Abstract
Viruses affect host physiology beyond causing acute disease, thereby giving rise to the concept that the virome is a component of the microbiome. However, the role of the enteric virome is understudied relative to the fast-paced research examining commensal bacteria in the intestine. In this article, I discuss our recent work on murine norovirus indicating that an animal virus in the intestine can provide many of the signals to the host that have been attributed to commensal bacteria. Our findings suggest that the surge in microbiome research should incorporate examination of the enteric virome.
INTRODUCTION
The human gut microbiome includes trillions of commensal bacteria that offer substantial benefits to the host, including aiding digestion, promoting the development of the immune system, and occupying niches that would otherwise be available to pathogens (1). Hence, the relationship between commensal bacteria and the host can be described as mutualism, a form of symbiosis in which both parties benefit. This mutualism may be conditional, because commensal bacteria are implicated in many chronic inflammatory disorders. In particular, mouse models of inflammatory bowel disease (IBD) have been helpful in establishing causality and have shown that intestinal bacteria that are otherwise innocuous or easily contained evoke disease in genetically susceptible hosts (2). The diversity, the volume, and this proposed role in disease have been used to justify the eruption of studies that catalog and characterize intestinal bacteria.
The enteric virome has received less attention, partly due to the technological barrier in identifying viruses and performing functional studies. The coding potential of the enteric virome is predicted to be immense, because it includes viruses that infect host cells, endogenous retroviruses, and viruses that infect the various microbial inhabitants of the gastrointestinal tract, such as bacteria, archaea, and fungi. Recent studies indicate that the most abundant members of the enteric virome, bacteriophages that infect commensal bacteria, are diverse and likely to have a substantial impact on the host (3). Here, I use our recent findings in a mouse model to propose that animal viruses in the gut not only are pathogens that infect host cells and cause gastroenteritis but also are symbiotic modulators of host physiology.
A VIRAL TRIGGER IN A MOUSE MODEL OF IBD
During my postdoctoral research in Skip Virgin's laboratory, we generated mice with decreased expression of the autophagy gene Atg16l1, with the intention of investigating the role of autophagy in viral pathogenesis. As we began characterizing these mice, a series of population genetics studies were published that linked a common polymorphism in Atg16l1 with IBD susceptibility. IBD is characterized by recurrent intestinal inflammation that is considered the consequence of a loss of tolerance toward commensal bacteria. The relationship between this potential microbial origin of the disease and genetic susceptibility factors, such as the polymorphism in Atg16l1, remains an active area of research. Motivated by the many unanswered questions surrounding the origin of IBD, we collaborated with pathologist Thad Stappenbeck to characterize these Atg16l1 mutant mice. We found that Atg16l1 mutation led to structural and functional abnormalities in Paneth cells, specialized epithelial cells found in small intestinal crypts that produce antimicrobial granules (4). We also observed similar Paneth cell abnormalities in IBD patients homozygous for the ATG16L1 risk allele, suggesting that this model may be useful in understanding disease pathogenesis.
The next part of the project was possible only because of a previous discovery made by Stephanie Karst and Christiane Wobus while they were postdoctoral fellows in the Virgin laboratory. Several years before my arrival, they discovered that laboratory mice harbor a positive-strand RNA virus with close sequence homology to human noroviruses, which was aptly named murine norovirus (MNV) (5). Similar to noroviruses that infect humans, MNV is readily detected in the gastrointestinal tract, and we were concerned about its widespread presence in animal facilities, including the facility in which Atg16l1 mutant mice were maintained at the time. Thus, we transferred Atg16l1 mutant mice to an advanced MNV-free barrier facility through embryo rederivation. We soon found that Atg16l1 mutant mice raised in this new facility did not display the Paneth cell abnormalities that previously appeared to develop spontaneously. It is not uncommon for the presence or degree of intestinal pathologies to display variability when mice raised in different facilities are compared, a phenomenon that is frequently attributed to differences in commensal bacteria or common pathogens, such as Helicobacter species (6). Indeed, we found that intentional MNV infection restored the appearance of Paneth cell abnormalities in Atg16l1 mutant mice. Additionally, MNV-infected Atg16l1 mutant mice, but not MNV-infected control mice or uninfected Atg16l1 mutant mice, developed IBD pathologies upon intestinal injury with the chemical dextran sodium sulfate (DSS) (7). Although Atg16l1 and autophagy are important in restricting the replication of intracellular pathogens, we did not observe increased MNV replication in Atg16l1 mutant mice compared with controls in this setting, suggesting that the intestinal abnormalities are due to a loss of tolerance to the virus. Thus, we observed a viral trigger of pathologies in a mouse model of IBD, a disease that has mainly been attributed to exacerbated immune responses directed toward commensal bacteria.
MNV CAN FUNCTIONALLY REPLACE THE BENEFICIAL CUES PROVIDED BY COMMENSAL BACTERIA
MNV has been used as a model to understand the biology of human noroviruses because of its ability to be propagated in cell culture and infect mice. A groundbreaking recent study suggests that MNV and human noroviruses may also share a dependence on B cells and commensal bacteria for efficient replication (8). However, rather than inducing an acute vomiting and diarrheal disease like human noroviruses, MNV frequently establishes persistent infection in immunocompetent mice without causing obvious disease. We were struck by the similarity between MNV and commensal bacteria—both can persist in the host without causing disease but induce intestinal abnormalities in IBD models—and this resemblance led us to examine whether MNV can mimic other features of commensal bacteria, perhaps even having a beneficial function for the host.
To test this idea, we monoassociated germfree mice with MNV by inoculating breeding pairs within a sterile gnotobiotic isolator and allowing the virus to be vertically transmitted to offspring (9). Due to the absence of detectable bacteria, germfree mice display structural and functional abnormalities in the intestine and associated mucosal immune system (10), some of which are listed in Fig. 1. As reported by other groups investigating enteric viral replication in mice deficient in bacteria (8, 11, 12), the amount of virus recovered from these monoassociated mice was reduced, but prolonged shedding was detectable. Remarkably, we found that MNV was able to partially, and in some cases completely, reverse many of the abnormalities that are present in germfree mice (Fig. 1) (9). MNV infection of mice treated with a cocktail of antibiotics, which also causes intestinal abnormalities, led to a similar restoration of intestinal morphology and lymphoid function. Depletion of commensal bacteria following antibiotic treatment is a health hazard in humans as well, as evidenced by antibiotic-associated diarrhea. We found that MNV provides substantial protection in antibiotic-treated mice in two models of intestinal damage, administration of dextran sodium sulfate (DSS) in drinking water and oral infection by Citrobacter rodentium (Gram-negative bacterium related to Escherichia coli) (9). When taken together, these observations indicate that one virus is able to perform many of the same functions as an entire community of bacteria that includes ∼1,000 species.
FIG 1.
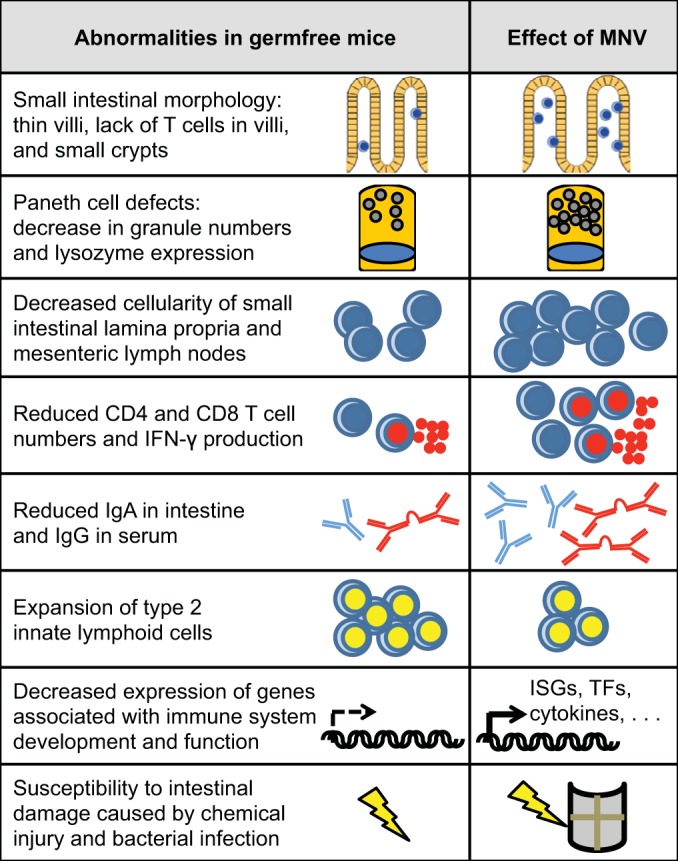
Abnormalities in germfree mice are reversed by murine norovirus infection. Due to the absence of detectable bacteria, germfree mice display several abnormalities in the intestine and associated immune compartments, a subset of which are listed in the left column. Monoassociating germfree mice with murine norovirus (MNV) is sufficient to partially or completely reverse many of these abnormalities, as indicated in the right column (9). IFN, interferon; ISG, interferon-stimulated gene; TF, transcription factor.
Although the array of responses elicited by a single viral infection is striking, MNV-monoassociated germfree mice are not exactly the same as conventional mice. For instance, MNV is unable to reduce the expansion of NK T cells observed in the colons of germfree mice (13). Also, when we compared three strains of MNV, we found that they evoked similar responses, but there were quantitative differences in their abilities to reverse intestinal abnormalities and provide protection following DSS injury. However, it is important to note that host responses evoked by individual commensal bacterial species also differ significantly from one another, particularly in their ability to induce T cell differentiation (14). A key question is whether MNV evokes only responses from the host that are redundant with those induced by commensal bacteria or whether there are unique benefits conferred by MNV infection that cannot be provided by commensal bacteria. Our previous observation that MNV triggers inflammatory pathologies in Atg16l1 mutant mice (when commensal bacteria are present) suggests that this virus and commensal bacteria evoke nonoverlapping responses from the host, at least under certain conditions.
PERSPECTIVES
The concept that viruses affect host trait and alter disease susceptibility is supported by pioneering studies demonstrating that lymphocytic choriomeningitis virus (LCMV) prevents diabetes in mice (15) and has subsequently been reinforced by many epidemiology observations and findings in animal models (16, 17). Our experiments with MNV build on this understanding of the virome by demonstrating that an enteric virus can function analogously to commensal bacteria that inhabit the gastrointestinal tract. Extending our results with one mouse virus to humans or other animals is premature, and enteric viruses such as noroviruses must continue to be treated as serious pathogens. Still, it is unlikely that MNV is the only virus that can evoke these responses from a host. The effect of MNV was dependent on type I interferon (IFN-I), which is a conserved response to viruses. A deeper understanding of how IFN-I functions in this setting, and what other pathways function downstream of MNV, may help determine what commensal viruses would look like if they exist in humans. Also, understanding why MNV is beneficial in antibiotic-treated mice but deleterious in Atg16l1 mutant mice could yield insight into how balanced immunity is maintained in the intestine in the presence of a diverse microbial flora.
Although viruses in human specimens remain difficult to identify and propagate in culture, existing technologies are able to detect sequences corresponding to prokaryotic and eukaryotic viruses in stool harvested from individuals that do not present disease at the time of collection. Our findings, as limited as they are, suggest that referring to the presence of these viruses as “asymptomatic infections” is much like ignoring commensal bacteria because they are found in healthy individuals. A similar argument can be made regarding other microbial inhabitants of the gastrointestinal tract, which include archaea, fungi, protozoa, and helminth parasites. This potential contribution of the enteric virome to human health is not always acknowledged. For instance, fecal transplants that are effective for treating Clostridium difficile colitis are being applied to other illnesses without much consideration for the presence of viruses. Innovations that facilitate detection of known and unknown enteric viruses, as well as techniques that allow their characterization, are an immediate need.
The MNV model is also revealing novel ways in which a viral infection can interact with other microbes in the intestine. The MNV-induced intestinal pathologies in DSS-treated Atg16l1 mutant mice are prevented by antibiotic treatment (7), which may be related to the observation that MNV replication is partially dependent on the presence of bacteria (8, 9). Additionally, two complementary studies showed that commensal bacteria suppress an IFN-λ response that would otherwise prevent persistent MNV infection (18, 19). Finally, helminth infection impairs immunity to MNV, which is unexpectedly independent of commensal bacteria (20). Therefore, the enteric virome is perhaps best described as a component of the gut microbiome that interacts with bacteria and other microbial agents.
ACKNOWLEDGMENTS
I thank Skip Virgin and Elisabeth Kernbauer for critically reading and providing helpful comments on the manuscript.
We are funded by the National Institutes of Health (R01 DK093668) and the American Heart Association (12GRNT12030041).
REFERENCES
- 1.Hooper LV, Gordon JI. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 2.Huttenhower C, Kostic AD, Xavier RJ. 2014. Inflammatory bowel disease as a model for translating the microbiome. Immunity 40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duerkop BA, Hooper LV. 2013. Resident viruses and their interactions with the immune system. Nat Immunol 14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW IV. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 6.Mizoguchi A. 2012. Animal models of inflammatory bowel disease. Prog Mol Biol Transl Sci 105:263–320. doi: 10.1016/B978-0-12-394596-9.00009-3. [DOI] [PubMed] [Google Scholar]
- 7.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kernbauer E, Ding Y, Cadwell K. 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature 516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda K, Littman DR. 2012. The microbiome in infectious disease and inflammation. Annu Rev Immunol 30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oldstone MB. 1988. Prevention of type I diabetes in nonobese diabetic mice by virus infection. Science 239:500–502. doi: 10.1126/science.3277269. [DOI] [PubMed] [Google Scholar]
- 16.Roossinck MJ. 2011. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol 9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 17.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. 27November2014. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 27November2014. Interferon lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, Tomov VT, Kobuley D, Tran SV, Bittinger K, Bailey AG, Laughlin AL, Boucher JL, Wherry EJ, Bushman FD, Allen JE, Virgin HW, Artis D. 2014. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 345:578–582. doi: 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]


