Abstract
Understanding the life cycle and pathogenesis of animal viruses requires that we have systems in which the viruses can replicate and cause disease. For the latter, we rely upon animal models or information that we can obtain from studying natural infections of humans and other animals. For the former, however, we are largely dependent on the availability of cell culture systems in which viruses can be propagated to investigate the molecular mechanisms of viral replication. For many years, it was assumed that replication in culture provided an accurate description of the life cycle of the organism. In this Gem, we will discuss two viruses, polyomavirus and cytomegalovirus, in which cell culture systems have accidentally provided unique potential insights into viral replication and persistence in their hosts.
INTRODUCTION
Ever since the development of eukaryotic cell culture systems in the early 1900s, virologists have taken advantage of tissue culture to isolate, identify, and characterize viruses (1). Viruses can easily be propagated to high titers in permissive host cells, which facilitates their isolation and purification. With rapid advances in molecular biology and cell biology, many basic aspects of virus infection and the interplay between viruses and their hosts have been elucidated. These include, but are not limited to, virus receptor identification, entry pathways, transcription, translation, replication mechanisms, and host innate immune responses to various virus infections. In addition, studies of viruses in cell culture have also greatly improved our understanding of how normal cells operate, and researchers continue to use viruses as tools to further our knowledge of many fundamental cellular processes.
The ability to persist for the life of the infected host is a common characteristic of many DNA viruses (2). Herpesviruses, adenoviruses, and polyomaviruses, for example, all establish lifelong persistent infections. In the persistent state, viral replication is either low or absent, and the virus has evolved strategies to evade the immune system, allowing it to coexist with the host. As such, our ability to study viruses in culture has generally allowed us to understand only a portion of viral biology, albeit an important aspect: what happens when a virus productively infects a cell. There are two major selective forces at play in these in vitro models. First, there is the lack of selection by the immune system. Adaptive immunity is nonexistent, and not all aspects of innate immunity are present. Second, there is selection for cytopathology and production of progeny virions. Indeed, before there were molecular biology tools, we relied on cell biological and biochemical assays to understand infection. The former force works to our advantage as researchers: we can propagate in culture viruses whose growth in an animal host may be otherwise inhibited by the immune response or whose host is not experimentally tractable or cannot be used for ethical reasons. The latter force, however, means that we are open to the possibility that we are examining rare variants that arose in the host prior to their addition to the cultured cells or that there has been selection in culture for replication ability that was not present in the initial isolate.
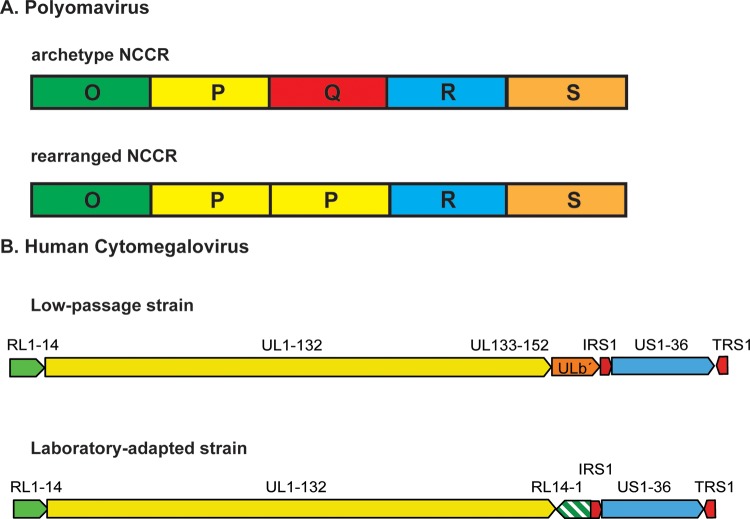
Polyomaviruses (PyVs) are small, nonenveloped viruses that contain an approximately 5-kb double-stranded circular DNA genome and have been isolated from a variety of species, from birds to humans (3). In the human host, these viruses are characterized by the fact that they rarely cause disease in individuals with a functional immune system. Rather, one usually sees disease in immunocompromised individuals, such as transplant patients being treated with immunosuppressive drugs, HIV-infected patients, patients being treated with certain monoclonal antibodies that block immune cell activities, or elderly people whose immune systems are waning (4). The PyV genome can be divided into three genetic regions: (i) the early region, which encodes the regulatory T antigens; (ii) the late region, which encodes the capsid proteins; and (iii) the noncoding control region (NCCR; sometimes called the regulatory region), which contains the viral origin of DNA replication and the transcriptional promoters for the early and late genes. For the two best-studied human PyVs, BKPyV and JCPyV, the structure of the NCCR depends upon the natural history of the virus (5). In the so-called archetype virus, which is the virus that circulates through the population and can be isolated from urine due to periodic, subclinical replication, there is a characteristic sequence that is largely invariant (Fig. 1A). In rearranged variants, there are large deletions and duplications within the NCCR. Interestingly, except under very artificial conditions, the archetype virus cannot be propagated in culture (6). In fact, attempts to culture archetype BKPyV result in the outgrowth of rearranged variants (7). Moreover, rearranged variants of BKPyV are associated with viremia in polyomavirus-associated nephropathy in renal transplant patients, and in contrast to the archetype virus, these variants can be propagated in culture (8). Similarly, rearranged variants of JCPyV can be isolated and cultured from patients with progressive multifocal leukoencephalopathy (9). Therefore, it is thought that the rearrangement of the NCCR is associated with enhanced viral replication and subsequent pathogenesis. Both BKPyV and JCPyV were initially isolated from patient specimens; if investigators had attempted to isolate the viruses from healthy individuals, they probably would have been unsuccessful unless they were lucky enough to get in vitro selection for rearrangements. Interestingly, none of the more recently isolated human PyVs have been efficiently propagated in culture (4). We speculate that this is a reflection of the fact that they are also archetype viruses whose natural mode of infection is persistence.
FIG 1.
Examples of genome rearrangements in PyV and HCMV. (A) Archetype and rearranged PyV NCCRs. The top shows the structure of the archetype BKPyV NCCR, which is arbitrarily divided into five blocks of DNA sequence named O (for origin of replication), P, Q, R, and S; the bottom shows a hypothetical rearranged NCCR in which the P block is duplicated and the Q block is deleted. The blocks are not drawn to scale. (B) HCMV low-passage-number and laboratory-adapted strain genome organization. The top shows the ORF organization of low-passage-number HCMV strains with a unique ULb′ region that is absent in laboratory strains (bottom). Additionally, the RL region is repeated in the laboratory strains (bottom). Block arrows depict the relative orientations of the RL1-14, UL1-152, ULb′, IRS1, US1-36, and TRS1 gene segments.
Cytomegaloviruses belong to the betaherpesvirus family and contain a large linear double-stranded DNA genome. The human cytomegalovirus (HCMV) establishes a lifelong infection in humans and, like PyVs, HCMV is an opportunistic pathogen that causes severe disease in immunocompromised people. Its genome includes two unique regions termed the unique long (UL) and unique short (US) regions. These two domains are flanked by terminal and internal repeat segments that can be recombined, resulting in genomic isomerization. Upon entry, the linear genome is circularized via unpaired bases at each end and the DNA is replicated by a rolling circle mechanism. The catenated DNA is subsequently cleaved into unit length molecules and packaged into virions. It is unclear whether during latency the viral DNA is replicated or not, but it is believed that the genome persists as an episome (10).
Clinical isolates of HCMV have been obtained from various settings, and some of them have been cloned and sequenced (11). These isolates have minimal passage history in culture and display similar genomic organization. In contrast, laboratory strains of HCMV have gone through multiple passages in fibroblasts, which resulted in selection for increased replication in fibroblasts and decreased ability to infect other cell types that are permissive to the clinical strains. Interestingly, a closer look revealed that in comparison to the clinical precursors, these cell culture-adapted strains display extensive genomic rearrangements, including large deletions, duplications, and inversions (11, 12) (Fig. 1B). The rearrangements are particularly prevalent in the two terminal regions, which are enriched in repeated sequences. Unlike PyVs in which genomic rearrangements are confined to the NCCRs, the reorganization in HCMV genomes can involve open reading frame (ORF)-containing sequences. It is inferred that the terminal regions contain genes that are not essential for HCMV replication in cell culture.
How these rearrangement events in the viral genomes lead to increased replication in culture is unclear. In PyVs, the NCCR controls both early and late transcription, and rearrangement is often observed within the late proximal side of the NCCR (13). This region contains many putative enhancer elements, and rearrangements within this region may certainly affect promoter activities. One possible outcome is a change in the expression of viral microRNAs (miRNAs), which regulate expression of early mRNAs, due to altered early and late promoter firing. For example, with BKPyV, the rearranged virus produces less viral miRNA than the archetype strain, thereby resulting in an enhancement of viral replication (14). It has been proposed that increased miRNA expression by the archetype strain may facilitate the establishment of persistence, as less early mRNA is being produced, therefore allowing the virus to be more likely to evade immune surveillance. This may not be generalizable to other PyVs, such as simian virus 40 (SV40), however (15). In HCMV, research suggests that disruption of a glycoprotein complex, gH-gL-pUL128-pUL130-pUL131, in laboratory strains may be responsible for increased replication (16, 17), although as is the case for PyVs, the exact mechanism is not well defined. In addition, all clinical strains of HCMV contain a unique region termed ULb′ that is absent in laboratory strains. This region has been shown to be necessary for viral latency establishment in hematopoietic progenitor cells, although it is dispensable for replication in fibroblasts (18). Taken together, these data suggest that rearrangements within these opportunistic DNA virus genomes may have profound impact on the ability of the viruses to either persist or replicate under different contexts of infection.
There is unfortunately a lack of knowledge on how genomic arrangements arise in these DNA viruses. Recombination appears to play a role in this process, as large deletions and duplications have been observed in both PyVs and HCMV (5, 16). It has been postulated that viral-replication-dependent recombination events might be important for generating observed sequence changes (19). Future studies focusing on host recombination pathways as well as a closer examination of a larger number of clinical isolates will reveal additional information on the underlying mechanisms of rearrangement.
SUMMARY
In summary, the selection for growth of viruses imposed by cell culture systems, while greatly enhancing our understanding of replication, has been a double-edged sword. On the one hand, we have uncovered basic strategies used by viruses to complete their life cycles. On the other hand, we now recognize that the genomes of these lab-adapted viruses do not always represent the viruses that circulate in the animal host. Therefore, what are considered wild-type strains in the laboratory may not necessarily be truly wild type in the host. Nonetheless, comparison of these two classes of genomic types has provided tremendous insight into viral biology and pathogenesis. Indeed, it appears that the primary strategy of some DNA viruses may be to express molecules that repress replication and allow the viruses to coexist with the host, rather than replicating robustly and destroying the infected cell. These persistent viruses can then sense the environment and switch to replicative mode when conditions are optimal for spread to a new host.
ACKNOWLEDGMENTS
We thank members of the Imperiale and Jiang labs for critical readings of the manuscript. We apologize to all our colleagues whose work could not be cited because of length restrictions.
This work was supported by NIH grant R01 AI060584 to M.J.I and by funding from the UAB startup fund and the Faculty Development Grant Program to M.J.
REFERENCES
- 1.Leland DS, Ginocchio CC. 2007. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldogh I, Albrecht T, Porter DD. 1996. Persistent viral infections. InBaron S. (ed), Medical microbiology, 4th ed.The University of Texas Medical Branch at Galveston, Galveston, TX. [PubMed] [Google Scholar]
- 3.DeCaprio JA, Imperiale MJ, Major EO. 2013. Polyomaviruses, p 1633–1661 InKnipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed.Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.DeCaprio JA, Garcea RL. 2013. A cornucopia of human polyomaviruses. Nat Rev Microbiol 11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moens U, Van Ghelue M. 2005. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology 331:209–231. doi: 10.1016/j.virol.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Broekema NM, Imperiale MJ. 2012. Efficient propagation of archetype BK and JC polyomaviruses. Virology 422:235–241. doi: 10.1016/j.virol.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinstein R, Schoonakker BC, Harley EH. 1991. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J Virol 65:1600–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosert R, Rinaldo CH, Funk GA, Egli A, Ramos E, Drachenberg CB, Hirsch HH. 2008. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med 205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosert R, Kardas P, Major EO, Hirsch HH. 2010. Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate. J Virol 84:10448–10456. doi: 10.1128/JVI.00614-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolovan-Fritts CA, Mocarski ES, Wiedeman JA. 1999. Peripheral blood CD14(+) cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood 93:394–398. [PubMed] [Google Scholar]
- 11.Murphy E, Shenk T. 2008. Human cytomegalovirus genome. Curr Top Microbiol Immunol 325:1–19. [DOI] [PubMed] [Google Scholar]
- 12.Wang A, Ren L, Abenes G, Hai R. 2009. Genome sequence divergences and functional variations in human cytomegalovirus strains. FEMS Immunol Med Microbiol 55:23–33. doi: 10.1111/j.1574-695X.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- 13.White MK, Safak M, Khalili K. 2009. Regulation of gene expression in primate polyomaviruses. J Virol 83:10846–10856. doi: 10.1128/JVI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broekema NM, Imperiale MJ. 2013. miRNA regulation of BK polyomavirus replication during early infection. Proc Natl Acad Sci U S A 110:8200–8205. doi: 10.1073/pnas.1301907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CJ, Burke JM, Kincaid RP, Azarm KD, Mireles N, Butel JS, Sullivan CS. 2014. Naturally arising strains of polyomaviruses with severely attenuated microRNA expression. J Virol 88:12683–12693. doi: 10.1128/JVI.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 70:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Shenk T. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol 79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umashankar M, Petrucelli A, Cicchini L, Caposio P, Kreklywich CN, Rak M, Bughio F, Goldman DC, Hamlin KL, Nelson JA, Fleming WH, Streblow DN, Goodrum F. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog 7:e1002444. doi: 10.1371/journal.ppat.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson EM, Wortman MJ, Dagdanova AV, Lundberg PS, Daniel DC. 2013. Polyomavirus JC in the context of immunosuppression: a series of adaptive, DNA replication-driven recombination events in the development of progressive multifocal leukoencephalopathy. Clin Dev Immunol 2013:197807. doi: 10.1155/2013/197807. [DOI] [PMC free article] [PubMed] [Google Scholar]