Abstract
Molecular-genetic imaging of cancer using nonviral delivery systems has great potential for clinical application as a safe, efficient, noninvasive tool for visualization of various cellular processes including detection of cancer, and its attendant metastases. In recent years, significant effort has been expended in overcoming technical hurdles to enable clinical adoption of molecular-genetic imaging. This chapter will provide an introduction to the components of molecular-genetic imaging and recent advances on each component leading to safe, efficient clinical applications for detecting cancer. Combination with therapy, namely, generating molecular-genetic theranostic constructs, will provide further impetus for clinical translation of this promising technology.
1. INTRODUCTION
In the context of cancer, the purpose of imaging is to visualize a target for diagnosis, staging, and/or therapeutic monitoring. Due to the noninvasive nature of medical imaging, a variety of imaging modalities have been developed and are continually being improved to enhance the sensitivity and specificity of detection. Conventional imaging methods, such as computed tomography (CT), ultrasound, and magnetic resonance imaging (MRI), provide sensitive detection of anatomic information at the macroscopic level. As opposed to those anatomic techniques, molecular imaging can provide visualization and quantification of biochemical processes at cellular and molecular levels. Initial approaches to molecular imaging involved directly targeting cell surface receptors, metabolic enzymes, and transporters using probe molecules that directly interact with their targets, e.g., antibodies, peptides, aptamers, and small molecules. The most widely used examples of such direct imaging is positron emission tomography with [18F]fluorodeoxyglucose (FDG-PET) for detecting cancer, which generally presents with elevated glucose metabolism (Blasberg & Tjuvajev, 2003). Although direct imaging in combination with conventional structural imaging, e.g., PET/CT and PET/MR, provides precise spatial, functional, and quantitative visualization of target diseases, it has limitations. First, there are a limited number of clinically viable, specific target-probe combinations for many diseases. Second, often targets for certain diseases are also present in normal cells causing high background noise. Actively inserting a target to cells of interest, via a reporter transgene, with subsequent detection of the target with already well-established imaging agents (probes) would provide a solution for some of the limitations of direct imaging. This indirect imaging approach, which is under vigorous preclinical development, requires the following components: (1) a promoter to drive the expression of a reporter gene in a target cell, (2) a reporter gene, (3) a mechanism, e.g., a viral or nonviral vector, to enable transfection of the target cells with the reporter transgene, and (4) an imaging probe that interacts with the reporter transgene in such a way as to enable visualization. Because genetic material is introduced to and later detected within target cells using a cognate probe, this indirect imaging method has been referred to as “molecular-genetic imaging.”
Through deliberate insertion of a customized transgene, the molecular-genetic imaging approach can provide versatile tools for imaging of many diseases that do not have unique or otherwise suitable targets for direct imaging. Despite its promise, there are many hurdles to overcome to bring this technology to the clinic. Perhaps the biggest challenge is the regulatory piece, as several of the components of the method noted earlier must be administered to the patient at different times, each potentially with a unique capacity to damage normal tissue. For example, due to the potential risk of causing malignant transformation of normal cells by random insertion of transfected DNA into chromosomes, clinical trials of gene therapy have been very closely scrutinized with approval for clinical use limited to very few lethal diseases for which there are no cures (Moran, 2012). Continuous effort from the gene therapy community resulting in recent promising and safe trials addressing diseases such as inherited blindness and immune deficiencies has led the US National Institutes of Health to announce on May 22, 2014 that they will no longer subject all proposed gene therapy trials to review by a special federal advisory committee. That will expedite the process for approval of clinical trials for molecular-genetic imaging as well. This chapter will introduce details regarding the components of molecular-genetic imaging, current limitations and efforts to resolve them, and criteria for successful clinical translation.
2. PROMOTERS
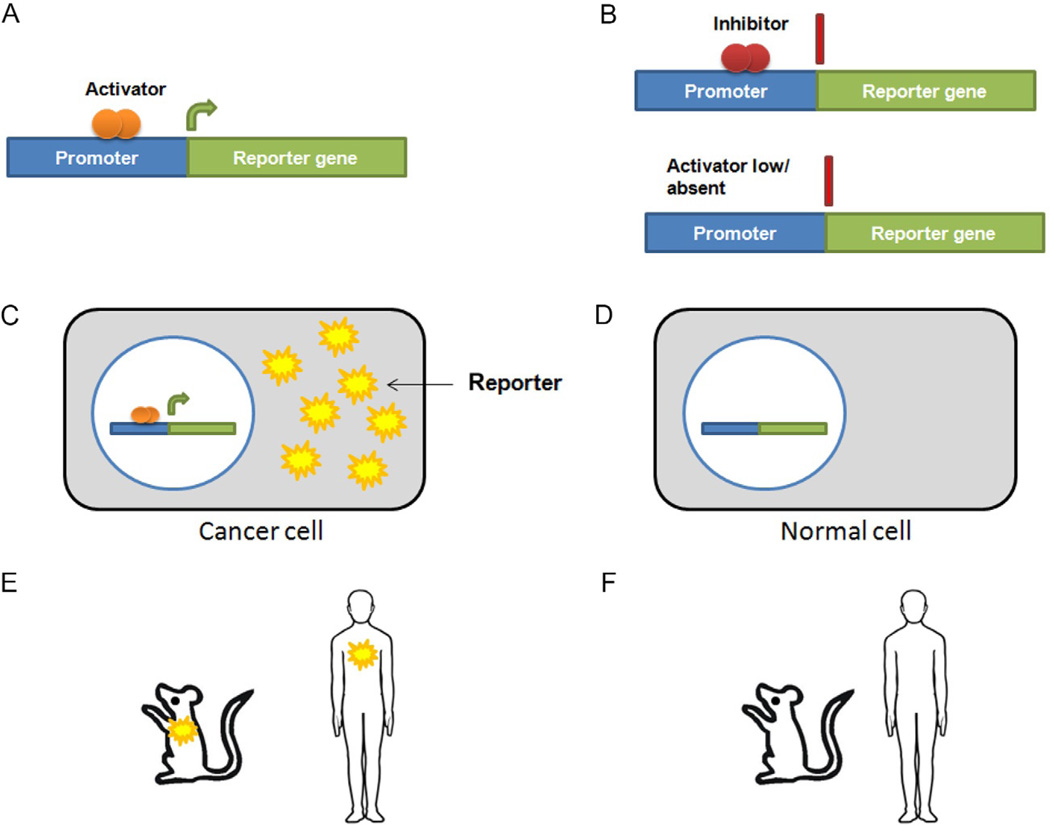
Promoters can broadly be described as regions of DNA located upstream of the transcriptional start site of a gene that serve as a binding site for the RNA polymerase complex and other factors. Several transcription factor-binding sites may be present along the length of the promoter, which can modulate transcription of the downstream gene. The binding of certain transcription factors (transcriptional activators/enhancers) enhances transcription of the downstream gene, while binding of other transcription factors (transcriptional repressors/silencers) blocks transcription of the downstream gene. Transcriptional targeting has been widely used to express specific reporter genes in particular tissues using tissue- or tumor-specific promoters (Bhatia et al., 2013). The reporter gene is only expressed in cells that have sufficient levels of transcription factors necessary to activate transcription through this specific promoter. Accordingly, promoters, enhancers, and repressors are critical components that regulate the duration and strength of a reporter gene’s expression in specific cells for optimal molecular-genetic imaging (Fig. 4.1).
Figure 4.1.
Schematic representation of the basic molecular-genetic imaging approach. (A) The tissue- or tumor-specific promoter is upstream of the reporter gene, and in the presence of an activator causes transcription of the reporter. (B) In the presence of an inhibitor or silencer or when the activator is low or absent, the reporter gene is not transcribed. (C) The cancer cell expresses the activator and is able to express the reporter. (D) Normal cells do not express the activator and accordingly do not express the reporter. (E) The reporter can be visualized in mice with tumors and potentially in patients with tumors. (F) Normal mice and individuals do not express the reporter.
The optimal promoters for molecular-genetic imaging for human cancer would be activated in cancer cells specifically and robustly while having minimal or no activity in normal tissues. In order to identify novel tumor-specific promoters, researchers evaluated promoters of genes that were frequently upregulated in cancers. Those included promoters of oncogenes; promoters of genes involved in modulating the cell cycle; and promoters of genes known to enhance cell survival, angiogenesis, cancer progression, and metastasis. Additionally, promoters that are both tumor-specific and tissue-specific have been identified that can potentially target reporter gene expression in a particular tissue type, for example, genes involved in melanin biosynthesis (Pleshkan, Alekseenko, Zinovyeva, Vinogradova, & Sverdlov, 2011).
Several types of promoters are utilized for tumor-specific reporter gene expression (Nettelbeck, Jerome, & Muller, 2000; Robson & Hirst, 2003). One type of promoter is the native or minimal active promoter of a specific gene, e.g., the progression elevated gene-3 (PEG-3) promoter. The PEG-3 promoter is a well-studied tumor-specific promoter used for molecular-genetic imaging. The PEG-3 gene was first identified in rodents as a gene upregulated upon malignant transformation and tumor progression in rat embryonic fibroblast cells (Su, Shi, & Fisher, 1997) and has no known human counterpart. Being a rodent gene promoter, utilizing the PEG-3 promoter is advantageous as it helps to avoid background expression of reporter signals, allowing for more accurate reporter readout. In addition, the PEG-3 promoter has no chance for chromosomal insertion to the human genome by homologous recombination (discussed in Section 7). The PEG-3 promoter is robustly activated in a number of human cancer cell lines including prostate, breast, and glioblastoma, with minimal activity in their normal counterparts (Su et al., 2005). Tumor-specific activation of the PEG-3 promoter was attributed to AP-1 and PEA-3 transcription factors, which are commonly upregulated in tumor cells. In fact, reporter genes under transcriptional control of the PEG-3 promoter were successfully used to detect micrometastatic disease in in vivo models of melanoma and breast cancer using both bioluminescent and radionuclide-based molecular-genetic imaging approaches (Bhang, Gabrielson, Laterra, Fisher, & Pomper, 2011).
Several other promoters that are specifically active in tumors as compared to normal tissues have also been identified and can potentially be utilized for targeting reporter gene expression specifically in tumors (Table 4.1). Those include human telomerase reverse transcriptase (hTERT), cyclooxygenase-2 (Cox-2), α-chemokine SDF-1 receptor (CXCR4), survivin (BIRC5), and α-fetoprotein (AFP) gene promoters. The hTERT promoter is highly active in human cancer cells but not in normal differentiated human cells (Wirth, Kuhnel, & Kubicka, 2005). The hTERT promoter was utilized to visualize tumors and metastasis in vivo in a colorectal tumor model (Kishimoto et al., 2006; Umeoka et al., 2004) and in a head and neck cancer model (Kurihara et al., 2009). Cox-2 is involved in prostaglandin biosynthesis. The Cox-2 promoter is active in intestinal neoplasias and tumors of epithelial origin but not in normal tissues. The Cox-2 promoter was utilized to visualize tumor xenografts using bioluminescent imaging (BLI) (Liang, Dmitriev, Kashentseva, Curiel, & Herschman, 2004; Liang, Yamamoto, Curiel, & Herschman, 2004). The CXCR4 promoter was found to be active in many cancer types but not in their normal counterparts and had low activity in the liver in vivo (Haviv, van Houdt, Lu, Curiel, & Zhu, 2004; Zhu et al., 2004a). Survivin belongs to the inhibitor of apoptosis protein family that is expressed in human cancers but is not detected in normal, differentiated tissues. The survivin promoter was identified to have high activity in melanoma cell lines and primary melanoma cells but not in human epithelial melanocytes. It also demonstrated extremely low activity in vivo in major mouse organs including the liver (Lu et al., 2005; Zhu et al., 2004b). AFP is a serum glycoprotein that decreases rapidly following birth and is repressed in adults. Interestingly, AFP expression is increased in about 80% of patients with hepatocellular carcinoma. The AFP promoter has been used to monitor hepatocellular carcinoma using BLI noninvasively in a transgenic mouse model (Park et al., 2011).
Table 4.1.
Tumor-selective promoters
| Promoter | Gene symbol |
Cancer |
|---|---|---|
| α-Fetoprotein | AFP | Hepatocellular carcinoma |
| Α-Lactalbumin | ALA | Breast cancer |
| Astrocyte elevated gene-1; Metadherin | AEG-1/MTDH | Breast cancer, prostate cancer, liver cancer |
| Cell division cycle 25C | Cdc25C | Melanoma |
| Carcinoembryonic antigen | CEA | Colorectal cancer, pancreatic cancer, breast cancer, lung cancer, adenocarcinomas |
| Cyclin A | CCNA | Melanoma |
| Calponin | CNN | Soft tissue and bone tumors |
| Cyclooxygenase-2 | Cox-2 | Ovarian cancer, pancreatic cancer, gastrointestinal cancer |
| Glucose-regulated protein 78 | Grp78 (BIP) | Solid tumors |
| Human epidermal growth factor receptor 2 | HER2/ErbB2 | Breast cancer, pancreatic cancer, ovarian cancer |
| Human telomerase reverse transcriptase | hTERT | Lung cancer, colorectal cancer, ovarian cancer, bladder cancer, cervical cancer, liver cancer, glioma, head and neck cancer |
| Human chorionic gonadotropin | hGC | Ovarian cancer |
| Lymphocyte cytosolic protein 1/l-plastin | LCP1 | Ovarian cancer, breast cancer, fibrosarcoma, carcinomas |
| Midkine | MK | Pancreatic cancer |
| Metallothionein | MT | Ovarian cancer |
| Mucin 1 | MUC1/DF3 | Adenocarcinomas |
| Osteocalcin | OC | Prostate cancer, ovarian cancer, lung cancer, brain cancer |
| Progression elevated gene-3 | PEG-3 | Prostate cancer, breast cancer, glioma |
| Probasin | ARR2PB | Prostate cancer |
| Prostate-specific antigen | PSA | Prostate cancer |
| Secretory leukoprotease inhibitor | SLPI | Lung cancer, breast cancer, oropharyngeal cancer, bladder cancer, endometrial cancer, ovarian cancer, colorectal cancer, cervical cancer, adenocarcinomas |
| Survivin | BIRC5 | Hepatocellular carcinoma, gastric cancer, glioma, gallbladder cancer, melanoma |
| Tyrosinase | TYR | Melanoma |
Another type of promoter is the composite promoter, which is designed to increase the strength of promoters that demonstrate suitable specificity but weaker activity. Composite promoters also help to reduce the promoter size while maintaining promoter activity. Small promoters are more convenient for introduction into various vectors, where space can be a limiting factor, and help to maintain the minimal size of a vector, which is critical for optimal transfection efficiency (discussed in Section 7). Composite promoters include the minimal active promoter along with enhancers from the same gene or different genes, e.g., human tyrosinase (TYR) promoter with multiple TYR enhancers (Siders, Halloran, & Fenton, 1996). Another example is a combination of promoter elements from multiple origins optimized for enhanced tumor-specific expression, e.g., a synthetic promoter generated by combining elements from the TYR and AFP promoters (Martinelli & De Simone, 2005).
Promoters can also be constitutive or inducible. Constitutive promoters can be tissue-specific, such as the TYR promoter for melanoma (Bentley, Eisen, & Goding, 1994) and the prostate-specific antigen (PSA) promoter for prostate cancer (Gotoh et al., 1998; Pang et al., 1995), while other promoters are active in a variety of tumor types, such as the hTERT promoter (Fujiwara, Urata, & Tanaka, 2007) and the PEG-3 promoter (Bhang et al., 2011). Inducible promoters are induced to express the reporter gene in response to exogenously provided hormones, radiation, heat, drugs, and small molecules, e.g., radiation-responsive elements (CArG) were identified in the early growth response 1 (Egr-1) promoter and were used in a clinical trial to express tumor necrosis factor α in patients with soft-tissue sarcomas receiving radiation (Hallahan et al., 2001). The HSP70B promoter has been used to activate genes in response to external heat/hyperthermia (Rome, Couillaud, & Moonen, 2005). An example of a composite inducible promoter is the probasin (ARR2PB) promoter combined with the bacterial chloramphenicol acetyltransferase reporter. That promoter could be induced by androgens and glucocorticoids and proved useful for targeting prostate cancer (Zhang, Thomas, Kasper, & Matusik, 2000).
Silencer elements are also being included in molecular-genetic approaches to limit gene expression to certain tissue types. For example, inclusion of a conserved neuron-restrictive silencer element caused repression of gene expression in all nonneuronal cells (Xie et al., 2013). Additionally, conditions associated with the tumor microenvironment are also being used to ensure tumor-specific targeting of reporter genes, for example, hypoxia-inducible factor (HIF)-responsive promoters that will be active in the hypoxic tumor microenvironment (Post et al., 2007). Some of the HIF-responsive promoters include the vascular endothelial growth factor (VEGF) promoter, the erythropoietin promoter, and the phosphoglycerate kinase-1 promoter (Robson & Hirst, 2003). Other promoters have multiple levels of specificity such as the p21WAF1/CIP1/MDA6 promoter, which is tumor-specific, radiation-induced, and hypoxia-specific (Robson & Hirst, 2003).
More information regarding tissue- and tumor-specific promoters can be found elsewhere (Nettelbeck et al., 2000; Papadakis, Nicklin, Baker, & White, 2004; Robson & Hirst, 2003; Saukkonen & Hemminki, 2004). As discussed earlier, the tissue- or tumor-specific promoter is placed upstream of a reporter gene to enable tissue- or tumor-specific expression of the reporter gene. Various reporters utilized for molecular-genetic imaging in combination with these promoters will be discussed in the next section.
3. REPORTERS
Molecular-genetic reporter genes under the transcriptional control of promoters, along with substrates or imaging agents, can be used noninvasively to track quantitatively various biological phenomena, including immune cell trafficking cancer development, progression, metastasis, molecular interactions, and delivery of macromolecules (genes, recombinant proteins) and cells. Molecular-genetic reporters improve the sensitivity and specificity of traditional imaging approaches, as the physiologic manifestations of molecular-genetic events precede anatomic changes (Bhang et al., 2011; Kang & Chung, 2008). Molecular-genetic reporters can broadly be classified as optical, radionuclide, and paramagnetic/MRI reporters. The most common molecular-genetic reporter genes based on optical probes are firefly luciferase (fLuc), renilla luciferase (rLuc), click beetle luciferase (cbLuc), and gaussia luciferase (gLuc) (Tannous, Kim, Fernandez, Weissleder, & Breakefield, 2005; Zhao et al., 2005; Table 4.2), and they have replaced traditional approaches using chloramphenicol-acetyl transferase (CAT) and β-galactosidase (β-gal) because of sensitivity, rapid acquisition or assay time, and the capacity for use in a high throughput mode (Wood, Lam, Seliger, & McElroy, 1989). fLuc is the most commonly used luciferase because of its robustness, reproducibility, tissue penetration, and sensitivity over the other luciferases for in vivo imaging (Bhang et al., 2011; Hayat, 2007). We showed that the fLuc reporter under the transcriptional control of a cancer-specific promoter (PEG-3 promoter) led to amplification and subsequent expression of fLuc in cancer cells, which permitted the detection of micrometastases in melanoma and breast cancer in mouse models (Bhang et al., 2011). Due to light scattering and absorption (Zhao et al., 2005), the luciferase-based reporters have limitations in deep tissue imaging as the emission range of wild-type fLuc is <600 nm, and may not enable visualization of internal organs of even small animals. The development of red or near-infrared (NIR)-emitting mutant luciferase (Ppy, Lcr, Ph-RED) (Branchini, Southworth, Khattak, Michelini, & Roda, 2005; Caysa et al., 2009; Mezzanotte et al., 2011) and fluorescent reporters along with codon optimization (Mezzanotte et al., 2011) for mammalian expression systems are constantly being updated with the primary aim of enhancing sensitivity and specificity of molecular-genetic reporters.
Table 4.2.
Reporters
| Reporters | Genes | Substrates | Modalities | Advantages | Limitations |
|---|---|---|---|---|---|
| Optical A. Luminescence B. Fluorescence |
fLuc, rLuc, cbLuc, gLuc gfp, rfp, Katuska | Luciferin, coelenterazine | Bioluminescence imaging Fluorescence imaging |
High sensitivity (50–100 cells) Specificity Quantitative Cost-effective Nonhazardous |
Limited tissue penetration Low spatial resolution Preclinical studies |
| Radionuclide | HSV1-tk, hD2R hSSTR2, hNaIS | [18F]FIAU, [18F]FEAU and analogues, [18F]FESP, 111In-DTPA- octreotide, 131I | PET, SPECT | Preclinical and clinical Extensive penetration High sensitivity |
Ionizing radiation Medium spatial resolution (several mm) |
| Paramagnetic agents | CK, β-gal Tyrosinase Ferritin Tf, hTR Tyrosinase | ATP, EgadMe, Fe, Tf-MIONs | MRI | Clinical Deep tissue penetration High spatial resolution (µm–mm) |
Low signal Low sensitivity Long acquisition time |
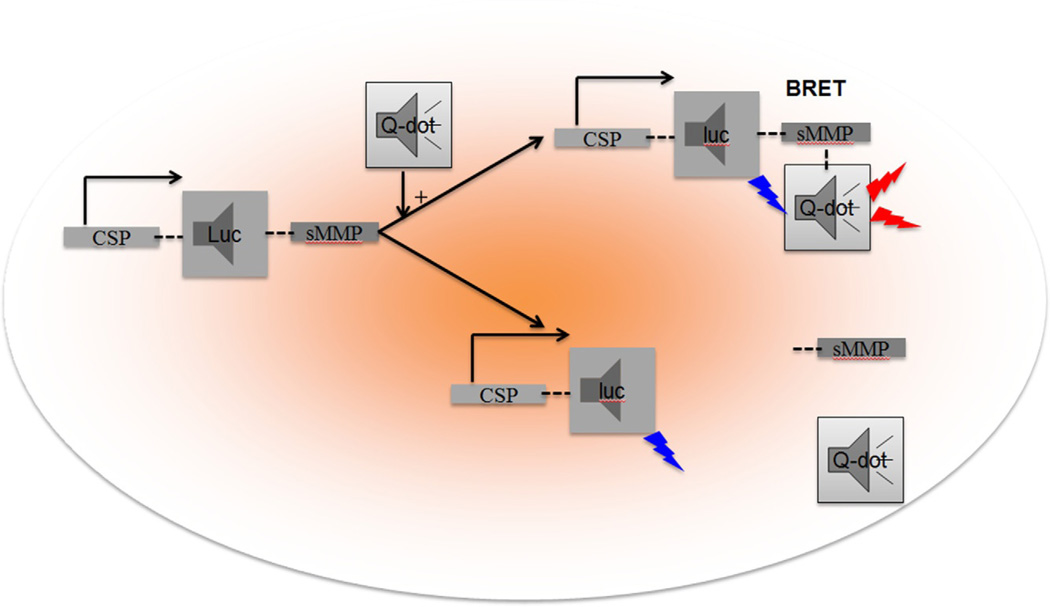
Although fluorescent reporter genes do not require any substrate to be irradiated, they do require an external light source, which further diminishes utility due to tissue scattering and endogenous fluorochromes (such as hemoglobin) at shorter wavelengths and water or aqueous solutions at longer wavelengths. Accordingly, NIR emission is the preferred wavelength for in vivo imaging. Based on that, progress has been made in developing engineered green fluorescent protein (GFP) reporter genes with NIR emission. Recently, Katuska and the monomeric mKate (Ex/Em max: 588/635) (Shcherbo et al., 2007) were developed, which showed promise as a reporter gene for whole body imaging of an animal, which can further be improved by developing far-red or NIR (650–700 nm) emission reporters with enhanced photostability. The availability of different fluorescent gene reporters in a vector or a living system will also help in studying molecular interactions utilizing the quantum physical phenomenon known as fluorescence resonance energy transfer (FRET) (Nagai & Miyawaki, 2004), which occurs between two fluorescent reporters with overlapping absorption and emission spectra lying in close proximity (~1000nm) to one another. The same principle can be applied to study protein interactions with bioluminescence resonance energy transfer (BRET) (Xu, Piston, & Johnson, 1999), where the donor is the luciferase gene and the acceptor is the fluorescent species (GFP, RFP, quantum, or Q-dots) with overlapping emission and absorption spectra, respectively. Q-dots might be the preferred acceptor fluorescent species over GFPs, because of their higher quantum yield and molar extinction coefficient, broader absorption spectra with narrower emission spectra (So, Xu, Loening, Gambhir, & Rao, 2006). The luciferase gene (luc) under the transcriptional control of a cancer-specific promoter (CSP) (Bhang et al., 2011) with either end conjugated with substrates for proteinase, mainly MMPs (sMMPs) (Yao, Zhang, Xiao, Xia, & Rao, 2007), i.e., CSP-luc-sMMP or ECM, or even by engineering a luciferase gene with an internal cleaved site for MMPs (Wigdal et al., 2008; Yao et al., 2007), can be introduced into living systems using plasmid/polyethyleneimine (PEI) nanocomplexes. They can also be utilized to generate transgenic mouse models that can serve as optical imaging reporters, where disease progression can be studied by luciferase expression and invasion can be studied by applying a Q-dot conjugated with chemical groups or metal ions. Those will form complexes with luciferase via cleavable sMMP (Yao et al., 2007) so that BRET can be utilized for observing fluorescence from Q-dots within a defined molecular distance (~1000 nm). With the change in tumor microenvironment or invasion due to the activity of MMPs, the substrate of the MMPs will be cleaved, BRET will be avoided, and fluorescence will not be detected (Fig. 4.2). That process will diminish the requirement for an external light source, required by fluorescent proteins, and is responsible for autofluorescence and photobleaching. As such, BRET with Q-dot acceptors may improve the sensitivity and specificity of the fluorescent reporter technique (Xu et al., 1999). That technique, along with the development of cooled charge-coupled device cameras with digital imaging modalities and high-resolution 3D diffuse light emission tomography (DLIT) (Cronin et al., 2012), has tremendous potential in developing nanosensors for detecting tumor progression in real time in animals and in patients in the near future (Cronin et al., 2012; Yao et al., 2007).
Figure 4.2.
Hypothetical scheme for molecular-genetic imaging utilizing bioluminescence resonance energy transfer (BRET): a cancer-specific or cancer-selective promoter (CSP) drives luciferase gene expression (luc) with the C-terminal end containing a cDNA sequence corresponding to amino acids for the substrate of a matrix metalloproteinase (sMMP) or collagen (i.e., CSP-luc-sMMP) forming Luc-sMMP. The construct generates bioluminescent signal by adding substrates for luciferin (D-luciferin or coelenterazine). The addition of Q-dots with overlapping absorption spectra with respect to the emission spectra of the version of luc chosen will undergo BRET enabling fluorescence detected in the NIR region to visualize the presence of tumor (upper sequence). As the tumor progresses, the tumor microenvironment changes, and the involvement of extracellular matrix proteins or MMPs lead to subsequent cleavage of sMMPs from Luc-sMMP, releasing the Q-dot. The Q-dot will no longer be activated by bioluminescence because the molecular distance (<1000 nm) between it and the bioluminescent construct will no longer enable BRET. BRET signal turns off during tumor invasion and metastasis, where MMPs abound. This is an example of an imaging reporter that can measure tumor growth while maintaining sensitivity to its microenvironment, via loss of BRET.
Although 3D-DLIT and 3D-fluorescence light imaging tomography (3D-FLIT) utilizing high-resolution BLI and fluorescence imaging coregistered with digital anatomic structures of animals can be utilized to quantify signal and depth, it is still limited to preclinical studies due to expected attenuation from deep tissues and internal organs. That led to the development of radionuclide-based molecular imaging reporters utilizing PET or single-photon emission computed tomography (SPECT), upon systemic administration of radiolabeled probes, such as nucleoside analogs or radionuclide itself. As noted earlier, we demonstrated that systemic administration of the imaging vector containing the cancer-specific PEG-3 promoter driving the expression of the HSV1-tk gene (i.e., pPEG-3-HSV-tk) led to cancer site-specific amplification of HSV1-tk, which subsequently accumulated 2′-fluoro-2′-deoxy-β-d-5-[125I] iodouracil-arabinofuranoside ([125I]FIAU) leading to signal amplification by ~30-fold in an experimental model of metastatic melanoma. We have also compared the [125I]FIAU-SPECT method with that of FDG in terms of its capacity to detected widespread metastases in experimental models. Although both molecular-genetic-based SPECT ([125I]FIAU) and FDG-PET imaging could detect lung nodules, the specificity of molecular-genetic-based imaging surpassed FDG-PET imaging in areas in close proximity to heart and brown fat of the mice, tissues that are FDG-avid in this model system (Bhang et al., 2011). HSV1-TK, being an enzyme, amplifies its signal on addition of the substrate and is therefore preferred over certain receptor-based reporter genes, which have a 1:1 relationship with their ligands (Kang & Chung, 2008).
Of current clinically relevant imaging modalities, MRI likely provides the best spatial resolution for deep tissues. It is also frequently used for functional studies and a variety of MR-based molecular imaging reporters are emerging (Airan, Li, Gilad, & Pelled, 2013). The most common MRI reporter genes used are enzyme-based, e.g., creatine kinase (CK), β-galactosidase (β-gal), and TYR, whereas nonenzyme-based iron (Fe) transporters or interacting partners such as transferrin (Tf), e.g., Tf-MION (monocrystalline iron oxide nanocompounds), can also be used to study the expression of human transferrin receptor utilizing MRI. Although MR-based molecular-genetic imaging is an emerging technology that can provide high spatial resolution in deep tissues, and the signal can be coregistered with functional and molecular tissue information, its primary limitation is sensitivity (Table 4.2).
Considering the advantages/applications and limitations of each modality, multimodality imaging modules may provide superior and complementary information on molecular events in vivo by leveraging the benefits of companion modalities. For example, the high spatial resolution of MRI can complement the high sensitivity of PET or BLI to provide an enhanced image that carries more information than any single modality can provide. Moreover, it has been shown that instead of using fused or different reporter genes for different imaging modalities, a single reporter gene that can be utilized for different imaging modalities, namely, a multimodality reporter gene, can also be engineered. For example, Qin et al. showed that the TYR reporter gene system, with melanin as multifunctional target, could be used to perform MRI by virtue of chelation of iron by melanin, PET ([18F]P3BZA), and Cerenkov luminescence imaging. Because of its compatibility with materials that can be administered to human subjects, this multimodality reporter gene has the potential to be translated into the clinic (Qin et al., 2013). Additional information on the various reporter genes used for molecular-genetic imaging can be found elsewhere (Doubrovin, Serganova, Mayer-Kuckuk, Ponomarev, & Blasberg, 2004; Gross & Piwnica-Worms, 2005; Kang & Chung, 2008).
4. SIGNAL ENHANCEMENT OF REPORTERS
One of the ways to achieve a higher target-to-background ratio for in vivo molecular-genetic imaging is to increase the expression level of the reporter transgenes. Such a strategy would particularly benefit many tissue-specific promoters, which are relatively weak compared with the strong viral promoters generally used to express transgenes (Bhang et al., 2011; Ribault et al., 2001). There are several factors that determine the expression level of a reporter, including strength of the promoter, the existence of enhancers, codon usage bias of a reporter gene, and presence of introns. There have been several attempts to enhance the expression of imaging reporter transgenes through genetic engineering, as summarized below.
4.1. Enhancers
An enhancer is a small (50–1500 bp) cis-acting DNA region to which gene-specific transcription factors can bind, enabling them to interact with basic transcription factors and RNA polymerase II (Maston, Evans, & Green, 2006; Pennacchio, Bickmore, Dean, Nobrega, & Bejerano, 2013). Generally those interactions increase the level of expression of the gene that they regulate. The location of enhancers can be either upstream or downstream of the gene and they perform their function of enhancement regardless of their orientation. Some enhancers have been found to be located at over 100,000 base pairs distant from the gene that they enhance or within the intron (Pennacchio et al., 2013). Strong enhancers have been successfully added to expression vectors, including the human cytomegalovirus (CMV) immediate-early enhancer (Boshart et al., 1985; Gruh et al., 2008) and simian virus 40 (SV40) enhancer (Haas, Ramanujam, Chandrasekharappa, & Subramanian, 1991; Luke et al., 2011). Enhancers from certain gene promoters have also been used to increase promoter-dependent, tissue-specific expression of a reporter. Examples include the SM22α promoter/creatine kinase enhancer for smooth muscle-specific expression (Ribault et al., 2001), PSA promoter/enhancer for prostate cancer-specific expression (Latham, Searle, Mautner, & James, 2000), endothelin-1 promoter/enhancer for endothelial cell-selective expression (Dronadula et al., 2011), and uroplakin II promoter/prostate stem cell antigen enhancer for bladder cancer-specific expression (Wang et al., 2010). Recently, Watanabe et al. developed the super gene expression (SGE) system to take full advantage of transcriptional enhancers for further enhancing expression. They inserted triple enhancer sequences composed of hTERT, SV40, and CMV downstream of the sequence of the bovine growth hormone poly-A under the control of the CMV promoter. They constructed that SGE system into an adenoviral expression system and compared the expression efficiency with CMV, Rous sarcoma virus, and CMV early enhancer/ chicken β-actin (CAG) promoters. The SGE system demonstrated superior expression efficiency over conventional promoters tested in vitro. In addition, the SGE system with a tumor suppressor protein (reduced expression in immortalized cells (REIC)/Dickkopf-3 (Dkk-3)) exhibited the highest anticancer effect on the subcutaneous tumor growth of RENCA mouse renal cell carcinoma cells in BALB/C mice (Watanabe et al., 2014).
4.2. Two-step transcriptional amplification
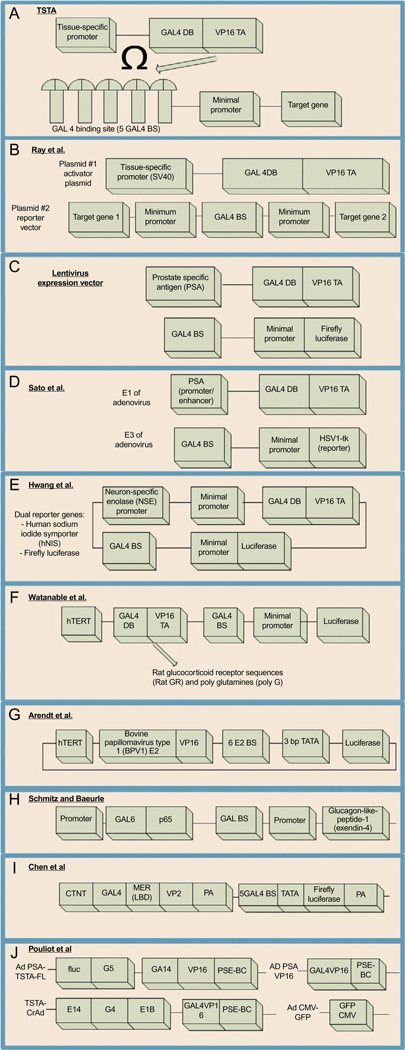
The concept of two-step transcriptional amplification (TSTA), where a weak promoter of interest, e.g., a tissue-specific promoter, drives the expression of a chimeric protein composed of a specific DNA-binding domain and a strong transcriptional activator, was tested by a German group in 1998 (Nettelbeck, Jerome, & Muller, 1998). In that study, Nettlebeck et al. fused the DNA-binding domain of the Lex A repressor to a strong VP16 activator of HSV1 to create a positive feedback loop of expression initiated by the endothelial cell-specific von Willebrand factor promoter. That system achieved an approximately 100-fold increase in expression while maintaining tissue specificity. A group at Stanford University further developed that new concept into the TSTA system for molecular-genetic imaging with modifications. The TSTA approach (Iyer et al., 2001) took advantage of the GAL4/upstream activation sequence (UAS) system (Brand & Perrimon, 1993) that has been used for studying gene expression and function in organisms such as D. melanogaster. The first step is to express a fusion protein composed of the GAL4 DNA-binding domain and VP16 transactivation domain (GAL4-VP16) via a promoter of interest. The GAL4-VP16 fusion protein, as a second step, will bind to a UAS located upstream of a reporter and express it. The strong transcriptional strength of VP16 and multiple copies of UAS upstream of the reporter gene enable a significant increase in the expression level of the corresponding reporter. The original TSTA attempt with the PSA promoter reported an approximately 50-fold increase in fLuc activity in a prostate cancer cell line while maintaining tissue selectivity (Iyer et al., 2001). The magnitude of enhanced signals using the TSTA strategy could strengthen the clinical utility of molecular-genetic imaging. Additionally, TSTA can be modified to equip the molecular-genetic imaging system with various additional features. We summarize recent efforts to enhance the TSTA system further below (Fig. 4.3).
Figure 4.3. See legend on next page.
Schematic diagrams of various TSTA approaches. (A) Original TSTA approach. A weak, tissue-specific promoter is used to drive the expression of a fusion protein that will bind to its binding sites and amplify the expression of a target gene. (B) Bidirectional vector with two target genes of interest that would be amplified when bound to the chi-meric protein created as a result of the tissue-specific promoter SV40. (C) Lenti-TSTA. The TSTA was inserted into a lentivirus expression system with the PSA promoter to amplify expression of a reporter gene such as fLuc. (D) Adeno-TSTA. The system was optimized to utilize the E1 and E3 segments of the adenovirus to enhance expression of the reporter gene HSV1-tk using PSA acting as a promoter for synthesis of the chimeric protein. (E) TSTA system for imaging cellular processes such as neuronal differentiation via the NSE promoter. (F) Addition of rat glucocorticoid receptor sequences (Rat GR) and poly-glutamines (polyG)in between the GAL4 and VP16 domains to increase efficiency of luciferase expression. (G) Replacing GAL4-UAS with a bovine papillomavirus 1 transcriptional activator. (H) Replacing VP-16 with p65. (I) Titrable TSTA using the cardiac troponin T (CTNT) promoter and the MER sequence responsive to raloxifene. (J) Dual TSTA system (dTSTA) for the expression of a reporter and a conditionally replicating Ad.
4.2.1 Bidirectional TSTA
Ray et al. developed a bidirectional vector system, where they constructed two independent UAS-reporter cassettes in the same vector to enable simultaneous expression of two reporters using one promoter that drives the expression of GAL4-VP16 (Ray et al., 2004). Interestingly, they used two separate plasmids, one for the UAS-reporter cassettes (reporter vector) and another for promoter-GAL4-VP16 (activator plasmid), and cotransfected these plasmids to target cells or animal models bearing xenograft tumors. When they cotransfected cells in vitro with varying doses of the activator plasmid they found a high correlation between the dose and the expression level of the reporters. Similar correlations were also observed in their two in vivo models (xenograft of cells cotransfected with vectors or hydrodynamic delivery of vectors to xenograft tumors). That approach will enable flexible applications for: (1) multimodal imaging when using two imaging reporters, (2) theranostic applications when using one imaging reporter and one therapeutic gene, and (3) dual-targeting therapy when using two different therapeutic genes. It will be important and interesting to compare the delivery and transfection efficiencies between cotransfecting two small vectors and transfecting one large vector harboring both the promoter-GAL4-VP16 and UAS-reporter cassettes.
4.2.2 Lentivirus-TSTA
The TSTA system has been successfully adapted to virus-mediated gene transfer tools. TSTA was cloned into a third-generation lentivirus (LV) expression vector with a promoter of PSA and fLuc as a reporter. This LV-TSTA efficiently infected xenograft prostate tumor when injected directly to the tumors and the luciferase activity lasted up to 3 months post-injection. LV-TSTA showed the possibility of prolonged and strong expression of a reporter driven by a weak, tissue-specific promoter. That will allow long-term monitoring of tumor development without invasive procedures (Iyer et al., 2004).
4.2.3 Adeno-TSTA
An approach similar to the LV-TSTA was taken for an adenoviral vector. Sato et al. applied the TSTA system with a PSA promoter/enhancer and HSV1-tk as a reporter. After successful introduction of the TSTA system to the adenoviral expression system, they tried to optimize the adeno-TSTA system for further enhancement of reporter expression. They explored various configurations of the TSTA system in adenoviral vectors and found that the physical separation of the two TSTA components via cloning PSA promoter/enhancer driven-GAL4-VP16 and UAS-HSV1-tk into E1 and E3 regions of adenovirus, respectively, enhanced the tissue-specific expression of the corresponding reporter. That adenoviral TSTA system proved capable of detecting lung metastasis of prostate cancer in animal models via PET (Sato et al., 2008).
4.2.4 Advanced TSTA system
Watanabe et al. (2011) tried to enhance further the transcriptional efficiency of a TSTA system by introducing poly-glutamines and rat glucocorticoid receptor sequences between the GAL4 and VP16 sequences. It has been reported that introduction of a homo-polymeric stretch of glutamine or proline could enhance the transcriptional activity of a transcription factor (Gerber et al., 1994). That “advanced TSTA system” with the cancer-selective hTERT promoter exhibited an approximately 17-fold increase in in vitro transfection efficiency compared with a conventional TSTA system. The authors created an adeno-associated virus harboring the advanced TSTA system and tested it in an orthotopic liver tumor model. in vivo BLI showed an approximately 16-fold increase in signal intensity for the advanced TSTA system compared with that from a conventional TSTA system.
4.2.5 Replacing components of the TSTA
Arendt et al. replaced GAL4 with the bovine papillomavirus 1 transcriptional activator E2 to create an E2-VP16 fusion protein. The UAS was also replaced with E2 binding sites followed by a minimal bovine papillomavirus 4 promoter containing three base pairs of the promoter sequence upstream from the TATA box (Arendt, Nasir, & Morgan, 2009). VP16-E2 proved to be superior to Gal4-VP16 as the transcriptional activator in a TSTA system driven by either of the two potentially cancer-specific promoters, telomerase RNA and hTERT, in several cell lines (Arendt et al., 2009). Another widely used transactivator is p65, a subunit of human NF-κB (Schmitz & Baeuerle, 1991). Gal6-p65 was tested for neuronal cell-selective expression using human synapsin-1 promoter (Liu et al., 2006). The TSTA system with p65 was also used to deliver a plasmid expressing glucagon-like peptide-1 (a.k.a. exendin-4) to diabetic mice (Kim, Lee, Nam, & Kim, 2013; Lee et al., 2006).
4.2.6 Titratable TSTA
In an effort to control further the tissue-specific expression of reporter genes, a titratable TSTA (tTSTA) system has been developed (Chen et al., 2014). tTSTA uses the cardiac troponin T promoter to drive the expression of the Gal4-mER(LBD)-VP2 and fLuc as the reporter. The Gal4-mER(LBD)-VP2 requires the binding of a mutant estrogen receptor (ERG521T) ligand binding domain (LBD) to an ER ligand such as raloxifene to act as a transactivator. Intraperitoneal injection of raloxifene after the intracardiac administration of the tTSTA plasmid led to an approximately 10-fold increase in luciferase activity selectively in cardiac tissues. That strategy can enhance benefit-to-risk ratio of future cardiac gene therapies.
4.2.7 Dual TSTA
Recently, Pouliot et al. have tried to augment further the expression efficiency of a TSTA system (Pouliot et al., 2013). They developed a dual TSTA system where the transactivator drives the expression of not only a reporter but also conditionally replicating adenovirus, which allows genomic amplification of infected adenoviral vectors. That approach showed an approximately 50-fold increase in reporter genome and 25-fold increase in the reporter activity in vivo compared with a conventional TSTA system. That would allow lowering the dose of viral administration while maintaining similar imaging or therapeutic efficacies in a clinical setting.
4.2.8 TSTA for imaging cellular differentiation
A combination of a TSTA system and a tissue-specific promoter was explored to track and image neuronal differentiation in vivo. Hwang et al. constructed an imaging vector composed of a TSTA system with a neuron-specific enolase (NSE) promoter and dual reporter genes [human sodium iodide symporter (hNIS) and fLuc]. The TSTA system enhanced signal intensity of fLuc driven by the NSE promoter by 130-fold. That system was able to detect differentiated neurons in vivo. This strategy can be used for tracking and imaging the differentiation of transplanted stem cells in vivo (Hwang et al., 2008). In addition, this study showed an example of the utility of the TSTA system for visualizing various cellular processes in vivo.
4.3. Codon optimization
The genetic code is degenerate as it has 64 different codons for 20 amino acids and transcriptional stop signs. Different species often have a preference for a particular codon for encoding an amino acid (Comeron & Aguade, 1998). That codon usage bias often makes it less efficient to express reporter genes from different species. For this reason, reporter genes of nonhuman origin have been optimized for their codon usage bias by replacing codons (DNA sequences) with the ones more frequently used in humans. An excellent example of that involves an attempt at the optimization of GFP as a reporter from Aequorea victoria (Yang, Cheng, & Kain, 1996). The humanized gLuc via codon optimization exhibited an approximately 1000-fold increase in signal intensity compared with its wild-type isolated from another marine organism, Gaussia princeps (Tannous et al., 2005). Recently, codon-optimized fLuc with a single mutation (S284T) has been shown to emit a red-shifted bioluminescent signal with enhanced intensity in human glioma cells (Caysa et al., 2009). Software is available online to aid in optimization of codons for desired species (Fox & Erill, 2010).
4.4. Posttranscriptional regulatory elements
Once messages are transcribed, their expression levels can be enhanced further at the translational level. Posttranscriptional regulatory elements (PTREs) are known to regulate the synthesis of polypeptides by modifying the efficiency of translation. PTREs include untranslated regions (UTRs) of mRNA, introns, poly-A signal, the Kozak sequence, and signal peptides (Luo & Reed, 1999; Makrides, 1999; Zhang, Leng, & Mixson, 2005; Zhao, Hyman, & Moore, 1999). Some of those PTREs have been tested for enhancing expression levels of genes of interest in mammalian expression systems. Mariati et al. have performed an informative study to compare the efficiencies of five different PTREs for enhancing the expression of target genes in human and Chinese hamster ovary cells (Mariati, Ho, Yap, Yap, & Yang, 2012). They tested: (1) 5′ UTR of the human heat shock protein 70, (2) 5′ UTR of the 163-bp splice variant of VEGF, (3) 5′ UTR of the tripartite leader sequence of human adenovirus linked with the major late promoter enhancer, (4) the first intron of human CMV immediate-early gene (intron A), and (5) woodchuck hepatitis virus PTRE. Among those, the tripartite leader sequence of human adenovirus 5′ UTR was found to provide the most universal and highest enhancement of the expression levels. Introduction of efficient PTRE into the expression vector will further enhance the efficiency per copy of the gene, especially when combined with transcriptional level mechanisms such as enhancers or TSTA. It will be critical to choose and optimize an appropriate PTRE for each specific application.
4.5. Synthetic super promoter
Another approach to accomplish enhancement of target gene expression was to create a synthetic promoter with elevated transcriptional ability. Two different methodologies have been attempted: (1) rational design of a synthetic promoter by combining known regulatory elements of strong promoters and (2) screening a library of short DNA sequences to identify strong enhancers and reconstituting these with known strong promoters. An example of the rational design method is the super core promoter 1 (SCP1) developed by Juven-Gershon, Cheng, and Kadonaga (2006). The SCP1 was created by combining four core promoter motifs: (1) the TATA box, (2) the initiator, (3) the motif ten element, and (4) the downstream promoter element. The SCP1 promoter exhibited elevated transcriptional activity compared with the CMV promoter and the adenovirus major late promoter both in vitro and in vivo, demonstrating the possibility of rational design of super promoters. A screening method has been developed by Schlabach et al., where they created a library of 52,429 unique 100-mer DNA pieces (Schlabach, Hu, Li, & Elledge, 2010). That library was built by printing all possible 10-mer DNA sequences on microarray as a 10 tandem repeats of same 10-mer sequence. They constructed a retroviral vector with the library upstream of the CMV minimal promoter and tested the promoter activity for expression of GFP in human cells. They found several 100-mer synthetic enhancers that were superior to the CMV promoter. These synthetic promoters demonstrated the proof-of-principle for development of synthetic target-specific and strong promoters for molecular-genetic imaging.
4.6. Introducing introns
Higher eukaryotic genes contain introns. Introns and their removal during transcription can significantly affect the efficiency of gene expression (Niu & Yang, 2011). Effects on gene expression efficiency could occur through many levels of gene expression including splicing, polyadenylation, mRNA export, translational efficiency, and mRNA stability. It has been reported that the presence of the intron could enhance the expression level of eukaryotic genes (Gruss, Lai, Dhar, & Khoury, 1979; Hamer, Smith, Boyer, & Leder, 1979). Nott et al. tested the effect of introducing an intron to an imaging vector with rLuc (Nott, Meislin, & Moore, 2003). They added an intron from human triose phosphate isomerase gene into either the 5′- or 3′-end of the rLuc gene and compared the expression levels. They found that the expression levels in HeLa cells were increased by 29-fold for 5′ insertion and 8-fold for 3′ insertion. They also found enhanced mRNA accumulation and higher translational yields.
5. PROLONGED EXPRESSION OF REPORTERS
The majority of nonviral expression systems provide only transient expression of a reporter gene, which is not suitable for situations that require long-term monitoring of targets or prolonged expression of therapeutic genes. Additionally, the rapid development of cell-based therapies for various diseases such as cancer, neurodegenerative disease, cardiovascular disease, and macular degeneration has necessitated noninvasive methods to track and monitor transplanted cells for relatively longer periods of time—longitudinally—than for the imaging methods described earlier. Conventional molecular-genetic imaging vectors that lack the machinery for self-replication will be gradually lost as the transfected cells divide and cannot be used for these indications. Expression vectors with a viral origin of replication, such as Epstein–Barr virus replication origin (OriP) and Epstein–Barr virus nuclear antigen 1 (EBNA-1), have enabled prolonged expression of genes (Jackson, Juranek, & Lipps, 2006; Stoll et al., 2001).
Another suggested mechanism of transient expression of transfected nonviral expression vectors involves rapid epigenetic changes such as methylation of promoters by host cells (Brooks et al., 2004). There have been several approaches to overcome that phenomenon. Among them, three approaches have been reported to enhance expression levels: (1) introducing ubiquitous chromatin opening elements (UCOEs), (2) introducing the stabilizing and antirepressor (STAR) element, and (3) introducing scaffold/ matrix attachment regions (S/MARs). The UCOEs are the regulatory elements of abundant housekeeping genes, which maintain a highly acetylated status of the genes for constitutive expression. Key examples of UCOEs are from HNRPA2B1 and CBX3 genes (Williams et al., 2005). Combining UCOE with the CMV promoter enhanced the expression efficiency up to 16-fold (Benton et al., 2002). STAR elements were identified through a screening for DNA sequences capable of blocking transcriptional repression (Kwaks et al., 2003). Those STAR elements enhanced the expression level when incorporated into expression vectors in different cell lines, and the expression levels were stable for up to 60 generations of cells transfected with the vector without any selection machinery, such as antibiotic-resistant genes. S/MARs are DNA regions that physically bind to nuclear matrix, creating loop-like structures of chromatin inside the nucleus (Boulikas, 1993). Although the molecular mechanisms accounting for the prevention of gene silencing by S/MARs have not been clearly identified, some S/ MARs, such as human β-interferon gene cluster S/MAR sequence, have been shown to increase the stability of the expression vector enabling maintenance of the vector as an episomal plasmid ( Jenke et al., 2002). In addition, being present at the matrix attachment regions increases the chance to interact with host replication machinery, enabling replication of the expression vectors as the host cell divides. The episomal vectors capable of replicating and maintaining the expression of reporters will enable longitudinal molecular-genetic imaging without multiple administrations of vectors to patients.
6. MACHINERY FOR GENE DELIVERY
Delivering plasmid vectors efficiently to target is the most important yet most challenging step toward successful application of molecular-genetic imaging. Viral vectors such as adenovirus, adeno-associated virus, LV, HSV, poxvirus, and Ebstein–Barr virus have been used to deliver imaging vectors. Those viral vectors generally have higher transfection efficiencies than nonviral strategies. The potential insertional mutagenesis, however, caused by residual viral elements, difficulty with systemic delivery and the possibility of immunogenicity make viral approaches largely unsuitable for clinical translation, particularly if systemic administration of the vectors is contemplated. In this section, we focus on nonviral delivery methods using biocompatible materials with various modifications to enhance the efficacy of gene delivery.
As polyanionic macromolecules, DNA vectors must overcome several barriers to be efficiently delivered into the nucleus of target cells, where the reporter genes are transcribed. First, vectors must cross the negatively charged plasma membrane of target cells. Second, endocytosed vectors must be released from endosomes into the cytoplasm. Third, released vectors should enter the nucleus. To facilitate transferring vectors through those barriers, several nanoparticle-based strategies have been employed including cationic polymers (polyplex), lipids with a positive charge (lipoplexes), and nanoparticles (nanoplex).
6.1. Cationic polymers (polyplexes)
DNA molecules interact with cationic polymers through electrostatic interactions resulting in formation of condensed polyplexes of nanosize range and increased permeability through plasma membranes. N/P ratio (charge ratio) between polymers and DNA can be optimized to enhance the transfection efficiency of polyplexes (Wolff & Rozema, 2008). PEI, cationic polypeptides such as poly-l-lysine and protamine, biodegradable cationic polysaccharides such as chitosan, and poly-β-amino ester (PBAE) are examples of the most widely used polycationic polymers (Sun & Zhang, 2010). Despite relative high transfection efficiency of cationic polymers, their clinical translation has been hampered by dose-dependent cytotoxicity (Grandinetti, Smith, & Reineke, 2012). In order to minimize the toxicity of cationic polymers, various surface modifications have been attempted. The most common approach has been to attach biocompatible polymers, such as polyethylene glycol (PEG) (Shim & Kwon, 2010). PEGylation may reduce the cytotoxicity of the cationic polymer and protect polyplexes from excessive protein binding and macrophage phagocytosis, but it also affects the formation of essential nonbilayer intermediates resulting in reduction of transfection efficiency (Drummond, Zignani, & Leroux, 2000). Jiang et al. controlled the shape of polyplexes by manipulating solvent polarity to enhance transcriptional efficiency with PEG ( Jiang et al., 2013). Other examples of modifications include pH-triggered deshielding of PEI-polyplexes to improve endosomal release (Walker et al., 2005). Another approach to reduce cytotoxicity and enhance the transcriptional efficacy is to modify the surface of the polyplex by attaching a targeting moiety. For example, Zugates et al. added aminogalactose to PBAE as a targeting ligand that binds to asialoglycoprotein receptor expressed on the surface of human hepatocellular carcinoma and liver cells (Zugates et al., 2007).
6.2. Positively charged lipids (lipoplexes)
Liposomes with positive charges (lipoplexes) form electrostatic interactions with negatively charged DNA vectors. Lipoplexes have been used heavily as in vitro experimental tools but also have been successfully used for efficient systemic delivery of DNA vectors in vivo (Kurosaki et al., 2008). Targeted lipoplexes have been tested by conjugating antibodies (Ab), sugars, and small ligands for surface receptors. Examples of Ab used to target liposomes are anti-HER-2 Ab-targeting HER-2/neu protein on breast cancer cells (Siwak, Tari, & Lopez-Berestein, 2002), anti-intracellular adhesion molecule (ICAM-1) Ab-targeting tumor endothelial cells (Almenarqueralt, Duperray, Miles, Felez, & Altieri, 1995), antivascular cell adhesion factor (VACAM-1) Ab-targeting tumor neovasculature (Gosk, Moos, Gottstein, & Bendas, 2008), and anti-RON receptor Ab-targeting acute hypoxic cancer cells (Guin et al., 2011). Sugar has been used to modify lipids to generate cationic glycolipids to target liver cancer cells expressing asialoglycoprotein (Srinivas, Samanta, & Chaudhuri, 2009). Ligand-targeting folate receptors on folic acid overexpressing cancer cells were used to formulate liposomes (Gorle, Ariatti, & Singh, 2014). Small peptide (RGDK) targeting integrin α5β1 has been implemented to create lipopeptide to deliver the p53 tumor suppressor gene to an in vivo model of tumor vasculature (Pramanik et al., 2008). In order to enhance further transfection efficiency, core–shell lipids have recently been developed for gene delivery, where liposome and other nucleic acid condensing materials have been used simultaneously. Harashima et al. created a multifunctional nanodevice (MEND) composed of DNA core condensed with cationic polymer surrounded by the liposome (Hatakeyama, Akita, & Harashima, 2011). The surface of the MEND liposome has been further modified by addition of protease-cleavable PEG for removal of PEG at the target site (Hatakeyama et al., 2007) and fusogenic peptide that forms alpha helix at acidic endosomal pH to improve endosomal release of the liposome (Sakurai et al., 2011). The long used calcium-phosphate principle of nucleic acid precipitation has been utilized for gene delivery, where the system has nanocalcium-phosphate-DNA as a core coated by cationic lipid shell (Zhou et al., 2010). Chitosan has also been used as a nucleic acid condensing core material for ocular therapy ( Jiang et al., 2012).
6.3. Nanoparticles (nanoplexes)
Various types of nanoparticles have recently been tested for in vivo delivery of genetic materials. The surface of gold nanorods has been modified to interact with double-stranded DNA. Since gold nanorods absorb NIR light and generate heat, irradiation of dsDNA-gold nanorod nanoplexes results in releasing the dsDNA, enabling controlled release of delivered vectors (Yamashita et al., 2011). Lee et al. created multilayered gold nanoplexes by layer-by-layer fabrication of poly-l-lysine and nucleic acid based on electrostatic interaction. That approach enabled slow, gradual release of the therapeutic gene (Lee, Han, Asokan, & Tung, 2011). Polyamidoamine (PAMAM) dendrimers have also been used for gene delivery utilizing their high solubility in aqueous solution, defined size and shape, and abundance of primary amino group on the surface (Navarro & Tros de Ilarduya, 2009). Liu et al. created disulfide crossed-linked dendrimers with high transfection efficiency and relatively low cytotoxicity (Liu, Wang, Yang, & Cheng, 2012). Other examples of dendrimer-based gene delivery are cyclodextrin–dendrimer conjugates (Arima et al., 2010), hyperbranched PAMAM dendrimers (Lee et al., 2003), dendrimers with a triethanolamine core (Zhou et al., 2006), and Jeffamine-cored PAMAM dendrimers (Aydin, Akbas, Senel, & Koc, 2012).
7. SIZE AND IMMUNOGENICITY
It has been tested and reported that the size of nonviral expression vectors can affect the transfection efficiency to mammalian cells. Kreisse et al. have tested the effect of plasmid DNA size on the ability to form lipoplex and transfection efficiency (Kreiss et al., 1999). They found that the size of the plasmid did not alter the formation of lipoplex but larger plasmids had lower transfection efficiencies. They also found that the smaller plasmid DNA had superior nuclear transportation efficiency. Later, Lukacs et al. discovered that plasmid DNA larger than 2000 base pairs was not capable of freely diffusing into the cytoplasm and virtually every size of DNA was immobile in the nucleus (Lukacs et al., 2000). A systematic study of the effect of the size of plasmid DNA on transfection efficiency was performed by Yin et al., where they discovered that the expression level of a reporter gene was inversely proportional to the size of plasmid DNA tested (Yin, Xiang, & Li, 2005). Interestingly, expression levels sharply declined with a plasmid size of 5100 base pairs or larger, providing a theoretical maximum reference size of expression vectors for efficient transfection.
In addition to the size of vectors, there are other elements that could affect the overall transfection efficiency. Once plasmid enters into the cytoplasm it is exposed to the attack of various exo- and/or endonucleases, resulting in rapid degradation. It has been reported that certain secondary structures with a single strand region, e.g., a poly-A site, is typically vulnerable to such attack (Azzoni, Ribeiro, Monteiro, & Prazeres, 2007). Replacing such regions with synthetic counterparts could enhance the stability of the vectors (Ribeiro, Monteiro, & Prazeres, 2004).
Although nonviral gene delivery vectors are generally considered to be safer compared with viral vectors for clinical applications, there are still remaining safety concerns that need to be avoided when designing such vectors. Chromosomal insertion of plasmid vectors via homologous recombination could happen (Ledwith et al., 2000; Manam et al., 2000; Nichols, Ledwith, Manam, & Troilo, 1995; Wang et al., 2004); therefore, it is strongly recommended to avoid any sequences homologous to human DNA. Sequences from bacterial origin such as antibiotic-resistant genes for selection and bacterial replication origin, and other unnecessary plasmid backbones can cause an immune response in humans, and should be avoided (Hartmann & Krieg, 1999; McMahon, Wells, Bamfo, Cartwright, & Wells, 1998; Yew et al., 1999). For example, the ampicillin resistance gene (AmpR), the most commonly used antibiotic-resistant gene, is not suitable for clinical use due to potential hyperreactivity of some patients to β-lactam antibiotics causing an allergic reaction to residual contamination in the final product (Carnes & Williams, 2007).
In order to avoid those safety issues and minimize the size of plasmid vectors, several strategies have been developed. One approach is to replace the relatively large antibiotic-resistant gene with a short essential gene or suppressor tRNA with an appropriate modification on the host bacterial chromosome. Soubrier et al. have developed a new vector (pCOR) for human gene therapy (Soubrier et al., 1999). pCOR has a R6K γ replication origin (400 base pairs), synthetic amber suppressor tRNA (200 base pairs) for selection, and ColE1 resolution fragment (400 base pairs) for preventing plasmid multimerization. The selection was done using arginine-deprived media as the synthetic amber suppressor tRNA is required for the synthesis of argE gene essential for arginine biosynthesis. Another approach was to excise unnecessary elements (e.g., antibiotic-resistant gene, bacterial replication origin, and prokaryote backbone) from the plasmid post-production by site-specific recombination (Chen, He, & Kay, 2005; Darquet, Cameron, Wils, Scherman, & Crouzet, 1997). The resultant minicircle plasmid possessed the minimum elements required for the expression of the gene of interest in human cells. The only drawback of that approach is the relatively high cost of production due to the additional steps of recombination and purification.
8. CONCLUDING REMARKS
Clinical translation of molecular-genetic imaging is challenging due to the multiple components that require concurrent optimization, namely, the imaging transgene, delivery vehicle, and imaging agent. Nevertheless, there are two strategies that could render molecular-genetic imaging viable in clinical practice. First, the imaging system should be developed for a broad spectrum of applications. Examples would include use of near-universal, cancer-specific (rather than tissue-specific) promoters, or promoters that show specific activity in cancer stem cells. Second, combining an imaging aspect with a therapeutic component to create a theranostic agent would provide substantial impetus to the development of in vivo molecular-genetic imaging systems. That could be accomplished using a reporter gene that can act as a therapeutic gene concurrently, e.g., HSV1-tk and hNIS, or by employing dual expression vectors, e.g., bidirectional TSTA, as discussed earlier. Other potential theranostic reporters are emerging. For example, we recently tested the prostate-specific membrane antigen (PSMA) as an imaging reporter and showed it performed as well or better than HSV1-tk or hNIS for imaging of animal models of human cancer (Castanares et al., 2014). PSMA would be a good candidate for a theranostic reporter as many imaging and therapeutic probes have already been developed for this human protein (Banerjee et al., 2008; Chen et al., 2009, 2008; Foss et al., 2005; Hillier et al., 2009; Mease et al., 2008; Pomper et al., 2002; Shallal et al., 2014), including several radiolabeled molecules that are in clinical trials (Barrett et al., 2013; Cho et al., 2012; Hillier et al., 2011).
In summary, criteria for clinical translation of molecular-genetic imaging include: (1) a nearuniversal promoter, (2) a translatable reporter-probe pair, (3) strong expression of the reporter, (4) an optimal size for the plasmid vector, (5) lack of homologous sequences to the human genome to avoid unwanted insertion, (6) lack of bacterial sequences to prevent unfavorable immune reactions, and (7) an efficient delivery vehicle with enhanced capacity for transcription (Table 4.3). Combination with a therapeutic aspect provides motivation to pursue imaging concurrently. As each of those criteria are currently under intensive optimization worldwide, and gene therapy is becoming more effective and less restricted, we anticipate that molecular-genetic imaging will soon see widespread clinical translation.
Table 4.3.
Criteria for clinical translation of molecular-genetic imaging
| Criteria for a translatable system | Approaches to meet the criteria |
|---|---|
| Universal promoter for target applications | Universal cancer-specific promoters |
| Translatable reporter-probe pair | Reporter-probe pairs already in clinical use |
| Reporter-probe pairs with better efficiency | |
| Strong expression of reporter | Enhancers |
| Two-step transcriptional amplification | |
| Codon optimization | |
| Posttranscriptional regulatory elements | |
| Introducing introns | |
| Synthetic promoters | |
| Mammalian replication origin | |
| Optimal size of plasmid vector | Minicircle vector |
| Lack of homologous sequence to human genome to avoid unwanted insertion | Replacing antibiotic-resistant gene |
| Minicircle vector | |
| Lack of bacterial sequence to prevent unfavorable immune reactions | Replacing antibiotic-resistant gene |
| Minicircle vector | |
| Efficient delivery vehicle with enhance transcription capability | Modified polyplex, lipoplex, and nanoplex |
ACKNOWLEDGMENTS
The authors thank U54 CA151838, R01 CA134675, P50 CA058236, R01 CA134721, P01 CA104177, DoD W81XWH-10-PCRP-SIDA, the A. David Mazzone Awards Program of the Prostate Cancer Foundation, the Goldman Foundation, the National Foundation for Cancer Research, and the Samuel Waxman Cancer Research Foundation for financial support. D. S. is a Harrison Scholar and P. B. F. holds the Thelma Newmeyer Corman Chair in Cancer Research in the VCU MCC.
REFERENCES
- Airan RD, Li N, Gilad AA, Pelled G. Genetic tools to manipulate MRI contrast. NMR in Biomedicine. 2013;26(7):803–809. doi: 10.1002/nbm.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almenarqueralt A, Duperray A, Miles LA, Felez J, Altieri DC. Apical topography and modulation of Icam-1 expression on activated endothelium. American Journal of Pathology. 1995;147(5):1278–1288. [PMC free article] [PubMed] [Google Scholar]
- Arendt ML, Nasir L, Morgan IM. A novel two-step transcriptional activation system for gene therapy directed toward epithelial cells. Molecular Cancer Therapeutics. 2009;8(12):3244–3254. doi: 10.1158/1535-7163.MCT-09-0543. [DOI] [PubMed] [Google Scholar]
- Arima H, Yamashita S, Mori Y, Hayashi Y, Motoyama K, Hattori K, et al. In vitro and in vivo gene delivery mediated by lactosylated dendrimer/alpha-cyclodextrin conjugates (G2) into hepatocytes. Journal of Controlled Release. 2010;146(1):106–117. doi: 10.1016/j.jconrel.2010.05.030. [DOI] [PubMed] [Google Scholar]
- Aydin Z, Akbas F, Senel M, Koc SN. Evaluation of Jeffamine(R)-cored PAMAM dendrimers as an efficient in vitro gene delivery system. Journal of Biomedical Materials Research. Part A. 2012;100(10):2623–2628. doi: 10.1002/jbm.a.34196. [DOI] [PubMed] [Google Scholar]
- Azzoni AR, Ribeiro SC, Monteiro GA, Prazeres DM. The impact of polyadenylation signals on plasmid nuclease-resistance and transgene expression. Journal of Gene Medicine. 2007;9(5):392–402. doi: 10.1002/jgm.1031. [DOI] [PubMed] [Google Scholar]
- Banerjee SR, Foss CA, Castanares M, Mease RC, Byun Y, Fox JJ, et al. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA) Journal of Medicinal Chemistry. 2008;51(15):4504–4517. doi: 10.1021/jm800111u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JA, Coleman RE, Goldsmith SJ, Vallabhajosula S, Petry NA, Cho S, et al. First-in-man evaluation of 2 high-affinity PSMA-avid small molecules for imaging prostate cancer. Journal of Nuclear Medicine. 2013;54(3):380–387. doi: 10.2967/jnumed.112.111203. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the microphthalmia gene product and role of the initiator. Molecular and Cellular Biology. 1994;14(12):7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton T, Chen T, McEntee M, Fox B, King D, Crombie R, et al. The use of UCOE vectors in combination with a preadapted serum free, suspension cell line allows for rapid production of large quantities of protein. Cytotechnology. 2002;38(1-2):43–46. doi: 10.1023/A:1021141712344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhang HE, Gabrielson KL, Laterra J, Fisher PB, Pomper MG. Tumor-specific imaging through progression elevated gene-3 promoter-driven gene expression. Nature Medicine. 2011;17(1):123–129. doi: 10.1038/nm.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Menezes ME, Das SK, Emdad L, Dasgupta S, Wang XY, et al. Innovative approaches for enhancing cancer gene therapy. Discovery Medicine. 2013;15(84):309–317. [PubMed] [Google Scholar]
- Blasberg RG, Tjuvajev JG. Molecular-genetic imaging: Current and future perspectives. The Journal of Clinical Investigation. 2003;111(11):1620–1629. doi: 10.1172/JCI18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M, Weber F, Jahn G, Dorschhasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalo-virus. Cell. 1985;41(2):521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Nature of DNA-sequences at the attachment regions of genes to the nuclear matrix. Journal of Cellular Biochemistry. 1993;52(1):14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- Branchini BR, Southworth TL, Khattak NF, Michelini E, Roda A. Red-and green-emitting firefly luciferase mutants for bioluminescent reporter applications. Analytical Biochemistry. 2005;345(1):140–148. doi: 10.1016/j.ab.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene-expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brooks AR, Harkins RN, Wang PY, Qian HS, Liu PX, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. Journal of Gene Medicine. 2004;6(4):395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- Carnes AE, Williams JA. Plasmid DNA manufacturing technology. Recent Patents on Biotechnology. 2007;1(2):151–166. doi: 10.2174/187220807780809436. [DOI] [PubMed] [Google Scholar]
- Castanares MA, Mukherjee A, Chowdhury WH, Liu M, Chen Y, Mease RC, et al. Evaluation of prostate-specific membrane antigen as an imaging reporter. Journal of Nuclear Medicine. 2014;55(5):805–811. doi: 10.2967/jnumed.113.134031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caysa H, Jacob R, Muther N, Branchini B, Messerle M, Soling A. A redshifted codon-optimized firefly luciferase is a sensitive reporter for bioluminescence imaging. Photochemical & Photobiological Sciences: Official Journal of the European Photochemistry Association and the European Society for Photobiology. 2009;8(1):52–56. doi: 10.1039/b814566k. [DOI] [PubMed] [Google Scholar]
- Chen Y, Dhara S, Banerjee SR, Byun Y, Pullambhatla M, Mease RC, et al. A low molecular weight PSMA-based fluorescent imaging agent for cancer. Biochemical and Biophysical Research Communications. 2009;390(3):624–629. doi: 10.1016/j.bbrc.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Foss CA, Byun Y, Nimmagadda S, Pullambhatla M, Fox JJ, et al. Radio halogenated prostate-specific membrane antigen (PSMA)-based ureas as imaging agents for prostate cancer. Journal of Medicinal Chemistry. 2008;51(24):7933–7943. doi: 10.1021/jm801055h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, He CY, Kay MA. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Human Gene Therapy. 2005;16(1):126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- Chen IY, Paulmurugan R, Nielsen CH, Wang DS, Chow V, Robbins RC, et al. A titratable two-step transcriptional amplification strategy for targeted gene therapy based on ligand-induced intramolecular folding of a mutant human estrogen receptor. Molecular Imaging and Biology. 2014;16(2):224–234. doi: 10.1007/s11307-013-0673-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SY, Gage KL, Mease RC, Senthamizhchelvan S, Holt DP, Jeffrey-Kwanisai A, et al. Biodistribution, tumor detection, and radiation dosimetry of 18F-DCFBC, a low-molecular-weight inhibitor of prostate-specific membrane antigen, in patients with metastatic prostate cancer. Journal of Nuclear Medicine. 2012;53(12):1883–1891. doi: 10.2967/jnumed.112.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Aguade M. An evaluation of measures of synonymous codon usage bias. Journal of Molecular Evolution. 1998;47(3):268–274. doi: 10.1007/pl00006384. [DOI] [PubMed] [Google Scholar]
- Cronin M, Akin AR, Collins SA, Meganck J, Kim JB, Baban CK, et al. High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS One. 2012;7(1):e30940. doi: 10.1371/journal.pone.0030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darquet AM, Cameron B, Wils P, Scherman D, Crouzet J. A new DNA vehicle for nonviral gene delivery: Supercoiled minicircle. Gene Therapy. 1997;4(12):1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- Doubrovin M, Serganova I, Mayer-Kuckuk P, Ponomarev V, Blasberg RG. Multimodality in vivo molecular-genetic imaging. Bioconjugate Chemistry. 2004;15(6):1376–1388. doi: 10.1021/bc0498572. [DOI] [PubMed] [Google Scholar]
- Dronadula N, Du L, Flynn R, Buckler J, Kho J, Jiang Z, et al. Construction of a novel expression cassette for increasing transgene expression in vivo in endothelial cells of large blood vessels. Gene Therapy. 2011;18(5):501–508. doi: 10.1038/gt.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DC, Zignani M, Leroux JC. Current status of pH-sensitive liposomes in drug delivery. Progress in Lipid Research. 2000;39(5):409–460. doi: 10.1016/s0163-7827(00)00011-4. [DOI] [PubMed] [Google Scholar]
- Foss CA, Mease RC, Fan H, Wang Y, Ravert HT, Dannals RF, et al. Radiolabeled small-molecule ligands for prostate-specific membrane antigen: In vivo imaging in experimental models of prostate cancer. Clinical Cancer Research. 2005;11(11):4022–4028. doi: 10.1158/1078-0432.CCR-04-2690. [DOI] [PubMed] [Google Scholar]
- Fox JM, Erill I. Relative codon adaptation: A generic codon bias index for prediction of gene expression. DNA Research. 2010;17(3):185–196. doi: 10.1093/dnares/dsq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Urata Y, Tanaka N. Telomerase-specific oncolytic virotherapy for human cancer with the hTERT promoter. Current Cancer Drug Targets. 2007;7(2):191–201. doi: 10.2174/156800907780058835. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263(5148):808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Gorle S, Ariatti M, Singh M. Novel serum-tolerant lipoplexes target the folate receptor efficiently. European Journal of Pharmaceutical Sciences. 2014;59:83–93. doi: 10.1016/j.ejps.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Gosk S, Moos T, Gottstein C, Bendas G. VCAM-1 directed immuno-liposomes selectively target tumor vasculature in vivo. Biochimica et Biophysica Acta-Biomembranes. 2008;1778(4):854–863. doi: 10.1016/j.bbamem.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Gotoh A, Ko SC, Shirakawa T, Cheon J, Kao C, Miyamoto T, et al. Development of prostate-specific antigen promoter-based gene therapy for androgen-independent human prostate cancer. The Journal of Urology. 1998;160(1):220–229. [PubMed] [Google Scholar]
- Grandinetti G, Smith AE, Reineke TM. Membrane and nuclear permeabilization by polymeric pDNA vehicles: Efficient method for gene delivery or mechanism of cytotoxicity? Molecular Pharmaceutics. 2012;9(3):523–538. doi: 10.1021/mp200368p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Piwnica-Worms D. Spying on cancer: Molecular imaging in vivo with genetically encoded reporters. Cancer Cell. 2005;7(1):5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Gruh I, Wunderlich S, Winkler M, Schwanke K, Heinke J, Blomer U, et al. Human CMV immediate-early enhancer: A useful tool to enhance cell-type-specific expression from lentiviral vectors. Journal of Gene Medicine. 2008;10(1):21–32. doi: 10.1002/jgm.1122. [DOI] [PubMed] [Google Scholar]
- Gruss P, Lai CJ, Dhar R, Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proceedings of the National Academy of Sciences of the United States of America. 1979;76(9):4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guin S, Ma Q, Padhye S, Zhou YQ, Yao HP, Wang MH. Targeting acute hypoxic cancer cells by doxorubicin-immunoliposomes directed by monoclonal antibodies specific to RON receptor tyrosine kinase. Cancer Chemotherapy and Pharmacology. 2011;67(5):1073–1083. doi: 10.1007/s00280-010-1408-8. [DOI] [PubMed] [Google Scholar]
- Haas MW, Ramanujam P, Chandrasekharappa SC, Subramanian KN. Sequence requirements for activation of replication by the SV40 transcriptional promoter or enhancer elements. Virology. 1991;180(1):41–48. doi: 10.1016/0042-6822(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Hallahan DE, Qu S, Geng L, Cmelak A, Chakravarthy A, Martin W, et al. Radiation-mediated control of drug delivery. American Journal of Clinical Oncology. 2001;24(5):473–480. doi: 10.1097/00000421-200110000-00012. [DOI] [PubMed] [Google Scholar]
- Hamer DH, Smith KD, Boyer SH, Leder P. SV40 recombinants carrying rabbit beta-globin gene coding sequences. Cell. 1979;17(3):725–735. doi: 10.1016/0092-8674(79)90279-4. [DOI] [PubMed] [Google Scholar]
- Hartmann G, Krieg AM. CpG DNA and LPS induce distinct patterns of activation in human monocytes. Gene Therapy. 1999;6(5):893–903. doi: 10.1038/sj.gt.3300880. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Akita H, Harashima H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: A strategy for overcoming the PEG dilemma. Advanced Drug Delivery Reviews. 2011;63(3):152–160. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Akita H, Kogure K, Oishi M, Nagasaki Y, Kihira Y, et al. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Therapy. 2007;14(1):68–77. doi: 10.1038/sj.gt.3302843. [DOI] [PubMed] [Google Scholar]
- Haviv YS, van Houdt WJ, Lu B, Curiel DT, Zhu ZB. Transcriptional targeting in renal cancer cell lines via the human CXCR4 promoter. Molecular Cancer Therapeutics. 2004;3(6):687–691. [PubMed] [Google Scholar]
- Hayat MA. Cancer imaging: Lung and breast carcinomas. Philadelphia, PA: Elsevier Science; 2007. [Google Scholar]
- Hillier SM, Kern AM, Maresca KP, Marquis JC, Eckelman WC, Joyal JL, et al. 123I-MIP-1072, a small-molecule inhibitor of prostate-specific membrane antigen, is effective at monitoring tumor response to taxane therapy. Journal of Nuclear Medicine. 2011;52(7):1087–1093. doi: 10.2967/jnumed.110.086751. [DOI] [PubMed] [Google Scholar]
- Hillier SM, Maresca KP, Femia FJ, Marquis JC, Foss CA, Nguyen N, et al. Preclinical evaluation of novel glutamate-urea-lysine analogues that target prostate-specific membrane antigen as molecular imaging pharmaceuticals for prostate cancer. Cancer Research. 2009;69(17):6932–6940. doi: 10.1158/0008-5472.CAN-09-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DW, Kang JH, Jeong JM, Chung JK, Lee MC, Kim S, et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. European Journal of Nuclear Medicine and Molecular Imaging. 2008;35(1):135–145. doi: 10.1007/s00259-007-0561-8. [DOI] [PubMed] [Google Scholar]
- Iyer M, Salazar FB, Lewis X, Zhang LQ, Carey M, Wu L, et al. Noninvasive imaging of enhanced prostate-specific gene expression using a two-step transcrip-tional amplification-based lentivirus vector. Molecular Therapy. 2004;10(3):545–552. doi: 10.1016/j.ymthe.2004.06.118. [DOI] [PubMed] [Google Scholar]
- Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir SS. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Molecular Therapy. 2006;14(5):613–626. doi: 10.1016/j.ymthe.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Jenke BHC, Fetzer CP, Stehle IM, Jonsson F, Fackelmayer FO, Conradt H, et al. An episomally replicating vector binds to the nuclear matrix protein SAF-A in vivo. EMBO Reports. 2002;3(4):349–354. doi: 10.1093/embo-reports/kvf070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Gan L, Zhu C, Dong Y, Liu J, Gan Y. Cationic core-shell liponanoparticles for ocular gene delivery. Biomaterials. 2012;33(30):7621–7630. doi: 10.1016/j.biomaterials.2012.06.079. [DOI] [PubMed] [Google Scholar]
- Jiang X, Qu W, Pan D, Ren Y, Williford JM, Cui HG, et al. Plasmid-templated shape control of condensed DNA-block copolymer nanoparticles. Advanced Materials. 2013;25(2):227–232. doi: 10.1002/adma.201202932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nature Methods. 2006;3(11):917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- Kang JH, Chung JK. Molecular-genetic imaging based on reporter gene expression. Journal of Nuclear Medicine. 2008;491(Suppl. 2):164S–179S. doi: 10.2967/jnumed.107.045955. [DOI] [PubMed] [Google Scholar]
- Kim PH, Lee M, Nam K, Kim SW. Enhanced incretin effects of exendin-4 expressing chimeric plasmid based on two-step transcription amplification system with dendritic bioreducible polymer for the treatment of Type 2 diabetes. Journal of Gene Therapy. 2013;1(1):7–15. [PMC free article] [PubMed] [Google Scholar]
- Kishimoto H, Kojima T, Watanabe Y, Kagawa S, Fujiwara T, Uno F, et al. In vivo imaging of lymph node metastasis with telomerase-specific replication-selective adenovirus. Nature Medicine. 2006;12(10):1213–1219. doi: 10.1038/nm1404. [DOI] [PubMed] [Google Scholar]
- Kreiss P, Cameron B, Rangara R, Mailhe P, Aguerre-Charriol O, Airiau M, et al. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Research. 1999;27(19):3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y, Onimatsu H, Kojima T, Shirota T, Hatori M, et al. Telomerase-specific virotheranostics for human head and neck cancer. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2009;15(7):2335–2343. doi: 10.1158/1078-0432.CCR-08-2690. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Kitahara T, Teshima M, Nishida K, Nakamura J, Nakashima M, et al. Exploitation of de novo helper-lipids for effective gene delivery. Journal of Pharmacy and Pharmaceutical Sciences. 2008;11(4):56–67. doi: 10.18433/j31s3b. [DOI] [PubMed] [Google Scholar]
- Kwaks THJ, Barnett P, Hemrika W, Siersma T, Sewalt RGAB, Satijn DPE, et al. Identification of anti-repressor elements that confer high and stable protein production in mammalian cells. Nature Biotechnology. 2003;21(7):822. doi: 10.1038/nbt814. (vol. 21, p. 553, 2003) [DOI] [PubMed] [Google Scholar]
- Latham JPF, Searle PF, Mautner V, James ND. Prostate-specific antigen promoter/enhancer driven gene therapy for prostate cancer: Construction and testing of a tissue-specific adenovirus vector. Cancer Research. 2000;60(2):334–341. [PubMed] [Google Scholar]
- Ledwith BJ, Manam S, Troilo PJ, Barnum AB, Pauley CJ, Griffiths TG, 2nd, et al. Plasmid DNA vaccines: Investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology. 2000;43(4-6):258–272. doi: 10.1159/000053993. [DOI] [PubMed] [Google Scholar]
- Lee SK, Han MS, Asokan S, Tung CH. Effective gene silencing by multilayered siRNA-coated gold nanoparticles. Small. 2011;7(3):364–370. doi: 10.1002/smll.201001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lim YB, Choi JS, Lee Y, Kim TI, Kim HJ, et al. Polyplexes assembled with internally quaternized PAMAM-OH dendrimer and plasmid DNA have a neutral surface and gene delivery potency. Bioconjugate Chemistry. 2003;14(6):1214–1221. doi: 10.1021/bc034095g. [DOI] [PubMed] [Google Scholar]
- Lee M, Oh S, Ahn CH, Kim SW, Rhee BD, Ko KS. An efficient GLP-1 expression system using two-step transcription amplification. Journal of Controlled Release. 2006;115(3):316–321. doi: 10.1016/j.jconrel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Liang Q, Dmitriev I, Kashentseva E, Curiel DT, Herschman HR. Non-invasive of adenovirus tumor retargeting in living subjects by a soluble adenovirus receptor-epidermal growth factor (sCAR-EGF) fusion protein. Molecular Imaging and Biology: MIB: The Official Publication of the Academy of Molecular Imaging. 2004;6(6):385–394. doi: 10.1016/j.mibio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Liang Q, Yamamoto M, Curiel DT, Herschman HR. Noninvasive imaging of transcriptionally restricted transgene expression following intratumoral injection of an adenovirus in which the COX-2 promoter drives a reporter gene. Molecular Imaging and Biology: MIB: The Official Publication of the Academy of Molecular Imaging. 2004;6(6):395–404. doi: 10.1016/j.mibio.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang H, Yang W, Cheng Y. Disulfide cross-linked low generation dendrimers with high gene transfection efficacy, low cytotoxicity, and low cost. Journal of the American Chemical Society. 2012;134(42):17680–17687. doi: 10.1021/ja307290j. [DOI] [PubMed] [Google Scholar]
- Liu BH, Yang Y, Paton JFR, Li F, Boulaire J, Kasparov S, et al. GAL4-NF-kappa B fusion protein augments transgene expression from neuronal promoters in the rat brain. Molecular Therapy. 2006;14(6):872–882. doi: 10.1016/j.ymthe.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Lu B, Makhija SK, Nettelbeck DM, Rivera AA, Wang M, Komarova S, et al. Evaluation of tumor-specific promoter activities in melanoma. Gene Therapy. 2005;12(4):330–338. doi: 10.1038/sj.gt.3302385. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. The Journal of Biological Chemistry. 2000;275(3):1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- Luke JM, Vincent JM, Du SX, Gerdemann U, Leen AM, Whalen RG, et al. Improved antibiotic-free plasmid vector design by incorporation of transient expression enhancers. Gene Therapy. 2011;18(4):334–343. doi: 10.1038/gt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrides SC. Components of vectors for gene transfer and expression in mammalian cells. Protein Expression and Purification. 1999;17(2):183–202. doi: 10.1006/prep.1999.1137. [DOI] [PubMed] [Google Scholar]
- Manam S, Ledwith BJ, Barnum AB, Troilo PJ, Pauley CJ, Harper LB, et al. Plasmid DNA vaccines: Tissue distribution and effects of DNA sequence, adjuvants and delivery method on integration into host DNA. Intervirology. 2000;43(4-6):273–281. doi: 10.1159/000053994. [DOI] [PubMed] [Google Scholar]
- Mariati Ho SCL, Yap MGS, Yap MGS, Yang YS. Post-transcriptional regulatory elements for enhancing transient gene expression levels in mammalian cells. Protein Expression in Mammalian Cells: Methods and Protocols. 2012;801:125–135. doi: 10.1007/978-1-61779-352-3_9. [DOI] [PubMed] [Google Scholar]
- Martinelli R, De Simone V. Short and highly efficient synthetic promoters for melanoma-specific gene expression. FEBS Letters. 2005;579(1):153–156. doi: 10.1016/j.febslet.2004.11.068. [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annual Review of Genomics and Human Genetics. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- McMahon JM, Wells KE, Bamfo JE, Cartwright MA, Wells DJ. Inflammatory responses following direct injection of plasmid DNA into skeletal muscle. Gene Therapy. 1998;5(9):1283–1290. doi: 10.1038/sj.gt.3300718. [DOI] [PubMed] [Google Scholar]
- Mease RC, Dusich CL, Foss CA, Ravert HT, Dannals RF, Seidel J, et al. N-[N-[(S)-1,3-Dicarboxypropyl]carbamoyl]-4-[18F]fluorobenzyl-L-cysteine, [18F]DCFBC: A new imaging probe for prostate cancer. Clinical Cancer Research. 2008;14(10):3036–3043. doi: 10.1158/1078-0432.CCR-07-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Lowik C. Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS One. 2011;6(4):e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. First gene therapy nears landmark European market authorization. Nature Biotechnology. 2012;30(9):807–809. doi: 10.1038/nbt0912-807. [DOI] [PubMed] [Google Scholar]
- Nagai T, Miyawaki A. A high-throughput method for development of FRET-based indicators for proteolysis. Biochemical and Biophysical Research Communications. 2004;319(1):72–77. doi: 10.1016/j.bbrc.2004.04.147. [DOI] [PubMed] [Google Scholar]
- Navarro G, Tros de Ilarduya C. Activated and non-activated PAMAM dendrimers for gene delivery in vitro and in vivo. Nanomedicine. 2009;5(3):287–297. doi: 10.1016/j.nano.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Nettelbeck DM, Jerome V, Muller R. A strategy for enhancing the transcriptional activity of weak cell type-specific promoters. Gene Therapy. 1998;5(12):1656–1664. doi: 10.1038/sj.gt.3300778. [DOI] [PubMed] [Google Scholar]
- Nettelbeck DM, Jerome V, Muller R. Gene therapy: Designer promoters for tumour targeting. Trends in Genetics: TIG. 2000;16(4):174–181. doi: 10.1016/s0168-9525(99)01950-2. [DOI] [PubMed] [Google Scholar]
- Nichols WW, Ledwith BJ, Manam SV, Troilo PJ. Potential DNA vaccine integration into host cell genome. Annals of the New York Academy of Sciences. 1995;772:30–39. doi: 10.1111/j.1749-6632.1995.tb44729.x. [DOI] [PubMed] [Google Scholar]
- Niu DK, Yang YF. Why eukaryotic cells use introns to enhance gene expression: Splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity. Biology Direct. 2011;6:24. doi: 10.1186/1745-6150-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9(5):607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Taneja S, Dardashti K, Cohan P, Kaboo R, Sokoloff M, et al. Prostate tissue specificity of the prostate-specific antigen promoter isolated from a patient with prostate cancer. Human Gene Therapy. 1995;6(11):1417–1426. doi: 10.1089/hum.1995.6.11-1417. [DOI] [PubMed] [Google Scholar]
- Papadakis ED, Nicklin SA, Baker AH, White SJ. Promoters and control elements: Designing expression cassettes for gene therapy. Current Gene Therapy. 2004;4(1):89–113. doi: 10.2174/1566523044578077. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim KI, Lee YJ, Lee TS, Kim KM, Nahm SS, et al. Non-invasive monitoring of hepatocellular carcinoma in transgenic mouse with bioluminescent imaging. Cancer Letters. 2011;310(1):53–60. doi: 10.1016/j.canlet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: Five essential questions. Nature Reviews. Genetics. 2013;14(4):288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleshkan VV, Alekseenko IV, Zinovyeva MV, Vinogradova TV, Sverdlov ED. Promoters with cancer cell-specific activity for melanoma gene therapy. Acta Naturae. 2011;3(2):13–21. [PMC free article] [PubMed] [Google Scholar]
- Pomper MG, Musachio JL, Zhang J, Scheffel U, Zhou Y, Hilton J, et al. 11C–MCG: Synthesis, uptake selectivity, and primate PET of a probe for glutamate carboxypeptidase II (NAALADase) Molecular Imaging. 2002;1(2):96–101. doi: 10.1162/15353500200202109. [DOI] [PubMed] [Google Scholar]
- Post DE, Sandberg EM, Kyle MM, Devi NS, Brat DJ, Xu Z, et al. Targeted cancer gene therapy using a hypoxia inducible factor dependent oncolytic adenovirus armed with interleukin-4. Cancer Research. 2007;67(14):6872–6881. doi: 10.1158/0008-5472.CAN-06-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot F, Sato M, Jiang ZK, Huyn S, Karanikolas BDW, Wu LL. A molecular imaging system based on both transcriptional and genomic amplification to detect prostate cancer cells in vivo. Molecular Therapy. 2013;21(3):554–560. doi: 10.1038/mt.2012.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik D, Majeti BK, Mondal G, Karmali PP, Sistla R, Ramprasad OG, et al. Lipopeptide with a RGDK tetrapeptide sequence can selectively target genes to proangiogenic alpha 5 beta 1 integrin receptor and mouse tumor vasculature. Journal of Medicinal Chemistry. 2008;51(22):7298–7302. doi: 10.1021/jm800915y. [DOI] [PubMed] [Google Scholar]
- Qin C, Cheng K, Chen K, Hu X, Liu Y, Lan X, et al. Tyrosinase as a multifunctional reporter gene for Photoacoustic/MRI/PET triple modality molecular imaging. Scientific Reports. 2013;3:1490. doi: 10.1038/srep01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Paulmurugan R, Hildebrandt I, Iyer M, Wu L, Carey M, et al. Novel bidirectional vector strategy for amplification of therapeutic and reporter gene expression. Human Gene Therapy. 2004;15(7):681–690. doi: 10.1089/1043034041361271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribault S, Neuville P, Mechine-Neuville A, Auge F, Parlakian A, Gabbiani G, et al. Chimeric smooth muscle-specific enhancer/promoters: Valuable tools for adenovirus-mediated cardiovascular gene therapy. Circulation Research. 2001;88(5):468–475. doi: 10.1161/01.res.88.5.468. [DOI] [PubMed] [Google Scholar]
- Ribeiro SC, Monteiro GA, Prazeres DM. The role of polyadenylation signal secondary structures on the resistance of plasmid vectors to nucleases. Journal of Gene Medicine. 2004;6(5):565–573. doi: 10.1002/jgm.536. [DOI] [PubMed] [Google Scholar]
- Robson T, Hirst DG. Transcriptional targeting in cancer gene therapy. Journal of Biomedicine & Biotechnology. 2003;2003(2):110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome C, Couillaud F, Moonen CT. Spatial and temporal control of expression of therapeutic genes using heat shock protein promoters. Methods. 2005;35(2):188–198. doi: 10.1016/j.ymeth.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Sakurai Y, Hatakeyama H, Sato Y, Akita H, Takayama K, Kobayashi S, et al. Endosomal escape and the knockdown efficiency of liposomal-siRNA by the fusogenic peptide shGALA. Biomaterials. 2011;32(24):5733–5742. doi: 10.1016/j.biomaterials.2011.04.047. [DOI] [PubMed] [Google Scholar]
- Sato M, Figueiredo ML, Burton JB, Johnson M, Chen M, Powell R, et al. Configurations of a two-tiered amplified gene expression system in adenoviral vectors designed to improve the specificity of in vivo prostate cancer imaging. Gene Therapy. 2008;15(8):583–593. doi: 10.1038/gt.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saukkonen K, Hemminki A. Tissue-specific promoters for cancer gene therapy. Expert Opinion on Biological Therapy. 2004;4(5):683–696. doi: 10.1517/14712598.4.5.683. [DOI] [PubMed] [Google Scholar]
- Schlabach MR, Hu JK, Li M, Elledge SJ. Synthetic design of strong promoters. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(6):2538–2543. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Baeuerle PA. The P65 subunit is responsible for the strong transcription activating potential of Nf-kappa-B. EMBO Journal. 1991;10(12):3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallal HM, Minn I, Banerjee SR, Lisok A, Mease RC, Pomper MG. Heterobivalent agents targeting PSMA and integrin-alphavbeta3. Bioconjugate Chemistry. 2014;25(2):393–405. doi: 10.1021/bc4005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, et al. Bright far-red fluorescent protein for whole-body imaging. Nature Methods. 2007;4(9):741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS Journal. 2010;277(23):4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- Siders WM, Halloran PJ, Fenton RG. Transcriptional targeting of recombinant adenoviruses to human and murine melanoma cells. Cancer Research. 1996;56(24):5638–5646. [PubMed] [Google Scholar]
- Siwak DR, Tari AM, Lopez-Berestein G. The potential of drug-carrying immunoliposomes as anticancer agents—Commentary re: J. W. Park et al., anti-HER2 immunoliposomes: Enhanced efficacy due to targeted delivery. Clin. Cancer Res. 2002;8:1172–1181. [PubMed] [Google Scholar]; Clinical Cancer Research. 2002;8(4):955–956. [PubMed] [Google Scholar]
- So MK, Xu C, Loening AM, Gambhir SS, Rao J. Self-illuminating quantum dot conjugates for in vivo imaging. Nature Biotechnology. 2006;24(3):339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- Soubrier F, Cameron B, Manse B, Somarriba S, Dubertret C, Jaslin G, et al. pCOR: A new design of plasmid vectors for nonviral gene therapy. Gene Therapy. 1999;6(8):1482–1488. doi: 10.1038/sj.gt.3300968. [DOI] [PubMed] [Google Scholar]
- Srinivas R, Samanta S, Chaudhuri A. Cationic amphiphiles: Promising carriers of genetic materials in gene therapy. Chemical Society Reviews. 2009;38(12):3326–3338. doi: 10.1039/b813869a. [DOI] [PubMed] [Google Scholar]
- Stoll SM, Sclimenti CR, Baba EJ, Meuse L, Kay MA, Calos MP. Epstein-Barr virus/human vector provides high-level, long-term expression of alpha1-antitrypsin in mice. Molecular Therapy. 2001;4(2):122–129. doi: 10.1006/mthe.2001.0429. [DOI] [PubMed] [Google Scholar]
- Su ZZ, Sarkar D, Emdad L, Duigou GJ, Young CS, Ware J, et al. Targeting gene expression selectively in cancer cells by using the progression-elevated gene-3 promoter. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(4):1059–1064. doi: 10.1073/pnas.0409141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su ZZ, Shi Y, Fisher PB. Subtraction hybridization identifies a transformation progression-associated gene PEG-3 with sequence homology to a growth arrest and DNA damage-inducible gene. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(17):9125–9130. doi: 10.1073/pnas.94.17.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XL, Zhang N. Cationic polymer optimization for efficient gene delivery. Mini-Reviews in Medicinal Chemistry. 2010;10(2):108–125. doi: 10.2174/138955710791185109. [DOI] [PubMed] [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Molecular Therapy: The Journal of the American Society of Gene Therapy. 2005;11(3):435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Umeoka T, Kawashima T, Kagawa S, Teraishi F, Taki M, Nishizaki M, et al. Visualization of intrathoracically disseminated solid tumors in mice with optical imaging by telomerase-specific amplification of a transferred green fluorescent protein gene. Cancer Research. 2004;64(17):6259–6265. doi: 10.1158/0008-5472.CAN-04-1335. [DOI] [PubMed] [Google Scholar]
- Walker GF, Fella C, Pelisek J, Fahrmeir J, Boeckle S, Ogris M, et al. Toward synthetic viruses: Endosomal pH-triggered deshielding of targeted polyplexes greatly enhances gene transfer in vitro and in vivo. Molecular Therapy. 2005;11(3):418–425. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Therapy. 2004;11(8):711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- Wang DG, Wang ZP, Tian JQ, He XD, Chowdhury WH, Zhang XB, et al. Prostate stem cell antigen enhancer and uroplakin II promoter based bladder cancer targeted tissue-specific vector. Urologic Oncology-Seminars and Original Investigations. 2010;28(2):164–169. doi: 10.1016/j.urolonc.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Sakaguchi M, Kinoshita R, Kaku H, Ariyoshi Y, Ueki H, et al. A novel gene expression system strongly enhances the anticancer effects of a REIC/Dkk-3-encoding adenoviral vector. Oncology Reports. 2014;31(3):1089–1095. doi: 10.3892/or.2013.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Ueki H, Ochiai K, Huang P, Kobayashi Y, Nasu Y, et al. Advanced two-step transcriptional amplification as a novel method for cancer-specific gene expression and imaging. Oncology Reports. 2011;26(4):769–775. doi: 10.3892/or.2011.1371. [DOI] [PubMed] [Google Scholar]
- Wigdal SS, Anderson JL, Vidugiris GJ, Shultz J, Wood KV, Fan F. A novel bioluminescent protease assay using engineered firefly luciferase. Current Chemical Genomics. 2008;2:16–28. doi: 10.2174/1875397300802010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S, Mustoe T, Mulcahy T, Griffiths M, Simpson D, Antoniou M, et al. CpG-island fragments from the HNRPA2B1/CBX3 genomic locus reduce silencing and enhance transgene expression from the hCMV promoter/enhancer in mammalian cells. BMC Biotechnology. 2005;5:17. doi: 10.1186/1472-6750-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Kuhnel F, Kubicka S. Telomerase-dependent gene therapy. Current Molecular Medicine. 2005;5(2):243–251. doi: 10.2174/1566524053586536. [DOI] [PubMed] [Google Scholar]
- Wolff JA, Rozema DB. Breaking the bonds: Non-viral vectors become chemically dynamic. Molecular Therapy. 2008;16(1):8–15. doi: 10.1038/sj.mt.6300326. [DOI] [PubMed] [Google Scholar]
- Wood KV, Lam YA, Seliger HH, McElroy WD. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244(4905):700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- Xie X, Mathias JR, Smith MA, Walker SL, Teng Y, Distel M, et al. Silencer-delimited transgenesis: NRSE/RE1 sequences promote neural-specific trans-gene expression in a NRSF/REST-dependent manner. BMC Biology. 2013;10:93. doi: 10.1186/1741-7007-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(1):151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Fukushima H, Akiyama Y, Niidome Y, Mori T, Katayama Y, et al. Controlled-release system of single-stranded DNA triggered by the photothermal effect of gold nanorods and its in vivo application. Bioorganic & Medicinal Chemistry. 2011;19(7):2130–2135. doi: 10.1016/j.bmc.2011.02.042. [DOI] [PubMed] [Google Scholar]
- Yang TT, Cheng LZ, Kain SR. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Research. 1996;24(22):4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Zhang Y, Xiao F, Xia Z, Rao J. Quantum dot/bioluminescence resonance energy transfer based highly sensitive detection of proteases. Angewandte Chemie International Edition in English) 2007;46(23):4346–4349. doi: 10.1002/anie.200700280. [DOI] [PubMed] [Google Scholar]
- Yew NS, Wang KX, Przybylska M, Bagley RG, Stedman M, Marshall J, et al. Contribution of plasmid DNA to inflammation in the lung after administration of cationic lipid:pDNA complexes. Human Gene Therapy. 1999;10(2):223–234. doi: 10.1089/10430349950019011. [DOI] [PubMed] [Google Scholar]
- Yin W, Xiang P, Li Q. Investigations of the effect of DNA size in transient transfection assay using dual luciferase system. Analytical Biochemistry. 2005;346(2):289–294. doi: 10.1016/j.ab.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Zhang L, Leng QX, Mixson AJ. Alteration in the IL-2 signal peptide affects secretion of proteins in vitro and in vivo. Journal of Gene Medicine. 2005;7(3):354–365. doi: 10.1002/jgm.677. [DOI] [PubMed] [Google Scholar]
- Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141(12):4698–4710. doi: 10.1210/endo.141.12.7837. [DOI] [PubMed] [Google Scholar]
- Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. Journal of Biomedical Optics. 2005;10(4):41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiology and Molecular Biology Reviews. 1999;63(2):405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chemical Communications (Cambridge, England) 2006;22:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- Zhou C, Yu B, Yang X, Huo T, Lee LJ, Barth RF, et al. Lipid-coated nano-calcium-phosphate (LNCP) for gene delivery. International Journal of Pharmaceutics. 2010;392(1-2):201–208. doi: 10.1016/j.ijpharm.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZB, Makhija SK, Lu B, Wang M, Kaliberova L, Liu B, et al. Tran-scriptional targeting of adenoviral vector through the CXCR4 tumor-specific promoter. Gene Therapy. 2004a;11(7):645–648. doi: 10.1038/sj.gt.3302089. [DOI] [PubMed] [Google Scholar]
- Zhu ZB, Makhija SK, Lu B, Wang M, Kaliberova L, Liu B, et al. Tran-scriptional targeting of tumors with a novel tumor-specific survivin promoter. Cancer Gene Therapy. 2004b;11(4):256–262. doi: 10.1038/sj.cgt.7700679. [DOI] [PubMed] [Google Scholar]
- Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, et al. Rapid optimization of gene delivery by parallel end-modification of poly(betaamino ester)s. Molecular Therapy. 2007;15(7):1306–1312. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]