ABSTRACT
Herpes simplex virus 1 (HSV-1) capsids are assembled in the nucleus, where they incorporate the viral genome. They then transit through the two nuclear membranes and are wrapped by a host-derived envelope. In the process, several HSV-1 proteins are targeted to the nuclear membranes, but their roles in viral nuclear egress are unclear. Among them, glycoprotein M (gM), a known modulator of virus-induced membrane fusion, is distributed on both the inner and outer nuclear membranes at the early stages of the infection, when no other viral glycoproteins are yet present there. Later on, it is found on perinuclear virions and ultimately redirected to the trans-Golgi network (TGN), where it cycles with the cell surface. In contrast, transfected gM is found only at the TGN and cell surface, hinting at an interaction with other viral proteins. Interestingly, many herpesvirus gM analogs interact with their gN counterparts, which typically alters their intracellular localization. To better understand how HSV-1 gM localization is regulated, we evaluated its ability to bind gN and discovered it does so in both transfected and infected cells, an interaction strongly weakened by the deletion of the gM amino terminus. Functionally, while gN had no impact on gM localization, gM redirected gN from the endoplasmic reticulum (ER) to the TGN. Most interestingly, gN overexpression stimulated the formation of syncytia in the context of an infection by a nonsyncytial strain, indicating that gM and gN not only physically but also functionally interact and that gN modulates gM's activity on membrane fusion.
IMPORTANCE HSV-1 gM is an important modulator of virally induced cell-cell fusion and viral entry, a process that is likely finely modulated in time and space. Until now, little was known of the proteins that regulate gM's activity. In parallel, gM is found in various intracellular locations at different moments, ranging from nuclear membranes, perinuclear virions, the TGN, cell surface, and mature extracellular virions. In transfected cells, however, it is found only on the TGN and cell surface, hinting that its localization is modulated by other viral proteins. The present study identifies HSV-1 gN as a binding partner for gM, in agreement with their analogs in other herpesviruses, but most excitingly shows that gN modulates gM's impact on HSV-1-induced membrane fusion. These findings open up new research avenues on the viral fusion machinery.
INTRODUCTION
Herpesviridae are among the most complex human viruses from the point of view of their large genomes and viral particle composition. Among them, herpes simplex virus 1 (HSV-1), the prototype of human alphaherpesviruses, incorporates its 152-kb genome into an icosahedral capsid surrounded by a multiprotein tegument layer and a cell-derived lipid layer containing over a dozen viral proteins (1, 2). Of the latter, those mediating viral entry, namely, gB, gD, and the gH/gL complex, are essential for the propagation of the virus (3). In contrast, the viral glycoprotein M (gM) is conserved throughout the family and typically critical for beta- and gammaherpesviruses but is not essential for most alphaherpesviruses, including HSV-1 (4–16). Consequently, when gM is depleted from HSV-1 or the related alphaherpesvirus pseudorabies virus, viral yields are minimally reduced by 3- to 50-fold. However, its impact is substantially increased when UL11 and gE/gI are codepleted in combination with gM, likely due to overlapping functions between these viral proteins (5, 17–19).
Despite its nonessential status in tissue culture, gM of several alphaherpesviruses has been associated with a number of functions throughout the viral life cycle (7, 10, 19–22). The glycoprotein is thus known to downregulate the surface expression of gD and the gH/gL complex, two key players in virus-induced membrane fusion, and facilitates the upstream incorporation of the gH/gL complex into mature virions (23, 24). Furthermore, gM has been shown to stimulate viral entry in the context of syncytial strains (22). However, despite its presence on nuclear membranes (see below), gM is seemingly not involved in the release of herpesviruses from the nucleus, where newly made viral capsids are initially assembled (17). In contrast, with the conserved gN viral protein, gM alters immunity against the virus by downregulating the transport and peptide loading of major histocompatibility complex class I in the endoplasmic reticulum (ER) (25–30). Finally, and perhaps most interestingly, gM has been reported to modulate virulence in animal models (31, 32). Thus, gM appears to exert important and diverse regulatory activities at potentially different intracellular localizations. In this context, targeting of gM to these distinct sites is likely one important means to regulate its function. It is therefore critical to understand this process and define its molecular players.
In several herpesviruses, gM, gN, and related homologues physically interact (16, 21, 33–36). In many cases, this association impacts their release from the ER and maturation at the Golgi apparatus. For Epstein-Barr virus (EBV) and infectious laryngotracheitis virus (ILTV), gM is required for the processing of the related gN (36, 37), while for bovine herpesvirus 1 (BHV-1), human herpesvirus 6 (HHV-6), and HHV-8, gN is a dominant determinant of gM transport and maturation (15, 21, 30). In contrast, the sole transfection of HSV-1 gM is sufficient to drive it to the trans-Golgi network (TGN) irrespective of the presence of gN or other viral genes (23, 38). However, a more complex and dynamic scenario takes place in infected cells with the early recruitment of HSV-1 gM to nuclear membranes at 4 h postinfection (hpi) and ultimately on perinuclear virions once they are produced (38, 39). Much later on, gM is redirected to the TGN, where it cycles back and forth with the plasma membrane and may ultimately be retargeted again to the ER at a time when the TGN is eventually disrupted by the infection (39). The glycoprotein is also incorporated on mature extracellular virions (6). HSV-1 gN localization is more problematic to access, as antibodies are currently unavailable to characterize it. It is thus unclear if it is complexed to gM, as are its counterparts in other herpesviruses, and how the two molecules depend on each other for their intracellular transport and maturation. Moreover, no function has yet been assigned to gN.
Given that gM expression is detectable as early as 2 hpi and that gN mRNAs are only discernible from 10 hpi onward, coincident with the release of gM from the ER (38), it was tempting to postulate that gN may be the molecular switch that releases gM from the nuclear membranes in infected cells. The present study examines this possibility using a yellow fluorescent protein (YFP)-tagged version of gN to monitor it and depicts an intricate situation where gM and gN do indeed form a complex in the absence of additional viral proteins, as the two proteins readily interacted in transfected and infected cells. While gM was targeted to the TGN independently of gN, the latter was exclusively at the ER when transfected alone. However, gN was significantly redirected to the TGN in the presence of gM. Importantly, the mutual coimmunoprecipitation (co-IP) of YFP-tagged gN with either endogenous or exogenous gM confirmed their physical interaction both in transfected and infected cells. Taking advantage of a previously reported mutant deleting the first two methionines at amino acids 1 and 19 (19), but still expressing lower levels of an amino-terminal truncated gene product starting from the third methionine residue at amino acid 133, we further showed that the formation of the gM/gN complex still occurs but is strongly disrupted by the deletion. Most excitingly, this physical interaction transcends into a functionally relevant one, as overexpression of gN in wild-type (WT)-infected cells leads to a significant increase of syncytium formation, a normally rare event unless syncytial mutants are used. All together, these results hint at a fine multiprotein modulation of cell-cell fusion that is deregulated by altering gN expression. Given the importance of cell-to-cell transmission to avoid immune detection by neutralizing antibodies, these findings likely have a significant impact for viral spread.
MATERIALS AND METHODS
Cells and viruses.
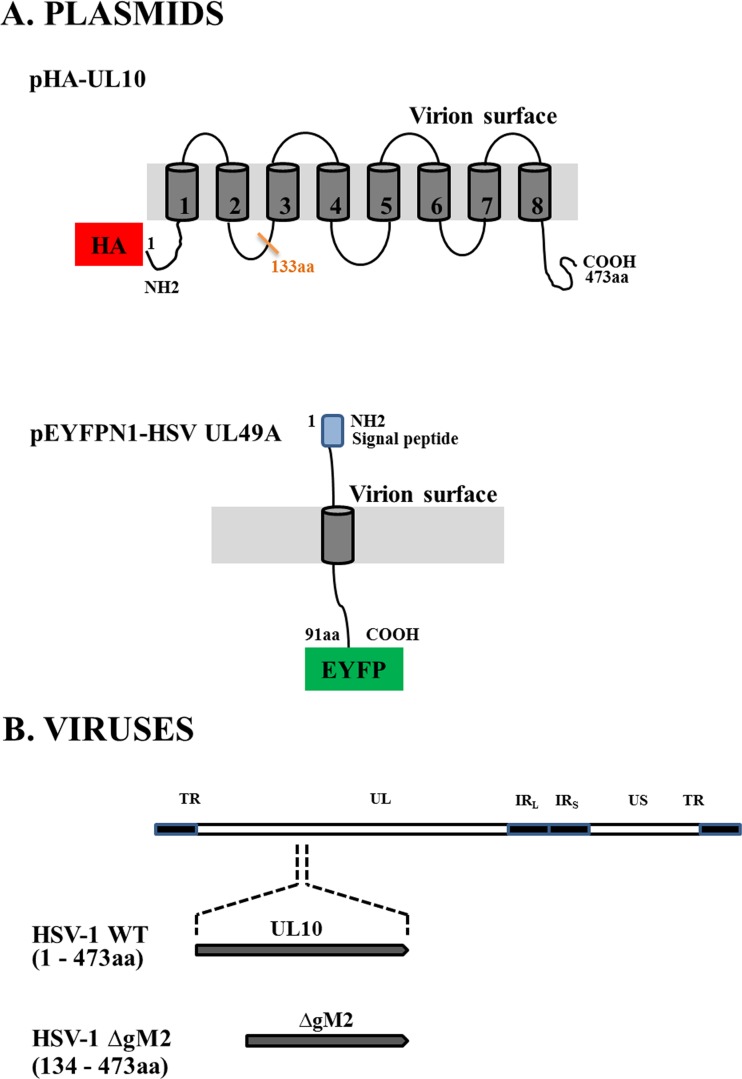
143B and HeLa (ATCC) cells were grown in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS; HyClone) and 2 mM l-glutamine (Invitrogen) in 5% CO2. 143B cells were also supplemented with 15 μg/ml 5-bromo-2 deoxyuridine (BrdU; Sigma) except prior to transfection or infection. The nonsyncytial wild-type HSV-1 strain 17+ (obtained from Beate Sodeik) and the HSV-1ΔgM2 mutant virus (provided by Konstantin Kousoulas [19]) were propagated on BHK cells, and their titers were determined by plaque assay on Vero cells. For clarity, the viruses and constructs used in the paper are shown in Fig. 1.
FIG 1.
Schematic view of plasmids and viruses used in this study. (A) Plasmid vectors expressing either the amino-terminal HA-tagged gM (pHA-UL10) or the carboxyl-terminal YFP-tagged gN (pEYFPN1-HSV UL49A) fusion proteins are shown. The gray areas indicate the predicted transmembrane domains. (B) The genomic map of the HSV-1 genome is showing the unique long (UL) and unique short (US) regions flanked by terminal (TR) and internal inverted (IRL and IRS) repeat sequences. In most experiments, wild-type virus coding for the full-length gM was used (473 amino acids; HSV-1 WT). Where indicated, a mutant virus coding for a truncated gM, in which the first 133 amino acids of gM were deleted (HSV-1 ΔgM2 mutant), was used.
Transfections and infections.
For cotransfection studies, 143B cells grown in 24-well plates were transfected with 0.8 μg/well of pHA-UL10 (expressing hemagglutinin [HA]-tagged HSV-1 gM; Mutagenex) and pEYFPN1-HSV UL49A (coding for HSV-1 gN; generously provided by Colin Crump) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours later, the cells were fixed and permeabilized for immunofluorescence microscopy. To monitor their subcellular localization in the context of an HSV-1 infection, the cells were first transfected with 0.8 μg/ml of pEYFPN1-HSV UL49A for 24 h and then absorbed with wild-type HSV-1 strain 17+ at a multiplicity of infection (MOI) of 2 for 1 h at 37°C and subsequently grown in standard medium at 37°C. At various times following postinfection, the cells were fixed and permeabilized for immunolabeling as detailed below.
Immunofluorescence microscopy.
For confocal microscopy, cells were fixed at 4°C for 30 min in 3% paraformaldehyde made in phosphate-buffered saline (PBS) and washed with PBS, and any remaining fixative was inactivated with 50 mM NH4Cl in PBS. The cells were permeabilized using 0.1% Triton X-100 for 4 min, and nonspecific protein binding sites were blocked with 10% FCS. The specimens were labeled for 1 h at room temperature with primary antibodies diluted in 10% FCS, washed, and incubated with secondary antibodies for 30 min. The samples were mounted on glass slides in Dako containing 0.1 μg/ml Hoechst 33342 (Sigma-Aldrich) to stain the nuclei. The following primary antibodies were used: anti-gM rabbit polyclonal 4c10 (courtesy of Joel Baines) to detect endogenous gM (infection) or a mouse monoclonal anti-HA antibody (Santa Cruz) to detect HA-tagged exogenous gM (transfection). We also used sheep anti-TGN46 (Serotec), rabbit anti-calnexin (Stressgen), and a mouse anti-green fluorescent protein (GFP; Roche). All secondary antibodies (Alexa 488, 568, and 674) were from Molecular Probes. Fluorescence microscopy was performed on an LSM510 confocal microscope and software (Carl Zeiss). The confocal sections were all acquired using a 100× objective, and contrast adjustments of immunofluorescence images were performed equally with Adobe Photoshop CS5 for all images within each experiment. Quantification of the subcellular localization of gM and gN was done by manually counting cells that were positive for both proteins. In all cases, at least 200 positive cells were compiled for each condition and independent experiment.
Syncytium formation.
143B cells grown in 24-well plates were transfected with 0.8 μg/well of pEYFPN1-HSV UL49A using Lipofectamine 2000 as described above. Twelve hours later, the cells were infected with the nonsyncytial wild-type HSV-1 strain 17+ or the HSV-1ΔgM2 mutant at an MOI of 2 for 1 h at 37°C and then grown in standard medium at 37°C. At various times postinfection, the cells were fixed and stained with 0.1 μg/ml Hoechst 33342 to identify the nuclei. The plasma membrane was additionally labeled with 5 μg/ml wheat germ agglutinin (WGA) conjugated to Alexa Fluor 647 (Molecular Probes) to delineate the cell boundaries. Syncytia, defined as fluorescent membranes containing two or more nuclei, were scored from randomly selected fields of view (200 cells per condition per independent experiment).
Immunoprecipitation.
For immunoprecipitation studies, HeLa cells were chosen because of their high rate of transfection. They were grown on 10-cm dishes and transfected at 80% confluence with either 24 μg/dish of pHA-UL10 or pEYFPN1-HSV UL49A or alternatively with 12 μg/dish of each plasmid. Twenty-four hours posttransfection, the cells were lysed for 30 min at 4°C with gentle agitation with RIPA buffer (0.15 M NaCl, 10 mM Tris-HCl [pH 7.4], 1% deoxycholic acid, 1% NP-40, 0.1% SDS, supplemented with protease inhibitor cocktail [Roche]). For immunoprecipitation studies in the context of infections, cells were transfected for 24 h with 24 μg/dish of pEYFPN1-HSV UL49A and subsequently infected with HSV-1 WT or the HSV-1 ΔgM2 mutant at an MOI of 5 for an additional 16 h. They were then lysed as described above. In both cases, cell debris was removed by centrifugation (14,000 × g, 15 min, 4°C), and the supernatants were incubated with preimmune serum for 1 h. These lysates were subsequently incubated with protein A agarose (Roche) for 1 h hour at 4°C with gentle agitation. After centrifugation at 10,000 × g for 5 min to remove the beads, these precleared lysates were incubated overnight at 4°C with mouse anti-HA (to detect exogenous gM), rabbit 4c10 polyclonal (to bring down endogenous gM; in our hands, the PAS980 polyclonal (see below) did not work well for this purpose), or mouse anti-GFP (which very efficiently binds enhanced YFP [EYFP] [data not shown]). Fresh protein A agarose beads were added for 60 min, and the immune complexes were washed three times with RIPA buffer and once with Tris-HCl (pH 7.4) to remove unbound proteins. The bead-bound material and total lysates were finally analyzed by Western blotting as detailed below. To evaluate the impact of the gM truncation on gN coimmunoprecipitation (co-IP), the quantification of the Western blot data was normalized to the level of gM and gN that could be immunoprecipitated by their respective antibodies. The ratio between the co-IP in the context of wild-type infection and the amount of co-IP with the HSV-1 ΔgM2 mutant was then calculated.
SDS-PAGE electrophoresis and Western blotting.
gM tends to aggregate near boiling temperatures (personal unpublished observation). Consequently, immunoprecipitated proteins and whole-cell lysates were heated prior to being loaded on denaturing 10% polyacrylamide gels with sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 2% β-mercaptoethanol) at 37°C for 1 h to analyze gN, 56°C for 2 min for gM, and 1 h at 37°C plus 2 min at 56°C when analyzing both proteins. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes. The membranes were immersed for 1 h in blocking buffer (5% nonfat dry milk, 13.7 mM NaCl, 0.27 mM KCl, 0.2 mM KH2PO4, 1 mM Na2HPO4, and 0.1% Tween 20) and subsequently incubated for 2 h with a rabbit polyclonal against HSV-1 gM (PAS980, kindly provided by Lynn Enquist) or rabbit anti-GFP antibodies diluted in blocking buffer to detect gM and gN, respectively. The blots were then washed and probed with secondary antibodies conjugated to horseradish peroxidase (anti-rabbit [Cedarlane]). When rabbit gM antibodies were used for both immunoprecipitations and Western blotting (for instance, to detect gM/gN complexes in infected cells), light-chain-specific antibodies (ImmunoResearch Johnson) were used as secondary antibodies at this stage to avoid the detection of the primary antibody. Final revealing was done with the Super Signal West Pico chemiluminescent substrate (Pierce) on Kodak BioMax Light film or by using a ChemiDoc MP system (Bio-Rad). Where indicated, the latter was used to quantify the expression of gM in wild-type and mutant strains using the onboard quantification tools (Image Lab version 5.0).
RESULTS
gM binds to and drives gN to the TGN in transfected cells.
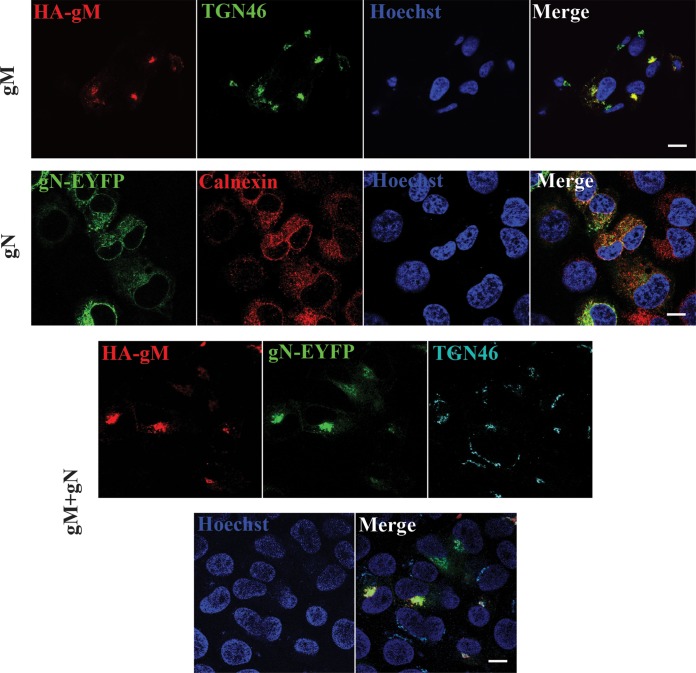
To directly probe the putative formation of an HSV-1 gM/gN complex, the two proteins were first examined by fluorescence microscopy in transfected cells. As anticipated, cells that were transfected with gM alone led to the accumulation of that protein at the TGN (Fig. 2), in agreement with previous reports (23, 38). In contrast, gN alone accumulated in the ER, as evidenced by its reticular staining and outer nuclear envelope pattern and overlapping with the calnexin marker (Fig. 2). When coexpressed with gM, both proteins strongly colocalized and were exclusively found at the TGN in doubly transfected cells (Fig. 2). Quantification of over 600 cells taken from multiple images and three independent experiments confirmed these findings with 100% of transfected gM alone at the TGN, 100% of gN alone in the ER, and 100% of cotransfected gM and gN at the TGN, respectively. Importantly, cotransfection of gN with an empty HA vector did not relocalize any gN to the TGN (data not shown). While gM targeting was independent of gN, the intracellular localization of gN was thus strongly influenced by gM. Taken together, these data strongly hinted at a functional interaction between gM and gN.
FIG 2.
gM drives gN to the TGN in transfected cells. 143B cells grown on coverslips were transfected with plasmids coding for HA-tagged gM and YFP-labeled gN or cotransfected with the two plasmids and processed as described in Materials and Methods. The coverslips were fixed and then reacted with antibodies against HA (to detect gM), TGN46 (to label the TGN), or calnexin (to identify the endoplasmic reticulum), while nuclei were labeled with Hoechst. Samples were analyzed by confocal laser scanning microscopy. Scale bar, 10 μm.
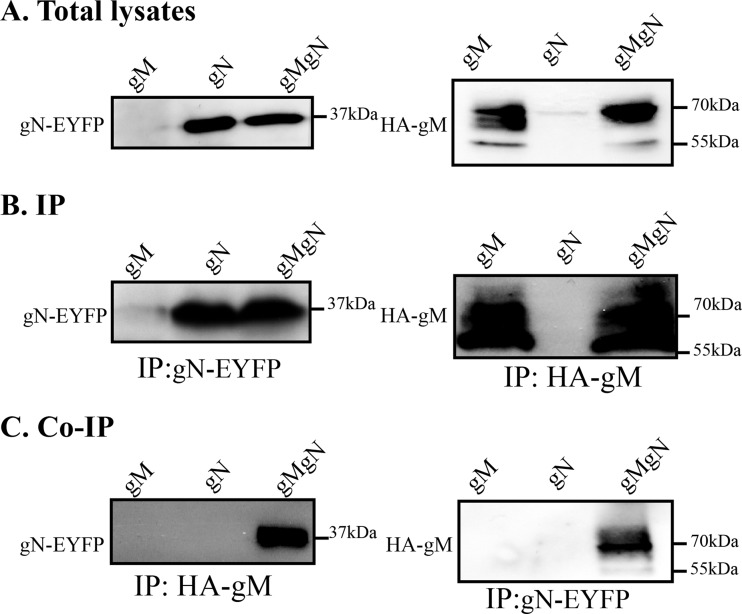
To assess whether the functional interaction seen in the cotransfected cells translated into direct physical interactions, cells were transfected with plasmids coding for either protein or cotransfected with both and analyzed by pulldown. Unfortunately, the initial gM polyclonal used (PAS980) was not suitable for immunoprecipitations in our hands, and a gN antiserum is unavailable as of yet; the two molecules were thus individually immunoprecipitated using an antibody against the HA tag present on gM or an anti-GFP antibody, which efficiently reacted with the related EYFP tag present on gN (data not shown). These samples were then analyzed by Western blotting to probe the association of the two proteins. As revealed in Fig. 3A and B, either antibody readily and specifically detected its intended target. Furthermore, the gM glycoprotein was, as expected, expressed in multiple forms, ranging from 55 to over 70 kDa, consistent with its previously documented post-translational processing (6, 38). Meanwhile, gN appeared as a single band near the 37-kDa marker, a mass consistent with the predicted 9.2-kDa gN when coupled to the 27-kDa YFP moiety (Fig. 3A). Most interestingly, the immunoprecipitation of gM brought down gN and vice versa, indicating that the two molecules did indeed interact and did so in the absence of any additional viral protein (Fig. 3C). Interestingly, the GFP immunoprecipitation appeared enriched in the higher-molecular-mass forms of gM and contained much less of the 55-kDa gM band seen in the total cell lysate or gM immunoprecipitation, suggesting the complex may preferentially be enriched for specific gM posttranslational forms (Fig. 3C).
FIG 3.
Physical interaction between gM and gN in transfected cells. HeLa cells were transfected with plasmids expressing HA-tagged gM (gM) or YFP-tagged gN (gN) or cotransfected with both constructs (gMgN). The cells were harvested at 24 h posttransfection and lysed with RIPA buffer. (A) As positive controls, 1/50th of the total lysate of each sample was also loaded onto the gels and analyzed. (B and C) The rest of the lysates were immunoprecipitated (IP) with mouse anti-HA antibodies for gM or mouse anti-GFP antibodies for gN and subjected to SDS-PAGE in 10% gel followed by Western blotting. The blots were reacted with the PAS980 anti-gM antibody or an anti-GFP antibody to detect gM and gN, respectively, as indicated to the left of each blot. The molecular masses of the molecular weight markers are indicated to the right of the panels.
The gM/gN complex exists in infected cells.
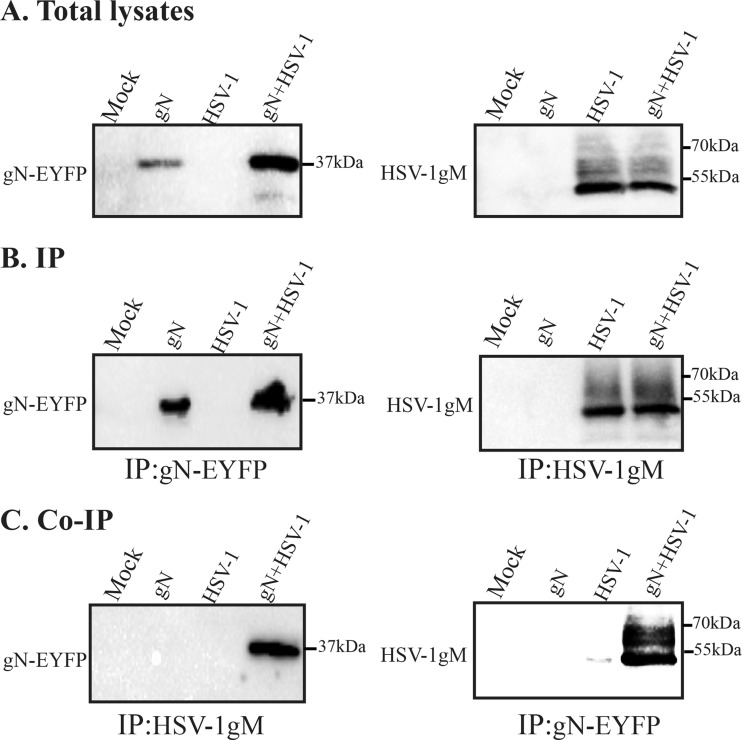
Given the differential behavior of gM in infected and transfected cells, it was crucial to evaluate if the above gM/gN complex could also be detected in the context of an infection. For this purpose, cells were transfected with pEYFPN1-HSV UL49A to circumvent the lack of gN antibody and then infected with the wild-type virus as a source of endogenous gM. The potential complex was probed by pulldown assays. The data confirm the specificity of the respective antibodies and their ability to bring down their intended targets (Fig. 4B). It also shows that a complex could indeed be readily detected in infected cells supplemented with exogenous YFP-tagged gN (Fig. 4C), in agreement with the aforementioned cotransfection results. As before, gN was detected as a single band, indicating that gN expression in the context of an infection was no different than in transfection experiments. Similarly, the complex appeared enriched in the higher-molecular-mass forms of gM (compare total or immunoprecipitated gM with that following an immunoprecipitation against the YFPP/gN protein). We thus concluded that gM does indeed form a complex with gN in both transfected and infected scenarios.
FIG 4.
Coimmunoprecipitation of the gM/gN complex in infected cells. HeLa cells were transfected with YFP-tagged gN for 24 h. They were then infected with wild-type virus at an MOI of 5, and the cells were harvested at 16 hpi and lysed. Two and a half percent (1/40th) of the lysates were directly loaded onto the gels (A), and the rest of the lysates were immunoprecipitated (IP) with the 4C10 anti-gM or anti-GFP antibody as indicated (B and C). All samples were analyzed by Western blotting. The blots were reacted with the PAS980 anti-gM (A to C, right side) or anti-GFP antibodies (A to C, left side). Uninfected cells were used as the negative control. The molecular masses of the molecular weight markers are indicated to the right of the panels.
Implication of the gM amino terminus in the gM/gN complex.
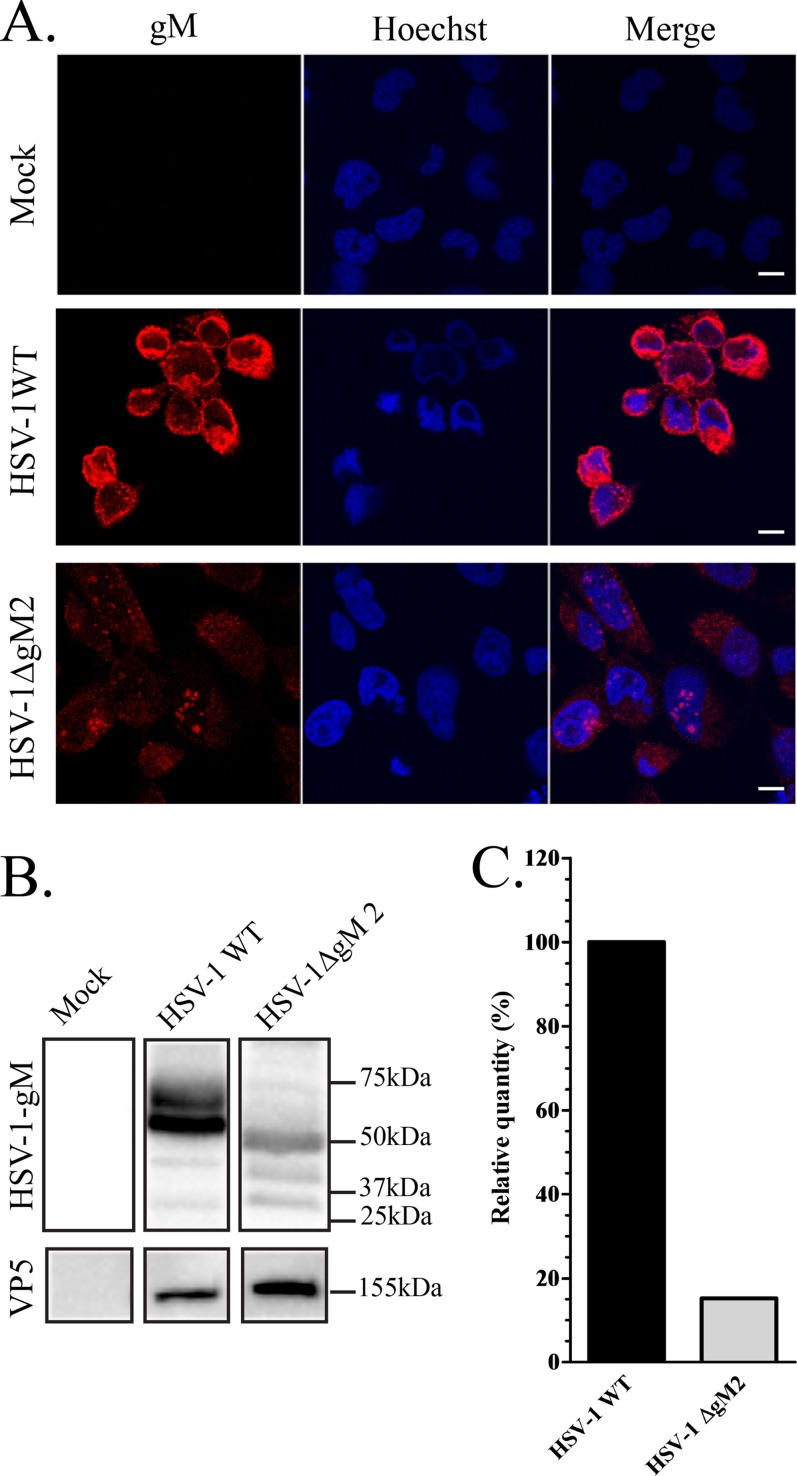
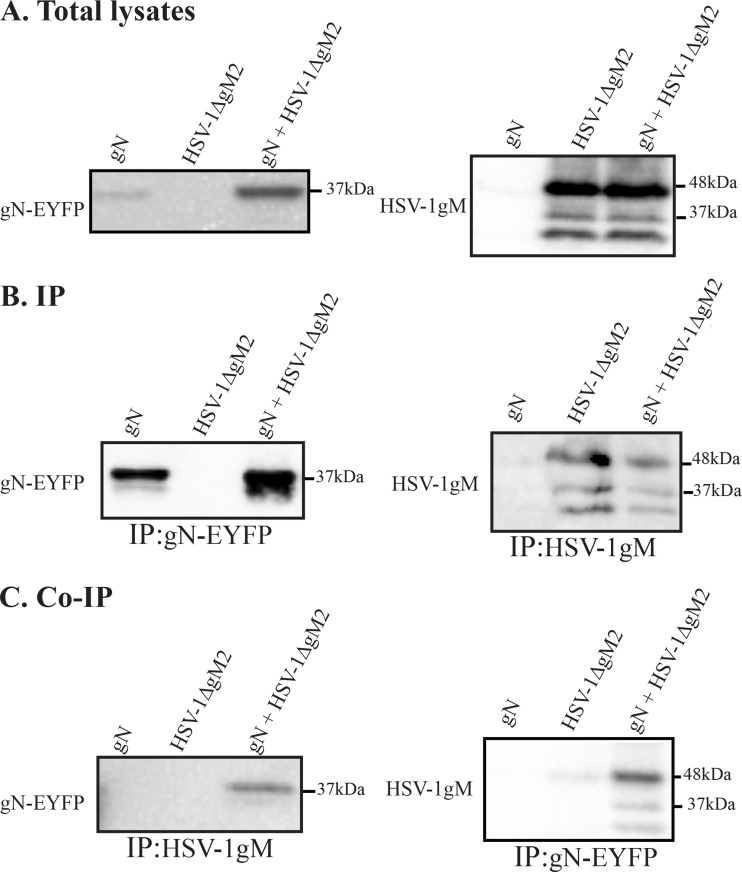
Knocking out the HSV-1 gM protein has proven an unusually difficult matter, as deleting its start site results in the expression of gM from internal methionines. To address this issue, Chouljenko and colleagues had previously reported the use of a double HSV-1 mutant strain in which the first two methionines were mutated to substantially reduce gM expression (19). To first confirm these findings, we probed this viral mutant with the carboxyl-terminal 4C10 gM antibody by both immunofluorescence and Western blotting. The data indicated that while the mutations were very effective at reducing gM expression, small residual amounts of the protein were nonetheless still detectable by immunofluorescence (Fig. 5A) and Western blotting (Fig. 5B). Quantification of the Western blot data indicated that the truncated mutant was expressed at approximately 15% of the level of wild-type gM (Fig. 5C). Far from being an impediment, this provided us with an opportunity to ask if the amino terminus of gM is important for its interaction with gN. Thus, cells transfected with exogenous pEYFP HSV1 UL49A and infected with the double gM (HSV-1ΔgM2) mutant were analyzed by coimmunoprecipitation as described above. The data showed that the gN still bound to the truncated gM (Fig. 6C). However, quantification of 3 independent experiments indicated that the detection of the gM/gN complex was reduced by 33-fold compared to full-length gM, taking into account the relative expression level of the two proteins and the ability of the antibodies to immunoprecipitate them. As noted before, the complex preferentially contained the higher-molecular-mass form of gM.
FIG 5.
Expression of gM in wild-type and mutant HSV-1 strains. (A) 143B cells grown on coverslips were infected with the HSV-1ΔgM2 mutant and, as a positive control, wild-type HSV-1. The cells were fixed at 16 hpi and stained with anti-gM (4C10) antibody (red) and Hoechst (blue). (B) HSV-1- or HSV-1ΔgM2 mutant-infected or mock-treated 143B cells were also harvested at 16 hpi, and cell lysates were analyzed by Western blotting. The blots were reacted with anti-gM (PAS980) antibodies or, as a loading control, antibodies against the major capsid protein VP5. Mock-infected cells were used as a negative control. Note that the predicted molecular mass for gM is 52 kDa, while that of the truncated HSV-1ΔgM2 mutant (lacking the first 132 amino acids) is 37.5 kDa. These predictions do not take into consideration the reported post-translational processing of the gM protein, clearly evident in the blots. (C) Quantification of the Western blot expression levels of gM in HSV-1 WT and the HSV-1ΔgM2 mutant. These estimates were normalized against VP5 (ratios of gM to VP5). For easier comparison, the data for wild-type virus was arbitrarily defined as 100%. Scale bar, 10 μm.
FIG 6.
gM/gN interacts less efficiently with the amino-truncated ΔgM2 protein. HeLa cells were transfected with YFP-tagged gN for 24 h. They were then infected with HSV-1ΔgM2 virus at an MOI of 5, and the cells were harvested at 16 hpi and lysed. Two and a half percent (1/40th) of the lysates were directly loaded onto the gels, and the rest of the lysates were immunoprecipitated (IP) with the 4C10 anti-gM or anti-GFP antibody (B and C) and analyzed by Western blotting. The blots were reacted with the PAS980 anti-gM (A to C, right side) or anti-GFP antibodies (A to C, left side). As before, the molecular masses of the molecular weight marker are indicated to the right of the panels.
Functional relevance of the complex.
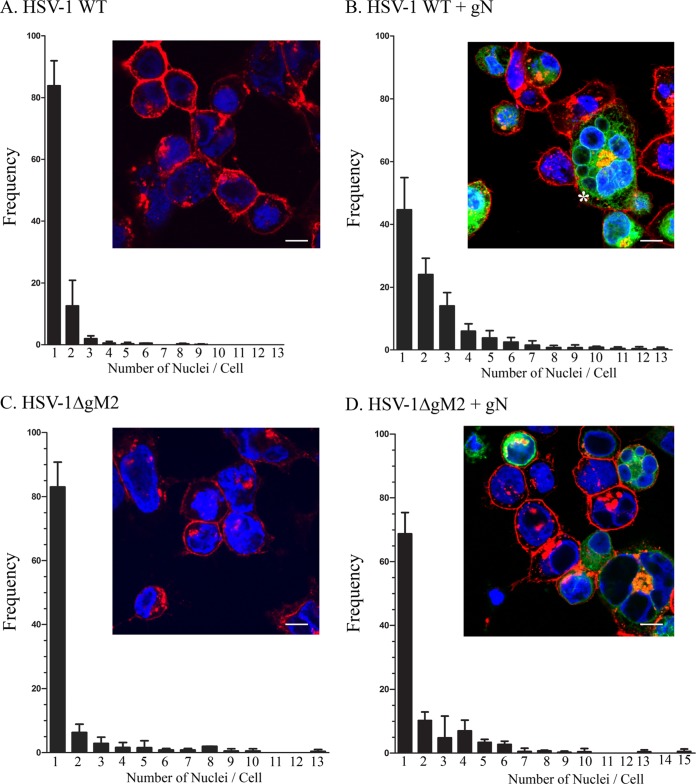
Various gM/gN complexes have been documented in distinct herpesviruses, hinting at a conserved role. Although the association of gM with gN typically causes a change of localization of one or both of the proteins, no function has been assigned to either gN or the gM/gN complex. However, gM is a negative modulator of cell-cell fusion when cotransfected with the HSV-1 fusion machinery constituted of the viral glycoproteins gB, gD, and gH/gL (23). In contrast, gM stimulates the entry by fusion of the virus in the context of strong syncytial strains (22), indicating that virus-induced fusion is normally very tightly controlled. Given this highly modulated fusion activity, we asked whether gN might also participate in this process. One clue came during the above-described studies when syncytia were noted upon gN overexpression in infected cells, something that is uncommon when using the 17+ wild-type HSV-1 strain. To address this more systematically, we quantified syncytium formation under various conditions using Hoechst to stain the nuclei and Alexa Fluor 647-labeled WGA to delineate the cell boundaries (see insets of Fig. 7). Not surprisingly, syncytia were never seen in gN-, gM-, or gM/gN-transfected cells in the absence of virus since the HSV-1 fusion apparatus is lacking in these scenarios (data not shown). Similarly, the sole presence of wild-type virus predominantly yielded a nonsyncytial phenotype, with over 80% of cells containing a unique nucleus or very few of them (Fig. 7A). In contrast, only 45% of cells contained a single nucleus when gN was overexpressed in wild-type-infected cells (Fig. 7B). This substantial increase in cell-cell fusion was statistically highly significant (P = 0.0039 when comparing the increase of cells with more than one nucleus; P = 0.0007 if one compares the decrease of cells with a single nucleus) and reached up to 13 nuclei per cell (Fig. 7B). Very interestingly, this syncytial phenotype depended on the presence of both gM and gN, as a viral strain depleted for gM (HSV-1ΔgM2 mutant) was statistically indistinguishable (P > 0.05) from wild-type virus in terms of syncytia, even upon the overexpression of gN (Fig. 7C and D). Moreover, the lack of any significant syncytia in the case of truncated gM plus YFP gN shows that the gN, and consequently the vector alone, does not induce cell-cell fusion per se. This was nonetheless confirmed by transfecting the empty vector and then infecting the cells with wild-type virus. In these experiments, 94% of cells had a single nuclei (n = 295 cells). This suggested that the gN overexpression deregulated the normally tightly gM-controlled virus-induced fusion.
FIG 7.
The HSV-1 gN viral protein induces syncytial formation in infected cells. 143B cells were transfected with (B and D) or without (A and C) YFP-tagged gN for 24 h and subsequently infected at an MOI of 2 with wild-type virus (A and B) or the HSV-1ΔgM2 mutant (C and D). At 12 hpi, the cells were fixed and stained without permeabilization for plasma membrane with Alexa 647-labeled wheat germ agglutinin to delineate the cell boundaries. Cells were then washed and examined by confocal laser scanning microscopy for the presence of syncytia, defined as single cells containing two or more nuclei. Quantification was done by counting 200 cells for each experiment. The reported values represent averages from three independent experiments. The error bars indicate standard deviations. Fluorescence microscopy insets show typical examples for each condition. The asterisk in panel B denotes an example of a syncytium. Scale bar, 10 μm.
DISCUSSION
From alphaherpesviruses to gammaherpesviruses, gM interacts with gN, and the complex typically leads to changes in intracellular targeting of either or both proteins (15, 21, 30, 36, 37, 40). The present study indicates that this extends to HSV-1 and further reveals that the amino terminal 132 amino acids of HSV-1 gM are important for an efficient interaction between the two proteins. At this point though, it is premature to conclude if the deletion of the amino gM terminus removes a binding domain or perhaps alternatively impacts the overall conformation of the protein. It may also be that the lower expression level of the truncated mutant lessens the probability of contact between the two proteins. In all cases, the complex was composed of a single gN molecular-mass band preferentially bound to, but not exclusively, higher-molecular-mass forms of gM, which are presumably the heavily glycosylated forms of the protein. This may mean the two proteins interact once gM is modified in the Golgi or, more likely, since gN is never TGN bound in the absence of gM, that the two proteins interact in the ER and the complex is then transported to the Golgi, where immature sugars on gM can be modified.
It is unfortunately not possible to address the intracellular localization of endogenously expressed and untagged gN, as no antibody is available and our efforts to produce such an antibody have failed (I. El Kasmi and R. Lippé, unpublished data). We thus cannot rule out that the ER localization of the overexpressed gN does not reflect that of gN expressed by the virus. However, as the YFP-tagged gN clearly physically and functionally interacted with both endogenous and exogenously expressed gM, we feel confident that the gM/gN complex reported here is biologically relevant. Moreover, the colocalization of the two proteins in intact cells indicated the immunoprecipitated complex is not an artifact based on a post-lysate event taking place in the test tube.
Interestingly, while gN is dominant and alters the localization and/or maturation of gM in BHV-1, equine herpesvirus 1 (EHV-1), and HHV-8 (21, 30, 40), the opposite seems true for HSV-1, EBV, and infectious ILTV, where gM is required for the transport and/or processing of gN (this study) (36, 37). While the reasons for this distinction among herpesviruses are not clear, that the complex formation has targeting consequences for gM or gN in all herpesviruses studied so far appears well conserved.
We previously reported the late targeting of gM from the nuclear membranes to the TGN in infected cells (38), but the mechanisms targeting gM to either compartment are not known. Our initial expectation was that gN might constitute the molecular switch that triggers HSV-1 gM relocalization since gN expression coincides with this change of targeting and is known to redirect gM in some herpesviruses. Unexpectedly, this was not the case for HSV-1. Given that gM is never nuclear when transfected alone but transits in different intracellular compartments during an infection, this indicates that one or more additional viral proteins controls gM intracellular targeting or that, alternatively, a host protein does so under the influence of the virus. How gM is targeted to the nucleus and escapes this compartment thus remains a mystery.
The impact of the gM/gN complex on virus-induced membrane fusion, a phenomenon normally under tight control, is of significant interest. Past studies have attributed both a negative and positive role for gM. Hence, gM cotransfection reduced cell-cell fusion mediated by the HSV-1 fusion machinery composed of gB, gD, and the gH/gL complex (23). In apparent contrast, gM was recently reported to stimulate viral entry, i.e., the fusion of the viral envelope with the cell surface in the context of strong syncytial strains (22). Interestingly, both cases involve “forced situations” where the fine controls of virally induced fusion were deregulated, as has been the case with the various syncytial strains studied in the past, all of which involve mutations (41–49). Despite this possible caveat, these experimental models are clearly very useful to unravel new fusion modulators that may otherwise be difficult to detect. It is in a similar context that gN′s role on syncytial formation was detected in the present study, in this case by overexpressing gN early during an infection with a nonsyncytial strain.
The apparent discrepancy between the transfection and infection scenarios must be reconciled somehow. It is worth noting that gM inhibition of fusion in the transfected model can occur in the absence of gN but is stimulated by it (23), while the stimulatory activity of gM on fusion during entry by syncytial strains happens in the presence of gN (22). In contrast, the sole coexpression of gM and gN in the absence of the gB, gD, and gH/gL is insufficient to drive cell-cell fusion. Our own present findings indicate that the sole overexpression of gN in transfection experiments or in the context of a virus expressing significantly reduced amounts of gM (HSV-1ΔgM2 viral mutant) does not induce syncytial formation. All together, these results suggest that gN′s activity first depends on the presence of gM and second that additional viral proteins temporarily and spatially modulate gM positive or negative regulation of fusion. Although speculative, it may be critical to identify additional modulators of fusion and finely manipulate this viral apparatus. This will surely be a significant challenge.
While the precise interplay between gM and gN remains elusive, it is tempting to speculate that gM may play a role early during the infection, when it is located on nuclear membranes and when gN is not yet expressed. At that stage, gM could regulate membrane fusion, perhaps preventing the unrestricted fusion of the inner and outer nuclear membrane at a time when new viral particles have yet to be assembled. The targeting of gM to nuclear membranes prior to the presence of the gB, gD, and gH fusogenic machinery would be consistent with such a concept (38, 50–53). As gM is subsequently detected on perinuclear virions at 14 hpi (39), it could then promote the fusion of the enveloped viral particles with the outer nuclear membrane, although data by Leege and colleagues argue otherwise, as the deletion of gM from the related pseudorabies virus does not lead to an accumulation of perinuclear virions (17). Further downstream, the partial release of gM from the nucleus (observed from 10 to 13 hpi) coincides with the expression of gN (mRNA detectable at 10 hpi) and targeting of gN to the TGN. The exact mechanism of action of gN is still unknown, but it may modulate gM by potentially sequestering it and/or preventing it from interacting with other proteins. This interaction of gN with gM in infected cells could thus prevent gM's negative modulation on membrane fusion and thus promote cell-cell fusion under perhaps yet-to-be-defined suitable conditions. This would enable the virus to evade neutralizing antibodies awaiting outside the cell and thus favor viral spreading.
ACKNOWLEDGMENTS
We thank the generosity of Konstantin Gus Kousoulas, Colin Crump, Joel Baines, Lynn Enquist, and Beate Sodeik for providing invaluable reagents. We particularly thank Kerstin Radtke for her help in the early stages of this work.
This work was financed by an NSERC grant to R.L. (RGPIN/342203-2010).
REFERENCES
- 1.Loret S, Guay G, Lippé R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henaff D, Radtke K, Lippé R. 2012. Herpesviruses exploit several host compartments for envelopment. Traffic 13:1443–1449. doi: 10.1111/j.1600-0854.2012.01399.x. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehner R, Meyer H, Mach M. 1989. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol 63:3792–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines JD, Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol 65:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines JD, Roizman B. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol 67:1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osterrieder N, Neubauer A, Brandmuller C, Braun B, Kaaden OR, Baines JD. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol 70:4110–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs W, Mettenleiter TC. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J Gen Virol 80:2173–2182. [DOI] [PubMed] [Google Scholar]
- 9.Cai JS, Jang HK, Izumiya Y, Tsushima Y, Kato K, Damiani AM, Miyazawa T, Kai C, Takahashi E, Mikami T. 1999. Identification and structure of the Marek's disease virus serotype 2 glycoprotein M gene: comparison with glycoprotein M genes of Herpesviridae family. J Vet Med Sci 61:503–511. doi: 10.1292/jvms.61.503. [DOI] [PubMed] [Google Scholar]
- 10.Dijkstra JM, Visser N, Mettenleiter TC, Klupp BG. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol 70:5684–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tischer BK, Schumacher D, Messerle M, Wagner M, Osterrieder N. 2002. The products of the UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J Gen Virol 83:997–1003 http://vir.sgmjournals.org/content/83/5/997.long. [DOI] [PubMed] [Google Scholar]
- 12.Konig P, Giesow K, Keil GM. 2002. Glycoprotein M of bovine herpesvirus 1 (BHV-1) is nonessential for replication in cell culture and is involved in inhibition of bovine respiratory syncytial virus F protein induced syncytium formation in recombinant BHV-1 infected cells. Vet Microbiol 86:37–49. doi: 10.1016/S0378-1135(01)00489-8. [DOI] [PubMed] [Google Scholar]
- 13.Ziegler C, Just FT, Lischewski A, Elbers K, Neubauer A. 2005. A glycoprotein M-deleted equid herpesvirus 4 is severely impaired in virus egress and cell-to-cell spread. J Gen Virol 86:11–21. doi: 10.1099/vir.0.80393-0. [DOI] [PubMed] [Google Scholar]
- 14.Yamagishi Y, Sadaoka T, Yoshii H, Somboonthum P, Imazawa T, Nagaike K, Ozono K, Yamanishi K, Mori Y. 2008. Varicella-zoster virus glycoprotein M homolog is glycosylated, is expressed on the viral envelope, and functions in virus cell-to-cell spread. J Virol 82:795–804. doi: 10.1128/JVI.01722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawabata A, Jasirwan C, Yamanishi K, Mori Y. 2012. Human herpesvirus 6 glycoprotein M is essential for virus growth and requires glycoprotein N for its maturation. Virology 429:21–28. doi: 10.1016/j.virol.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Mach M, Kropff B, Dal Monte P, Britt W. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J Virol 74:11881–11892. doi: 10.1128/JVI.74.24.11881-11892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leege T, Fuchs W, Granzow H, Kopp M, Klupp BG, Mettenleiter TC. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J Virol 83:896–907. doi: 10.1128/JVI.01842-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brack AR, Dijkstra JM, Granzow H, Klupp BG, Mettenleiter TC. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol 73:5364–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chouljenko DV, Kim IJ, Chouljenko VN, Subramanian R, Walker JD, Kousoulas KG. 2012. Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J Virol 86:4262–4270. doi: 10.1128/JVI.06766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp BG, Nixdorf R, Mettenleiter TC. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol 74:6760–6768. doi: 10.1128/JVI.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koyano S, Mar EC, Stamey FR, Inoue N. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J Gen Virol 84:1485–1491. doi: 10.1099/vir.0.18941-0. [DOI] [PubMed] [Google Scholar]
- 22.Kim IJ, Chouljenko VN, Walker JD, Kousoulas KG. 2013. Herpes simplex virus 1 glycoprotein M and the membrane-associated protein UL11 are required for virus-induced cell fusion and efficient virus entry. J Virol 87:8029–8037. doi: 10.1128/JVI.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crump CM, Bruun B, Bell S, Pomeranz LE, Minson T, Browne HM. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J Gen Virol 85:3517–3527. doi: 10.1099/vir.0.80361-0. [DOI] [PubMed] [Google Scholar]
- 24.Ren Y, Bell S, Zenner HL, Lau SY, Crump CM. 2012. Glycoprotein M is important for the efficient incorporation of glycoprotein H-L into herpes simplex virus type 1 particles. J Gen Virol 93:319–329. doi: 10.1099/vir.0.035444-0. [DOI] [PubMed] [Google Scholar]
- 25.Barker DE, Roizman B. 1992. The unique sequence of the herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J Virol 66:562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnett BC, Dolan A, Telford EA, Davison AJ, McGeoch DJ. 1992. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J Gen Virol 73:2167–2171. doi: 10.1099/0022-1317-73-8-2167. [DOI] [PubMed] [Google Scholar]
- 27.Liang X, Tang M, Manns B, Babiuk LA, Zamb TJ. 1993. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5. Virology 195:42–50. doi: 10.1006/viro.1993.1344. [DOI] [PubMed] [Google Scholar]
- 28.Said A, Azab W, Damiani A, Osterrieder N. 2012. Equine herpesvirus type 4 UL56 and UL49.5 proteins downregulate cell surface major histocompatibility complex class I expression independently of each other. J Virol 86:8059–8071. doi: 10.1128/JVI.00891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deruelle MJ, Van den Broeke C, Nauwynck HJ, Mettenleiter TC, Favoreel HW. 2009. Pseudorabies virus US3- and UL49.5-dependent and -independent downregulation of MHC I cell surface expression in different cell types. Virology 395:172–181. doi: 10.1016/j.virol.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Lipinska AD, Koppers-Lalic D, Rychlowski M, Admiraal P, Rijsewijk FA, Bienkowska-Szewczyk K, Wiertz EJ. 2006. Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M. J Virol 80:5822–5832. doi: 10.1128/JVI.02707-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dijkstra JM, Gerdts V, Klupp BG, Mettenleiter TC. 1997. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J Gen Virol 78:2147–2151. [DOI] [PubMed] [Google Scholar]
- 32.MacLean CA, Robertson LM, Jamieson FE. 1993. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J Gen Virol 74(Part 6):975–983. [DOI] [PubMed] [Google Scholar]
- 33.Jons A, Dijkstra JM, Mettenleiter TC. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol 72:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X, Chow B, Raggo C, Babiuk LA. 1996. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J Virol 70:1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu SX, Zhu XP, Letchworth GJ. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the U(L)49.5 protein. J Virol 72:3029–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs W, Mettenleiter TC. 2005. The nonessential UL49.5 gene of infectious laryngotracheitis virus encodes an O-glycosylated protein which forms a complex with the non-glycosylated UL10 gene product. Virus Res 112:108–114. doi: 10.1016/j.virusres.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Lake CM, Molesworth SJ, Hutt-Fletcher LM. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J Virol 72:5559–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Nagel CH, Sodeik B, Lippé R. 2009. Early, active, and specific localization of herpes simplex virus type 1 gM to nuclear membranes. J Virol 83:12984–12997. doi: 10.1128/JVI.01180-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baines JD, Wills E, Jacob RJ, Pennington J, Roizman B. 2007. Glycoprotein M of herpes simplex virus 1 is incorporated into virions during budding at the inner nuclear membrane. J Virol 81:800–812. doi: 10.1128/JVI.01756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph J, Seyboldt C, Granzow H, Osterrieder N. 2002. The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J Virol 76:2952–2963. doi: 10.1128/JVI.76.6.2952-2963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baines JD, Ward PL, Campadelli-Fiume G, Roizman B. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol 65:6414–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melancon JM, Foster TP, Kousoulas KG. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J Virol 78:7329–7343. doi: 10.1128/JVI.78.14.7329-7343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders PG, Wilkie NM, Davison AJ. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J Gen Virol 63:277–295. doi: 10.1099/0022-1317-63-2-277. [DOI] [PubMed] [Google Scholar]
- 44.Bzik DJ, Fox BA, DeLuca NA, Person S. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 45.Bond VC, Person S. 1984. Fine-structure physical map locations of alterations that affect cell-fusion in herpes-simplex virus type-1. Virology 132:368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- 46.Debroy C, Pederson N, Person S. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 47.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham FL, Johnson DC. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol 66:5603–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pogue-Geile KL, Lee GT, Shapira SK, Spear PG. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100–109. doi: 10.1016/0042-6822(84)90251-4. [DOI] [PubMed] [Google Scholar]
- 49.Ruyechan WT, Morse LS, Knipe DM, Roizman B. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol 29:677–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torrisi MR, Di Lazzaro C, Pavan A, Pereira L, Campadelli-Fiume G. 1992. Herpes simplex virus envelopment and maturation studied by fracture label. J Virol 66:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stannard LM, Himmelhoch S, Wynchank S. 1996. Intra-nuclear localization of two envelope proteins, gB and gD, of herpes simplex virus. Arch Virol 141:505–524. doi: 10.1007/BF01718314. [DOI] [PubMed] [Google Scholar]
- 52.Farnsworth A, Wisner TW, Webb M, Roller R, Cohen G, Eisenberg R, Johnson DC. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci U S A 104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson DC, Wisner TW, Wright CC. 2011. Herpes simplex virus glycoproteins gB and gD function in a redundant fashion to promote secondary envelopment. J Virol 85:4910–4926. doi: 10.1128/JVI.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]