ABSTRACT
Epstein-Barr virus (EBV) is a well-established B-cell-tropic virus associated with various lymphoproliferative diseases of both B-cell and non-B-cell origin. EBV is associated with a number of T-cell lymphomas; however, in vitro studies utilizing prototypical EBV type 1 (EBV-1) laboratory strains have generally failed to readily infect mature T cells in culture. The difficulties in performing in vitro T-cell experiments have left questions regarding the role of EBV in the pathogenesis of EBV-positive T-cell lymphoproliferative diseases largely unresolved. We report here that the EBV type 2 (EBV-2) strain displays a unique cell tropism for T cells. In remarkable contrast to EBV-1, EBV-2 readily infects primary T cells in vitro, demonstrating a propensity for CD8+ T cells. EBV-2 infection of purified T cells results in expression of latency genes and ultimately leads to T-cell activation, substantial proliferation, and profound alteration of cytokine expression. The pattern of cytokine production is strikingly skewed toward chemokines with roles in lymphocyte migration, demonstrating that EBV-2 has the ability to modulate normal T-cell processes. Collectively, these novel findings identify a previously unknown cell population potentially utilized by EBV-2 to establish latency and lay the foundation for further studies to elucidate the role of EBV in the pathogenesis of T-cell lymphoproliferative diseases.
IMPORTANCE The ability of EBV to infect T cells is made apparent by its association with a variety of T-cell lymphoproliferative disorders. However, studies to elucidate the pathogenic role of EBV in these diseases have been limited by the inability to conduct in vitro T-cell infection experiments. Here, we report that EBV-2 isolates, compromised in the capacity to immortalize B cells, infect CD3+ T cells ex vivo and propose a working model of EBV-2 persistence where alteration of T-cell functions resulting from EBV-2 infection enhances the establishment of latency in B cells. If indeed EBV-2 utilizes T cells to establish a persistent infection, this could provide one mechanism for the association of EBV with T-cell lymphomas. The novel finding that EBV-2 infects T cells in culture will provide a model to understand the role EBV plays in the development of T-cell lymphomas.
INTRODUCTION
While Epstein-Barr virus (EBV) establishes lifelong latency in B cells and is associated with B-cell malignancies, it is also associated with malignancies and diseases that originate from T cells, including NK/T-cell lymphomas (1), hemophagocytic lymphohistiocytosis (2), hydroa vacciniforme (HV) (3), and chronic active EBV (CAEBV) (4, 5). In these diseases, EBV can be detected in CD4+ T cells, CD8+ T cells, or γδ T cells (6, 7), with the virus predominantly existing as a latent infection (8, 9). The etiology of these T-cell diseases, and in particular whether EBV infection of T cells is an aberrancy in a virus known for its B-cell tropism in vitro and in vivo, remains unknown.
Based on genetic differences in the Epstein-Barr nuclear antigen 2 (EBNA-2), EBNA-3a, and EBNA-3c latent genes, EBV has been classified into two major strains (10–14), which are referred to as EBV type 1 (EBV-1) and type 2 (EBV-2) (14). The two strains not only differ in their genotypes, they also have functional differences in their transforming capacities. EBV-1 readily transforms B cells in culture, leading to the outgrowth of immortalized lymphoblastoid cell lines (LCL), while EBV-2 is poorly transforming (11, 14). The genetic differences in EBNA-2, -3A, and -3C are thought to lead to these phenotypic differences (11, 15–18). The potential of EBV to cause proliferation and ultimately to immortalize B cells in culture is thought to be the in vitro manifestation of the ability of EBV to establish latency in vivo (19). Thus, the fact that EBV-2 transformation of B cells is inefficient is contradictory to the observation that EBV-2 persists in the human population (20–22), suggesting that EBV-2 could utilize unique mechanisms to establish a persistent infection in vivo.
In this study, we show that, in contrast to EBV-1, EBV-2 readily infected primary T cells in culture with the expression of the latent genes. In addition, we show that CD8+ T cells were the predominant cell type infected, which resulted in their activation and in the induction of several cytokines known to alter lymphocyte trafficking. These results provide a possible mechanism by which EBV-2 may utilize the T-cell compartment during the primary stage of infection to enhance the establishment of latency, thus allowing EBV-2 to be retained in the human population.
MATERIALS AND METHODS
Production of EBV.
The EBV cell lines B95.8, Akata, Jijoye, and BL5 were used in this study for production of virus. The B95.8 and Akata cell lines have previously been typed and found to carry EBV-1, and the Jijoye cell line carries EBV type 2 (10). The BL5 cell line, a gift from W. Harrington, University of Miami, was typed by PCR for a region of EBNA-3c as previously described (23) and was determined to carry EBV-2. EBV stocks were generated as previously described (24). Briefly, the B95.8, Jijoye, BL5, and Ramos cell lines were treated with sodium butyrate and tetradecanoyl phorbol acetate (TPA) (at 4 mM and 24 ng/ml, respectively), and the Akata cell line was treated with anti-human Ig (50 μg/ml). Supernatants were centrifuged at 4,000 × g for 10 min and passed over a 0.7-μm filter to remove cellular debris. Viral particles were pelleted by ultracentrifugation at 16,000 × g for 90 min and resuspended in 1/100 the initial volume using complete RPMI. Virus stocks were quantified following DNase treatment by quantitative PCR (qPCR) using a method previously described to amplify EBV BALF5 (25). For some experiments, virus derived from the Jijoye cell line was UV irradiated in a Bio-Rad GS Gene linker for 5 min. The Ramos cell line is an EBV-negative Burkitt's lymphoma (BL) cell line and was used to generate mock-infected supernatants.
T-cell purification and infection.
After obtaining informed consent, peripheral blood was obtained from healthy U.S. adult donors as approved by the Institutional Review Board of SUNY Upstate Medical University and according to the Declaration of Helsinki. The blood was layered over Ficoll-Paque (GE Healthcare, Little Chalfont, United Kingdom) to isolate peripheral blood mononuclear cells (PBMCs). T cells were isolated from peripheral blood mononuclear cells by negative enrichment using the human Pan T-cell Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) or, where indicated, by depleting PBMCs of B cells using a CD19 MicroBead kit (Miltenyi Biotec, Bergisch Gladbach, Germany). T-cell subsets were isolated by negative enrichment using CD4+ or CD8+ T-cell isolation kits (Miltenyi Biotec, Bergisch Gladbach, Germany). Following magnetic enrichment, purity analysis was performed via flow cytometry with CD3-allophycocyanin (APC), CD19-peridinin chlorophyll protein (PerCp)-Cy5.5, CD8-phycoerythrin (PE)-Cy7, and CD4-APC-Cy7 antibodies. At the time of isolation, all T-cell cultures were found to have a purity of >94%, with <0.07% B-cell contamination. Notably, the majority of the T-cell cultures had a purity of >97%. In all experiments, cells were plated at 106 cells/ml in complete RPMI containing 1 μg/ml cyclosporine to inhibit the T-cell receptor-mediated activation of EBV-specific T cells and infected at a multiplicity of infection (MOI) of 10 genomes per cell. Cell cultures were maintained at 37°C and supplemented with 5% CO2.
Cell imaging. (i) Immunofluorescent staining.
LMP-1 (clone SC 1-4) antibody was biotinylated utilizing an EZ-Link Sulfo-NHS-LC Biotinylation kit (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. Mock- or Jijoye-infected T-cell cultures were harvested at 7 days postinfection (p.i.) and washed in staining buffer containing phosphate-buffered saline (PBS), 1% bovine serum albumin, and 0.1% sodium azide. The cells were Fc blocked (human Fc binding inhibitor; eBiosciences, San Diego, CA) in staining buffer for 20 min. After washing, the cells were incubated at 37°C for 1 h with the primary antibodies CD3-fluorescein isothiocyanate (FITC) (clone BW264/56) and LMP-1–biotin (clone SC 1-4). Following incubation with primary antibodies, the cells were washed 3 times and incubated with streptavidin-APC for 1 h at room temperature. After washing, the cells were fixed in 4% paraformaldehyde for 10 min at room temperature, transferred to slides with a cytospin centrifuge, and stained with DAPI (4′,6-diamidino-2-phenylindole). Cell images were acquired using a Nikon Eclipse Ti inverted microscope and imaging system, using NIS-Elements Br software (Nikon, Tokyo, Japan). Cell images were taken at ×100 magnification.
(ii) White-light images.
Cell aggregation was measured microscopically using a Nikon Eclipse Ti inverted microscope and imaging system, using NIS-Elements Br software (Nikon, Tokyo, Japan). Cell images were taken at ×10 magnification, and the autoexposure function was used to select the exposure time for each image.
Flow cytometry and cell proliferation and activation.
Cells were stained prior to infection with e450 Cell Proliferation Dye (eBioscience, San Diego, CA) according to the manufacturer's protocol. Cultures were harvested at the indicated times and washed twice in flow cytometry buffer containing phosphate-buffered saline, 1% bovine serum albumin, and 0.1% sodium azide. The cells were Fc blocked in flow buffer for 20 min. After washing, the cells were stained for 20 min with the following antibodies: CD3-APC, CD19-PerCp-Cy5.5, CD8-PE-Cy7, CD4-APC-Cy7, and CD69-FITC (Biolegend, San Diego, CA). All experiments were run on a BD LSR-Fortessa flow cytometer equipped with FACS Diva software (BD Biosciences, San Jose, CA). Flow analysis was performed in FlowJo v9.5.2 (Tree Star, Ashland, OR).
Quantitative PCR to determine EBV copy numbers.
Following magnetic purification, T cells were placed in culture and infected as stated above with no drugs or with the addition of acyclovir (50 μM) or phosphonoacetic acid (PAA) (1 mM). Cells were harvested at the indicated time points and subsequently washed three times in PBS to remove any cell-free virus. DNA was extracted from the cells using a Qiagen DNeasy kit (Qiagen) according to the manufacturer's protocol. EBV DNA levels were determined as previously described using primers and probes designed to detect a 70-bp region of the EBV BALF5 gene and the β-actin gene as a control for DNA input (25, 26).
Immunoblotting of EBV proteins.
Cell lysates were made using a 1:1 ratio of PBS and Laemmli buffer with 2-mercaptoethanol and heating for 5 min in boiling water, and 80 μg of total cellular protein from each sample was separated using SDS-PAGE on 10% acrylamide gels, followed by immunoblotting. Membranes were blocked in a 10% milk–Tris-buffered saline (TBS)–Tween 20 solution, followed by overnight incubation with primary antibody (anti-EBNA-1 [clone 1B5], anti-EBNA-2 [clone PE2], anti-LMP-1 [clone SC 1-4], anti-LMP-2 [clone 14B7], or anti-β-actin [clone mAbcam8224] [Abcam, Cambridge, MA]). The blots were then incubated with the secondary antibody goat anti-rat IgG horseradish peroxidase (HRP) or goat anti-rabbit IgG HRP. Antibody-labeled protein bands were detected using an ECL chemiluminescence substrate (GE Healthcare Life Sciences).
Limiting-dilution PCR.
For limiting-dilution PCR analyses, T cells were harvested at 7 days p.i. and serially 10-fold diluted in a background of uninfected PBMCs. A total of 10,000 cells were plated in a 96-well PCR plate at 12 wells per dilution. Samples containing 10, 1, 0.1, or no copies of EBV-2 EBNA3c DNA were included as controls. Following lysis with proteinase K, real-time PCR was performed using primers for EBNA-3c (forward, 5′ AGAAGGGGAGCGTGTGTTG 3′; reverse, 5′ GGCTCGTTTTTGACGTCGG 3′) and an EBV-2-specific probe (CCGTGTGACTGGAAGACATGGCACTCC). The real-time PCR conditions were as follows: 95°C for 3 min, followed by 45 cycles of 95°C for 10 s and 55°C for 30 s. All reactions were performed with the iTaq Universal Probes Supermix (Bio-Rad). Wells were scored for the presence of the EBV-2 genome by the threshold cycle (CT) value. The frequency of T cells positive for the viral genome was calculated by Poisson distribution analysis using mean data from tests of infection on 3 independent PBMC donors.
Cytokine measurement.
Cultures were harvested at the indicated times, and cells were removed from the supernatants by centrifugation at 300 × g for 7 min. Cytokines were measured using a 25-plex cytokine Bio-Plex assay (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions and run on a Bio-Rad Bio-Plex 200 system (Bio-Rad, Hercules, CA). Cytokines were quantified using a standard curve of known concentrations of all the cytokines tested, and the background fluorescence was subtracted.
Statistical analysis.
All statistics were performed using GraphPad Prism v5.0b (GraphPad, La Jolla, CA). Multiple groups were compared using one- or two-way analysis of variance (ANOVA), as appropriate, with Bonferroni's posttest correction. Significance was determined as a P value of <0.05.
RESULTS
EBV-2 induces T-cell proliferation in cultured peripheral blood mononuclear cells.
EBV-1 induces substantial proliferation of B cells following primary infection in vitro, while EBV-2 induces only a moderate level of B-cell proliferation (16). To confirm this early observation, PBMCs were labeled with a proliferation dye (e450) and then infected with either EBV-1 or EBV-2 at an MOI of 10 genome copies per cell. EBV-1 was isolated from either the B95-8 cell line or the Akata cell line, while EBV-2 was isolated from the Jijoye cell line or the BL5 cell line. After 7 days, cells were harvested and stained with fluorescence-labeled anti-CD19 or anti-CD3 monoclonal antibodies to assess proliferation in B cells or T-cell compartments, respectively. We observed that, as expected, all EBV strains induced proliferation of CD19+ B cells (Fig. 1A and B). Additionally, the CD19+ B cells infected with B95-8- or Akata-derived EBV-1 had undergone multiple rounds of replication, as indicated by the dilution of the e450 dye. In contrast, no more than 2 rounds of replication were observed in CD19+ B cells infected with Jijoye- or BL5-derived EBV-2 (Fig. 1A). No proliferation of CD3+ T cells was observed following infection with either B95-8- or Akata-derived EBV-1. However, infection with either Jijoye- or BL5-derived EBV-2 resulted in induction of proliferation in over 30% of the CD3+ T cells, suggesting that EBV-2 can infect T cells and that the infection of T cells is not dependent on the source of EBV-2 (Fig. 1A and B). Mock treatment of CD19+ B cells or CD3+ T cells did not result in proliferation, indicating that any residual TPA or Na-butyrate in the media following isolation of EBV from cell lines was not responsible for cell proliferation.
FIG 1.
EBV type 2 induces T-cell proliferation in cultured peripheral blood mononuclear cells. PBMCs were labeled with e450 proliferation dye and infected with EBV-1 (B95.8- or Akata-derived virus) or EBV-2 (Jijoye- or BL5-derived virus) or not infected (medium). At 7 days postinfection, cells were harvested and stained for flow cytometry. Lymphoblasts and lymphocytes were gated initially based on FSC/SSC, followed by CD19+ B and CD3+ T cells, and subsequently, proliferated cells were determined by e450 dilution. (A) Representative histograms of CD19+ B-cell and CD3+ T-cell proliferation measured by fluorescence intensity of the e450 proliferation dye from 3 to 9 replicates of 4 independent experiments. (B) Mean frequencies of B- and T-cell proliferation. The error bars represent means ± standard errors. ***, P < 0.001, and ns, P > 0.05, using a two-way ANOVA with Bonferroni's posttest.
EBV-2 latently infects T cells.
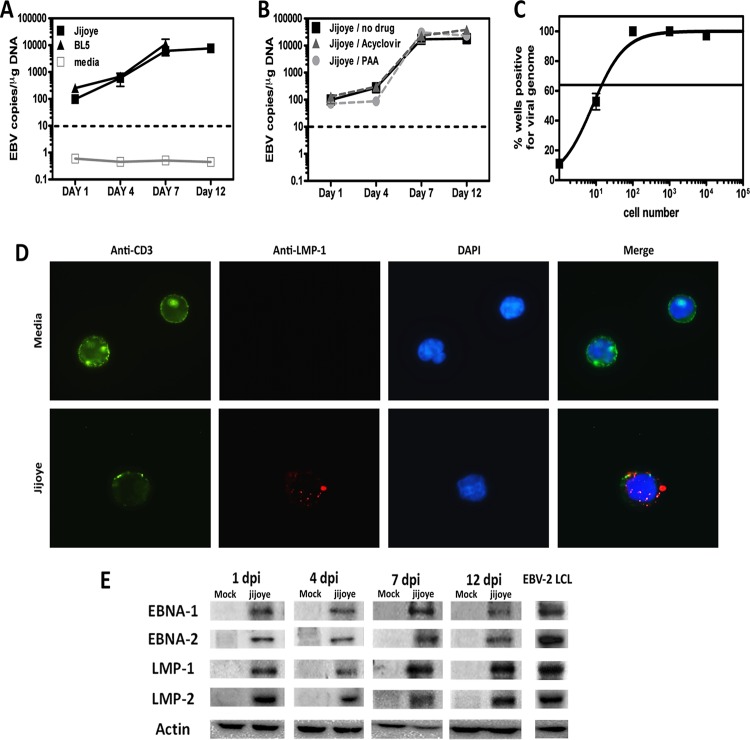
To further characterize the infection of T cells by EBV-2, the EBV genome load was measured by qPCR following infection of purified CD3+ T cells over 12 days. EBV-2 derived from either Jijoye or BL5 cells, was found to infect T cells, with the viral load logarithmically increasing through 7 days p.i. and stabilizing at later time points (Fig. 2A). In contrast, the viral genome was not detected at concentrations above those in noninfected cells at any time point in EBV-1-infected T-cell samples (data not shown), suggesting infection of T cells is unique to EBV-2.
FIG 2.
EBV type 2 infects T cells. T cells were purified from PBMCs of 3 or 4 independent donors. (A) T cells were infected with EBV-2 isolated from the Jijoye or BL5 cell line or were cultured in medium only. (B) T cells were infected with EBV-2 (Jijoye derived) in the absence of drugs or in the presence of acyclovir or PAA. (A and B) T cells were harvested 1, 4, 7, and 12 days postinfection for DNA extraction, and multiplex qPCR for the EBV genome and actin was performed to determine the EBV copy number per microgram of DNA (n = 3 for each time point). (C) Real-time limiting-dilution PCR for the viral genome was utilized to determine the frequency of T cells that harbored EBV-2. Primers and probes specific for EBV-2 EBNA-3c were used to detect the EBV-2 genome. The frequency of cells positive for the viral genome was calculated by Poisson distribution analysis of mean data (n = 3). Samples containing 10, 1, 0.1, or no copies of EBNA3c DNA were included as controls. The line at 63.2% indicates the point at which one viral genome-positive cell per reaction was predicted to occur. The x axis shows the numbers of cells per reaction; the y axis shows the percentages of 12 reactions positive for the viral genome. The error bars represent means ± standard errors. (D) Immunofluorescent staining of T cells was performed at 7 days p.i. Mock- and Jijoye-infected T cells were stained to visualize CD3 (green) and LMP-1 (red) on the cell surface and with DAPI to visualize nuclei (blue). The images were captured with a 100× objective using a Nikon Eclipse Ti inverted microscope. (E) Whole-cell lysates from EBV-2- and mock-infected T cells were prepared at 1, 4, 7, and 12 days postinfection (dpi) and subjected to immunoblotting to detect LMP-1 (42 kDa), LMP-2 (50 kDa), EBNA-1 (70 kDa), EBNA-2 (75 kDa), and actin (42 kDa). The data shown are for noninfected (media) and EBV-2 (Jijoye)-infected samples from 1 of 4 individual donors for each time point. As shown, an EBV-2 LCL was used as a positive-staining control.
The increase in the viral genome load could be due to lytic viral replication. To test this possibility, purified CD3+ T cells were infected with EBV-2 or EBV-2 in the presence of acyclovir or PAA, two well-known inhibitors of gammaherpesvirus lytic replication (27, 28). Interestingly, the increase in the viral load over time was not diminished by the presence of acyclovir or PAA (Fig. 2B), strongly suggesting that increased EBV-2 genome copy numbers over time occur via expansion of latently infected cells. Although we infected the CD3+ T cells at an MOI of 10 genome copies/cell, it is unlikely that all the cells in the culture were infected. We performed limiting-dilution PCR at 7 days p.i. to assess the frequency of latently infected cells following infection of CD3+ T cells with EBV-2 (Fig. 2C). We found approximately 1 in 14 EBV-2-infected CD3+ T cells, indicating that approximately 70,000 cells/106 total CD3+ T cells in the culture were infected with EBV-2.
Although minimal B-cell contamination was detected in the purified T-cell cultures (day 0, <0.07%, and day 7, <0.95%) and the viral genome was not detected in T-cell cultures infected with EBV-1, we wanted to confirm that these results were due to the direct infection of T cells by EBV-2 and not to infection of contaminating cells present in the cultures. To accomplish this, we performed immunofluorescence microscopy on mock- and EBV-2-infected T-cell cultures to visualize the pan-T-cell marker CD3 and the EBV latency protein LMP-1, which is known to be expressed in EBV-positive T cell lymphomas (29). At 7 days post-EBV-2 infection, we detected LMP-1 and CD3+ double-positive cells, demonstrating that EBV-2 clearly infects T cells. Notably, we also observed a marked decrease in the concentration of CD3 on the surfaces of EBV-2-infected T cells compared to T cells from mock-infected cultures (Fig. 2D). Downregulation of CD3 is a characteristic of activated T cells, suggesting that subsequent to infection, EBV-2 induces the activation of T cells.
As the data so far were suggestive of a latent EBV infection, we next confirmed the presence of the viral latency proteins EBNA-1, EBNA-2, LMP-1, and LMP-2 during primary infection of T cells. Western blot analysis was performed on EBV-2-infected T cells from 4 different donors. Shown in Fig. 2E is a representative analysis for a single donor. Even at 1 day p.i., all four latency proteins could be detected. All four latency proteins were detected through 12 days post-EBV-2 infection, further suggesting that EBV-2 initiates a latent infection following primary infection of CD3+ T cells. Notably, samples from different time points were run on separate gels and, with the exception of actin, were visualized using different exposure times; thus, it is currently unknown how the concentrations of the latency proteins differ over time. As controls, Western blots of mock-infected CD3+ T cells showed no viral proteins, while analysis of an EBV-2 LCL showed that the antibodies used were able to detect EBV-2-derived latent proteins (Fig. 2E).
EBV-2 induces proliferation of CD8+ T cells.
We next wanted to determine whether EBV-2 infected the CD8+ or CD4+ T-cell subset by first measuring T-cell subset proliferation following EBV-2 infection. CD3+ T cells were isolated from PBMCs, stained with the e450 proliferation dye, and infected with EBV-1 (derived from B95-8 cells), EBV-2 (derived from Jijoye cells), or medium alone; mock infected; or infected with UV-inactivated EBV-2 (UV Jijoye). At 7 days p.i., CD4+ and CD8+ T-cell subsets were analyzed via flow cytometry for cell proliferation, as indicated by dilution of the e450 proliferation dye (Fig. 3A shows a representative analysis, and Fig. 3B, C, and D shows summarized results from 3 experiments with 3 [mock infected and UV Jijoye] to 8 [medium, B95.8, and Jijoye] donors). EBV-2-infected CD4+ T cells showed minimal proliferation at 7 days postinfection, with less than 14% of the total CD4+ T-cell population undergoing only 1 round of cell division (Fig. 3B and C). In contrast, infection of CD8+ T cells with EBV-2 resulted in 50% of the cells undergoing up to 4 rounds of division by 7 days postinfection (Fig. 3B and D). Both mock-infected and UV-inactivated EBV-2-infected CD8+ T cells showed a modest one round of proliferation, which was significantly less than the proliferation induced by infection with EBV-2, suggesting that infection with EBV-2 and not binding to the cells or residual TPA was inducing the cellular proliferation.
FIG 3.
EBV type 2 induces CD8 T-cell proliferation. T cells were labeled with e450 proliferation dye and subsequently infected with EBV-1 (B95.8) or EBV-2 (Jijoye). T cells cultured with medium only, mock infected, or exposed to UV-irradiated Jijoye-derived virus were included as controls. T-cell populations were analyzed at 7 days postinfection for proliferation, as measured by dilution of the fluorescence intensity of the e450 proliferation dye. (A) Representative histograms of CD4+ and CD8+ T-cell proliferation from 5 independent donors. (B) The percentages of proliferated CD4+ and CD8+ T cells were compared for all experimental groups. (C and D) Percentages of CD4+ and CD8+ T cells that underwent 0, 1, 2, 3, and ≥4 rounds of proliferation. The data are presented as means ± standard errors. CD4+ T cells, Jijoye versus medium, P < 0.05; Jijoye versus mock, P > 0.05; Jijoye versus B95.8, P < 0.05; Jijoye versus UV Jijoye, P > 0.05. CD8+ T cells, Jijoye versus medium, P < 0.001; Jijoye versus mock, P < 0.001; Jijoye versus B95.8, P < 0.001; Jijoye versus UV Jijoye, P < 0.001.
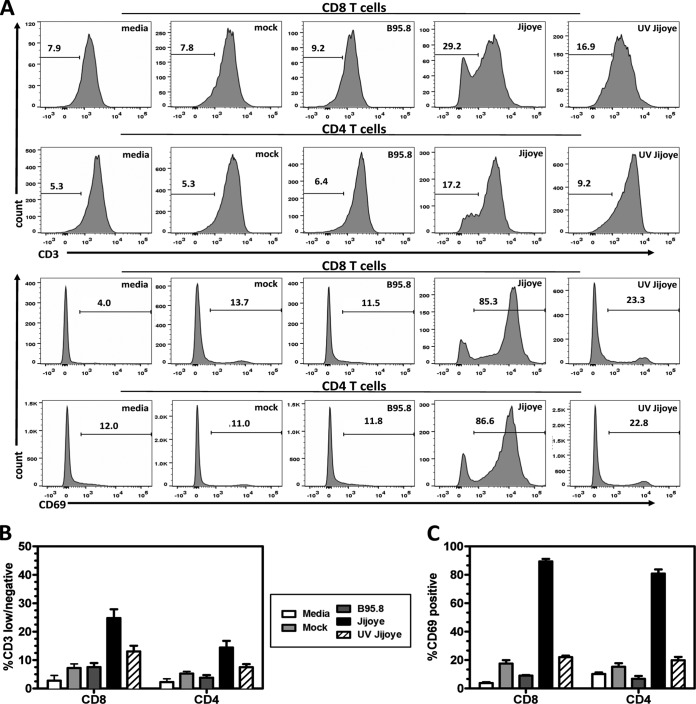
EBV-2 induces T-cell activation.
Primary infection of B cells by EBV is known to drive the cells to become activated lymphoblasts. EBV accomplishes this by expression of nine latent genes referred to as the growth program (30). The pattern of EBV protein production observed in EBV-2-infected T cells suggests that the growth program is also initiated following infection of T cells. T-cell activation is accompanied by increased CD69 expression, decreased CD3 expression, and increased cell size. To determine whether EBV-2 infection induces T-cell activation, we assessed cell size and expression of CD3 and CD69 in both CD4+ and CD8+ T cells by flow cytometry following infection. At 7 days p.i., EBV-2-infected CD4+ and CD8+ T cells displayed significant increases in the percentages of CD69-positive and CD3-low or -negative cells relative to EBV-1-infected T cells (Fig. 4A to C). This finding is consistent with our earlier observation of decreased amounts of CD3 on the surfaces of infected T cells (Fig. 2D). Moreover, based on forward (FSC) and side (SSC) scatter analysis, EBV-2-infected CD8+ and CD4+ T cells increased in size by 7 days p.i. (data not shown). Collectively, these data demonstrate that EBV-2 induces activation of CD8+ and CD4+ T cells. These effects were observed for both the CD4+ and CD8+ T cells (Fig. 4A to C), demonstrating that EBV-2 activates both T-cell subsets, in contrast to cell proliferation, where only CD8+ T cells were significantly induced to proliferate. These data establish that upon primary infection, EBV-2 significantly induces the activation of T cells.
FIG 4.
EBV type 2 induces T-cell activation. T cells purified from PBMCs were infected with B95.8-derived EBV-1 or Jijoye-derived EBV-2. T cells cultured with medium only, mock infected, or exposed to UV-irradiated Jijoye-derived EBV-2 were included as controls. At 7 days postinfection, cells were harvested and analyzed using flow cytometry. (A) Representative histograms of CD3 and CD69 surface expression on CD4+ and CD8+ T cells for all experimental groups. (B) The percentages of CD3-negative and -low CD4 and CD8 T cells at day 7 postinfection were plotted. (C) The percentages of CD69-positive CD4 and CD8 T cells at 7 days postinfection were plotted. The data are shown as means ± standard errors from 3 independent donors (n = 3). CD8+, CD69, Jijoye versus medium, P < 0.001; CD3, Jijoye versus medium, P < 0.001. CD4+, CD69, Jijoye versus medium, P < 0.001; CD3, Jijoye versus medium, P < 0.001. CD8+, CD69, Jijoye versus mock, P < 0.001; CD3, Jijoye versus mock, P < 0.001. CD4+, CD69, Jijoye versus mock, P < 0.001; CD3, Jijoye versus mock, P < 0.01. CD8+, CD69, Jijoye versus B95.8, P < 0.001; CD3, Jijoye versus B95.8, P < 0.001. CD4+, CD69, Jijoye versus B95.8, P < 0.001; CD3, Jijoye versus B95.8, P < 0.01. CD8+, CD69, Jijoye versus UV Jijoye, P < 0.001; CD3. Jijoye versus UV Jijoye, P < 0.001. CD4+, CD69, Jijoye versus UV Jijoye, P < 0.001; CD3, Jijoye versus UV Jijoye, P < 0.05.
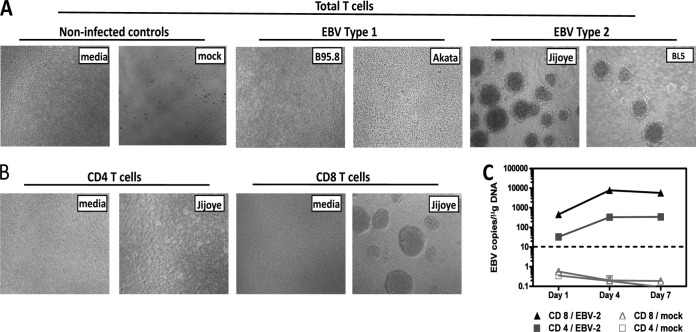
EBV-2 preferentially infects and induces aggregation of CD8+ T cells.
In vitro, EBV-induced activation of B cells results in aggregation and proliferation of the infected cells. Thus, purified CD4+ or CD8+ T-cell cultures were infected with B95-8-derived EBV-1 or Jijoye or BL5-derived EBV-2 or with medium alone and subsequently microscopically observed for aggregation. Consistent with the finding that EBV-2 uniquely induces T-cell activation, EBV-2-treated T cells, but not EBV-1- or medium-treated T cells, induced an aggregated cell phenotype (Fig. 5A). Cell aggregation was observed following infection with either Jijoye- or BL5-derived EBV-2, indicating that this effect was common to EBV-2 and not unique to a particular viral isolate. Moreover, observation of the CD4+ and CD8+ T-cell subsets revealed that the aggregation phenotype was induced only in the CD8+ T-cell population (Fig. 5B). Because we observed different effects of EBV-2 infection on CD4+ and CD8+ T cells, we wanted to determine whether the increases we observed in EBV genome loads in CD3+ T cells represented infection of CD4+ or CD8+ T-cell subsets. Purified CD4+ or CD8+ T cells were infected with EBV-2, and the viral load was determined by qPCR over 7 days p.i. The EBV genome could be detected in both CD4+ and CD8+ T cells at all time points. However, a 13-fold-higher viral load was detected in the CD8+ T-cell population than in the CD4+ T-cell population at 1 day p.i., with the differences increasing over time to 37-fold-higher viral loads in the CD8+ T cells by 7 days p.i. (Fig. 5C). Collectively, these data demonstrate that EBV-2 can infect both CD8+ and CD4+ T cells but preferentially infected CD8+ T cells.
FIG 5.
EBV type 2 induces T-cell aggregation and preferentially infects CD8 T cells. T cells were purified using an untouched pan-T-cell, untouched CD4, or untouched CD8 isolation kit. (A and B) T cells were infected with EBV-1 strain B95.8 or Akata or EBV-2 strain Jijoye or BL5. Noninfected controls were T cells treated with medium only and mock-infected T cells. EBV-induced T-cell aggregation was observed microscopically at 7 days p.i. Representative images of T-cell aggregation are shown for total T cells (A) and T-cell subsets (B). (C) CD4+ and CD8+ T cells were harvested 1, 4, and 7 days postinfection for DNA extraction. Multiplex qPCR for the EBV genome and β-actin was performed to determine the EBV-2 copy number per μg of DNA (n = 2 for each time point).
EBV-2 causes dysregulated cytokine production during CD8+ T-cell infection.
The finding that EBV-2 infects T cells raises another important question: does infection cause functional changes in T cells? To address this question, CD8+ T cells were purified and cytokine production was analyzed following mock or EBV-2 infection using a multiplex Luminex assay. Virtually no increase in cytokines was observed following mock infection of CD8+ T cells, with the exception of a very low level of CCL5 (RANTES) and interleukin-1RA (IL-1RA) at 7 days p.i. (Fig. 6A and B). In supernatants from EBV-2-infected CD8 T cells, large increases in several chemokines, including CCL5 (RANTES), CCL3 (MIP-1α), CCL4 (MIP-1β), CCL2 (MCP-1), and IL-1RA were observed at 1, 4, and 7 days p.i., with mean levels reaching between 1 and 4 ng/ml during the course of infection (Fig. 6). EBV-2 infection also induced increased expression of IL-2R and IL-8, to levels ranging from 0.092 ng/ml to 0.555 ng/ml. High-level production of these chemotactic cytokines suggests that EBV-2 infection drives the expression of specific cytokines in infected T cells.
FIG 6.
EBV type 2 causes dysregulated cytokine production during CD8+ T-cell infection. CD8+ T cells were purified and infected with Jijoye (EBV-2) or not infected (media). Supernatants were collected from replicate wells for Bio-Plex cytokine measurement at 1, 4, and 7 days postinfection. Cytokine production was standardized for cell proliferation and compared to a standard curve of known cytokine concentrations for all 25 cytokines analyzed. All the cytokines analyzed were plotted against each other for the medium control and Jijoye-infected T cells and compared using one-way ANOVA with Bonferroni's posttest at each time point studied. The data are representative of 3 separate donors (n = 3). *, P < 0.05; ***, P < 0.001; and no asterisk, P > 0.05 for EBV-2 versus medium control. The error bars represent means ± standard errors.
DISCUSSION
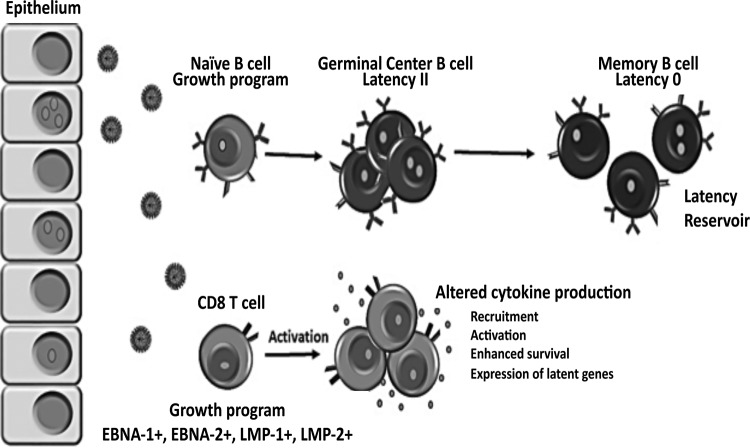
EBV is known to persist in the human population by establishing a latent infection in B cells. The immortalization of B cells in culture is thought to reflect the ability of EBV to establish latency in vivo (19). Thus, the impaired ability of EBV-2 to immortalize B cells is contradictory to the observation that EBV-2 persists in the human population, suggesting EBV-2 could be utilizing additional cell types and alternative mechanisms to establish a persistent infection. EBV is able to infect T cells in vivo, as evidenced by its association with a variety of T-cell lymphoproliferative disorders; however the role EBV plays in the pathogenesis of these diseases is currently unknown. Additionally, whether EBV infection of T cells is intentional or aberrant remains uncertain. In this study, we found that EBV-2 isolates preferentially infected CD8+ T cells, induced their proliferation and activation, and altered their cytokine profiles. In contrast, while EBV-1 derived from the B95.8 or Akata strain readily infects B cells, these EBV-1 viruses did not infect T cells ex vivo. Based on these observations, we propose a working model of EBV-2 persistence where alteration of T-cell functions resulting from EBV-2 infection, such as production of lymphotactic cytokines, enhances the establishment of latency in B cells (the model is shown in detail in Fig. 7). Although not addressed in this report, as we focused here on characterizing the primary infection of T cells, our finding that EBV-2 latently infects T cells suggests that T cells could also enhance EBV-2 persistence by acting as an additional latency reservoir. If indeed EBV-2 utilizes T cells at any stage of infection to establish a persistent infection, this could provide one mechanism for the association of EBV with T-cell lymphoproliferative diseases. The discovery that EBV-2 readily infects T cells in culture will enable studies to better understand the pathogenic role EBV plays in the development of these lymphomas.
FIG 7.
Working model of EBV type 2 latency establishment and persistence based on our in vitro T-cell infection studies. Following primary exposure to EBV-2, CD8+ T cells, in addition to B cells, in the tonsil are infected, and the growth program is initiated, resulting in the activation and proliferation of the infected lymphocytes. At this time, EBV-2-infected CD8+ T cells could enhance the ability of the virus to establish latency in the B-cell compartment. EBV-2 may accomplish this by inducing alteration of T-cell cytokine production that could hamper the antiviral response, recruit additional target cells to the site of infection, enhance the activation and survival of lymphocytes, and/or induce expression of critical viral genes. Another, non-mutually exclusive possibility is that, in addition to memory B cells, T cells serve as a latency reservoir.
Distinct patterns of EBV latent gene expression are observed in different EBV-associated lymphoproliferative disorders and are thought to reflect the latency programs utilized by EBV to establish a persistent infection. The current paradigm suggests that following primary infection of naive B cells, EBV initiates the growth program (also known as latency III). At this time, all latency-associated genes are expressed, providing the surrogate signals that naive B cells normally receive upon antigen encounter. This results in lymphoblast formation and establishment of germinal centers, where latent viral gene expression is restricted to EBNA-1 and LMP-1 and -2 (latency II). These cells subsequently differentiate to become memory B cells, where the virus latently resides long term (31, 32). Our results suggest that EBV-2 also expresses the growth program following primary infection of T cells in vitro, as EBNA-2, the latency protein that initiates transcription of the growth program (33), was expressed throughout the first 12 days of infection. Notably, this is the first observation of the growth program in cells of non-B-cell origin. EBV-associated T-cell lymphomas have previously been shown to exhibit viral gene expression restricted to EBNA-1, LMP-1, and LMP-2 (latency II) (29). However, studies of primary EBV T-cell infection, including the pattern of viral gene expression, have been severely limited due to previous difficulties in infecting primary T cells in culture with EBV-1. Some success has been achieved following infection of immature thymocytes with EBV-1, which resulted in expression of EBNA-2 transcripts and the lytic transcript BZLF1 (34), as well as inducing their proliferation (35). An important additional question is whether EBNA-2 expression is sustained in T cells beyond 12 days p.i. Long-term T-cell culture experiments are ongoing to elucidate the roles EBNA-2 and the growth program play at different stages of EBV-2 T-cell infection. Addition of two well known lytic cycle inhibitors, acyclovir and PAA, showed no difference in the increase in EBV genome copy numbers over time, suggesting that the infection of T cells is primarily a latent infection. As a second measure, we also stained EBV-2-infected CD3+ T cells with an anti-gp350 antibody, and no cells were seen to be positive (data not shown). Notably, previous studies of EBV primary B-cell infection have established a temporal pattern of latent gene expression spanning the first 2 weeks of infection, with EBNA-2 transcripts detected within 24 h, EBNA-1 transcripts reaching peak levels by 2 to 3 days, and LMP-1 and LMP-2 transcripts peaking at approximately 2 weeks postinfection (36–38). Following initial infection of T cells, we detected consistent levels of all four of these latent proteins from 24 h through 12 days postinfection, demonstrating a unique pattern of EBV gene expression during primary T-cell infection.
Attempts to generate an appropriate ex vivo system to study EBV infection of T cells have been hampered by the use of EBV-1 strains, such as the prototypical laboratory strain, B95.8, to perform in vitro studies. Thus, the discovery that EBV-2 readily infects T cells provides an in vitro system to study EBV infection of T cells. The data we present here suggest that infection of T cells ex vivo could be unique to the EBV-2 subtype, as two independent EBV-2 strains were found to readily infect primary T cells in culture. However, the cellular origin of EBV has previously been shown to have an impact on viral tropism (39). The EBV-2 strains used in this study were all raised in human Burkitt's lymphoma cell lines, whereas the classic EBV-1, B95.8, was raised in tamarin monkey B cells. To control for this, we also tested EBV-1 derived from the Akata BL cell line and found that, similar to B95.8 virus, Akata-derived EBV-1 also failed to infect T cells. Alternatively, EBV-1 and EBV-2 isolates also differ in their glycoprotein genes (40), which have been shown to affect cellular tropism. EBV infects B cells through binding of the viral gp350 to CD21 (41, 42). We and others have observed that peripheral blood T cells do not normally express the EBV receptor CD21 (43, 44; E. M. Wohlford and R. Rochford, unpublished data). EBV infection of epithelial cells differs radically from B-cell infection, as there is no requirement for viral gp350 (45) and infection is not dependent on CD21 (46). This suggests that EBV likely uses alternative viral glycoproteins and cellular receptors to infect T cells. The observation that EBV-2 is able to infect and activate both CD4+ and CD8+ T cells suggests that the receptor used by EBV-2 to infect T cells is expressed on both T-cell subsets; however, the finding that EBV-2 induces proliferation of only the CD8+ T-cell subset and of an ∼1-log-unit-higher viral copy number demonstrates a fundamental difference in the course of infection following viral entry.
Infection of T cells by EBV-2 caused profound differences in chemokine expression. CD8+ T cells infected with EBV-2 produced significantly higher levels of MIP-1α, MIP-1β, MCP-1, IL-1Rα, IL-2R, RANTES, and IL-8 than mock-infected T cells. These cytokines have been shown to be simultaneously produced by activated CD8+ T cells (47). This is consistent with our finding that EBV-2 preferentially infects CD8+ T cells. Notably, the majority of the cytokines detected at elevated concentrations in supernatants of EBV-2-infected T cells have lymphotactic properties (48–50). Production of lymphotactic cytokines has been hypothesized to be a strategy used by EBV to recruit additional lymphocyte targets during infection to enhance the establishment of a persistent infection (51). Indeed, EBV LMP-1, which was detected at all time points studied during EBV-2 T-cell infection, has been shown to upregulate production of IL-8, RANTES, and MIP-1α (52–55). Whether LMP-1 is directly responsible for the high levels of these cytokines observed in EBV-2-infected T-cell cultures and how expression of the cytokines affects the infectious process in vivo have yet to be determined.
To generate the virus stocks, we needed to use TPA and Na-butyrate, two compounds that could also indirectly induce some of the phenotypes we observed following EBV-2 infection. To control for this, we generated a mock infection using an EBV-negative BL cell line, Ramos, which was treated exactly the same as the EBV-infected cell lines were to generate virus. Treatment of T cells with mock-infected supernatant did not result in significant induction of proliferation, activation, or aggregation, providing further support for the idea that the effects on T cells we were seeing were due to infection with EBV-2. In addition, UV-inactivated virus did not induce significant cell proliferation or activation in CD4+ or CD8+ T cells, indicating that this is not due to binding of the virus to T cells. However, we cannot rule out the possibility that infected cells were inducing changes in noninfected cells, as not all cells in CD3+ cultures were infected.
Overall, our results provide the first evidence that EBV-2 efficiently activates and causes proliferation of T cells during primary infection. We also show that EBV-2 infection of T cells alters cytokine production. The inability of EBV-2 to efficiently transform B cells in vitro suggests that, unlike EBV-1, EBV-2 is impaired in the ability to independently drive B-cell differentiation and establish latency in the memory B-cell compartment. Our results provide a possible mechanism by which EBV-2 could be maintained in the human population, through T-cell infection and immune disruption (Fig. 7). In vivo studies are needed to determine whether EBV-1 and EBV-2 have different effects during primary infection of humans and, importantly, if EBV-2 is predominant in EBV-associated T-cell lymphomas and lymphoproliferative diseases.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health, NCI CA102667 (R.R.) and F30 CA168358 (E.M.W.).
We thank Gary Chan and Megan Peppenelli for their assistance with Western blot experiments, Jennifer Moffat for the acyclovir and PAA reagents and protocols, and Karen Gentile in the SUNY MAC core faculty for assistance with the Luminex assay.
REFERENCES
- 1.Park S, Ko YH. 2014. Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disorders. J Dermatol 41:29–39. doi: 10.1111/1346-8138.12322. [DOI] [PubMed] [Google Scholar]
- 2.Kasahara Y, Yachie A. 2002. Cell type specific infection of Epstein-Barr virus (EBV) in EBV-associated hemophagocytic lymphohistiocytosis and chronic active EBV infection. Crit Rev Oncol Hematol 44:283–294. doi: 10.1016/S1040-8428(02)00119-1. [DOI] [PubMed] [Google Scholar]
- 3.Iwatsuki K, Satoh M, Yamamoto T, Oono T, Morizane S, Ohtsuka M, Xu ZG, Suzuki D, Tsuji K. 2006. Pathogenic link between hydroa vacciniforme and Epstein-Barr virus-associated hematologic disorders. Arch Dermatol 142:587–595. doi: 10.1001/archderm.142.5.587. [DOI] [PubMed] [Google Scholar]
- 4.Fujiwara S, Kimura H, Imadome K, Arai A, Kodama E, Morio T, Shimizu N, Wakiguchi H. 2014. Current research on chronic active Epstein-Barr virus infection in Japan. Pediatr Int 56:159–166. doi: 10.1111/ped.12314. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JI, Jaffe ES, Dale JK, Pittaluga S, Heslop HE, Rooney CM, Gottschalk S, Bollard CM, Rao VK, Marques A, Burbelo PD, Turk S-P, Fulton R, Wayne AS, Little RF, Cairo MS, El-Mallawany NK, Fowler D, Sportes C, Bishop MR, Wilson W, Straus SE. 2011. Characterization and treatment of chronic active Epstein-Barr virus disease: a 28-year experience in the United States. Blood 117:5835–5849. doi: 10.1182/blood-2010-11-316745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawabe S, Ito Y, Gotoh K, Kojima S, Matsumoto K, Kinoshita T, Iwata S, Nishiyama Y, Kimura H. 2012. Application of flow cytometric in situ hybridization assay to Epstein-Barr virus-associated T/natural killer cell lymphoproliferative diseases. Cancer Sci 103:1481–1488. doi: 10.1111/j.1349-7006.2012.02305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu W-W, Hsieh P-P, Chuang S-S. 2013. Cutaneous EBV-positive γδ T-cell lymphoma vs. extranodal NK/T-cell lymphoma: a case report and literature review. J Cutan Pathol 40:310–316. doi: 10.1111/cup.12066. [DOI] [PubMed] [Google Scholar]
- 8.Chen CL, Sadler RH, Walling DM, Su IJ, Hsieh HC, Raab-Traub N. 1993. Epstein-Barr virus (EBV) gene expression in EBV-positive peripheral T-cell lymphomas. J Virol 67:6303–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groux H, Cottrez F, Montpellier C, Quatannens B, Coll J, Stehelin D, Auriault C. 1997. Isolation and characterization of transformed human T-cell lines infected by Epstein-Barr virus. Blood 89:4521–4530. [PubMed] [Google Scholar]
- 10.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A 81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowe M, Young LS, Cadwallader K, Petti L, Kieff E, Rickinson AB. 1989. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol 63:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sculley TB, Apolloni A, Stumm R, Moss DJ, Mueller-Lantczh N, Misko IS, Cooper DA. 1989. Expression of Epstein-Barr virus nuclear antigens 3, 4, and 6 are altered in cell lines containing B-type virus. Virology 171:401–408. doi: 10.1016/0042-6822(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 13.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol 64:4084–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan A, Addison C, Gatherer D, Davison AJ, McGeoch DJ. 2006. The genome of Epstein-Barr virus type 2 strain AG876. Virology 350:164–170. doi: 10.1016/j.virol.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Farrell PJ. 1995. Epstein-Barr virus immortalizing genes. Trends Microbiol 3:105–109. doi: 10.1016/S0966-842X(00)88891-5. [DOI] [PubMed] [Google Scholar]
- 16.Rickinson AB, Young LS, Rowe M. 1987. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol 61:1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancian L, Bosshard R, Lucchesi W, Karstegl CE, Farrell PJ. 2011. C-Terminal region of EBNA-2 determines the superior transforming ability of type 1 Epstein-Barr virus by enhanced gene regulation of LMP-1 and CXCR7. PLoS Pathog 7:e1002164. doi: 10.1371/journal.ppat.1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucchesi W, Brady G, Dittrich-Breiholz O, Kracht M, Russ R, Farrell PJ. 2008. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J Virol 82:7456–7466. doi: 10.1128/JVI.00223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorley-Lawson DA, Gross A. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med 350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 20.Young LS, Yao QY, Rooney CM, Sculley TB, Moss DJ, Rupani H, Laux G, Bornkamm GW, Rickinson AB. 1987. New type B isolates of epstein-Barr virus from Burkitt's lymphoma and from normal individuals in endemic areas. J Gen Virol 68:2853–2862. doi: 10.1099/0022-1317-68-11-2853. [DOI] [PubMed] [Google Scholar]
- 21.Goldschmidts WL, Bhatia K, Johnson JF, Akar N, Gutiérrez MI, Shibata D, Carolan M, Levine A, Magrath IT. 1992. Epstein-Barr virus genotypes in AIDS-associated lymphomas are similar to those in endemic Burkitt's lymphomas. Leukemia 6:875–878. [PubMed] [Google Scholar]
- 22.Sixbey JW, Shirley P, Chesney PJ, Buntin DM, Resnick L. 1989. Detection of a second widespread strain of Epstein-Barr virus. Lancet ii:761–765. [DOI] [PubMed] [Google Scholar]
- 23.Tomkinson B, Robertson E, Kieff E. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol 67:2014–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutt-Fletcher LM, Turk SM. 2001. Virus isolation. Methods Mol Biol 174:119–123. doi: 10.1385/1-59259-227-9:119. [DOI] [PubMed] [Google Scholar]
- 25.Moormann AM, Chelimo K, Sumba OP, Lutzke ML, Ploutz-Snyder R, Newton D, Kazura J, Rochford R. 2005. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J Infect Dis 191:1233–1238. doi: 10.1086/428910. [DOI] [PubMed] [Google Scholar]
- 26.Moormann AM, Chelimo K, Sumba PO, Tisch DJ, Rochford R, Kazura JW. 2007. Exposure to holoendemic malaria results in suppression of Epstein-Barr virus-specific T cell immunosurveillance in Kenyan children. J Infect Dis 195:799–808. doi: 10.1086/511984. [DOI] [PubMed] [Google Scholar]
- 27.Coen N, Duraffour S, Topalis D, Snoeck R, Andrei G. 2014. Spectrum of activity and mechanisms of resistance of various nucleoside derivatives against gammaherpesviruses. Antimicrob Agents Chemother 58:7312–7323. doi: 10.1128/AAC.03957-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summers WC, Klein G. 1976. Inhibition of Epstein-Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J Virol 18:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieff E, Rickinson A. 2001. Fields virology, 4th ed, p 2511–2574 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 30.Thorley-Lawson DA, Duca KA, Shapiro M. 2008. Epstein-Barr virus: a paradigm for persistent infection—for real and in virtual reality. Trends Immunol 29:195–201. doi: 10.1016/j.it.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Thorley-Lawson DA. 2005. EBV the prototypical human tumor virus—just how bad is it? J Allergy Clin Immunol 116:251–261. doi: 10.1016/j.jaci.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Tsang SF, Kurilla MG, Cohen JI, Kieff E. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol 64:3407–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito Y, Kawamura Y, Iwata S, Kawada J-I, Yoshikawa T, Kimura H. 2013. Demonstration of type II latency in T lymphocytes of Epstein-Barr Virus-associated hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 60:326–328. doi: 10.1002/pbc.24319. [DOI] [PubMed] [Google Scholar]
- 34.Paterson RL, Kelleher CA, Streib JE, Amankonah TD, Xu JW, Jones JF, Gelfand EW. 1995. Activation of human thymocytes after infection by EBV. J Immunol 154:1440–1449. [PubMed] [Google Scholar]
- 35.Todd SC, Tsoukas CD. 1996. EBV induces proliferation of immature human thymocytes in an IL-2-mediated response. J Immunol 156:4217–4223. [PubMed] [Google Scholar]
- 36.Price AM, Luftig MA. 2014. Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation. Adv Virus Res 88:279–313. doi: 10.1016/B978-0-12-800098-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price AM, Tourigny JP, Forte E, Salinas RE, Dave SS, Luftig MA. 2012. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation. J Virol 86:11096–11106. doi: 10.1128/JVI.01069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alfieri C, Birkenbach M, Kieff E. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595–608. doi: 10.1016/0042-6822(91)90893-G. [DOI] [PubMed] [Google Scholar]
- 39.Borza CM, Hutt-Fletcher LM. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med 8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 40.Kwok H, Tong AHY, Lin CH, Lok S, Farrell PJ, Kwong DLW, Chiang AKS. 2012. Genomic sequencing and comparative analysis of Epstein-Barr virus genome isolated from primary nasopharyngeal carcinoma biopsy. PLoS One 7:e36939. doi: 10.1371/journal.pone.0036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT. 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci U S A 81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemerow GR, Wolfert R, McNaughton ME, Cooper NR. 1985. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2). J Virol 55:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braun M, Melchers I, Peter HH, Illges H. 1998. Human B and T lymphocytes have similar amounts of CD21 mRNA, but differ in surface expression of the CD21 glycoprotein. Int Immunol 10:1197–1202. doi: 10.1093/intimm/10.8.1197. [DOI] [PubMed] [Google Scholar]
- 44.Calattini S, Sereti I, Scheinberg P, Kimura H, Childs RW, Cohen JI. 2010. Detection of EBV genomes in plasmablasts/plasma cells and non-B cells in the blood of most patients with EBV lymphoproliferative disorders by using Immuno-FISH. Blood 116:4546–4559. doi: 10.1182/blood-2010-05-285452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janz A, Oezel M, Kurzeder C, Mautner J, Pich D, Kost M, Hammerschmidt W, Delecluse H-J. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J Virol 74:10142–10152. doi: 10.1128/JVI.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imai S, Nishikawa J, Takada K. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol 72:4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorner BG, Scheffold A, Rolph MS, Huser MB, Kaufmann SHE, Radbruch A, Flesch IEA, Kroczek RA. 2002. MIP-1a, MIP-1b, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc Natl Acad Sci U S A 99:6181–6186. doi: 10.1073/pnas.092141999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy WJ, Taubu DD, Anveru M, Conlonu K, Oppenheimu JJ, Kelvinu DJ, Longof DL. 1994. Human RANTES induces the migration of human T lymphocytes into the peripheral tissues of mice with severe combined immune deficiency. Eur J Immunol 24:1823–1827. doi: 10.1002/eji.1830240815. [DOI] [PubMed] [Google Scholar]
- 49.Schall TJ, Bacon K, Camp RD, Kaspari JW, Goeddel DV. 1993. Human macrophage inflammatory protein alpha (MIP-1 alpha) and MIP-1 beta chemokines attract distinct populations of lymphocytes. J Exp Med 177:1821–1826. doi: 10.1084/jem.177.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang JM, Xu L, Murphy WJ, Taub DD, Chertov O. 1996. IL-8-induced T-lymphocyte migration: direct as well as indirect mechanisms. Methods 10:135–144. doi: 10.1006/meth.1996.0087. [DOI] [PubMed] [Google Scholar]
- 51.McColl SR, Roberge CJ, Larochelle B, Gosselin J. 1997. EBV induces the production and release of IL-8 and macrophage inflammatory protein-1 alpha in human neutrophils. J Immunol 159:6164–6168. [PubMed] [Google Scholar]
- 52.Yoshizaki T, Horikawa T, Qing-Chun R, Wakisaka N, Takeshita H, Sheen T-S, Lee S-Y, Sato H, Furukawa M. 2001. Induction of interleukin-8 by Epstein-Barr virus latent membrane protein-1 and its correlation to angiogenesis in nasopharyngeal carcinoma. Clin Cancer Res 7:1946–1951. [PubMed] [Google Scholar]
- 53.Buettner M, Meyer B, Schreck S, Niedobitek G. 2007. Expression of RANTES and MCP-1 in epithelial cells is regulated via LMP1 and CD40. Int J Cancer 121:2703–2710. doi: 10.1002/ijc.23018. [DOI] [PubMed] [Google Scholar]
- 54.Lai H-C, Hsiao J-R, Chen C-W, Wu S-Y, Lee C-H, Su I-J, Takada K, Chang Y. 2010. Endogenous latent membrane protein 1 in Epstein-Barr virus-infected nasopharyngeal carcinoma cells attracts T lymphocytes through upregulation of multiple chemokines. Virology 405:464–473. doi: 10.1016/j.virol.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 55.Tsai S-C, Lin S-J, Lin C-J, Chou Y-C, Lin J-H, Yeh T-H, Chen M-R, Huang L-M, Lu M-Y, Huang Y-C, Chen H-Y, Tsai C-H. 2013. Autocrine CCL3 and CCL4 induced by the oncoprotein LMP1 promote Epstein-Barr virus-triggered B cell proliferation. J Virol 87:9041–9052. doi: 10.1128/JVI.00541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]