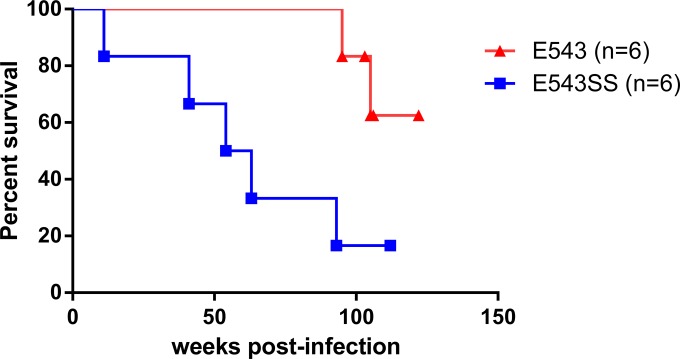
The clinical outcomes for these macaques are summarized in Table 1. Macaques were euthanized when they showed AIDS-related symptoms as described in Materials and Methods. Five of the macaques infected with the TRIM5α-resistant mutant SIVsmE543-3 S37S98 progressed to AIDS within 2 years after infection. Among these five macaques, one, Rh826, was a rapid progressor and was euthanized at 11 weeks p.i. The other four macaques in this group were euthanized around 1 to 2 years postinfection. All of these five macaques showed significant weight loss and severe, recurrent diarrhea unresponsive to medical treatment. One macaque (Rh833) also developed progressive multifocal leukoencephalitis (PML), tentatively ascribed to simian virus 40 (SV40) infection. Opportunistic infections (Brachyspira, Candida, and cytomegalovirus infections) and SIV-related enterocolitis were also detected in these macaques. In contrast, only two macaques infected with the TRIM5α-sensitive virus SIVsmE543-3 progressed to AIDS within 2 years after infection. One progressor (Rh831) was euthanized at 95 weeks p.i., and the other (Rh824) was euthanized at 105 weeks p.i. due to neurologic symptoms. Opportunistic SV40 infection was also indicated in macaque Rh824. The survival rates of these two groups were compared by a log rank test and are shown as a Kaplan-Meier plot in Fig. 2. Macaques infected with SIVsmE543-3 survived significantly longer than those infected with the TRIM5α-resistant mutant (P = 0.0141). The median survival time for the group infected with the TRIM5α-resistant mutant SIVsmE543-3 S37S98 was 58.5 weeks p.i., whereas the median survival time for the SIVsmE543-3-infected group could not be calculated due to the survival of four animals in this group but is more than 120 weeks. These results indicate that TRIM5α restriction delays disease progression and improves the survival rate after SIV infection.
FIG 2.
Survival curves of macaques infected with SIVsmE543-3 or SIVsmE543-3 S37S98. The cumulative survival rates of macaques infected with SIVsmE543-3 (red) or SIVsmE543-3 S37S98 (blue) are shown as Kaplan-Meier curves and were compared by a log rank test (P = 0.0141).
TRIM5α restriction resulted in the preservation of central memory CD4 T cell subsets in SIV-infected macaques.
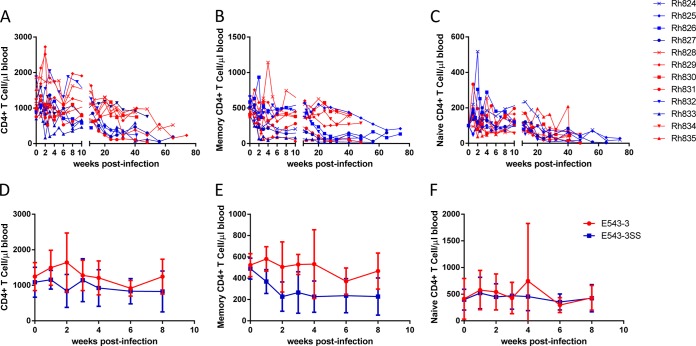
To further investigate the impact of TRIM5α restriction on disease progression, we analyzed CD4+ T cell kinetics in these infected macaques. As shown in Fig. 3A, declines in the levels of peripheral CD4+ T cells were observed in all of the infected macaques. This decline was due primarily to depletion of both central memory (CD28high CD95high) (Fig. 3B) and naïve (CD28high CD95low) (Fig. 3C) subsets of CD4+ T cells. The macaques that controlled viremia (Rh828, Rh829, and Rh830 in the SIVsmE543-3-infected group and Rh834 in the SIVsmE543-3 S37S98-infected group) showed significantly better preservation of peripheral central memory CD4+ T cells than the macaques that progressed to AIDS (Rh824 and Rh831 in the SIVsmE543-3-infected group and Rh826, Rh827, Rh832, Rh833, and Rh835 in the SIVsmE543-3 S37S98-infected group) (Fig. 3B), indicating a correlation between the loss of central memory CD4+ T cells and disease progression. To investigate the impact of TRIM5α restriction, the CD4+ T cell kinetics in macaques infected with SIVsmE543-3 and in those infected with the TRIM5α-resistant mutant SIVsmE543-3 S37S98 were compared by two-way ANOVA. The decline in the level of memory CD4+ T cells during the acute phase of infection was significantly delayed in SIVsmE543-3-infected macaques compared to that in SIVsmE543-3 S37S98-infected macaques (Fig. 3E) (P = 0.0126). The declines in the levels of total peripheral CD4+ T cells (Fig. 3D) and naïve subsets (Fig. 3F) did not differ significantly between these two groups (P > 0.05). These results indicate that the delay in disease progression and the improvement of the survival rate by TRIM5α restriction are associated with preservation of the central memory CD4+ T cell subset.
FIG 3.
Peripheral CD4+ T cell kinetics. (A to C) Peripheral CD4+ T cells (A), central memory CD4+ T cells (CD28+ CD95+) (B), and naïve CD4+ T cells (CD28+ CD95−) (C) were quantified by FACS and are shown as the absolute numbers per microliter of blood. (D to F) The kinetics of total CD4+ T cells (D), central memory CD4+ T cells (E), and naïve CD4+ T cells (F) at the acute phase of infection for groups infected with SIVsmE543-3 (red line) or SIVsmE543-3 S37S98 (blue line) were compared by two-way ANOVA.
TRIM5α restriction selected escape mutations.
As described above, three macaques (Rh824, Rh825, and Rh831) in the SIVsmE543-3-infected group failed to control virus replication. Two of them (Rh824 and Rh831) progressed to AIDS, although they survived longer than progressors in the SIVsmE543-3 S37S98-infected group. Furthermore, we also observed transient viremia suppression below 103 RNA copies per ml in macaques Rh824 and Rh825 before the failure of viremia control. These results suggest that SIV had escaped from TRIM5α restriction at the later stages of infection. To address this question, we collected plasma samples from these macaques at the chronic and end stages of infection and amplified gag regions by RT-PCR. The capsid regions of these PCR products were sequenced and were aligned to the SIVsmE543-3 capsid sequence. The nucleotide sequence alignments are shown in Fig. 4. A nonsynonymous C-to-T substitution was found at codon 37 in capsids amplified from each of these three macaques at both the chronic and the end stage of infection. This substitution resulted in the replacement of capsid amino acid (aa) 37, proline, by serine, a substitution we have observed in previous studies (24). This is the same substitution that we introduced into the parental virus to generate our TRIM-resistant variant SIVsmE543-3 S37S98. Several nonsynonymous substitutions were also observed at codon 98, encoding the other amino acid that we had mutated to achieve resistance to TRIM5TFP in SIVsmE543-3 S37S98. In our previous study (24), escape at position 98 was associated exclusively with an arginine-to-serine substitution. In contrast, the substitutions in the present study resulted in the replacement of aa 98 (arginine) variously by glycine, threonine, isoleucine, or serine (Table 2). The combination of mutations at codons 37 and 98 conferred escape from TRIM5α TFP restriction in a single-cycle infectivity assay (data not shown). In contrast, mutations were not observed at these two codons in virus sequences from macaques infected with the TRIM5α-resistant mutant SIVsmE543-3 S37S98, indicating that the mutations introduced were stable and did not result in a major fitness cost to the virus. The spontaneous appearance of mutations at these two critical capsid residues in viruses from macaques Rh824, Rh825, and Rh831 is consistent with their selection by TRIM5α and explains the eventual lack of viremia control in these animals.
FIG 4.
Variance of the SIV capsid sequence at chronic and late stages of infection. The nucleotide sequences encoding the capsid domains of SIV Gag cloned from the plasma of macaques Rh824, Rh825, and Rh831 (SIVsmE543-3 infected) and from the plasma of macaques Rh826, Rh827, Rh832, Rh833, and Rh835 (SIVsmE543-3 S37S98 infected) were aligned to the SIVSmE543-3 capsid sequence. The sequences encoding the capsid N-terminal domains are shown. Codons 37 and 98, which are associated with TRIM5α sensitivity, are highlighted in yellow. w.p.i., weeks postinfection.
TABLE 2.
Summary of amino acid substitutions in progressors
| Virus and macaque | Time point (weeks p.i.)a | Capsid amino acid at position: |
|
|---|---|---|---|
| 37 | 98 | ||
| SIVsmE543-3 | P | R | |
| Rh824 | 40 | S | G |
| 105* | S | T | |
| Rh825 | 60 | S | R/I |
| 106* | S | T | |
| Rh831 | 48 | S | R |
| 95* | S | R/S | |
| SIVsmE543-3 S37S98 | S | S | |
| Rh826 | 11* | S | S |
| Rh827 | 36 | S | S |
| 63* | S | S | |
| Rh832 | 35 | S | S |
| 93* | S | S | |
| Rh833 | 54* | S | S |
| Rh835 | 41* | S | S |
*, necropsy time point.
DISCUSSION
In this paper, we demonstrate that TRIM5α restriction affects the clinical outcomes of SIV-infected rhesus macaques. Using SIVsm clones that were either TRIM5α resistant or TRIM5α sensitive in macaques with the same restrictive TRIM5α genotype, we observed a 100- to 1,000-fold difference in viral load (24). In the present paper, we evaluated clinical outcome in this cohort and found that macaques infected with the TRIM5α-sensitive SIVsm clone survived longer and progressed to AIDS more slowly than those infected with the TRIM5α-resistant virus. A previous study by Lim et al. using SIVmac251-infected rhesus cohorts demonstrated similar effects of the TRIM5α genotype on viral load and survival (27), which was unexpected, since the capsid sequences of SIVmac251/239 appear to have TRIM5 escape mutations. Although the differences observed in the latter study were statistically significant, they were considerably less profound than those in our cohort infected with SIVsmE543-3, in agreement with the less effective restricting activity of TRIM5 on SIVmac251/239 strains (23, 27). Indeed, other studies have failed to observe differences in viral load or disease outcome due to the TRIM genotype in SIVmac251-infected macaques (28). While studies in macaques infected with SIV cannot be directly extrapolated to humans with HIV-1, these results nevertheless support the idea that the introduction of similar capsid mutations into human TRIM5 would have a significant effect on HIV replication, particularly since rhesus TRIM5 alleles strongly restrict HIV-1. This supports the feasibility of exploiting TRIM5α as a target for cell-based gene modification therapy for HIV-1. Furthermore, we also found that the preservation of central memory CD4+ T cells was associated with the protection by TRIM5α restriction, suggesting central memory CD4+ T cells or their progenitors as ideal targets for gene modification. It is also clear that despite a significant effect on survival, SIV was capable of evolving to escape TRIM restriction. Consequently, this may not be an effective stand-alone strategy and may require combination with other targets.
For several reasons, TRIM5α was considered a potential target for gene therapy when it was first identified as a protein restricting HIV replication in macaque cells. First, TRIM5α efficiently blocks HIV replication at a postentry stage before reverse transcription (6). Mathematical modeling has predicted that inhibitors that block replication before reverse transcription, as opposed to postintegration, would confer therapeutic benefit by promoting the survival and expansion of protected cells (29). Second, TRIM5α blocks virus replication by interaction with the HIV capsid protein. The sequence of the capsid protein is highly conserved, and therefore, TRIM5α can restrict across many different HIV clades and subtypes. Third, TRIM5α restriction is not antagonized by any viral accessory proteins (30). Furthermore, restriction proteins APOBEC3G and SAMHD1 have been reported to increase virus fitness by accelerating virus mutation and recombination (31–35), which has not been found during TRIM5α restriction.
The immunogenicity of TRIM5α may be an important issue when cell-based TRIM5α gene modification therapy goes into clinical trials. In humans, modification of human TRIM5α, usually in its SPRY/B30.2 domain, has been suggested for gene therapy, since natural human TRIM5α does not restrict HIV replication. It is possible, however, that the modified TRIM5α protein could stimulate an unexpected immune response and result in the clearance of transplanted gene-modified cells. In recent studies, lentiviral vectors were used to transfect transplanted progenitor cells in which modified TRIM5α protein was stably expressed (11, 13, 36). However, natural expression of TRIM5α protein was modulated by interferon responses. Furthermore, TRIM5α was reported to be involved in innate immune signaling and activation of the AP1 and NF-κB proinflammatory signaling pathways (37). Thus, the effect of overexpression of TRIM5α protein on the activation of the innate immune response should be evaluated. A possible solution would be to directly modify human TRIM5α into restrictive isoforms in the human genome using zinc finger nucleases or CRISPR/Cas9 gene-editing strategies (38–41) instead of introducing exogenous expression vectors. However, the safety of TRIM5α modification by this method should also be evaluated before clinical trials.
Our results suggest that virus mutation and escape may be another important issue when cell-based TRIM5α gene modification therapy goes into clinical trials. In the present study, mutants escaping TRIM5α restriction were detected in half of the macaques infected with TRIM5α-sensitive SIVsm. All of these macaques failed to control virus replication, and two of them progressed to AIDS. The efficacy of these modified TRIM5α isoforms in blocking HIV replication should be evaluated in vitro and in vivo. The combination of TRIM5α modification with other inhibitory therapy may be a possible way to reduce the appearance of escape mutants. For example, a recent study by Anderson et al. evaluated a combination lentiviral vector that encoded a CCR5 short hairpin RNA (shRNA), a human/rhesus macaque chimeric TRIM5α, and a transactivation response element (TAR) decoy. Transduction of this combination vector inhibited HIV replication and blocked the generation of escape mutants in both cultured and primary CD34+ hematopoietic progenitor cell (HPC)-derived macrophages in vitro (36). In the present study, we also found that one macaque (Rh834), expressing the restrictive MHC I allele Mamu-B17, successfully controlled virus replication despite inoculation with a TRIM5α-resistant virus. Although we did not evaluate the effect of MHC restriction on clinical outcome in this paper, these results suggest that multiple factors influence clinical outcome and that the SIVsm-infected rhesus macaque will be a relevant model with which to study the mechanisms and potential efficacy of such an approach.
ACKNOWLEDGMENTS
We thank Heather Cronise-Santis, Joanne Swerczek, and Richard Herbert at NIHAC for excellent care of the study animals.
This work was supported with federal funds from the intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Dybul M, Fauci AS, Bartlett JG, Kaplan JE, Pau AK, Panel on Clinical Practices for Treatment of HIV . 2002. Guidelines for using antiretroviral agents among HIV-infected adults and adolescents. Ann Intern Med 137:381–433. doi: 10.7326/0003-4819-137-5_Part_2-200209031-00001. [DOI] [PubMed] [Google Scholar]
- 2.Lewden C, Chene G, Morlat P, Raffi F, Dupon M, Dellamonica P, Pellegrin JL, Katlama C, Dabis F, Leport C, Agence Nationale de Recherches sur le Sida et les Hépatites Virales CO8 APROCO-COPILOTE and CO3 AQUITAINE Study Groups . 2007. HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J Acquir Immune Defic Syndr 46:72–77. doi: 10.1097/QAI.0b013e318134257a. [DOI] [PubMed] [Google Scholar]
- 3.Corbeau P, Reynes J. 2011. Immune reconstitution under antiretroviral therapy: the new challenge in HIV-1 infection. Blood 117:5582–5590. doi: 10.1182/blood-2010-12-322453. [DOI] [PubMed] [Google Scholar]
- 4.Hütter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. 2009. Long-term control of HIV by CCR5 delta32/delta32 stem-cell transplantation. N Engl J Med 360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 5.Hoxie JA, June CH. 2012. Novel cell and gene therapies for HIV. Cold Spring Harb Perspect Med 2:a007179. doi: 10.1101/cshperspect.a007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 7.Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 8.Yap MW, Nisole S, Stoye JP. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr Biol 15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Li X, Stremlau M, Lee M, Sodroski J. 2006. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J Virol 80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grutter C, Martinetti G, Mazzucchelli L, Grutter M, Manz MG, Luban J. 2009. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Invest 119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham QT, Bouchard A, Grutter MG, Berthoux L. 2010. Generation of human TRIM5α mutants with high HIV-1 restriction activity. Gene Ther 17:859–871. doi: 10.1038/gt.2010.40. [DOI] [PubMed] [Google Scholar]
- 13.Anderson J, Akkina R. 2008. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5α (TRIM 5α) in CD34+ cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum Gene Ther 19:217–228. doi: 10.1089/hum.2007.108. [DOI] [PubMed] [Google Scholar]
- 14.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy PE, Kumaraswami V, Giorgi JV, Detels R, Hunter J, Chopek M, Berger EA, Fauci AS, Nutman TB, Murphy PM. 1997. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med 3:23–36. [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyer SL, Wu LI, Akey JM, Emerman M, Malik HS. 2006. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5α in humans. Curr Biol 16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 18.Javanbakht H, An P, Gold B, Petersen DC, O'Huigin C, Nelson GW, O'Brien SJ, Kirk GD, Detels R, Buchbinder S, Donfield S, Shulenin S, Song B, Perron MJ, Stremlau M, Sodroski J, Dean M, Winkler C. 2006. Effects of human TRIM5α polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology 354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 19.Speelmon EC, Livingston-Rosanoff D, Li SS, Vu Q, Bui J, Geraghty DE, Zhao LP, McElrath MJ. 2006. Genetic association of the antiviral restriction factor TRIM5α with human immunodeficiency virus type 1 infection. J Virol 80:2463–2471. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldschmidt V, Bleiber G, May M, Martinez R, Ortiz M, Telenti A, Swiss HIV Cohort Study . 2006. Role of common human TRIM5α variants in HIV-1 disease progression. Retrovirology 3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama EE, Carpentier W, Costagliola D, Shioda T, Iwamoto A, Debre P, Yoshimura K, Autran B, Matsushita S, Theodorou I. 2007. Wild type and H43Y variant of human TRIM5α show similar anti-human immunodeficiency virus type 1 activity both in vivo and in vitro. Immunogenetics 59:511–515. doi: 10.1007/s00251-007-0217-7. [DOI] [PubMed] [Google Scholar]
- 22.van Manen D, Rits MA, Beugeling C, van Dort K, Schuitemaker H, Kootstra NA. 2008. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog 4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O'Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol 8:e1000462. doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F, Kirmaier A, Goeken R, Ourmanov I, Hall L, Morgan JS, Matsuda K, Buckler-White A, Tomioka K, Plishka R, Whitted S, Johnson W, Hirsch VM. 2013. TRIM5α drives SIVsmm evolution in rhesus macaques. PLoS Pathog 9:e1003577. doi: 10.1371/journal.ppat.1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed.National Academies Press, Washington, DC. [Google Scholar]
- 26.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins WR, Montefiori DC. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol 71:1608–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim SY, Rogers T, Chan T, Whitney JB, Kim J, Sodroski J, Letvin NL. 2010. TRIM5α modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog 6:e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenizia C, Keele BF, Nichols D, Cornara S, Binello N, Vaccari M, Pegu P, Robert-Guroff M, Ma ZM, Miller CJ, Venzon D, Hirsch V, Franchini G. 2011. TRIM5α does not affect simian immunodeficiency virus SIVmac251 replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J Virol 85:12399–12409. doi: 10.1128/JVI.05707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Laer D, Hasselmann S, Hasselmann K. 2006. Impact of gene-modified T cells on HIV infection dynamics. J Theor Biol 238:60–77. doi: 10.1016/j.jtbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 30.McCarthy KR, Johnson WE. 2014. Plastic proteins and monkey blocks: how lentiviruses evolved to replicate in the presence of primate restriction factors. PLoS Pathog 10:e1004017. doi: 10.1371/journal.ppat.1004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadler HA, Stenglein MD, Harris RS, Mansky LM. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol 84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulder LC, Harari A, Simon V. 2008. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A 105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wolinsky SM. 2010. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol 84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monajemi M, Woodworth CF, Benkaroun J, Grant M, Larijani M. 2012. Emerging complexities of APOBEC3G action on immunity and viral fitness during HIV infection and treatment. Retrovirology 9:35. doi: 10.1186/1742-4690-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen LA, Kim DH, Daly MB, Allan KC, Kim B. 2014. Host SAMHD1 protein promotes HIV-1 recombination in macrophages. J Biol Chem 289:2489–2496. doi: 10.1074/jbc.C113.522326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson JS, Javien J, Nolta JA, Bauer G. 2009. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5α protein, a CCR5 shRNA, and a TAR decoy. Mol Ther 17:2103–2114. doi: 10.1038/mt.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll D. 2011. Genome engineering with zinc-finger nucleases. Genetics 188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 40.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu F, Ourmanov I, Kuwata T, Goeken R, Brown CR, Buckler-White A, Iyengar R, Plishka R, Aoki ST, Hirsch VM. 2012. Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J Virol 86:8835–8847. doi: 10.1128/JVI.00923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]