ABSTRACT
Coevolution of herpesviruses with their respective host has resulted in a delicate balance between virus-encoded immune evasion mechanisms and host antiviral immunity. BILF1 encoded by human Epstein-Barr virus (EBV) is a 7-transmembrane (7TM) G-protein-coupled receptor (GPCR) with multiple immunomodulatory functions, including attenuation of PKR phosphorylation, activation of G-protein signaling, and downregulation of major histocompatibility complex (MHC) class I surface expression. In this study, we explored the evolutionary and functional relationships between BILF1 receptor family members from EBV and 12 previously uncharacterized nonhuman primate (NHP) lymphocryptoviruses (LCVs). Phylogenetic analysis defined 3 BILF1 clades, corresponding to LCVs of New World monkeys (clade A) or Old World monkeys and great apes (clades B and C). Common functional properties were suggested by a high degree of sequence conservation in functionally important regions of the BILF1 molecules. A subset of BILF1 receptors from EBV and LCVs from NHPs (chimpanzee, orangutan, marmoset, and siamang) were selected for multifunctional analysis. All receptors exhibited constitutive signaling activity via G protein Gαi and induced activation of the NF-κB transcription factor. In contrast, only 3 of 5 were able to activate NFAT (nuclear factor of activated T cells); chimpanzee and orangutan BILF1 molecules were unable to activate NFAT. Similarly, although all receptors were internalized, BILF1 from the chimpanzee and orangutan displayed an altered cellular localization pattern with predominant cell surface expression. This study shows how biochemical characterization of functionally important orthologous viral proteins can be used to complement phylogenetic analysis to provide further insight into diverse microbial evolutionary relationships and immune evasion function.
IMPORTANCE Epstein-Barr virus (EBV), known as an oncovirus, is the only human herpesvirus in the genus Lymphocryptovirus (LCV). EBV uses multiple strategies to hijack infected host cells, establish persistent infection in B cells, and evade antiviral immune responses. As part of EBV's immune evasion strategy, the virus encodes a multifunctional 7-transmembrane (7TM) G-protein-coupled receptor (GPCR), EBV BILF1. In addition to multiple immune evasion-associated functions, EBV BILF1 has transforming properties, which are linked to its high constitutive activity. We identified BILF1 receptor orthologues in 12 previously uncharacterized LCVs from nonhuman primates (NHPs) of Old and New World origin. As 7TM receptors are excellent drug targets, our unique insight into the molecular mechanism of action of the BILF1 family and into the evolution of primate LCVs may enable validation of EBV BILF1 as a drug target for EBV-mediated diseases, as well as facilitating the design of drugs targeting EBV BILF1.
INTRODUCTION
Epstein-Barr virus (EBV) is a human gammaherpesvirus classified as Human herpesvirus 4 in the genus Lymphocryptovirus (LCV). Worldwide, over 90% of adults are infected with EBV (1). Although EBV infections are normally asymptomatic, primary encounter with the virus can present as infectious mononucleosis (2). Under appropriate conditions (immunosuppression or Plasmodium falciparum coinfection), EBV can also function as an oncovirus, and EBV-driven transformation of B cells and epithelial cells is strongly associated with tumors (3).
Herpesviruses have coevolved with their respective hosts over millions of years, resulting in the development of strategies to promote their lifelong persistence within the host. Many of these strategies involve sophisticated immune evasion mechanisms to modulate the host immune response. Normal EBV infection therefore represents the establishment of a balance between host immune response and multiple viral immune evasion strategies. Despite the activation of a robust EBV-directed primary T-cell response and establishment of EBV-specific memory, acute EBV infection always results in lifelong persistence of the virus, even in immunocompetent individuals (4). Similar to the case with other herpesviruses, this ability of EBV to persist is presumably due, at least in part, to its ability to enter a stage of latency in which protein expression is minimized, thereby limiting viral antigen display to the immune system. However, in order to spread to a new host, infectious virus particles must be produced by entering the replicative or lytic phase (5).
EBV can be classified into two strains (types 1 and 2) based on sequence variation of the EBV-borne nuclear antigen (EBNA) gene (6). EBV type 1 is the more prevalent strain globally and is frequently detected in Caucasian and Asian people. In contrast, all EBV type 2 isolates originate from Central Africa, La Réunion, and New Guinea (7). Coinfection with both EBV strains is also possible but is more commonly detected in immunocompromised individuals (6). A recent study showed that EBV belongs to a single group of hominoid LCVs that arose by interspecies transfer from an Old World monkey (8), consistent with the occurrence of cross-species LCV transmission over evolutionary time. The restricted geographical localization of EBV type 2 to the Southern Hemisphere has also fostered the suggestion that the 2 viral strains may have evolved by a subsequent recombination event between EBV and a second, related Old World monkey LCV (7).
Aside from EBV, only two nonhuman primate (NHP) LCVs have been characterized: rhesus LCV (RhLCV; taxonomic name: Macacine herpesvirus 4), which infects Old World rhesus monkeys, and callitrichine LCV (CalHV3; Callitrichine herpesvirus 3), which infects New World marmosets. All 3 LCVs appear to share similar immune evasion strategies and have similar biological characteristics, with promotion of lymphocyte growth and transformation in vitro and persistent infection of B cells with the potential for oncogenic transformation in vivo (9, 10). Even though it is separated by an evolutionary distance of approximately 6 million years, RhLCV has a repertoire of viral genes identical to that of EBV, with an especially high degree of homology among the lytic-cycle proteins (range, 49 to 98% amino acid similarity) (8, 10). Although it shares a colinear genome, CalHV3 is more distantly related, with the absence of a number of genes present within RhLCV and EBV. Due to genetic and biological similarities with EBV, RhLCV infection in rhesus monkeys has been used as an animal model to study EBV infection. Similar to EBV infection in humans, persistent RhLCV infection appears to rely on a balanced host-virus relationship, as experimental infection with RhLCV can lead to the induction of B-cell lymphomas in immunosuppressed rhesus monkeys (11).
Several herpes- and poxviruses have captured and maintained a number of immune modulatory 7-transmembrane (7TM)-spanning G-protein-coupled receptor (GPCR) genes (12–14). These ancient events of molecular piracy have presumably been essential for the survival of LCVs during coevolution with their hosts over evolutionary time. Viral 7TM receptors have been shown to serve many functions. These functions include chemokine scavenge through broad-spectrum chemokine binding, as shown for the two most thoroughly characterized herpesvirus-encoded 7TM receptors: unique short region US28 from human cytomegalovirus (HCMV) (15) and open reading frame 74 (ORF74) from Kaposi's sarcoma-associated herpesvirus (KSHV) (16). In addition to scavenging host chemokines (17, 18), these receptors have additional functions and have been shown to reprogram intracellular signaling networks (19, 20), modulate cellular motility, and stimulate cytokine and growth factor secretion (21–24). Consistent with their immune evasion functions, most viral 7TM receptors characterized to date are dispensable for viral replication in vitro but are critical for normal viral growth in vivo (25–27).
EBV encodes a 7TM receptor (EBV BILF1) that is constitutively active through multiple signaling pathways (28, 29), similar to HCMV US28 (20, 30, 31) and KSHV ORF74 (16, 19, 31, 32). However, in contrast to most herpesvirus-encoded 7TM receptors, EBV BILF1 does not function as a chemokine receptor. Instead, EBV BILF1 is the first 7TM receptor shown to downregulate cell surface major histocompatibility (MHC) class I molecules (33–35). EBV BILF1 affects both the exocytic and the endocytic pathways of MHC class I molecules, contributing to impaired antigen presentation of MHC class I/peptide targets to CD8+ T cells (34). EBV-BILF1 receptor signaling and conformation also seem to play a central role. However, the precise molecular mechanism by which the BILF1 receptor targets MHC class I molecules is still unclear. RhLCV and CalHV3 are also known to encode BILF1 orthologues (33, 35). However, only BILF1 from RhLCV appears to share the MHC class I targeting function of EBV BILF1 (33, 35).
In the current study, we investigated BILF1 orthologues from an additional 12 recently identified NHP LCVs. As EBV has been suggested to originate from transmission of an ancient Old World monkey LCV to humans, we searched for putative BILF1 receptors encoded by these uncharacterized LCVs of NHPs and then explored their evolution, sequence conservation, and functional properties. Based on high amino acid sequence conservation, we identified putative BILF1 orthologues in all investigated LCVs at the same genomic position as the EBV BILF1. These BILF1 orthologues were identified in LCVs from diverse NHPs, ranging from great and lesser apes to Old and New World monkeys. BILF1 proteins clustered the identified LCVs into 3 phylogenetic groups (clades A to C). Selected receptors from each group were characterized further in terms of receptor signaling, cellular localization, and internalization in comparison to EBV BILF1.
MATERIALS AND METHODS
Ethics statement.
The samples analyzed in the present study originated from previous studies (8, 36). A detailed ethics statement is available in the publication of Scuda et al. (36).
Sample collection and processing.
Blood and tissue samples from 12 nonhuman primate species, previously collected from deceased wild and captive apes, Old World monkeys, and New World monkeys with PCR-confirmed presence of LCV (8), were included in the present study. The primate species are listed in Table 1, and details of the samples are available upon request. The DNA was prepared with the QiaAmp tissue kit according to the manufacturer's instructions.
TABLE 1.
Host species, viruses, abbreviations, and GenBank accession numbers of novel and known BILF1 sequences
| Host species | Virus name | Virus abbreviation | Complete receptor-encoding sequencea (bp) | Partial receptor-encoding sequence (bp) | Receptor nameb | GenBank accession no. |
|---|---|---|---|---|---|---|
| Novel BILF1 orthologues | ||||||
| Apes | ||||||
| Chimpanzee | Pan troglodytes lymphocryptovirus 1 | PtroLCV1 | 927 | PtroLCV1 BILF1c | KM091910 | |
| Gorilla | Gorilla gorilla lymphocryptovirus 1 | GgorLCV1 | 939 | GgorLCV1 BILF1c | KM091912 | |
| Gorilla gorilla lymphocryptovirus 2 | GgorLCV2 | 142 | GgorLCV2 BILF1 | —d | ||
| Orangutan | Pongo pygmaeus lymphocryptovirus 1 | PpygLCV1 | 948 | PpygLCV1 BILF1c | KM091909 | |
| Pongo pygmaeus lymphocryptovirus 2 | PpygLCV2 | 607 | PpygLCV2-BILF1 | KM091916 | ||
| Siamang | Symphalangus syndactylus lymphocryptovirus 1 | SsynLCV1 | 1041 | SsynLCV1-BILF1c | KM091911 | |
| Symphalangus syndactylus lymphocryptovirus 2 | SsynLCV2 | 607 | SsynLCV2 BILF1 | KM091915 | ||
| Old World monkeys | ||||||
| Long-tailed macaque | Macaca fascicularis lymphocryptovirus 1 | MfasLCV1 | 607 | MfasLCV1 BILF1 | KM091917 | |
| Patas monkey | Erythrocebus patas lymphocryptovirus 1 | EpatLCV1 | 957 | EpatLCV1 BILF1c | KM091913 | |
| Red colobus | Piliocolobus badius lymphocryptovirus 1 | PbadLCV1 | 975 | PbadLCV1 BILF1c | KM091907 | |
| New World monkeys | ||||||
| Black spider monkey | Ateles paniscus lymphocryptovirus 1 | ApanLCV1 | 924 | ApanLCV1 BILF1c | KM114261 | |
| White-faced saki | Pithecia pithecia lymphocryptovirus 1 | PpitLCV1 | 918 | PpitLCV1 BILF1c | KM091908 | |
| Known BILF1 orthologues | ||||||
| Primates | ||||||
| Human | Epstein-Barr virus | EBV | 939 | EBV BILF1 | NC_009334 | |
| Rhesus monkey (Old World primate) | Rhesus lymphocryptovirus | RhLCV | 939 | RhLCV BILF1 | AY037858 | |
| Marmoset (New World primate) | Callitrichine herpesvirus 3 | CalHV3 | 918 | CalHV3 BILF1 | KM091914 | |
| Ungulates | ||||||
| Domestic pig | Porcine lymphotropic herpesvirus 1 | PLHV1 | 978 | PLHV1 BILF1 | AF478169 | |
| Porcine lymphotropic herpesvirus 2 | PLHV2 | 915 | PLHV2 BILF1 | AY170317 | ||
| Porcine lymphotropic herpesvirus 3 | PLHV3 | 1017 | PLHV3 BILF1 | AY170315 | ||
| Wildebeest | Alcelaphine herpesvirus 1 | AlHV1 | 909 | AlHV1 BILF1 | NC_002531 | |
| Sheep | Ovine herpesvirus 2 | OvHV2 | 1254 | OvHV2 BILF1 | NC_007646 | |
| Horse | Equine herpesvirus 2 | EHV2 | 621 | EHV2 BILF1 | NC_001650 |
Including stop codon.
Original annotations of known BILF1 orthologues ORF in GenBank: ORF6 (CalHV3), vGPCR (PLHV1, -2, and -3), A5 (AIHV1), Ov5 (OvHV-2), and E6 (EHV2).
Detected by generic PCR using degenerate primers.
—, nucleotide sequence too short for GenBank submission; sequence information available on request.
PCR methods and sequencing.
Details of the different PCR methods are given below. All PCR products were purified by using a PCR purification kit (Qiagen) and directly sequenced with a BigDye Terminator cycle sequencing kit (Applied Biosystems, United Kingdom) in a 377 DNA automated sequencer (Applied Biosystems).
Generic PCR for amplification of LCV LF2 sequences.
For generic detection of partial LF2 sequences of LCV, PCR was carried out with a set of degenerate, nested primers with deoxyinosine substitutions (see Table S1 in the supplemental material) under conditions described previously (37). The primer-binding sites were placed in regions conserved among the gammaherpesviruses and only minimally degenerate in order to avoid amplification of gamma 2 herpesviruses.
For selective amplification of ORF LF2 of PpygLCV1 from samples doubly infected with Pongo pygmaeus lymphocryptovirus 1 (PpygLCV1) and PpygLCV2, an oligonucleotide with locked nucleic acid (LNA) substitutions (5′-cc+T+T+Acc+T+Gc+T+G+Gc+G-3′; TIB MOLBIOL GmbH, Germany) was used to selectively inhibit amplification of PpygLCV2 LF2 sequence under conditions described previously (38).
Specific LD PCR for amplification of sequences spanning from the BALF5 gene to the LF2 gene.
Nested nondegenerate primers (see Table S1) were designed on the basis of previously identified LCV BALF5 sequences (8, 39) and the LCV LF2 sequences determined in this study. Specific long-distance (LD) PCR was performed for each LCV in a nested format with the TaKaRa-Ex PCR system (TaKaRa Bio Inc., France) according to the manufacturer's instructions.
BILF1 PCR.
Partial BILF1 sequences were amplified with nested primer pairs, either degenerate (primer pairs 4141 and 4142) or nondegenerate (primer pairs 4176 and 4177) (see Table S1). They were deduced from an alignment of known BILF1 sequences and those determined in this study with BALF5-to-LF2 LD PCR. EBV BILF1 and CalHV3 BILF1 were amplified from primary sample material (8) using specific primers (not shown) based on the sequences published in GenBank (NCBI). PCR was carried out under the following conditions: 1 cycle at 95°C for 12 min and 45 cycles at 95°C for 30 s (annealing temperature [Tm] primer according to the manufacturer's instructions) and 72°C for 180 s, and 1 cycle at 72°C for 10 min.
Phylogenetic analysis and identification of conserved BILF1 receptor motifs.
For phylogenetic analysis of the BILF1 orthologues, amino acid sequences were aligned using the MAFFT (40) plug-in of the software Geneious Pro, version 6.1.6 (Biomatters Ltd.). The phylogenetic tree was constructed with the neighbor-joining module, and the reliability of the tree was analyzed by bootstrap analysis (100-fold resampling).
The alignments of the discovered BILF1 sequences with those from GenBank were done in Geneious 6.0.5 using MAFFT v6.814b. The BLOSUM62 matrix was applied with gap open penalty and offset values of 1.53 and 0.123, respectively. The distinction between extra- and intracellular regions was done using the Phobius webserver as a prediction program (41). A manual correction was implemented in the used prediction, extending TM3 so it began with CysIII:01. The sequence logo was generated using the web-based program WebLogo (http://weblogo.berkeley.edu).
Receptor cloning and recombinant G-protein plasmid.
The cloning of EBV BILF1, Pan troglodytes lymphocryptovirus 1 (PtroLCV1) BILF1, PpygLCV1 BILF1, Symphalangus syndactylus lymphocryptovirus 1 (SsynLCV1) BILF1, and CalHV3 BILF1 was done after amplification of the ORF with standard PCR techniques using Expand high-fidelity polymerase (Roche, Switzerland). The ORF was inserted into the vectors pCMV-HA and pCMV-c-myc (TaKaRa Bio Europe, France) by cohesive end ligation and transformed into Escherichia coli. All experiments were performed with the pCMV-HA constructs, except for the colocalization studies in which both pCMV-HA and pCMV-c-myc constructs were used. The recombinant G protein GαΔ6qi4myr was kindly provided by Evi Kostenis (Institute for Pharmaceutical Biology, University of Bonn, Bonn, Germany). GαΔ6qi4myr has switched receptor specificity from wild-type Gαq-interacting receptors to Gαi-interacting receptors and lacks the first 6 N-terminal amino acids, replacing the 4 C-terminal amino acids of wild-type Gαq with the corresponding 4 C-terminal amino acids from Gαi and including an N-terminal myristoylation site (42, 43).
Cell culture and transfection.
HEK-293 and COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Germany) plus 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. During luciferase-based experiments, the cell medium (DMEM) was modified to include heat-inactivated FBS and no penicillin-streptomycin. The HEK-293 cells were transfected using Lipofectamine 2000 (Invitrogen, Germany) according to the manufacturer's protocol for the transcription factor and the enzyme-linked immunosorbent assay (ELISA) experiments. For the [3H]inositol phosphate (IP3) turnover experiments, COS-7 cells were transfected by the calcium precipitation method (44). For the immunohistochemistry experiments, HEK-293 cells were transfected using the Effectene transfection reagent (Qiagen, Germany) according to the manufacturer's recommendations.
NFAT (nuclear factor of activated T cells), NF-κB, and CREB reporting experiments.
HEK-293 cells were seeded in 96-well culture plates and were transfected with 50 ng/well of the (cis-)reporter plasmids pNFAT-LUC and pNF-κB-LUC (both from Stratagene, USA) and various amounts of receptor DNA for CalHV3 BILF1 (5 ng/well). For the trans-reporting CREB (cyclic AMP response element binding protein) system, 50 ng/well of trans-activator plasmid (pFR-LUC) was cotransfected with 6 ng/well of the trans-reporter plasmid (pFA2-CREB) (both from Stratagene) and various amounts of receptor DNA as well as with and without 30 ng/well of the GαΔ6qi4myr plasmid DNA. Twenty-four hours after transfection, the cells were washed twice in phosphate-buffered saline (PBS), and 100 μl of PBS supplemented with 1 mg MgCl2 together with 100 μl of luciferase assay reagent (SteadyLite; PerkinElmer, USA) was added. Luminescence was measured in a Top-Counter (Top Count NXT, Packard, USA). For the forskolin stimulation, HEK-293 cells were transfected with various amounts of receptor DNA and the above-mentioned concentrations of CREB reporter plasmids. Twenty-four hours following transfection, cells were treated with 10 μM forskolin (Sigma, USA) in 100 μl of assay medium. The assay was terminated 6 h later and luminescence measured as described above.
Inositol phosphate production.
The inositol phosphate turnover experiment was performed as described previously. Briefly, 1 × 106 COS-7 cells were transfected with 5 μg of receptor DNA in the presence or absence of 5 μg of GαΔ6qi4myr. One day after transfection, the COS-7 cells were transferred to a 96-well plate (3.5 × 104 cells/well) and incubated with 4 μCi of myo-[3H]inositol in 100 μl of growth medium per well for 24 h. Cells were washed twice in PBS and were incubated in 0.1 ml Hanks' balanced salt solution (Invitrogen, United Kingdom) supplemented with 10 mM LiCl at 37°C for 45 min. Cells were extracted by addition of 40 μl of 10 mM formic acid to each well, followed by incubation on ice for 30 min. Twenty microliters of the extract was transferred to a white 96-well plate, and 80 μl of 1:8-diluted YSi poly-d-lysine-coated beads (PerkinElmer, USA) was added. Plates were sealed and shaken at maximum speed for at least 30 min and centrifuged for 5 min at 400 × g, and gamma radiation was counted in the Top-Counter mentioned above.
ELISA.
HEK-293 cells were transiently transfected with the receptor constructs at 15 ng/well. Twenty-four hours after transfection, the cells were fixed with 3.7% formaldehyde for 20 min, washed with PBS, and blocked in PBS containing 0.1% Tween 20 and 2% bovine serum albumin (BSA) for 1 h at room temperature (RT). The cells were subsequently incubated with a primary anti-hemagglutinin (anti-HA) antibody (anti-H11, clone 16B12, mouse; HISS Diagnostics, Germany) for 1 h at RT, washed (PBS), and incubated with a secondary goat anti-mouse horseradish peroxidase-conjugated antibody (Dianova, Denmark) for 1 h at RT. The peroxidase activity was determined by adding 3,3′-5,5′-tetramethyl benzidine substrate (Kem-En-Tec, Denmark), and the optical density (OD) at 450 nm was measured using a Victor2 multitask plate reader (PerkinElmer, USA).
Immunocytochemistry and confocal microscopy. (i) Colocalization of the BILF1 receptor orthologues with eGFPF.
HEK-293 cells were cultured on glass coverslips in 24-well plates and transiently transfected with the receptor DNA (400 ng/well) together with eGFPF plasmid DNA (200 ng/well) (Clontech). The farnesylated enhanced green fluorescent protein (eGFPF) vector contains the 20-amino-acid farnesylation signal from c-HA-Ras fused to a modified form of eGFPF. The cells were fixed 48 h after the transfection with 3.7% formaldehyde, washed with PBS, and permeabilized in 0.1% Triton-X for 10 min at RT. The cells were subsequently blocked in PBS containing 0.1% Tween 20 and 2% BSA for 1 h at RT and incubated with a primary antihemagglutinin antibody (anti-H11, clone 16B12, mouse; HISS Diagnostics) for 1 h at RT. After washing with PBS, the cells were incubated with a secondary Rhodamine Red-X-conjugated goat anti-mouse antibody (Dianova) for 1 h at RT. Cells were mounted with Moviol (Sigma) and analyzed using an inverted confocal microscope with a 63× water immersion plan-apochromatic objective with a 1.4 numerical aperture (Zeiss, Germany).
(ii) Colocalization of the BILF1 receptors.
HEK-293 cells were seeded and cotransfected with the constructs EBV BILF1 (HA tagged) and PtroLCV1 BILF1 (c-myc tagged), PpygLCV1 BILF1 (HA tagged) and PtroLCV1 BILF1 (c-myc tagged), and SsynLCV1 BILF1 (HA tagged) and PtroLCV1 BILF1 (c-myc tagged) (200 ng/well of receptor DNA). Cells were fixed and permeabilized as described for the colocalization studies of the BILF1 receptors with eGFPF. PtroLCV1 BILF1 cellular distribution was detected by incubation with a primary anti-c-myc antibody (rabbit; Sigma, USA) and a secondary Cy5-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch, Germany) for 1 h at RT. EBV BILF1 and SsynLCV1 BILF1 receptors were detected by direct immune staining for 1 h at RT with an anti-HA fluorescein isothiocyanate (FITC)-conjugated antibody (clone HA-7 FITC; Sigma). The washing, mounting, and confocal microscopy steps were performed as described for colocalization studies of the BILF1 receptor orthologues with eGFPF.
(iii) Antibody uptake studies.
HEK-293 cells were seeded and transfected as described for the colocalization studies. Forty-eight hours after the transfection, the cells were incubated in cold DMEM containing an anti-HA antibody (anti-H11, clone 16B12, mouse; HISS Diagnostics) in saturation for 1 h at 4°C. After 3 washes in cold DMEM, the cells were incubated in prewarmed medium at 37°C for 30 min and then fixed in 3.7% formaldehyde. The cells were washed with PBS and incubated with a FITC-conjugated goat anti-mouse antibody (Sigma) for 1 h at RT. The cells were permeabilized in PBS with 2% BSA and 0.2% saponin for 20 min, followed by incubation with a Rhodamine Red-X-conjugated goat anti-mouse antibody (Dianova) for 1 h at RT. Ultimately, the cells were analyzed by confocal microscopy.
Statistical analyses.
The data comparison between groups was performed by an unpaired two-tailed t test. The means ± SEM and statistics were calculated with GraphPad Prism software.
Nucleotide sequence accession numbers.
The novel BILF1 sequences determined in this study were deposited in GenBank under the accession numbers given in Table 1.
RESULTS
Identification of BILF1 receptor orthologues from uncharacterized LCVs of NHPs.
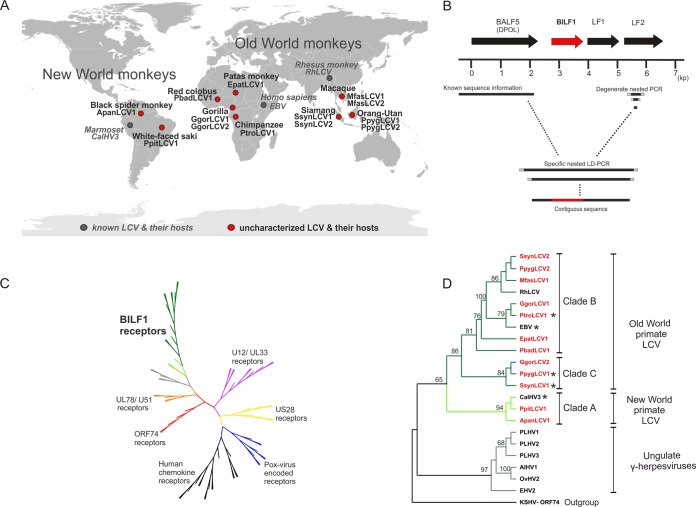
We searched for BILF1 receptor homologues in 12 novel LCVs identified in NHP hosts from Africa, Asia, and South America (Fig. 1A). As the genomes of RhLCV and CalHV3 have open reading frames (ORFs) similar in sequence, genomic position, and orientation to those of BALF5 (DNA polymerase [DPOL]), BILF1, LF1, and LF2 of EBV, we assumed that the same ORF organization is present in all NHP LCVs. Samples from 9 different NHP species had been identified earlier as LCV positive (3 NHP species hosted 2 LCVs) with generic PCR assays targeting the highly conserved glycoprotein B and DPOL genes (BALF4 and BALF5 in the EBV genome) (8). Using these samples, the viral genome region from BALF5 (DPOL) to LF2 containing the BILF1 orthologous ORF was amplified in two steps. First, partial LF2 sequences were amplified from the 12 LCVs by using generic nested PCR with degenerate primers that target conserved motifs in LF2 (Fig. 1B). Based on the published DPOL sequences of LCVs (8) and the newly determined LF2 sequences, specific DPOL sense and LF2 antisense primers were selected for each virus. Using nested long-distance (LD) PCR, the intervening sequence (approximately 3.5 kb) was then amplified. Eight LCV sequences were successfully amplified with LD PCR, sequenced, and determined with BLAST analysis to span a region encoding a BILF1 orthologue (Table 1). In cases where either generic LF2 PCR or LD PCR failed, partial BILF1-homologous sequences were amplified with generic nested PCR using primers that had been deduced from an alignment of known and novel BILF1 nucleic acid sequences. This approach was successful in the 4 remaining LCVs, of which partial BILF sequence of Gorilla gorilla lymphocryptovirus 2 (GgorLCV2) was amplified with the nested BILF1 primer set 4176/4177, whereas those of Macaca fascicularis lymphocryptovirus 1 (MfasLCV1), Pongo pygmaeus lymphocryptovirus 2 (PpygLCV2), and Symphalangus syndactylus lymphocryptovirus 2 (SsynLCV2) were amplified with the nested BILF1 primer set 4141/4142 (see Table S1 in the supplemental material). As revealed by BLAST analysis of GenBank, BILF1 sequences were amplified from all 12 LCVs, including 8 complete ORF and 4 partial sequences (Table 1). The analysis revealed also that although they are more distantly related to EBV BILF1, members of the genera Macavirus and Percavirus (ungulate gammaherpesviruses) also encode putative 7TM receptors at the homologous genomic position: porcine lymphotropic herpesviruses 1, 2, and 3, alcelaphine herpesvirus 1 and ovine herpesvirus 2 (genus Macavirus), and equine herpesvirus 2 (genus Percavirus) (Table 1).
FIG 1.
Identification of novel BILF1 receptor sequences from uncharacterized LCVs of nonhuman primates and phylogenetic studies. (A) World map of primate hosts harboring novel and known LCVs. The viruses are indicated in red and gray circles (novel and known LCVs, respectively); names of viruses and their hosts are written in bold blac (novel viruses) and italic gray (known viruses). Three ape hosts (gorilla, siamang, and orangutan) harbor 2 distinct LCVs. (B) Map of amplified genes and diagram of PCR strategy. EBV ORF BALF5 (DPOL), BILF1, LF1, and LF2 are symbolized as arrows (BILF1 ORF in red). A scale (in kb) oriented on the EBV genome is given below. The right side shows degenerate nested primers (light gray squares) used to amplify part of the LF2 gene. A short solid black line represents the amplified fragment. The left side represents published sequence information for the DNA polymerase gene of the investigated LCV (solid black line). Based on both DPOL and LF2 sequences, specific primers (gray squares) were selected and long-distance PCR was performed. A final contiguous sequence of approximately 3.5 kb was obtained, including the BILF1 ORF (red). (C) Phylogenetic tree of viral and endogenous 7TM receptors. The amino acid sequences were aligned with MAFFT and an unrooted tree was constructed using the neighbor-joining algorithm. The published U12/UL33 (purple), US28 (yellow), poxvirus-encoded receptor (blue), human chemokine receptor (black), ORF74 (red), and UL78/U51 (orange branches) sequences were aligned with the published and newly detected BILF1 sequences (green branches) and subjected to phylogenetic analyses. (D) Phylogenetic tree of BILF1 receptor family. BILF1 amino acid sequences were aligned with MAFFT and a rooted tree was constructed, using the phyml/bootstrap algorithm and the KSHV ORF74 sequence as an outgroup. The BILF1 sequences of New World primate LCVs (clade A; bright green branches) separate from those of Old World primate LCVs (clades B and C; dark green branches). The BILF1-homologous sequences from gammaherpesviruses of ungulates (gray branches) cluster separately from the primate BILF1 sequences. Newly discovered BILF1 sequences are highlighted in red, and BILF1 receptors used for functional studies are marked with a star.
Phylogenetic analysis of novel BILF1 receptors.
We performed phylogenetic analysis based on an alignment of a total of 63 7TM receptors comprising the 12 novel BILF1 sequences, 33 other virus-encoded 7TM ORF sequences, and 18 human chemokine receptors currently available in GenBank. The phylogenetic tree revealed that the BILF1 family constitutes the largest among the virus-encoded 7TM receptors, with closest relationship to the UL78/U51 and the most distant relationship to the human chemokine receptors (Fig. 1C). A more detailed phylogenetic analysis (PHYML algorithm) of the 12 novel and 3 previously known BILF1 sequences resulted in division of these primate LCVs into 3 major sister clades (A, B, and C) (Fig. 1D), comparable to results from viral glycoprotein B-based phylogeny (8). Clade A contains BILF1 orthologues from 3 New World monkey LCVs (Ateles paniscus lymphocryptovirus 1 [ApanLCV1; from black spider monkey], Pithecia pithecia lymphocryptovirus 1 [PpitLCV1; from white-faced saki], and CalHV3 [from marmoset]). Clade B is highly populated and comprises 4 novel BILF1 orthologues of great and lesser ape LCV (PtroLCV1 [from chimpanzee], GgorLCV1 [from gorilla], PpygLCV2 [from orangutan], and SsynLCV2 [from siamang]) and 3 novel BILF1 orthologues from Old World monkey LCVs (Erythrocebus patas lymphocryptovirus 1 [EpatLCV1], MfasLCV1, and Piliocolobus badius lymphocryptovirus 1 [PbadLCV1] [from patas monkey, long-tailed macaque, and red colobus, respectively]), besides the 2 known BILF1 receptors of EBV and rhesus macaque (EBV BILF1 and RhLCV BILF1). Clade C contains novel BILF1 orthologues from orangutan LCV (PpygLCV1), gorilla LCV (GgorLCV2), and siamang LCV (SsynLCV1).
Identification of conserved motifs in BILF receptor orthologues.
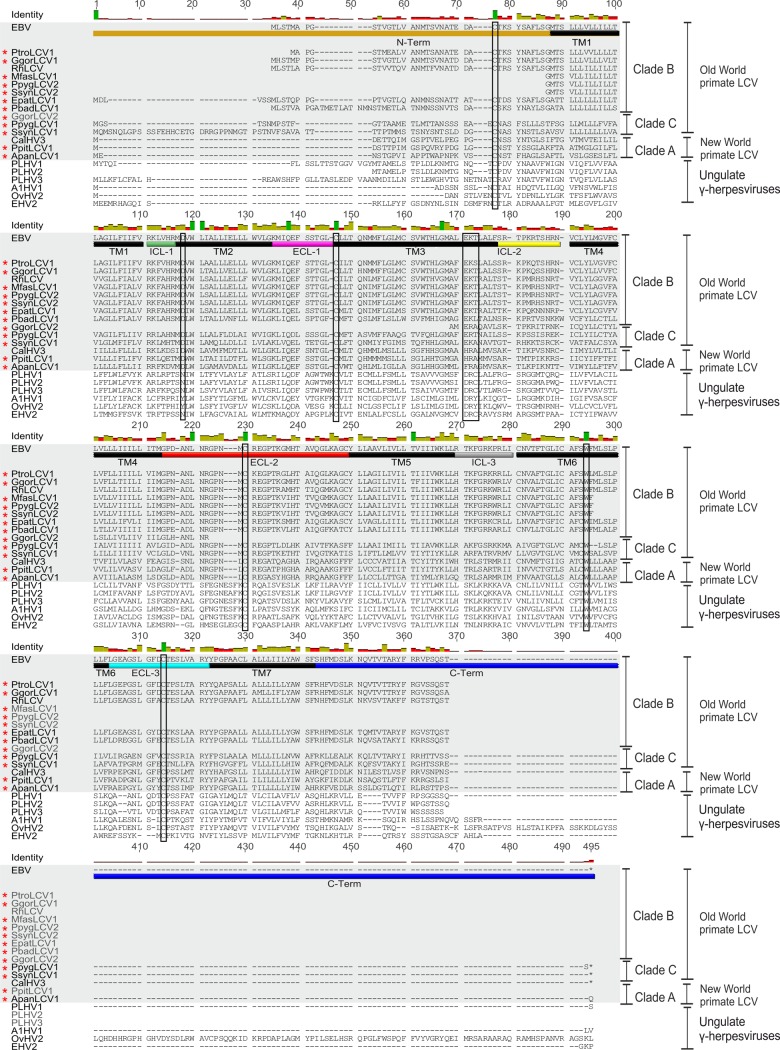
Structural analysis within the BILF1 family has so far been challenging due to the limited sequence information represented by only 3 primate-derived BILF1 and 6 more distantly related sequences from nonprimate ungulate gammaherpesviruses (Fig. 2). With identification of the 12 new NHP LCV BILF1 sequences, we were able to perform multiple sequence analysis of 15 closely related primate LCV BILF1 sequences and 6 sequences from the ungulate gammaherpesviruses (Fig. 2). BILF1 receptors are classified as class A 7TM receptors (45, 46). As expected, an overall high degree of conservation was observed within the predicted transmembrane (TM) helices of the BILF1 molecules. However, only a few residues were completely conserved. In TM3, CysIII:01 (3.25) (see next paragraph) was 100% conserved. This was also the case for residues putatively equivalent to AspII:10 (2.50) in TM2 and TrpVI:13 (6.48) in TM6 (45, 46). In contrast, prolines in TM5, -6, and -7 (ProV:16 [5.50], ProVI:15 [6.50], and ProVII:17 [7.50]) and AsnI:18 (1.50) in TM1, which are highly conserved among endogenous 7TM receptors, were absent from all sequences. The functionally important and highly conserved E/DRY (Glu/Asp-Arg-Tyr) motif (47) at the bottom of TM3 was only partially conserved. All BILF1 receptors display a positively charged amino acid (Arg or Lys) in position 2, and LCVs from apes and Old World monkeys (clades B and C) display a negatively charged amino acid (Glu) in position 1. Interestingly, all New World monkey clade A LCVs show a nonconserved change in position 1 (D to H/F). The third position is not conserved (Fig. 2).
FIG 2.
Multiple-sequence alignment of primate BILF1 receptors. An alignment of 21 BILF1 amino acid sequences was performed using MAFFT (Geneious 6.16). TM1 to TM7 of the BILF1 receptors are indicated with black bars. The intracellular loop regions (ICL-1, -2, and -3) are indicated with green, yellow, and gray bars, and the extracellular loop regions (ECL-1, ECL-2, and ECL-3) are indicated with bars in magenta, red and light blue. N and C termini are highlighted with bars in brown and dark blue, respectively. The alternative DRY motif and important conserved amino acid residues are marked with rectangles. The primate BILF1 amino acid sequences are highlighted with gray boxes and newly detected BILF1 amino acid sequences with red stars.
(The generic numbering system suggested by Schwartz and Rosenkilde (48, 49) is followed by the numbering system of Ballesteros and Weinstein (50). In the first system, the helices are numbered by roman numbers (I to VII) and the residues by arabic numbers. A helix is estimated to contain 26 residues, and the residue in the middle has number 13. Residues located to the N terminus of this have numbers below 13, and residues in the C terminus are counted upwards from 13. The second system assigns the most conserved residues in each helix with the number 50. A downward count towards the N terminus and an upward count towards the C terminus are applied for the remaining residues.)
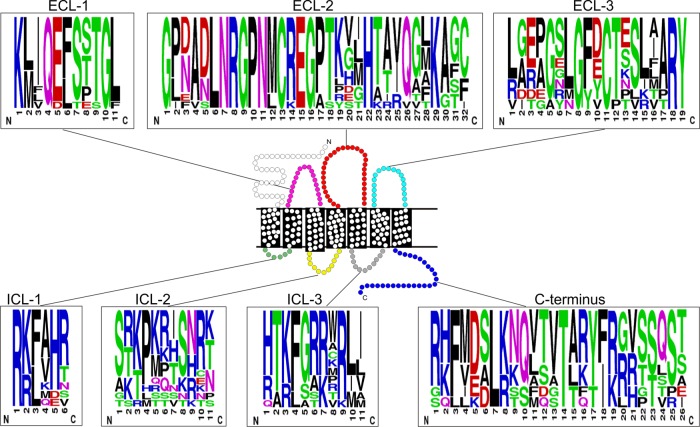
We next analyzed extracellular receptor regions (Fig. 2 and 3) and found a surprisingly high degree of conservation in particular in extracellular loop 2 (ECL-2)—a level of conservation that exceeded even that of the TM regions. Disulfide bridges between 2 Cys residues are common structural traits of extracellular loops of class A 7TM receptors. The Cys residues on the top of TM3 and in ECL-2, which are known to be involved in a disulfide bridge in class A 7TM receptors (51), were conserved in the BILF1 family (Fig. 2 and 3). All BILF1 receptors also contained a Cys in the N terminus and in ECL-3, consistent with the conservation of a second disulfide bridge between these two residues (52). Similar to the extracellular regions (ECL-1 to -3), the intracellular regions were also conserved in length and contained several conserved positively charged amino acid residues, in addition to an enrichment of serine and threonine residues, particularly in the C terminus.
FIG 3.
Sequence logo of the ICL and ECL domains as well as from the C termini of the BILF1 receptors. In the schematic picture of a 7TM receptor, the ECL and the ICL regions are depicted with the same colors as in Fig. 2. In the sequence logo, the chemical properties of the amino acids are represented in color (polar, green; neutral, purple; basic, blue; acidic, red; and hydrophobic, black). The figure was created using the web-based program WebLogo (http://weblogo.berkeley.edu).
Functional domain analysis of the BILF1 receptor orthologues.
BILF1 sequences from ape and Old World primate LCVs clustered within 2 phylogenetic clades (B and C) (Fig. 1D). EBV is believed to have been derived by transmission of an LCV from an Old World monkey of the subfamily Cercopithecinae to humans or an early hominoid lineage (8). Consistent with this derivation of EBV, BILF1 from RhLCV and EBV were both located within clade B (Fig. 1D and Table 1) and have been shown to share similarities in signaling (28, 29, 33). To expand on these earlier studies, we searched for putative conserved functional traits within the 7 novel BILF1 receptors in clade B and the 2 novel receptors in clade C. Two clade B members (EBV BILF1, and PtroLCV1 BILF1 from the chimpanzee) and 2 members from clade C (PpygLCV1 BILF1 and SsynLCV1 BILF1 from the orangutan and siamang, respectively) were selected for these comparative studies. PtroLCV1 BILF1 showed the highest sequence identity to EBV BILF1 (83.8%), followed by RhLCV BILF1 (80.4%) (Fig. 4). Despite the close evolutionary relationship of their hosts (orangutan and siamang) to humans (53), PpygLCV1 BILF1 and SsynLCV1 BILF1 showed relatively low identity to EBV BILF1 (46.7% and 45.7%, respectively) (Fig. 4).
FIG 4.
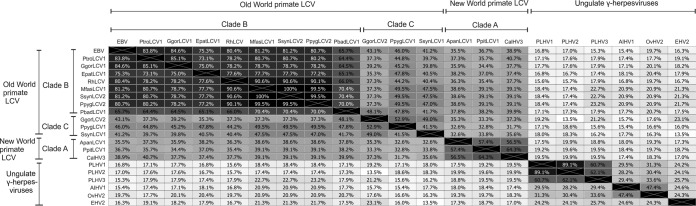
BILF1 amino acid sequence identities. Amino acid sequence identity was estimated based on an alignment of 21 BILF1 sequences and is displayed as a heat map.
Conserved constitutive signaling through Gαi and NF-κB-driven gene activation.
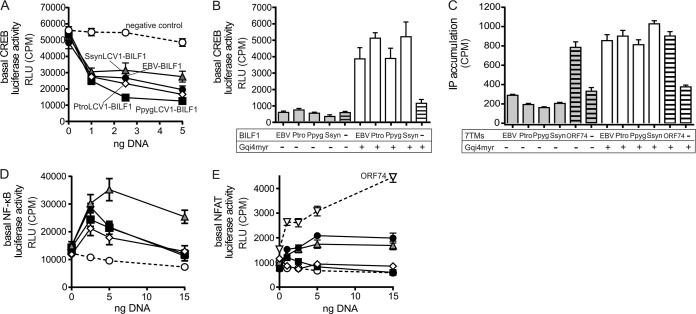
The selected BILF1 ORFs were inserted into the mammalian expression vector pCMV-HA for receptor functional studies. The pCMV-HA enables target protein expression to be confirmed based on the N-terminally localized hemagglutinin epitope tag. HEK-293 cells are commonly used for 7TM receptor signaling studies and were accordingly used for the molecular characterization of the primate BILF1 receptors, as the cells naturally targeted by the primate LCVs are in most instances not available. To determine whether the BILF1 receptor orthologues signal through Gαi as previously shown for EBV and RhLCV BILF1 (28, 29), receptor-mediated inhibition of forskolin-induced CREB (cyclic AMP response element binding protein) activation was investigated in transiently transfected HEK-293 cells (Fig. 5A). All ape-derived BILF1 receptors significantly inhibited the forskolin-induced increase of CREB in a dose-dependent manner, indicating that they all activate Gαi. No activity was observed in the absence of forskolin, indicating the absence of signaling through Gαs (Fig. 5B), again consistent with previous findings for EBV and RhLCV BILF1 (28, 29). Gαi signaling was supported by a robust activation of CREB following cotransfection of ape BILF1 receptor with the chimeric G protein GαΔ6qi4myr (Fig. 5B). This chimeric G protein is recognized as Gαi by 7TM receptors that utilize Gαi, but it functions as a Gαq subunit, leading to activation of CREB through phospholipase C. To examine signaling through Gαq, we measured the accumulation of [3H]inositol phosphate (IP3) in transiently transfected HEK-293 cells. For these studies, the constitutively active KSHV ORF74 7TM receptor served as a positive control (19, 31, 54). None of the 4 ape BILF1 receptors resulted in any basal activity through this pathway. KSHV ORF74 expression resulted in IP3 accumulation, as expected (Fig. 5C). Cotransfection with GαΔ6qi4myr resulted in accumulation of IP3, again confirming the activation of Gαi by the BILF1 receptors (Fig. 5C).
FIG 5.
CREB-mediated transcription regulation and activation of the transcription factors NF-κB and NFAT by BILF1 receptors. (A) Inhibition of forskolin (10 μM)-induced CREB activation with increasing doses of BILF1 receptor DNA in transiently transfected HEK-293 cells. The negative control is displayed in white circles, SsynLCV1 BILF1 in gray triangles, EBV-BILF1 in black circles, PtroLCV1 BILF1 in white squares, and PpygLCV1 BILF1 in black squares. (B) CREB activation by the BILF1 receptors in the absence (gray bars) and presence (white bars) of the promiscuous chimeric G protein GαΔ6qi4myr. The negative controls are shown as bars with black horizontal lines. (C) IP3 turnover in HEK-293 cells transfected with receptor DNA and in the absence (gray) or presence (white columns) of GαΔ6qi4myr. Positive controls (KSHV-ORF74) and negative controls (empty vector) are shown as bars with black lines. (D/E) Activation of the NF-κB (D) and the NFAT (E) transcription factors in transiently transfected HEK-293 cells by the different BILF1 receptors, with negative (vector) and positive (KSHV ORF74) controls. The controls and the BILF1 receptors are displayed as described for Fig. 3A (results of all experiments are shown as means ± SEM; n = 3).
EBV and RhLCV BILF1 have previously been shown to mediate NF-κB activation (29, 34, 35). This transcription factor plays a central role in inflammation through its ability to induce transcription of proinflammatory genes (55). An NF-κB reporter-based system in transiently transfected HEK-293 cells was used to study NF-κB activation of BILF1 receptors. As shown in Fig. 5D, the 3 novel ape BILF1 receptors activated NF-κB in a gene-dose-dependent manner, similar to EBV BILF1. Of these, the SsynLCV1-BILF1 showed the highest activity, whereas the BILF1 from EBV, PpygLCV1, and PtroLCV1 had similar activities.
An additional transcription factor, NFAT (nuclear factor of activated T cells), was analyzed due to its crucial role in the development and function of the immune system (56), as well as cell proliferation and induction of apoptosis (57). Activation of NFAT has never been tested for any of the BILF1 receptors, including EBV. We tested all 4 ape BILF1 receptors together with KSHV ORF74 as a positive control (Fig. 5E) (31). In contrast to the similar activation of CREB and NF-κB mediated by the 4 ape BILF1 receptors, only EBV and SsynLCV1 BILF1 receptors induced NFAT activation (at a level of approximately 40% activity compared to that of KSHV ORF74). Taken together, these results show that signaling profiles are not entirely conserved within the BILF1 family, yet there is a strong conservation of the Gαi signaling and the NF-κB activation, indicating that the latter pathways may play important roles in crucial aspects of LCV replication in its primate host.
Cellular expression, localization, and internalization of primate BILF1 receptors.
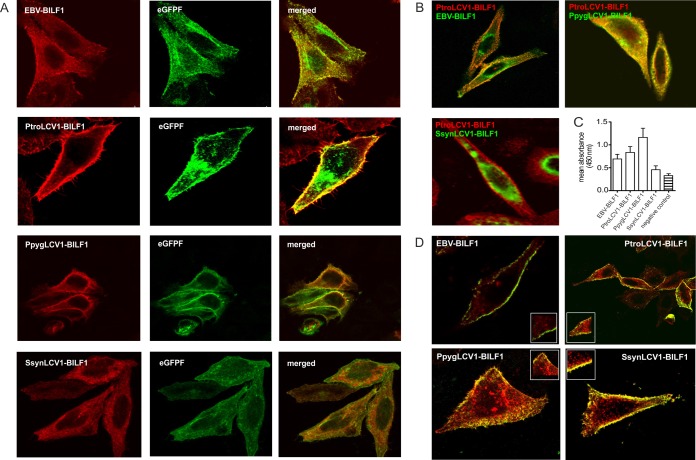
EBV-BILF1 mainly localizes to the cell membrane (28). To determine whether the other 3 ape BILF1 receptors showed a similar cellular localization pattern, HEK-293 cells were cotransfected with the EBV BILF1, PtroLCV1 BILF1, PpygLCV1 BILF1, or SsynLCV1 BILF1 expression plasmids together with a plasmid expressing a farnesylated enhanced green fluorescent protein (eGFPF) (a marker for cell membrane localization) followed by confocal microscopy analysis. As shown in Fig. 6A, 2 of the BILF1 receptors, PtroLCV1 BILF1 and PpygLCV1 BILF1, were predominantly localized to the cell membrane. Consistent with previously reported data (28), EBV BILF1 was also localized to the cell membrane, but it also displayed an additional intracellular accumulation. In contrast, SsynLCV1 BILF1 was primarily located within the cell.
FIG 6.
Distinct cellular localization patterns of BILF1 receptors. (A) HEK-293 cells were cotransfected with EBV BILF1, PtroLCV1 BILF1, PpygLCV1 BILF1, and SsynLCV1 BILF1 and the farnesylated enhanced green fluorescent protein (eGFPF). The representative pictures show the localization of the BILF1 receptors detected with a primary HA antibody against the HA tag expressed at the N termini of the receptors (red signal, left side), the signal of eGFPF (green signal, middle), and the merge signals of the BILF1 receptors with eGFPF (orange signal, right side). (B) Representative pictures of EBV BILF1- and PpygLCV1 BILF1-expressing HEK-293 cells (top) and SsynLCV1 BILF1-expressing HEK-293 cells (bottom), cotransfected with PtroLCV1 BILF1. EBV, PpygLCV1, and SsynLCV1 BILF1 molecules were detected with an anti-HA FITC-conjugated antibody using an N-terminal HA tag (green signal). PtroLCV1-BILF1was detected with a primary antibody against the c-myc tag (N terminal) and a secondary Cy5-conjugated antibody (red signal). (C) The graph shows cell surface expression levels of the BILF1 receptors as estimated in ELISA using an N-terminal HA tag (means ± SEM; n = 3). (D) Representative pictures of the constitutive internalization of the BILF1 receptors determined by antibody uptake studies. HEK-293 cells were transfected with HA-tagged EBV BILF1, PtroLCV1 BILF1, PpygLCV1 BILF1, and SsynLCV1 BILF1. Forty-eight hours after the transfection, receptors present at the cell surface were stained with a hemagglutinin antibody for 1 h at 4°C. Subsequently, cells were incubated at 37°C for 30 min to induce internalization and fixed afterwards. Labeled receptors still residing at the cell surface were detected with a FITC-conjugated antibody (green signal) prior to permeabilization, while internalized receptors were detected with a rhodamine-conjugated antibody (red signal).
To control for differences in subcellular localization due to different protein expression levels, we performed colocalization studies using cells that have been cotransfected with different BILF1 receptors expressing distinct epitope tags (c-myc and HA). HEK-293 cells were transfected with either HA-tagged EBV BILF1, PpygLCV1 BILF1, or SsynLCV1 BILF1, together with myc-tagged PtroLCV1 BILF1 (Fig. 6B). Consistent with the single-receptor expression studies (Fig. 6A), EBV BILF1 and, to a greater extent, SsynLCV1 BILF1 displayed intracellular accumulation in addition to the cell surface expression, in contrast to the primarily cell surface-localized PtroLCV1 BILF1and PpygLCV1 BILF1. Finally, to quantitatively assess the relative cell surface expression levels of the BILF1 proteins, we used an HA-specific ELISA to measure cell surface expression of HA-tagged versions of the 4 BILF1 receptors. Consistent with the results from the immunofluorescence studies, PpygLCV BILF1 and PtroLCV1 BILF1 showed higher cell surface localization than did EBV BILF1 and SsynLCV1 BILF1 (which had the lowest surface expression levels) (Fig. 6C). Overall, these results indicate that similar to NFAT activation, cellular localization patterns are not conserved within the BILF1 family.
Despite a suggested link between receptor endocytosis and MHC class I downregulation (33), uptake from the membrane has never been assessed for any BILF1 receptor. We therefore used transiently transfected HEK-293 cells combined with antibody uptake and confocal microscopy to assess whether the BILF1 receptors undergo constitutive internalization. For all 4 receptors, internalization was induced into endocytic vesicles after 30 min at 37°C (Fig. 6D).
The BILF1 receptor from an LCV of a New World monkey (marmoset CalHV3) displays properties similar to those of great ape BILF1 and EBV BILF1.
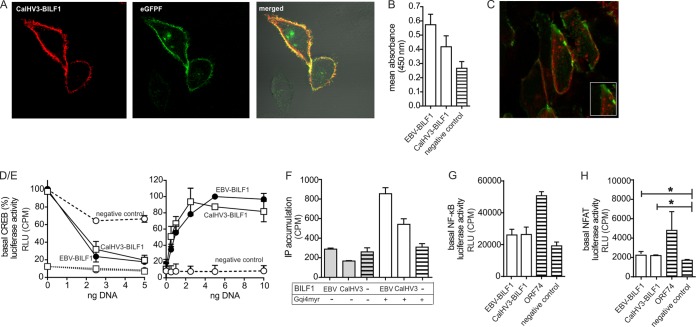
Phylogenetic analysis places the BILF1 receptor of the New World monkey (marmoset) LCV CalHV3 within clade A together with the BILF1 receptors of the New World monkey LCVs from PpitLCV1 and ApanLCV1 (Fig. 1D). However, CalHV3 BILF1 has previously been reported to differ from EBV and RhLCV BILFI through its inability to target MHC class I molecules (35). CalHV3 BILF1 also shows low identity (38.9%) to EBV BILF1 (Fig. 4). We therefore determined whether CalHV3 BILF1 receptor display the same characteristics of expression, signaling, and internalization as BILF1 from Old World primate LCVs. The CalHV3 BILF1 ORF was cloned into pCMV-HA and used for expression analysis and functional studies. CalHV3 BILF1 was found to be expressed mainly at the cell surface (Fig. 7A), with slightly lower expression levels than EBV BILF1 (Fig. 7B). Intriguingly, despite the lack of MHC class I targeting properties, this receptor undergoes constitutive endocytosis (Fig. 7C), similar to other BILF1 receptors (Fig. 6D). CalHV3 BILF1 also signaled constitutively via Gαi (determined by activation of CREB upon cotransfection of GαΔ6qi4myr) to the same level as EBV BILF1 (Fig. 7D/E). CalHV3 BILF1 also inhibited forskolin-induced CREB activation and mediated CREB activation only in the presence of GαΔ6qi4myr, thereby indicating the absence of signaling through Gαs (Fig. 7D/E). Similar to the other BILF1 receptors, CalHV3 BILF1 did not signal through Gαq, as IP accumulation was initiated only in the presence of GαΔ6qi4myr (Fig. 7F). With regard to transcription factor activation, CalHV3-BILF1 activated NF-κB and NFAT to the same extent as EBV-BILF1 (Fig. 7G and H). In summary, the New World marmoset LCV-derived CalHV3 BILF1 is a functional 7TM receptor with pharmacological properties similar to those of BILF1 receptors of clades B and C, but without the ability to downmodulate MHC class I molecules.
FIG 7.
Functional studies of CalHV3 BILF1. (A and B) To estimate cell surface expression levels of CalHV3 BILF1 in comparison to EBV BILF1, confocal microscopy (A) and ELISA (B) studies were performed as described for Fig. 6A and C. (C) Internalization studies were performed for CalHV3 BILF1 as described for Fig. 6D. (D/E) As shown in Fig. 5A and B for the ape and Old World monkey BILF1 receptors, CalHV3 BILF1-mediated CREB activity was estimated by cotransfection of CalHV3 BILF1 with GαΔ6qi4myr and in the presence of forskolin (D), as well as by cotransfection of CalHV3 BILF1 with GαΔ6qi4myr (E). EBV BILF1 was included in both experiments as a positive control. (F) IP3 turnover for CalHV3 BILF1 and EBV BILF1 is shown in the presence and in the absence of. GαΔ6qi4myr as described for Fig. 5C. (G and H) NF-κB (G) and NFAT (H) activation by CalHV3 BILF1 and EBV BILF1 was estimated by cotransfecting HEK-293 cells with receptor and reporter plasmid DNA. The significance of the NFAT activity is proven by Student's 2-tailed unpaired t test (for EBV BILF1, P < 0.011, and for CalHV3 BILF1, P < 0.021; significance is indicated with an asterisk for both). Representative pictures from the confocal microscopy studies (A and C) are shown, and the ELISA (B) and signaling (D to H) studies were performed 3 times (values are means ± SEM).
DISCUSSION
The frequent identification of virus-encoded 7TM receptors within pox- and herpesvirus genomes suggests that these receptors play important roles in the virus life cycle. With 7-membrane-spanning α-helices and mechanistic coupling to G proteins, these viral receptors all bear the structural hallmarks of classical endogenous 7TM receptors. With myriad identified pharmacological inhibitors, cellular 7TM receptors are accepted as highly druggable targets, and these virus-encoded counterparts may similarly represent putative drug targets for amelioration of virus-mediated diseases (58).
The most extensively studied viral 7TM receptors (KSHV ORF74 and HCMV US28) function as chemokine receptor homologues that interact with host (human) chemokines (16–18). The capacity of these receptors to promiscuously bind (and scavenge) human chemokines suggests their function in manipulation of the host immune system presumably to enhance survival as part of a viral immune evasion strategy (14). In contrast to KHSV ORF74 and HCMV US28, the LCV BILF1 receptors are not close homologues to any known cellular 7TM receptor and do not appear to bind host chemokines. However, the capacity of EBV and RhLCV BILF1 to modulate cellular signaling pathways and target a wide range of MHC class I molecules suggests that these receptors may similarly play a central role for immune evasion (28, 29, 33–35). The importance of BILF1 is underscored by the observation that all primate LCVs investigated in this study encode an intact BILF1 receptor ORF. Even within the ungulate gammaherpesviruses of the pig, wildebeest, sheep, and horse that have low sequence identity to primate LCVs, a putative 7TM receptor-encoding ORF has been identified (but not characterized yet) at a similar genomic position (59–61). However, not all gammaherpesviruses encode a BILF1 orthologue. Murine gammaherpesvirus 68 (MHV-68), a member of the related Rhadinovirus (RHV) genus, contains approximately 80 ORFs, 80% of which are homologues to genes of KSHV (genus RHV) and EBV (genus LCV) (62). MHV-68 is also closely related in its biological function to both viruses (63). Despite this high similarity to LCVs, neither MHV-68 nor KSHV encodes a BILF1 receptor, neither at the expected genomic position nor elsewhere. This exclusivity of BILF1 receptors to LCVs, ungulate percaviruses, and macaviruses suggests that a 7TM receptor-encoding ORF was captured once by a common ancestor of members of these genera, after divergence from the rhadinovirus lineage, which then evolved into BILF1 and its orthologues.
New and Old World monkey and great ape LCVs have a number of differences. Compared to EBV, the CalHV3 genome lacks 11 genes, some of which have known immune modulatory roles. This marked difference in gene repertoire makes New World LCVs interesting from an evolutionary perspective (10). Serological studies showed that LCV infection in New World primates may not be as ubiquitous as in humans (64), and the absence of these immune modulatory genes could feasibly have impacted the capacity for CalHV3 transmission or persistence within its host (65). Regarding receptor function, CalHV3 BILF1 is unable to downregulate MHC class I cell surface expression levels (35), which contrasts with LCVs of the Old World. In the present study, we found no clear differences between CalHV3 BILF1, and those of the other primate LCVs, suggesting that the lack of MHC class I targeting may rely more on subtle structural differences than on functional differences in the BILF1 receptors—knowledge that may facilitate an understanding of the molecular mechanism of how EBV BILF1 targets MHC class I molecules.
It has been suggested that both the exocytosis and the endocytosis of MHC class I are affected by EBV BILF1 (34). However, it has so far not been determined whether EBV BILF1 undergoes internalization from the cell surface. Herein, we show that EBV BILF1 does undergo recycling from the cell surface and that this property is conserved for all investigated New and Old World monkey and ape BILF1 molecules. This internalization may be necessary for the immune evasive mechanism of EBV BILF1, as it could occur in complex with MHC class I, thereby reducing MHC class I cell surface expression levels. A similar immune evasion strategy has been described for HCMV US28, which removes (scavenges) chemokines from the surface of HCMV-infected cells by constitutive internalization and recycling (17, 18). However, as CalHV3 BILF1 is clearly recycled from the cell surface but does not downregulate MHC class I, receptor recycling may be necessary but is obviously not sufficient for this immune evasion function. At present, the specific internalization and sorting motifs and signal-transducing adaptor proteins in EBV BILF1 remain to be identified, though it has been shown that the C terminus and the intracellular portion of the MHC class I molecules are essential for BILF1-mediated downregulation of MHC class I (35). Our sequence analysis of novel BILF1 receptors allowed the identification of several highly conserved amino acids in the C terminus, which may be required for internalization. Mapping experiments as the requirement of receptor cycling for MHC class I downregulation are the focus of ongoing experiments.
Our sequence analysis of the BILF1 receptors showed the conserved signaling motif (E/DRY motif at the bottom of TM3) to be present in a partially modified version. Other motifs were completely absent (for example, the PIF motif [in TM3] and the NPxxY motif [in TM7]) (45, 66). This modified version of the E/DRY motif has also been observed among other herpesvirus-encoded 7TM receptors (47). In spite of these differences, all BILF1 receptors signaled through Gαi and induced upregulation of NF-κB-controlled gene expression. For EBV BILF, G-protein signaling is known to be important for MHC class I targeting, as its disruption (by an inactivating point mutation in the EKT modified E/DRY motif) (67) abolished MHC class I downmodulation (33). Whether NF-κB signaling also plays a role for the immune evasion function of EBV BILF1 remains to be elucidated. Many viruses are believed to modulate NF-κB due to its role as a central mediator of the immune response via regulation of the expression of inflammatory cytokines, chemokines, and cell adhesion molecules (68). NF-κB also stimulates the transcription of MHC class I genes (69, 70), a cellular defense mechanism to help the host immune system to eradicate the virus-infected cell. This raises the question as to why viruses would activate NF-κB. EBV contains several immune evasive genes involved in targeting MHC class I molecules (71). As too-low MHC class I expression levels could feasibly alert the immune system (for natural killer cell-mediated removal), a counteraction could be needed to enable fine-tuning of MHC-I expression through virus-regulated NF-κB activation. Alternatively, the observed NF-κB activation could be irrelevant for the immune evasion function of EBV BILF1 and merely impact virus replication by altering cellular signaling pathways in the lytic phase.
In contrast to the overall similarity in the functional properties of the BILF, we observed differences in their subcellular distribution, with SsynLCV1 and EBV BILF1 differing from the predominant cell membrane localization of PpygLCV1, PtroLCV1, and CalHV3 BILF1. Variations in cellular expression patterns are not uncommon for virus-encoded 7TM receptors. HCMV US28 is predominantly intracellular (72), while KSHV ORF74 mainly locates to the cell membrane (73). Interestingly, only SsynLCV1, CalHV3, and EBV BILF1 mediated NFAT activity. Thus, SsynLCV1 BILF1 (clade C) and CalHV3 BILF1 (clade A), with low sequence identity to EBV BILF1 (clade B), have a higher degree of functional similarity to EBV BILF1, indicating that sequence similarity does not necessarily correlate with function. These differences could also be in part due to the use of HEK-293 cells as the default cell line, as these are not the natural target cells of primate LCVs. As viral receptors coevolve with their host, it is also possible that these receptors gain specific functions beneficial for host-virus interaction, thereby developing certain minor functional differences.
As an auxiliary effect, the constitutive signaling of EBV BILF1 has been shown to lead to the release of vascular endothelial growth factor (VEGF), cell transformation, and tumor formation in a xenograft tumor model (67). EBV BILF1 is an early lytic gene, yet lytic genes have been shown to exert an effect on EBV-associated tumorigenesis (74). Similar to the case with KSHV ORF74 and HCMV US28 (32, 54, 75), the proliferative and tumorigenic properties of EBV BILF1 have been linked to its constitutive activity (67). Consequently, EBV BILF1 may represent a potential druggable target for the treatment of EBV-mediated proliferative diseases.
We identified novel BILF1 sequences in 12 recently identified NHP LCVs with high or moderate sequence identity to EBV BILF1. This allowed identification of conserved motifs within the BILF1 receptors and revealed a large level of sequence variation and absence of common structural motifs compared to endogenous host cellular 7TM receptors. The absence of the 3 highly conserved prolines in TM5 to -7 represents such changes and would be expected to result in substantial structural changes as prolines commonly provide kinks in α-helices (45). This lack of conservation indicates overall major structural differences between the BILF1 receptor family and endogenous host 7TM receptors. In contrast, extracellular loop 2 (ECL-2) was conserved. This domain is generally important for receptor function by contributing to protein folding and conformational constraining in addition to forming the ligand-binding sites (76–78). ECL-2 is, on average, the longest of the 3 ECLs and encompasses the most divergent loop with sequence variations even within otherwise conserved class A 7TM receptor families (52, 79). The length and sequence of ECL-2 are surprisingly conserved for the BILF1 receptors, indicating an essential role of this receptor region. This could be as part of a ligand-binding site (MHC class I or yet unidentified ligand), as a regulator of ligand entry into the receptor, or as essential for conformational changes in the BILF receptors. A more thorough investigation of the role of conserved receptor parts could pave the path for the development of drug-like substances for EBV BILF1 and thereby a step further into opportunities for potential treatment of EBV-mediated diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fabian H. Leendertz, Kerstin Mätz-Rensing, and Andreas Ochs for supply with samples from wild and captive nonhuman primates, Nezlisah Yasmum, Petra Kurzendörfer, Sonja Liebmann, and Cornelia Walter for excellent technical support, and Tim Finsterbusch, Bettina Müller, Valentina Kubale, and Jacek Mokrosinski for their scientific advice.
This work was supported by the Lundbeck Foundation (R19-2008-2376), the Novo Nordisk Foundation (11-32538), and the Danish Research Council (12-127384).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02716-14.
REFERENCES
- 1.Küppers R. 2003. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat Rev Immunol 3:801–812. doi: 10.1038/nri1201. [DOI] [PubMed] [Google Scholar]
- 2.Henle G, Henle W, Diehl V. 1968. Relation of Burkitt's tumor-associated herpes-ytpe virus to infectious mononucleosis. Proc Natl Acad Sci U S A 59:94–101. doi: 10.1073/pnas.59.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutok JL, Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 4.Hislop AD, Ressing ME, van Leeuwen D, Pudney VA, Horst D, Koppers-Lalic D, Croft NP, Neefjes JJ, Rickinson AB, Wiertz EJHJ. 2007. A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J Exp Med 204:1863–1873. doi: 10.1084/jem.20070256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford DH. 2001. Biology and disease associations of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci 356:461–473. doi: 10.1098/rstb.2000.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. 2009. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res 143:209–221. doi: 10.1016/j.virusres.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimber U, Adldinger HK, Lenoir GM, Vuillaume M, Knebel-Doeberitz MV, Laux G, Desgranges C, Wittmann P, Freese UK, Schneider U. 1986. Geographical prevalence of two types of Epstein-Barr virus. Virology 154:56–66. doi: 10.1016/0042-6822(86)90429-0. [DOI] [PubMed] [Google Scholar]
- 8.Ehlers B, Spiess K, Leendertz F, Peeters M, Boesch C, Gatherer D, McGeoch DJ. 2010. Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus. J Gen Virol 91:630–642. doi: 10.1099/vir.0.017251-0. [DOI] [PubMed] [Google Scholar]
- 9.Cho Y, Ramer J, Rivailler P, Quink C, Garber RL, Beier DR, Wang F. 2001. An Epstein-Barr-related herpesvirus from marmoset lymphomas. Proc Natl Acad Sci U S A 98:1224–1229. doi: 10.1073/pnas.98.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F. 2013. Nonhuman primate models for Epstein-Barr virus infection. Curr Opin Virol 3:233–237. doi: 10.1016/j.coviro.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orlova N, Wang F, Fogg MH. 2011. Persistent infection drives the development of CD8+ T cells specific for late lytic infection antigens in lymphocryptovirus-infected macaques and Epstein-Barr virus-infected humans. J Virol 85:12821–12824. doi: 10.1128/JVI.05742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcami A, Lira SA. 2010. Modulation of chemokine activity by viruses. Curr Opin Immunol 22:482–487. doi: 10.1016/j.coi.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raftery M, Müller A, Schönrich G. 2000. Herpesvirus homologues of cellular genes. Virus Genes 21:65–75. doi: 10.1023/A:1008184330127. [DOI] [PubMed] [Google Scholar]
- 14.Rosenkilde MM, Smit MJ, Waldhoer M. 2008. Structure, function and physiological consequences of virally encoded chemokine seven transmembrane receptors. Br J Pharmacol 153(Suppl):S154–S166. doi: 10.1038/sj.bjp.0707660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kledal TN, Rosenkilde MM, Schwartz TW. 1998. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett 441:209–214. doi: 10.1016/S0014-5793(98)01551-8. [DOI] [PubMed] [Google Scholar]
- 16.Rosenkilde MM, Kledal TN, Bräuner-Osborne H, Schwartz TW. 1999. Agonists and inverse agonists for the herpesvirus 8-encoded constitutively active seven-transmembrane oncogene product, ORF-74. J Biol Chem 274:956–961. [DOI] [PubMed] [Google Scholar]
- 17.Billstrom MA, Lehman LA, Scott Worthen G. 1999. Depletion of extracellular RANTES during human cytomegalovirus infection of endothelial cells. Am J Respir Cell Mol Biol 21:163–167. doi: 10.1165/ajrcmb.21.2.3673. [DOI] [PubMed] [Google Scholar]
- 18.Bodaghi B, Jones TR, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier J-L, Michelson S. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J Exp Med 188:855–866. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn MC, Cesarman E. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 20.Casarosa P, Bakker RA, Verzijl D, Navis M, Timmerman H, Leurs R, Smit MJ. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J Biol Chem 276:1133–1137. doi: 10.1074/jbc.M008965200. [DOI] [PubMed] [Google Scholar]
- 21.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gershengorn MC, Mesri EA, Gerhengorn MC. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 22.Sodhi A, Montaner S, Patel V, Zohar M, Bais C, Mesri EA, Gutkind JS. 2000. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res 60:4873–4880. [PubMed] [Google Scholar]
- 23.Pati S, Cavrois M, Guo HG, Foulke JS, Kim J, Feldman RA, Reitz M. 2001. Activation of NF-kappaB by the human herpesvirus 8 chemokine receptor ORF74: evidence for a paracrine model of Kaposi's sarcoma pathogenesis. J Virol 75:8660–8673. doi: 10.1128/JVI.75.18.8660-8673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maussang D, Langemeijer E, Fitzsimons CP, Stigter-van Walsum M, Dijkman R, Borg MK, Slinger E, Schreiber A, Michel D, Tensen CP, van Dongen GAMS, Leurs R, Smit MJ. 2009. The human cytomegalovirus-encoded chemokine receptor US28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res 69:2861–2869. doi: 10.1158/0008-5472.CAN-08-2487. [DOI] [PubMed] [Google Scholar]
- 25.Davis-Poynter NJ, Lynch DM, Vally H, Shellam GR, Rawlinson WD, Barrell BG, Farrell HE. 1997. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol 71:1521–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beisser PS, Grauls G, Bruggeman CA, Vink C. 1999. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J Virol 73:7218–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira J, Schall TJ, Corey L, Geballe AP. 1998. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J Virol 72:8158–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen SJ, Rosenkilde MM, Eugen-Olsen J, Kledal TN. 2005. Epstein-Barr virus-encoded BILF1 is a constitutively active G protein-coupled receptor. J Virol 79:536–546. doi: 10.1128/JVI.79.1.536-546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beisser PS, Verzijl D, Gruijthuijsen YK, Beuken E, Smit MJ, Leurs R, Bruggeman CA, Vink C. 2005. The Epstein-Barr virus BILF1 gene encodes a G protein-coupled receptor that inhibits phosphorylation of RNA-dependent protein kinase. J Virol 79:441–449. doi: 10.1128/JVI.79.1.441-449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldhoer M, Kledal TN, Farrell H, Schwartz TW. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J Virol 76:8161–8168. doi: 10.1128/JVI.76.16.8161-8168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLean KA, Holst PJ, Martini L, Schwartz TW, Rosenkilde MM. 2004. Similar activation of signal transduction pathways by the herpesvirus-encoded chemokine receptors US28 and ORF74. Virology 325:241–251. doi: 10.1016/j.virol.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 32.Holst PJ, Rosenkilde MM, Manfra D, Chen SC, Wiekowski MT, Holst B, Cifire F, Lipp M, Schwartz TW, Lira SA. 2001. Tumorigenesis induced by the HHV8-encoded chemokine receptor requires ligand modulation of high constitutive activity. J Clin Invest 108:1789–1796. doi: 10.1172/JCI13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo J, Currin A, Griffin BD, Shannon-Lowe C, Thomas WA, Ressing ME, Wiertz EJHJ, Rowe M. 2009. The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by targeting MHC class I molecules for degradation. PLoS Pathog 5:e1000255. doi: 10.1371/journal.ppat.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo J, Quinn LL, Tamblyn J, Thomas WA, Feederle R, Delecluse H-J, Hislop AD, Rowe M. 2011. The Epstein-Barr virus-encoded BILF1 protein modulates immune recognition of endogenously processed antigen by targeting major histocompatibility complex class I molecules trafficking on both the exocytic and endocytic pathways. J Virol 85:1604–1614. doi: 10.1128/JVI.01608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin BD, Gram AM, Mulder A, Van Leeuwen D, Claas FHJ, Wang F, Ressing ME, Wiertz E. 2013. EBV BILF1 evolved to downregulate cell surface display of a wide range of HLA class I molecules through their cytoplasmic tail. J Immunol 190:1672–1684. doi: 10.4049/jimmunol.1102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scuda N, Madinda NF, Akoua-Koffi C, Adjogoua EV, Wevers D, Hofmann J, Cameron KN, Leendertz SAJ, Couacy-Hymann E, Robbins M, Boesch C, Jarvis MA, Moens U, Mugisha L, Calvignac-Spencer S, Leendertz FH, Ehlers B. 2013. Novel polyomaviruses of nonhuman primates: genetic and serological predictors for the existence of multiple unknown polyomaviruses within the human population. PLoS Pathog 9:e1003429. doi: 10.1371/journal.ppat.1003429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chmielewicz B, Goltz M, Lahrmann K-H, Ehlers B. 2003. Approaching virus safety in xenotransplantation: a search for unrecognized herpesviruses in pigs. Xenotransplantation 10:349–356. doi: 10.1034/j.1399-3089.2003.02074.x. [DOI] [PubMed] [Google Scholar]
- 38.Prepens S, Kreuzer K-A, Leendertz F, Nitsche A, Ehlers B. 2007. Discovery of herpesviruses in multi-infected primates using locked nucleic acids (LNA) and a bigenic PCR approach. Virol J 4:84. doi: 10.1186/1743-422X-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehlers B, Ochs A, Leendertz F, Goltz M, Boesch C, Mätz-Rensing K. 2003. Novel simian homologues of Epstein-Barr virus. J Virol 77:10695–10699. doi: 10.1128/JVI.77.19.10695-10699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Käll L, Krogh A, Sonnhammer ELL. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res 35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostenis E, Martini L, Ellis J, Waldhoer M, Heydorn A, Rosenkilde MM, Norregaard PK, Jorgensen R, Whistler JL, Milligan G. 2005. A highly conserved glycine within linker I and the extreme C terminus of G protein alpha subunits interact cooperatively in switching G protein-coupled receptor-to-effector specificity. J Pharmacol Exp Ther 313:78–87. [DOI] [PubMed] [Google Scholar]
- 43.Conklin BR, Farfel Z, Lustig KD, Julius D, Bourne HR. 1993. Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature 363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 44.Kissow H, Hartmann B, Holst JJ, Viby N-E, Hansen LS, Rosenkilde MM, Hare KJ, Poulsen SS. 2012. Glucagon-like peptide-1 (GLP-1) receptor agonism or DPP-4 inhibition does not accelerate neoplasia in carcinogen treated mice. Regul Pept 179:91–100. doi: 10.1016/j.regpep.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. 2009. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci 30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Rosenkilde MM, Benned-Jensen T, Frimurer TM, Schwartz TW. 2010. The minor binding pocket: a major player in 7TM receptor activation. Trends Pharmacol Sci 31:567–574. doi: 10.1016/j.tips.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Jensen A-SM, Sparre-Ulrich AH, Davis-Poynter N, Rosenkilde MM. 2012. Structural diversity in conserved regions like the DRY-motif among viral 7TM receptors—a consequence of evolutionary pressure? Adv Virol 2012:231813. doi: 10.1155/2012/231813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz TW. 1994. Locating ligand-binding sites in 7TM receptors by protein engineering. Curr Opin Biotechnol 5:434–444. doi: 10.1016/0958-1669(94)90054-X. [DOI] [PubMed] [Google Scholar]
- 49.Baldwin JM, Schertler GF, Unger VM. 1997. An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol 272:144–164. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 50.Ballesteros JA, Weinstein H. 1995. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors, p 366–428 InSealfon SC. (ed), Receptor molecular biology. Academic Press, San Diego, CA. [Google Scholar]
- 51.Mirzadegan T, Benkö G, Filipek S, Palczewski K. 2003. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry 42:2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rummel PC, Thiele S, Hansen LS, Petersen TP, Sparre-Ulrich AH, Ulven T, Rosenkilde MM. 2013. Extracellular disulfide bridges serve different purposes in two homologous chemokine receptors, CCR1 and CCR5. Mol Pharmacol 84:335–345. doi: 10.1124/mol.113.086702. [DOI] [PubMed] [Google Scholar]
- 53.Sibley CG, Ahlquist JE. 1984. The phylogeny of the hominoid primates, as indicated by DNA-DNA hybridization. J Mol Evol 20:2–15. doi: 10.1007/BF02101980. [DOI] [PubMed] [Google Scholar]
- 54.Rosenkilde MM, Waldhoer M, Lüttichau HR, Schwartz TW. 2001. Virally encoded 7TM receptors. Oncogene 20:1582–1593. doi: 10.1038/sj.onc.1204191. [DOI] [PubMed] [Google Scholar]
- 55.Baldwin AS. 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 56.Macian F. 2005. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 57.Robbs BK, Cruz ALS, Werneck MBF, Mognol GP, Viola JPB. 2008. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol 28:7168–7181. doi: 10.1128/MCB.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gruber CW, Muttenthaler M, Freissmuth M. 2010. Ligand-based peptide design and combinatorial peptide libraries to target G protein-coupled receptors. Curr Pharm Des 16:3071–3088. doi: 10.2174/138161210793292474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goltz M, Ericsson T, Patience C, Huang CA, Noack S, Sachs DH, Ehlers B. 2002. Sequence analysis of the genome of porcine lymphotropic herpesvirus 1 and gene expression during posttransplant lymphoproliferative disease of pigs. Virology 294:383–393. doi: 10.1006/viro.2002.1390. [DOI] [PubMed] [Google Scholar]
- 60.Hart J, Ackermann M, Jayawardane G, Russell G, Haig DM, Reid H, Stewart JP. 2007. Complete sequence and analysis of the ovine herpesvirus 2 genome. J Gen Virol 88:28–39. doi: 10.1099/vir.0.82284-0. [DOI] [PubMed] [Google Scholar]
- 61.Boudry C, Markine-Goriaynoff N, Delforge C, Springael J-Y, de Leval L, Drion P, Russell G, Haig DM, Vanderplasschen AF, Dewals B. 2007. The A5 gene of alcelaphine herpesvirus 1 encodes a constitutively active G-protein-coupled receptor that is non-essential for the induction of malignant catarrhal fever in rabbits. J Gen Virol 88:3224–3233. doi: 10.1099/vir.0.83153-0. [DOI] [PubMed] [Google Scholar]
- 62.Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71:5894–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong-Ho E, Wu T-T, Davis ZH, Zhang B, Huang J, Gong H, Deng H, Liu F, Glaunsinger B, Sun R. 2014. Unconventional sequence requirement for viral late gene core promoters of murine gammaherpesvirus 68. J Virol 88:3411–3422. doi: 10.1128/JVI.01374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fogg MH, Carville A, Cameron J, Quink C, Wang F. 2005. Reduced prevalence of Epstein-Barr virus-related lymphocryptovirus infection in sera from a New World primate. J Virol 79:10069–10072. doi: 10.1128/JVI.79.15.10069-10072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rivailler P, Cho Y-G, Wang F. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a New World primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J Virol 76:12055–12068. doi: 10.1128/JVI.76.23.12055-12068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. 2006. Molecular mechanism of 7TM receptor activation—a global toggle switch model. Annu Rev Pharmacol Toxicol 46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 67.Lyngaa R, Nørregaard K, Kristensen M, Kubale V, Rosenkilde MM, Kledal TN. 2010. Cell transformation mediated by the Epstein-Barr virus G protein-coupled receptor BILF1 is dependent on constitutive signaling. Oncogene 29:4388–4398. doi: 10.1038/onc.2010.173. [DOI] [PubMed] [Google Scholar]
- 68.Baeuerle PA, Henkel T. 1994. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol 12:141–179. [DOI] [PubMed] [Google Scholar]
- 69.Israël A, Le Bail O, Hatat D, Piette J, Kieran M, Logeat F, Wallach D, Fellous M, Kourilsky P. 1989. TNF stimulates expression of mouse MHC class I genes by inducing an NF kappa B-like enhancer binding activity which displaces constitutive factors. EMBO J 8:3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plaksin D, Baeuerle PA, Eisenbach L. 1993. KBF1 (p50 NF-kappa B homodimer) acts as a repressor of H-2Kb gene expression in metastatic tumor cells. J Exp Med 177:1651–1662. doi: 10.1084/jem.177.6.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spiess K, Rosenkilde MM. 2014. Functional properties of virus-encoded and virus-regulated G protein-coupled receptors. In Methods Pharmacol Toxicol 2014:45–65. doi: 10.1007/978-1-62703-779-2_3. [DOI] [Google Scholar]
- 72.Fraile-Ramos A, Pelchen-Matthews A, Kledal TN, Browne H, Schwartz TW, Marsh M. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218–232. doi: 10.1034/j.1600-0854.2002.030307.x. [DOI] [PubMed] [Google Scholar]
- 73.Schwarz M, Murphy PM. 2001. Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor constitutively activates NF-kappa B and induces proinflammatory cytokine and chemokine production via a C-terminal signaling determinant. J Immunol 167:505–513. doi: 10.4049/jimmunol.167.1.505. [DOI] [PubMed] [Google Scholar]
- 74.Halder S, Murakami M, Verma SC, Kumar P, Yi F, Robertson ES. 2009. Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One 4:e7214. doi: 10.1371/journal.pone.0007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, Lira SA, Söderberg-Nauclér C, Smit MJ. 2010. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal 3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 76.Woolley MJ, Watkins HA, Taddese B, Karakullukcu ZG, Barwell J, Smith KJ, Hay DL, Poyner DR, Reynolds CA, Conner AC. 2013. The role of ECL2 in CGRP receptor activation: a combined modelling and experimental approach. J R Soc Interface 10:20130589. doi: 10.1098/rsif.2013.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A. 2007. Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J Biol Chem 282:25677–25686. doi: 10.1074/jbc.M702311200. [DOI] [PubMed] [Google Scholar]
- 78.Peeters MC, van Westen GJP, Guo D, Wisse LE, Müller CE, Beukers MW, Ijzerman AP. 2011. GPCR structure and activation: an essential role for the first extracellular loop in activating the adenosine A2B receptor. FASEB J 25:632–643. doi: 10.1096/fj.10-164319. [DOI] [PubMed] [Google Scholar]
- 79.Schwartz TW, Rosenkilde MM. 1996. Is there a “lock” for all “keys” in 7TM receptors? Trends Pharmacol Sci 17:213–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.