Abstract
The specificity of DNA hybridization allows for the modular design of 2D and 3D shapes with wide-ranging applications including sensors, actuators, and even logic devices. The inherent biocompatibility of DNA and the ability to produce monodisperse structures of controlled shape and size make DNA nanostructures of interest as potential drug and gene delivery vehicles. In this review, we discuss several new approaches for the assembly of DNA nanostructures, advances in the modeling of these structures, and we highlight recent studies on the use of DNA nanotechnology for therapeutic applications such as drug delivery in tumor models.
Introduction
The unique properties of DNA have enabled the design and fabrication of intricate three-dimensional structures, molecular actuators, and rudimentary computational devices. The so-called “origami” approach of Rothemund in particular [1], combined with powerful open-source software tools [2], has allowed for great advances in the complexity of shapes, and of increasing size. Nevertheless, DNA nanostructures of relatively simple “wireframe” structure are still actively used in studies due to their rapid assembly and less demanding material requirements.
The well-defined size, shape, and hybridization properties of DNA nanostructures have positioned them to be investigated in applications such as sensing and delivery. Other characteristics of DNA sequences, such as non-Watson-Crick base pairing (e.g., i-motifs), protein binding (e.g., aptamers), and enzymatic activity (e.g., DNAzymes) provide a rich toolbox for the development of complex multi-functional nanostructures. While this shift into applications is of interest to the broader bionanotechnology community, it is also proceeding in parallel with new approaches to assemble, and modulate, structural properties. In this review we will focus on recent developments in the assembly of DNA nanostructures, their characterization, and works relevant to the field of drug and gene delivery.
Assembly
Assembly of DNA nanostructures be categorized by the method used to achieve the final structure. In the first category, single-stranded DNA (ssDNA) is assembled through a thermal annealing process. In the second category, assembly occurs through a reaction of strands; this could be an enzymatic reaction used to create a backbone strand or a non-enzymatic reaction, such as the hybridization chain reaction (HCR) [3].
The architectural features of DNA nanostructures provide yet another categorization scheme. For example, simple “wireframe” structures are defined by objects having helical double-stranded DNA along their edges. Such objects may be topologically open, as in dendrimer-like structures [4, 5], or closed, as in polyhedral structures. Regardless of their topology, wireframe objects tend to be relatively sparse and flexible as compared to “origami” structures. In origami structures, a long strand of ssDNA is used as a scaffold and shorter strands are used as “staples” to assemble a complex 2D or 3D structure. These structures typically make use of DNA crossovers [6, 7], whereby a single strand participates in several DNA helices. As a consequence, origami structures can be quite dense and rigid.
One barrier to the origami approach is the need for a long scaffold strand, which is generally obtained by purification from M13 bacteriophage (and associated bacterial cultures). Nickels et al. have reported the creation of origami structures from intact bacteriophages, circumventing the need for purification of the desired scaffold strand [8•]. By mixing staple strands, proteases, and denaturing agents with the bacteriophage particles at elevated temperatures in buffer containing MgCl2, several origami structures were produced from with yields comparable to conventional thermal annealing with purified scaffold. Assembly of these origami structures was also achieved inside M13-infected bacterial liquid culture. This assembly scheme was also attempted on much larger λ bacteriophage, and although origami structures were successfully assembled, the yield was very low.
Despite the success of DNA nanostructure assembly from intact bacteriophages, purification is still necessary for biomedical applications where endotoxin is a serious concern. Mathur and Henderson have reported the creation of similar complex origami structures solely from short ssDNA strands (i.e., oligonucleotides) [9••]. By “breaking” the scaffold strand into smaller strands, complex structures could still be formed through single-pot assembly by thermal annealing, although the yield is lower than the conventional origami method. This method thus eliminates concerns over bacterial and endotoxin contamination when using scaffold strands isolated from bacteriophages. However, the exclusive use of oligonucleotides introduces significantly more nicks into the resulting nanostructure, which may adversely affect mechanical properties and thermal stability.
Nanotubes
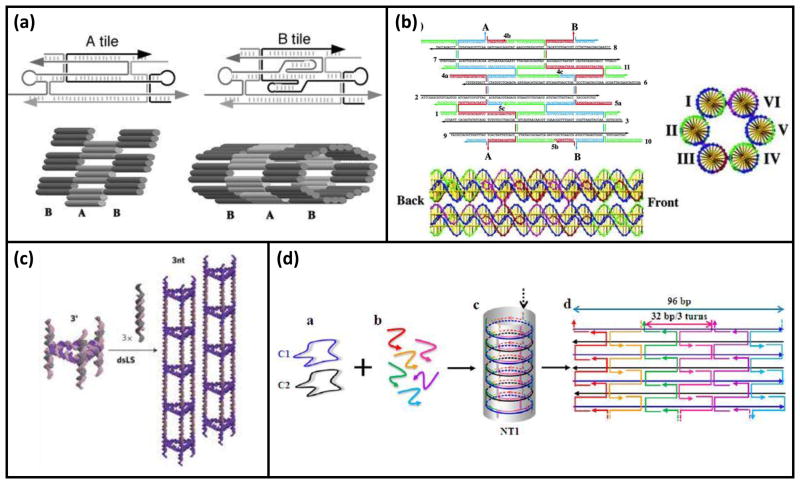
DNA nanotubes are of interest due to their high aspect ratio and potential applications for patterning. Distinct assembly strategies have been developed for assembling DNA nanotubes, reviewed in detail by Sleiman and co-workers [10]. In one strategy, 2D arrays of DNA tiles are formed, and then are caused to roll up into a tube by directed disulfide bond formation between tiles [11] (Figure 1a). In a second strategy, helical bundles of DNA having crossovers with specifically designed curvature are assembled and connected to each other by overhang hybridization to elongate them into tubes [12] (Figure 1b). Essentially, these bundles are intrinsically curved tiles. A third strategy involved the assembly of cyclic building blocks into nanotubes [13] (Figure 1c).
Figure 1.
Examples strategies for DNA nanotube assembly. (a–c) Representative schemes of the three methods of nanotube assembly described by Lo et al. [10]. (a) Example of nanotubes created by folding a 2D tile array upon itself (Reprinted with permission from [11]. Copyright 2004 Proceedings of the National Academy of Sciences of the United States of America.) (b) Example of helical bundles assembled to form nanotubes by crossover junctions (Reprinted with permission from [12]. Copyright 2005 American Chemical Society.) (c) Examples of nanotubes created by assembled cyclic building blocks longitudinally (Reprinted with permission from [13]. Copyright 2009 Nature Publishing Group.) (d) Nanotubes assembled from circular oligonucleotides connected by short staple strands. (Reprinted with permission from [14]. Copyright 2014 American Chemical Society.)
In a novel approach to nanotube assembly, Xiao and co-workers reported the use of small circular DNA (Figure 1d) [14]. This strategy allowed for the creation of rigid nanotubes with helices perpendicular to the direction of the tube, as opposed to most nanotubes whose helices run along the backbone of the tube. Such increased structural rigidity could be an important feature for future applications such as lithography. While this assembly strategy bears some resemblance to the strategy of Mathieu et al. [12], it is not a wireframe structure and would be much more rigid as a result of its crossovers.
Assembly of DNA nanotubes using an enzymatic reaction has also been reported [15]. By using linker strands as rungs, the backbone of the nanotube is produced by rolling circle amplification (RCA), an enzymatic reaction that uses a circular DNA template and DNA polymerase to amplify a short DNA primer to form a long strand. Through design of the rungs, variable addressable regions are presented at each rung, which allow for the incorporation of cargo. This addressability was tested by the incorporation of uorescent dyes, displaying the strength of this assembly method. This assembly takes place rapidly at room temperature, allowing for the incorporation of delicate cargo. An advantage of this strategy is the modular design; the addressable regions are not part of the structural design, allowing for various such regions to be easily incorporated.
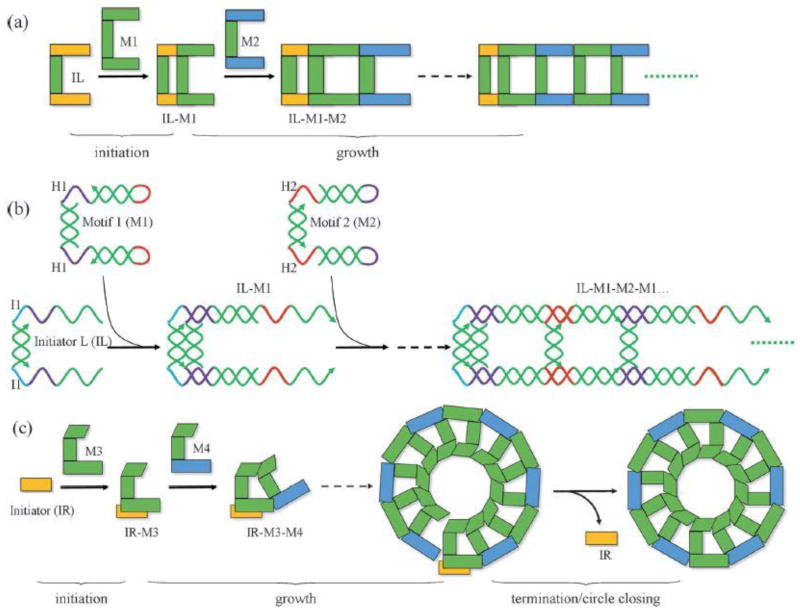
Although self-assembly of complex DNA nanostructures through thermal annealing is fairly straightforward, it is very different from the isothermal conditions of many biological systems. In order to accommodate this biological constraint, Mao and co-workers have reported a clever scheme in which two assembly processes, HCR + HCR or HCR + T-junction cohesion, synchronize to yield complex DNA nanostructures under isothermal conditions [16••]. In an HCR scheme, two ssDNA strands display stable hairpins; these strands are designed to hybridize with the addition of an initiator strand. With the addition of the initiator, one of the hairpin strands will hybridize with the initiator, freeing the rest of the strand to hybridize with the other hairpin strand and creating a hybridization cascade [3]. This reaction scheme is essentially a strand-displacement reaction that is self-perpetuating. Indeed, HCR is analogous to chain polymerization of synthetic polymers. The initiator molecule reacts with a “monomer” to produce an active monomer that can itself react with another monomer to repeat the process in a chain reaction. Using two cascading hybridization chain reactions and symmetric motifs, ladder-like structures were created. By using asymmetric motifs and an HCR process plus a T-junction cohesion, nanorings were created. The synchronization of the processes was controlled by proximity of the reaction sites to each other. In the case of the nanorings, the proximity of the HCR reaction site and the T-junction cohesion site stabilizes the latter reaction which would otherwise be unstable under the experimental conditions. While this method does not allow for control over the length of the structures (a termination reaction was necessary to close the nanorings), it does provide a powerful scheme for assembly that neither requires enzymatic reactions nor thermal annealing. One can envision the possibility of using this scheme to assemble nanostructures within a living cell.
Zhu et al. have also used HCR to assemble so-called “nanotrains” [17], analogous to the ladder-like structures described by Nie et al. [16••]. In this study, the initiator strand had aptamer functionality to increase the binding of the nanostructures to the surfaces of target cells. The hairpin strands involved in the HCR cascade allowed for the inclusion, in this case, of fluorescent tags. While this method allows for the relatively simple assembly of nanostructures including targeting moieties, their size cannot be controlled unless a termination reaction is included. Therefore such nanotrains, specifically those without termination, could prove of limited utility for drug and gene delivery.
It is well known that DNA nanostructures can be reconfigured by strand-displacement reactions. An interesting application of this strategy was demonstrated by Wei et al., where by adding specifically designed “carving” strands to an assembled tile-based DNA canvas or cube, sections of the structure were removed to create a new structure [18]. Due to the modular design, the carving of the structure was highly controlled, allowing for the development of a library of carving strands to produce complex, arbitrary shapes. Reconfiguration of the structure by carving was possible without external overhangs, but adding external overhangs led to higher completion of the carving. Carving a strand o3 of the canvas exposes overhangs in the resulting structure, so that carving proceeds as a cascade of strand-displacement reactions without external overhangs; when external overhangs are part of the structure, this cascade does not occur, as all of the strands being carved are accessible from the beginning. This is a powerful and reversible method for creating different complex structures from one canvas, however many carving strands are necessary to create each structure. Furthermore, without external overhangs, the cascade of strand displacements leads to a slow response time.
Zhu et al. have reported the creation of DNA “nanoflowers” by self-assembly that does not rely on base pairing [19]. Instead, the liquid crystallization of DNA strands drives the dense assembly of strands into aggregates termed nanoflowers. The DNA building blocks were synthesized from primer and template strands by rolling circle replication (RCR). Assembly through liquid crystallization is suggested by the authors due to the concentration and time dependency of structure formation. While this assembly method is a interesting approach to the creation of DNA nanostructures, it does not provide the same degree of size and shape control as conventional DNA assembly.
Mechanical properties
While the mechanical properties of ssDNA and dsDNA have been extensively studied, the mechanical properties of DNA nanostructures remain relatively unexplored. In terms of mechanical modeling, there has been some work to create a model for the mechanical properties of 2D DNA crossover structures by building on a previous ds-DNA model called the stack of plates [20]. In the stack of plates model, each base pair is modeled as an elliptical plate and the plates are connected by sugar-phosphate bonds modeled as elastic springs. However, the original model was not applicable for DNA crossovers, as it did not account for neighboring strands (which become important due to the proximity of strands required by crossover junctions). By including electrostatic interaction considerations in the model, Monte Carlo sampling was used to sample local and global moves of the structure, yielding the characteristic separation between strands. Using both this data and the experimental characteristic separation determined by AFM, the persistence length computed by the new model was shown to agree with that computed from the experimental data. The new model was also able to describe the helical deformation as a result of crossover points in the structure.
In addition to crossovers, the mechanical properties of DNA nanotubes assembled by tile arrays have also been studied [21]. By measuring thermal deformations of fluorescently labeled nanotubes, the persistence lengths of nanotubes of varying circumference were calculated. The persistence length as a function of circumference was used to model the tubes as bundles of elastic cylinders. This model accurately predicts the untwisting of the supertwists designed into the nanotubes due to shearing of cylinders relative to each other. This model also predicts that nanotube bending is not a simple compression and extension of the cylinders, but also includes shearing of the cylinders against each other.
Modeling of origami structures has also received some interest in order to predict 3D structures in solution [22]. By modeling dsDNA strands as isotropic elastic rods and taking nicks and single-strand linkers into account, finite element analysis was used to accurately predict the structure of complex DNA origami. This model can even predict non-intuitive out-of-plane bending, which cannot be seen by electron microscopy due to imaging limitations. Further work in this area may ultimately lead to the ability to design mechanical properties into DNA nanostructures with the same ease, and predictability, as structural features.
Stability against degradation
Independent of how they are assembled, the resulting nanostructures need to be sufficiently stable within their environment to achieve intended outcomes. Biological applications present challenges with respect to fouling (non-specific protein deposition), degradation (nucleases), phagocytic cell uptake, and (renal) elimination. Steps towards addressing these challenges have mainly focused on understanding how DNA nanostructures are degraded by nucleases found in blood, or serum (see Table 1). Our group was the first to explore this issue [23] and we discovered a surprising stability of wireframe tetrahedra in 10% fetal bovine serum. Hamblin et al. examined the serum stability of wireframe nanotubes and found that those constructed by RCA were significantly more stable than either linear dsDNA or non-RCA nanotubes [24]. However, the mechanistic origin of this difference was not identified. More recently, Conway et al. introduced subtle changes to their triangular prisms by labeling oligonucleotide termini with different functional groups [25]. Specifically, either hexanediol (C6) or hexaethylene glycol (HEG) groups dramatically improved the stability of triangular prisms over their unmodified analogues. While the mechanism for this increased stabilization is not clear, native gel electrophoresis revealed that only the HEG-modified prisms retain their structure in serum, suggesting a role for steric factors. We should note that native gel electrophoresis typically gives half-lives than are smaller than denaturing gel electrophoresis, due to the difference between monitoring assembly versus strand cleavage.
Table 1.
Characteristic sizes (in applicable units) of various DNA nanostructures and their half-lives τs in 10% fetal bovine serum, as determined by denaturing gel electrophoresis.
| Sample | Size | τs (h) | Reference |
|---|---|---|---|
| Linear dsDNA | 14 bp | 0.5 | [24] |
| Linear dsDNA | 63 bp | 0.6 | [23] |
| Linear dsDNA | 80 bp | 0.5 | [24] |
| Triangular prisms | 5.3 nm | 12.5 | [25] |
| Triangular prisms-C6 | 5.3 nm | 38 | [25] |
| Triangular prisms-HEG | 5.3 nm | 43 | [25] |
| Triangular prisms, ligated | 5.3 nm | 200 | [25] |
| Tetrahedra | 7 nm | 29 | [23] |
| Nanotubes | 1.2 μm | 0.8 | [24] |
| RCA-Nanotubes | 0.7 μm | 2.5 | [24] |
Hahn et al. systematically investigated the effects of magnesium ion Mg2+ concentration, serum levels, and various treatments to improve the stability of several model DNA origami structures in cell culture conditions [26]. At physiological Mg2+ levels, only one of their model structures remained assembled, and this potential problem was circumvented by the deliberate addition of Mg2+ to cell culture media. It should be noted, however, that nuclease activity strongly depends on (divalent) cation concentration [27], and therefore adjustment of these concentrations beyond physiological levels may have undesired effects in vivo. Hahn et al. also found that due to its animal-derived nature, the effect of serum (and its changes upon heat-inactivation) are quite difficult to control, and therefore partly explain the variation in Table 1. Nevertheless, such detailed experiments are important steps to improve the correlation of cell culture data with in vivo performance.
Two groups have taken a biomimetic approach towards improving stability of DNA nanostructures. Perrault et al. encapsulated their DNA nanostructures (origami octahedra) within lipid vesicles [28•]. Their strategy entailed presenting lipid-conjugated DNA strands on the external face of the octahedra, which presumably served as nucleation sites for the formation of lipid vesicles. The resulting liposome-DNA particles exhibited significant protection from DNase I activity, even in the presence of a large excess of this nuclease. Taking a slightly different inspiration, Mikkilä et al. combined DNA origami rectangles with capsid proteins from the cowpea chlorotic mottle virus (CCMV) to yield hybrid protein-DNA particles [29]. These authors did not explicitly test for stability against degradation, but it is likely that the hybrid nature of these particles will pose barriers to degradation by nucleases. Of course, because CCMV is a plant virus, it is to be expected that future works will attempt to repeat the strategy of Mikkilä et al. but instead using capsid proteins from mammalian viruses such as adenovirus or adeno-associated viruses.
While protecting cargo from degradation is an important goal of delivery vehicles, they are ideally able to efficiently release their cargo at the proper time and in the proper location. Thus over-protection of cargo is possible and would be detrimental to the overall efficacy of delivery vehicles. Indeed, such a difficulty has been identified for polycation-DNA complexes [30], which are similar to the hybrid particles developed by Mikkilä et al. [29]. Nevertheless, at the present time the major effort has been to increase DNA nanostructure stability, towards the ultimate goal of increasing the vehicle’s ability to remain in circulation. Stabilization strategies could also include covalent crosslinking (e.g., ligation), which has been shown to dramatically improve resistance to degradation by serum [25]. The introduction of partial synthetic character, as done in the DNA nanotube work by Hamblin et al. [24], can also improve stability against nuclease degradation but such an approach must be weighed against possible changes in bioactivity [31].
Delivery into cells and tissues
In vitro/cell culture
Several efforts have examined the uptake of DNA nanostructures into cells, and a consensus has to yet to emerge. Part of the difficulty appears to stem from the widely varying DNA nanostructure shapes and sizes. At both the micronand nano-scale it has already been well established that shape and size (among other properties) can dramatically affect cell uptake [32, 33]. A more subtle difficulty in comparing separate works is the variation in cell lines and conditions (e.g., dosing frequency, incubation time, serum levels).
In spite of variation among different works, there is evidence for non-specific uptake of DNA nanostructures For example, non-specific uptake of wireframe DNA tetrahedra has been observed in a few cases, notably in human cervical cancer cells (HeLa) [34•], in human breast cancer cells [35] and in human embryonic kidney cells (HEK) [36]. The high aspect ratio of wireframe nanotubes is likely to contribute to the non-specific uptake by HeLa cells [24], and may occur through a mechanism distinct from tetrahedra. With the exception of professional phagocytic cells, non-specific uptake is expected to be relatively low due to electrostatic repulsion between the DNA nanostructure and the cell surface [37].
Towards understanding the mechanisms underlying the non-specific uptake of DNA nanostructures, Liang et al. used sophisticated microscopy to track the internalization of wireframe tetrahedra. They found evidence for receptor-mediated endocytosis with active transport to lysosomes in HeLa cells [34•]. Independently, Kim et al. used pharmacological inhibitors of endocytosis pathways to identify contributions of caveolae-mediated endocytosis and macropinocytosis in MCF-7 cells [35]. Because of the lack of targeting ligands, we feel that these collective results are probably best explained by macropinocytotic uptake.
The non-specific uptake of complex hybrid particles has also been investigated. Protein-DNA particles were found to be internalized by human embryonic kidney (HEK) cells, and this uptake could be driven in a concentration-dependent manner [29]. Curiously, this study found higher uptake than a commercially available transfection reagent, although no mechanistic explanation was proposed for this outcome. Liposome-DNA particles showed negligible uptake in primary mouse splenocytes, comparable to control liposomes [28•]. This result is not surprising due to (i) the presence of a neutral polymer, polyethylene glycol, on the surface of the liposomes and (ii) the absence of any ligands to interact with the cell surfaces. A next step in such work will presumably be to add targeting ligands that direct binding and uptake to specific cell types.
Indeed, in contrast to non-specific uptake, the display of specific ligands can mediate receptor-mediated internalization, as demonstrated by two separate works using DNA aptamers on wireframe DNA nanostructures [38, 39]. In both these studies, the aptamers provide cell-selective targeting and uptake, an important feature for future delivery vehicles. The use of aptamers as ligands on DNA nanostructures is also attractive due to its facile incorporation and avoidance of chemical modifications to the DNA itself.
The anticancer drug doxorubicin (Dox) has now been extensively used as a model cargo to test the potential of DNA nanostructures as therapeutic carriers. This choice is a logical one given that Dox non-covalently binds to DNA by an intercalation mechanism. Zhao et al. used DNA origami nanotubes with differing “twist” to study non-targeted delivery of Dox to three breast cancer cell lines [40]. Twisted nanotubes were able to both carry more drug and release it more slowly than straight nanotubes, interpreted as a physical consequence of the intercalation of Dox into the nanostructures. Twisted nanotubes loaded with Dox significantly reduced cell viability as compared to the free drug in all three cell lines studied. Kim et al. also studied non-targeted delivery Dox to breast cancer cells, but their nanostructures were wireframe tetrahedra [35]. The Dox-loaded tetrahedra and free drug showed similar effects on the MCF-7 breast cancer cell line, reducing cell viability in a concentration-dependent manner. However, the Dox-loaded tetrahedra were able to reduce cell viability in a drug-resistant breast cancer cell line, demonstrating a clear advantage over the free drug. These in vitro results demonstrate the proof-of-concept ability of DNA nanostructures to deliver therapeutic cargo into cancer cells.
In vivo/animal models
Moving beyond cell culture into animal models is a necessary step in the translation from basic science to clinical use. These studies have been used to determine the biodistribution and circulation half-lives of DNA nanostructures, their efficacy in tumor models, and to assess their immunogenicity.
To our knowledge, Lee et al. were the first to study the fate of DNA nanostructures in an animal model [41•]. Using a (cervical cancer) xenograft tumor mouse model, biodistribution after 12 h showed accumulation primarily in the tumor and kidney, attributed to the folate ligands displayed by the tetrahedra. Zhang et al. ranked three different DNA origami structures (triangle, square, and tube) on the basis of their biodistribution in a (breast cancer) xenograft tumor mouse model [42]. After 24 h, all the structures had predominantly accumulated in the tumor and liver, with triangles showing the highest relative tumor accumulation. Noting the hydrodynamic radii of the structures: triangles (Rh = 59 nm); squares (Rh = 81 nm); tubes (Rh = 99 nm), the EPR effect would play a role in the biodistribution results, with aspect ratio also being important.
Lee et al. determined the circulation half-life τc of their wireframe tetrahedra to be approximately 24 min, which is significantly longer than the corresponding siRNA (Table 2). Perrault et al. also determined the circulation half-lives of bare and liposome-encapsulated origami octahedra, finding that a liposomal coating confers a dramatic improvement in circulation [28•]. While these steps are important, it remains unclear how much further improvement in both circulation and targeting will be necessary to make DNA-based delivery vehicles competitive with existing technologies.
Table 2.
Hydrodynamic radii Rh of various DNA nanostructures and their circulation half-lives τc, as determined by animal models.
The ability of DNA nanostructures to passively accumulate in tumor tissue motivated Zhang et al. to load their triangles with Dox and test efficacy in their animal tumor model. They found that Dox-loaded triangles gave the best reduction in tumor volume (as compared to free Dox, unloaded triangles, and saline [42]. Using folate ligands, Lee et al. took a targeted approach to delivery and furthermore demonstrated functionality, using siRNA-mediated downregulation of gene expression in their animal tumor model [41•]. Zhu et al. used aptamer-targeted DNA “nanotrains” to deliver several drugs, including Dox, in an animal tumor model [17]. Although the sizes of such objects are not well-defined, they were nevertheless able to reduce tumor volume slightly better than free Dox, and to increase survival times.
As a test for immune response, Lee et al. measured interferon-α (IFN-α) levels in blood samples at 6 h post-injection, and found no significant increase in IFN-α compared to levels in untreated mice [41•]. Zhang et al. also measured IFN-α levels at 6 h post-injection and found levels similar to negative controls [42]. These early results indicate a minimal immune response to DNA nanostructures and provide optimism regarding safety.. Nevertheless more in-depth studies are needed to establish an absence of an adaptive immune response (as might occur after repeated administrations).
Conclusions
The current literature shows significant advances in assembly strategies, particularly in clever design methods to control size/shape and to achieve assembly under isothermal conditions. The field is now shifting to applications such as sensing and delivery, with the latter efforts showing promise for drug and gene delivery. Finally, hybrid structures of DNA and other biomolecules are intriguing examples of future research directions in designer nanoscale particles.
Figure 2.
DNA nanoladders and nanorings assembled by two synchronized reactions. (a,c) Schematic of nanoladder and nanoring assembly, respectively. (b) Detailed illustration of HCR reaction used to assemble nanoladders. (Reprinted with permission from [16••]. Copyright 2014 Wiley.)
Highlights.
Review of recent advances in DNA nanostructure self-assembly and delivery.
Nanostructures assembled by synchronized hybridization chain reactions.
No consensus on uptake of nanostructures into cells due to experimental variation.
Encapsulation of DNA octahedra in lipid vesicles protects structure from nucleases.
Acknowledgments
The authors thank P. Charoenphol for helpful discussions. This work was supported by the National Institutes of Health (R21 CA158977), the National Science Foundation (DMR-0847558) and, in part, by the NSF Center for Hierarchical Manufacturing (CMMI-1025020).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37(15):5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A. 2004;101(43):15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Tseng YD, Kwon SY, D’Espaux L, Bunch JS, McEuen PL, Luo D. Controlled assembly of dendrimer-like DNA. Nat Mater. 2004;3(1):38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 5.Um SH, Lee JB, Kwon SY, Li Y, Luo D. Dendrimer-like DNA-based fluorescence nanobarcodes. Nat Protoc. 2006;1(2):995–1000. doi: 10.1038/nprot.2006.141. [DOI] [PubMed] [Google Scholar]
- 6.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99(2):237–247. doi: 10.1016/0022-5193(82)90002-9. (Print) [DOI] [PubMed] [Google Scholar]
- 7.Seeman NC. DNA in a material world. Nature. 2003;421(6921):427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 8•.Nickels PC, Ke Y, Jungmann R, Smith DM, Leichsenring M, Shih WM, Liedl T, Högberg B. DNA origami structures directly assembled from intact bacteriophages. Small. 2014;10(9):1765–1769. doi: 10.1002/smll.201303442. This paper reports the successful assembly of DNA origami structures through the addition of short staple strands directly to unpurified bacteriophages and even M13-infected bacterial liquid cultures. [DOI] [PubMed] [Google Scholar]
- 9••.Mathur D, Henderson ER. Complex DNA nanostructures from oligonucleotide ensembles. ACS Synth Biol. 2013;2(4):180–185. doi: 10.1021/sb3000518. This paper reports the assembly of DNA origami by exclusively using oligonucleotides, therefore eliminating the need for phage-derived scaffold strands and concerns of biological contamination. [DOI] [PubMed] [Google Scholar]
- 10.Lo PK, Metera KL, Sleiman HF. Self-assembly of three-dimensional DNA nanostructures and potential biological applications. Curr Opin Chem Biol. 2010;14(5):597–607. doi: 10.1016/j.cbpa.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Park SH, Reif JH, LaBean TH. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc Natl Acad Sci U S A. 2004;101(3):717–722. doi: 10.1073/pnas.0305860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathieu F, Liao S, Kopatscht J, Wang T, Mao C, Seeman N. Six-helix bundles designed from DNA. Nano Lett. 2005;5(4):661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aldaye FA, Lo PK, Karam P, McLaughlin CK, Cosa G, Sleiman HF. Modular construction of DNA nanotubes of tunable geometry and singleor double-stranded character. Nat Nanotechnol. 2009;4(6):349–52. doi: 10.1038/nnano.2009.72. [DOI] [PubMed] [Google Scholar]
- 14.Zheng H, Xiao M, Yan Q, Ma Y, Xiao SJ. Small circular DNA molecules act as rigid motifs to build DNA nanotubes. J Am Chem Soc. 2014;136(29):10194–10197. doi: 10.1021/ja504050r. [DOI] [PubMed] [Google Scholar]
- 15.Hamblin GD, Hariri AA, Carneiro KMM, Lau KL, Cosa G, Sleiman HF. Simple design for DNA nanotubes from a minimal set of unmodified strands: rapid, room-temperature assembly and readily tunable structure. ACS Nano. 2013;7(4):3022–8. doi: 10.1021/nn4006329. [DOI] [PubMed] [Google Scholar]
- 16••.Nie Z, Wang P, Tian C, Mao C. Synchronization of two assembly processes to build responsive DNA nanostructures. Angew Chem Int Ed Engl. 2014;53(32):8402–8405. doi: 10.1002/anie.201404307. This paper reports the assembly of DNA nanoladders and nanorings using synchronized hybridization chain reactions, eliminating the need for enzymes or thermal annealing. [DOI] [PubMed] [Google Scholar]
- 17.Zhu G, Zheng J, Song E, Donovan M, Zhang K, Liu C, Tan W. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci U S A. 2013;110(20):7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei B, Ong LL, Chen J, Jaffe AS, Yin P. Complex reconfiguration of DNA nanostructures. Angew Chem Int Ed Engl. 2014;53(29):7475–7479. doi: 10.1002/anie.201402437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu G, Zhang S, Song E, Zheng J, Hu R, Fang X, Tan W. Building fluorescent DNA nanodevices on target living cell surfaces. Angew Chem Int Ed Engl. 2013;52(21):5490–5496. doi: 10.1002/anie.201301439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbona JM, Aimé JP, Elezgaray J. Modeling the mechanical properties of DNA nanostructures. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;86(5 Pt 1):051912. doi: 10.1103/PhysRevE.86.051912. [DOI] [PubMed] [Google Scholar]
- 21.Schiffels D, Liedl T, Fygenson DK. Nanoscale structure and microscale stiffness of DNA nanotubes. ACS Nano. 2013;7(8):6700–6710. doi: 10.1021/nn401362p. [DOI] [PubMed] [Google Scholar]
- 22.Kim DN, Kilchherr F, Dietz H, Bathe M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012;40(7):2862–8. doi: 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keum JW, Bermudez H. Enhanced Resistance of DNA Nanostructures to Enzymatic Digestion. Chem Commun. 2009;(45):7036–7038. doi: 10.1039/B917661F. [DOI] [PubMed] [Google Scholar]
- 24.Hamblin GD, Carneiro KMM, Fakhoury JF, Bujold KE, Sleiman HF. Rolling circle amplification-templated DNA nanotubes show increased stability and cell penetration ability. J Am Chem Soc. 2012;134(6):2888–91. doi: 10.1021/ja2107492. [DOI] [PubMed] [Google Scholar]
- 25.Conway JW, McLaughlin CK, Castor KJ, Sleiman H. DNA nanostructure serum stability: greater than the sum of its parts. Chem Commun. 2013;49(12):1172–1174. doi: 10.1039/c2cc37556g. [DOI] [PubMed] [Google Scholar]
- 26.Hahn J, Wickham SFJ, Shih WM, Perrault SD. Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano. 2014;8(9):8765–8775. doi: 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charoenphol P, Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Molecular Pharmaceutics. 2014;11(5):1721–1725. doi: 10.1021/mp500047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8(5):5132–5140. doi: 10.1021/nn5011914. This paper reports on how DNA nanostructures can be encapsulated into liposomes, improving stability against nucleases and increasing circulation time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkilä J, Eskelinen AP, Niemelä EH, Linko V, Frilander MJ, Törmä P, Kostiainen MA. Virus-encapsulated DNA origami nanostructures for cellular delivery. Nano Lett. 2014;14(4):2196–2200. doi: 10.1021/nl500677j. [DOI] [PubMed] [Google Scholar]
- 30.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 31.Juliano RL, Bauman J, Kang H, Ming X. Biological Barriers to Therapy with Antisense and siRNA Oligonucleotides. Mol Pharm. doi: 10.1021/mp900093r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champion JA, Mitragotri S. Role of target geometry in phagocytosis. Proc Natl Acad Sci U S A. 2006;103(13):4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 34.Liang L, Li J, Li Q, Huang Q, Shi J, Yan H, Fan C. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew Chem Int Ed Engl. 2014;53(30):7745–7750. doi: 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- 35.Kim KR, Kim DR, Lee T, Yhee JY, Kim BS, Kwon IC, Ahn DR. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells. Chem Commun. 2013;49(20):2010–2. doi: 10.1039/c3cc38693g. [DOI] [PubMed] [Google Scholar]
- 36.Walsh AS, Yin H, Erben CM, Wood MJA, Turberfield AJ. DNA cage delivery to Mammalian cells. ACS Nano. 2011;5(7):5427–32. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 37.Segura T, Shea LD. Materials for non-viral gene delivery. Annu Rev Mater Res. 2001;31:25–46. [Google Scholar]
- 38.Chang M, Yang CS, Huang DM. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5(8):6156–63. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]
- 39.Charoenphol P, Bermudez H. Design and application of multifunctional DNA nanocarriers for therapeutic delivery. Acta Biomater. 2014;10(4):1683–1691. doi: 10.1016/j.actbio.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao YX, Shaw A, Zeng X, Benson E, Nyström AM, Högberg B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012;6(10):8684–8691. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7(6):389–93. doi: 10.1038/nnano.2012.73. • This paper reports the first in vivo study using DNA nanostructures, demonstrating delivery of siRNA, establishing circulation time, and measurement of initial immune response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Jiang Q, Li N, Dai L, Liu Q, Song L, Wang J, Li Y, Tian J, Ding B, Du Y. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8(7):6633–6643. doi: 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]