Abstract
Background
Measurement of anti-malarial drug efficacy and resistance relies mainly on in vivo clinical trials, in vitro/ex vivo assays and molecular markers detection. The existing in vitro/ex vivo assays, in particular those that are using non-radioactive devices, need to be standardized and adapted to field conditions. SYBR Green assay offers a rapid and cheap alternative to other in vitro assays, but it requires tools not commonly available in field laboratories. Here is described a modified SYBR green I protocol to perform the parasite growth test with blood samples in endemic areas, followed later by the SYBR green fluorescence assay performed at a specialized laboratory level.
Methods
In vitro susceptibility of Plasmodium falciparum clones HB3, 3D7, W2 and 7G8 to chloroquine (CQ), dihydroartemisinin (DHA), pyronaridine (PYD) and piperaquine (PPQ) was tested. Fresh isolates of P. falciparum from imported malaria cases were collected for ex vivo assays. The parasite suspension was added in 96-well plates predosed with anti-malarial drugs and incubated for 72 hours at 37°C, 5% CO2. SYBR green I protocol was modified to dry the plates after freeze-thawed process to mimic storage and shipping conditions. The plates were rehydrated with 200 μl of complete RPMI medium for fluorescence assay.
Results
There were no significant differences in IC50 values of CQ, DHA, PYD and PPQ, determined by the modified protocol, compared to standard protocol. Longer storage did not affect the IC50 values.
Conclusion
The SYBR green I modified protocol produced reliable results and could be a suitable method for in vitro/ex vivo assays in field.
Keywords: Malaria, SYBR green, In vitro, Ex vivo, Drug resistance
Background
Malaria kills many children annually and is endemic in more than 100 countries [1,2]. The spread of resistant malaria parasites to anti-malarial drugs, including artemisinin-based combination therapy, is a major concern for malaria control in endemic countries [3]. In the absence of effective vaccine, chemotherapy remains one of the strategies to control malaria. Continuous monitoring of current anti-malarial drugs is needed to detect the spread of resistant parasites strains and estimate the risk of therapeutic failure.
Three main approaches including in vivo trials, in vitro/ex vivo assays and molecular markers of drug resistance are currently used to monitor anti-malarial drug efficacy and drug resistance [4]. In vivo studies are considered the gold standard for measuring the efficacy of anti-malarial drugs [5]. They allow the measurement of clinical and parasitological efficacy of anti-malarials drugs and the detection of changes in treatment outcomes. Carrying out in vivo efficacy trials requires time, qualified medical staff and financial resources, which can represent a challenge in resource-limited malaria-endemic countries. In addition, in vivo trials measure therapeutic efficacy rather than drug resistance [6,7].
In vitro/ex vivo assays are indispensable means for measuring the intrinsic susceptibility of malaria parasites to anti-malarial drugs, and establishing baseline susceptibility of local parasite isolates to newly introduced drugs. Several techniques for in vitro/ex vivo tests have been developed for decades. These techniques include the WHO microtest, the isotopic based tests, the enzyme-linked immunosorbent assay (ELISA) and the SYBR green-based tests [8,9]. However, the availability of these techniques is limited in low-income countries due to financial cost, time consumption and problems related to the management of radioactive waste. The existing in vitro/ex vivo assays, in particular those that are non-radioactive, need to be standardized [8]. SYBR Green I in vitro/ex vivo assay offers a rapid, reproducible and cheap alternative to radioisotopic methods [4,10,11]. This also requires specific material and expertise commonly not available in field laboratories.
Here is the report of a modified SYBR green I protocol to perform in vitro/ex vivo tests in two steps: doing parasite culture on 96-well plate containing anti-malarial drugs in standard conditions, and postponing the step of SYBR green fluorescence assay for outsourcing with specialized laboratories. Such a standardized protocol for conducting in vitro/ex vivo drug sensitivity assays for field monitoring of drug-resistant malaria would allow direct comparisons of in vitro/ex vivo results from different laboratories involved in networks of anti-malarial drugs testing. It would also allow collecting more data during field studies.
Methods
Anti-malarial drugs
The following drugs were tested for ex vivo and in vitro assay: chloroquine diphosphate salt (CQ, RMCQ20120605-03 [20 mg]); dihydroartemisinin (DHA, RMDHA20120511-04 [20 mg]); pyronaridine tetraphosphate (PYD, RMPD20130114-01 [200 mg]) and piperaquine tetraphosphate tetrahydrate (PPQ, RMPQ20130114-01 [200 mg]). These drugs were provided by the WorldWide Antimalarial Resistance Network (WWARN). The stock solutions of CQ, DHA and PPQ were prepared in sterile distilled water and DHA in methanol. A two point five–fold serial dilution of stock solutions was made to obtain seven different concentrations. The final concentrations ranged from 6.55 nM to 1600 nM for CQ, 0.07 nM to 16 nM for DHA, 0.33 nM to 80 nM for PYD and 1.31 to 320 nM for PPQ (Table 1). Drugs were distributed at 25 μl per well in 96-wells plates. Each drug was distributed in triplicate in the plate. The plates for ex vivo tests were dried overnight at room temperature in sterile conditions and sealed with an adhesive plastic, then stored at +4°C until use.
Table 1.
The final concentrations of drugs in serial dilutions for in vitro / ex vivo assay
| Dilution (nM) of Drugs | D1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 |
|---|---|---|---|---|---|---|---|---|
| CQ | 1600 | 640 | 256 | 102 | 41 | 16.4 | 6.55 | 0 |
| DHA | 16 | 6.4 | 2.56 | 1.02 | 0.41 | 0.16 | 0.07 | 0 |
| PYD | 80 | 32 | 12.8 | 5.12 | 2.05 | 0.82 | 0.33 | 0 |
| PPQ | 320 | 128 | 51.2 | 20.5 | 8.19 | 3.28 | 1.31 | 0 |
CQ = chloroquine; DHA = dihydroartemisinin; PPQ = piperaquine; PYD = pyronaridine. nM = nanomolar. All drugs concentrations are in nM.
SYBR green I in vitro/ex vivo modified protocol
In vitro assay
Two chloroquine-sensitive (HB3, 3D7) and two chloroquine-resistant (7G8, W2) clones of Plasmodium falciparum were tested for in vitro susceptibility against chloroquine, dihydroartemisinin, pyronaridine and piperaquine. 3D7 and W2 were provided by WWARN; HB3 and 7G8 were from The Malaria Research and Reference Reagent Resource Center (MR4). The clones were cultured in complete RPMI 1640 medium, with 0.5% Albimax + hypoxanthine and gentamicin. They were incubated at 37°C, 5%CO2, 5%O2, 90%N2. An aliquot of the culture was diluted to reduce the parasitaemia to 0.5%, and the haematocrit was adjusted to 1.5%. This suspension was then added (175 μl per well) to microplates predosed with 25 μl of drugs and incubated for 72 hours at 37°, 5%C02, 5%O2, 90%N2. After freeze-thawed process, the test was performed according to the SYBR Green I standard and modified protocols (Figure 1). For the standard protocol, the test was performed as described by Bacon et al. [10]. Briefly, after homogenization of the medium, lysis buffer and SYBR Green I were added and fluorescence was read with the fluorimeter Tristar2 (Multimode Reader LB 942 Bertold Technologies) at 485 nm wave length using Microwin 2000 software. The standard protocol was then modified by drying the plates at 50°C for 10 hours after freeze-thawed process to evaporate the culture medium (Figure 1). To assess the impact of storage period on the values of IC50, the dried plates were stored at room temperature during 24 hours and nine days. The plates were then rehydrated with 200 μl of complete RPMI 1640 medium and incubated at room temperature for at least 1 hour. Lysis buffer and SYBR Green I were added and fluorescence were read with the fluorimeter Tristar2 (Multimode Reader LB 942 Bertold Technologies) at 485 nm wave length using Microwin 2000 software. One millilitre of the suspension was frozen at -20°C as control for parasite growth.
Figure 1.
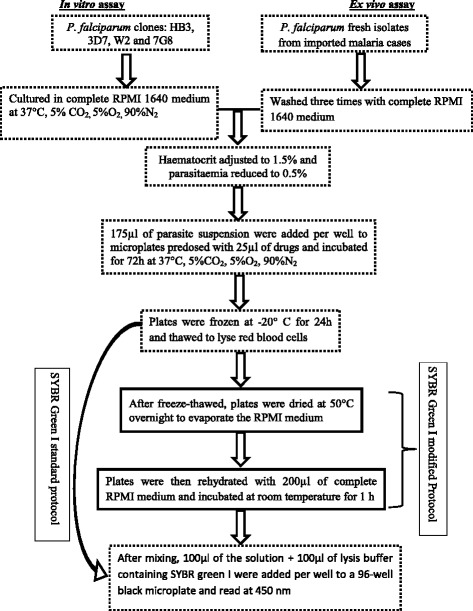
SYBR Green I modified protocol for ex vivo/in vitro assay.
Ex vivo assay
Fresh isolates were collected from nine imported clinical malaria cases with parasitaemia ≥ 0.5% at the Lyon teaching hospital, France. Tests were done within 48 hours after bleeding, without culture adaptation. Blood samples were collected before any anti-malarial treatment, washed three times in complete RPMI 1640 medium and adjusted to 1.5% haematocrit and 0.5% parasitaemia by adding fresh red blood cells. 175 μl suspension was added per well to the plates containing 25 μl of the anti-malarial drugs. Plates were incubated for 72 hours at 37°C in 5% CO2 and O2 controlled atmosphere. After the incubation time, plates were frozen and thawed for red blood cells lysis before SYBR Green I analysis. SYBR Green I analysis was performed according to the standard and modified protocols as already described above for in vitro tests (Figure 1).
Statistical analysis
The results from the Tristar2 were given in fluorescence units (FU). The concentration at which the drugs were able to inhibit 50% of parasite growth (IC50) was calculated through non-linear regression software. IC50 values were validated if there was sufficient growth of parasites with the ratio of FU (FU at concentration 0/FU at maximum concentration) ≥ 2, and a sigmoid curve generated by the software. IC50 means were compared using the t-test (α = 0.05) and the Ficher-Snedecor F-test for variances.
Results
Overall, three in vitro tests were performed for each clone. The IC50 means calculated respectively by standard and modified protocols are summarized in Table 2. No significant differences were seen in IC50s generated for in vitro tests between the standard and modified protocols.
Table 2.
The means of CI 50 of the SYBR green based in vitro anti-malarial drugs assay according to protocols
| Clone/drugs | Protocol standard | New Protocol (test) | P | ||
|---|---|---|---|---|---|
| Mean CI 50 (nM) | 95% CI | Mean CI 50 (nM) | 95% CI | ||
| 3D7 | |||||
| CQ | 31.10 ± 6.02 | 24.28-37.91 | 32.43 ± 4.31 | 26.22-35.97 | 0.77 |
| DHA | 2.30 ± 0.68 | 1.53-3.06 | 1.93 ± 0.43 | 1.81-2.78 | 0.47 |
| PYD | 8.82 ± 1.17 | 7.49-10.14 | 8.89 ± 1.54 | 7.07-10.56 | 0.97 |
| PPQ | 26.33 ± 1.71 | 21.38-28.27 | 25.66 ± 6.82 | 15.61-34.4 | 0.88 |
| W2 | |||||
| CQ | 467.66 ± 51.18 | 409.74-525.57 | 500.66 ± 87.22 | 368.96-566.35 | 0.60 |
| DHA | 1.94 ± 1.03 | 0.77-3.10 | 1.81 ± 0.99 | 0.81-3.06 | 0.81 |
| PYD | 5.35 ± 1.27 | 3.90-6.79 | 4.33 ± 1.21 | 3.98-6.71 | 0.37 |
| PPQ | 19.50 ± 2.91 | 16.20-22.79 | 17.20 ± 3.14 | 15.94-23.05 | 0.40 |
| HB3 | |||||
| CQ | 37.92 ± 6.66 | 30.38-45.47 | 50.28 ± 16.77 | 31.30-69.26 | 0.33 |
| DHA | 2.10 ± 0.97 | 0.99-3.20 | 1.99 ± 0.41 | 1.53-2.46 | 0.87 |
| PYD | 11.78 ± 2.36 | 9.10-14.46 | 12.51 ± 3.40 | 8.65-16.36 | 0.70 |
| PPQ | 21.70 ± 1.76 | 19.70-23.70 | 18.51 ± 2.24 | 15.97-21.06 | 0.12 |
| 7G8 | |||||
| CQ | 416.98 ± 92.57 | 311.82-521.33 | 420.55 ± 96.04 | 311.87-529.24 | 0.18 |
| DHA | 0.65 ± 0.17 | 0.45-0.85 | 0.73 ± 0.12 | 0.58-0.87 | 0.56 |
| PYD | 4.43 ± 1.39 | 2.84-6.01 | 5.05 ± 0.34 | 4.66-5.44 | 0.52 |
| PPQ | 17.82 ± 3.57 | 13.77-21.87 | 15.98 ± 1.22 | 14.59-17.38 | 0.47 |
CQ = Chloroquine; DHA = Dihydroartemisinin; PPQ = Piperaquine; PYD = Pyronaridine; SD = Standard deviation, nM = nanomolar.
Ex vivo susceptibility of P. falciparum was tested for nine samples from patients. The average culture success rate was 61.5% (ranged from 55.5-66.6%). The IC50 were calculated respectively by standard and modified protocols and the results are summarized in Table 3. No significant differences were seen in IC50s generated for ex vivo samples between the standard and modified protocols.
Table 3.
Ex vivo susceptibility of Plasmodium falciparum isolates to chloroquine, dihydroartemisinin, pyronaridine and piperaquine (standard and modified protocols)
| Culture success rate (n/N) | Standard protocol | Modified protocol | P | |
|---|---|---|---|---|
| IC 50 mean (nM) [95% CI] | IC 50 mean (nM) [95% CI] | |||
| CQ | 66.6% (6/9) | 484.08 [176-791] | 730.66 [595-995] | 0.35 |
| DHA | 66.6% (6/9) | 1.67 [1.21-2.12] | 1.44 [0.88-2] | 0.55 |
| PYD | 55.5% (5/9) | 14.23 [2.83-25.63] | 16.85 [1.69-32] | 0.79 |
| PPQ | 55.5% (5/9) | 78.22 [36.56-119.87] | 74 [24.61-123.38] | 0.90 |
CQ = Chloroquine; DHA = Dihydroartemisinin; PPQ = Piperaquine; PYD = Pyronaridine, nM = nanomolar.
Discussion
Among the different in vitro/ex vivo assay methods, the use of fluorescent labeling of DNA with SYBR Green offers some advantages compared to methods that use tritiated hypoxanthine or ELISA [4,10-12]. However, the fluorescence determination requires material not commonly available in field laboratories. Postponing the step of fluorescence determination allows the use of SYBR Green I protocol in field laboratories with limited resources. It also offers opportunity (i) to do simultaneously ex vivo and in vivo tests during field studies, (ii) to standardize protocols for data collection during multicenter clinical studies, (iii) to store data for future use, (iv) and to share original data with collaborators. The SYBR Green I modified protocol described in this study enables to dry and store the plates, which will be analysed later by well-equipped laboratories. Networks devoted to anti-malarial drugs resistance assay [13-17], may use this modified protocol for multicentre clinical studies, that could result in reduction of instrumental and systematic bias and facilitate the compilation, analysis and interpretation of data from different study sites. The risks of environment pollution are scarce and the technique does not require specific protection of manipulator since the SYBR Green is not radioactive [8] while its toxic for DNA, and will be added to the culture in the laboratories where the fluorescence will be read.
The plates were dried at 50°C to minimize the risk of contamination of the culture with bacteria or fungi (higher temperatures could eliminate the risk of contamination, but they could denature the DNA of the parasite). The modified protocol was first tested with laboratory clones in vitro, then with field fresh isolates ex vivo. Samples were collected from nine imported malaria cases (six from Cameroon and three from the Democratic Republic of Congo). Four patients have declared the use of chemoprophylaxis against malaria (with chloroquine or chloroquine-proguanil) prior to admission. The average of culture success rate (interpretable tests) of ex vivo assays was 61.5%, slightly lower than this described by Tinto el al. (85%) [18], probably due to the small sample size of the study reported here.
There was no significant difference between the means of IC50 determined by the standard and modified protocols. The values of IC50 determined in vitro by the modified protocol were broadly similar to those calculated using the standard protocol (p > 0.10). The duration of storage did not affect the IC50 value (IC50 of CQ against 7G8 ranged from 676 nM in standard protocol to 502 nM after 24 hours and 9 days of storage of dried plates in modified protocol). A slight but not significant decrease was observed in IC50 values of dried plates. These findings suggest that the dried plates can be stored at room conditions and be sent without special transportation requirements since biological material is stable and non-infectious.
The means of IC50 of chloroquine against sensitive clones 3D7 (32.43 nM, 95% CI: 26.22-35.97) and HB3 (50.28 nM, 95% CI: 31.3-69.2) determined in vitro by the modified SYBR Green I protocol are broadly similar to those already described in vitro by Garbi et al. in Senegal, Mali, Cameroon and Côte d’Ivoire [19] and Issaka et al. in Niger [20]. For ex vivo assays, our IC50 values of CQ (484.08 and 730.66 nM) are also comparable to the maximum values described by Tinto et al. (using the semiautomated microdilution technique) in Burkina Faso (8.3-595.9 nM) [18]. The lowers values (8.3 nM) obtained by Tinto et al. could be the fact that resistant and sensitive strains are circulating simultaneously in populations, and the large sample size included in this study.
The means of IC50 of the chloroquine were high in ex vivo assays (484.08 and 730.66 nM respectively in standard and modified protocols). Malaria cases were imported from chloroquine-resistance areas. High resistance rates of P. falciparum to chloroquine have been described in Cameroon since 2000 [21], even though recent studies have shown decreases in resistance due to changes in malaria treatment policies in these areas [19].
Conclusion
Drying and storing the plates did not affect the means of IC50 values in a modified SYBR Green I protocol to assess anti-malarial drugs. The modified protocol allows performing in vitro/ex vivo tests in two steps. By postponing the step that requires SYBR Green methodology, the tests could be done in field conditions and the dried plates could be transported in ambient conditions. Field staff involved in multicentre studies may use this protocol to standardize the collect and share of data. It offers the possibility to do simultaneously clinical in vivo and ex vivo/in vitro assays in field. More in vitro/ex vivo studies with large sample size are necessary for the validation of this protocol.
Abbreviations
- CQ
Chloroquine
- DHA
Dihydroartemisinin
- ELISA
Enzyme-linked immunosorbent assay
- FU
Fluorescence units
- GIS
Geographic information system
- MR4
Malaria research and reference reagent resource center (MR4)
- IC50
The 50% inhibitory concentration
- nM
Nanomolar
- PPQ
Piperaquine
- PYD
Pyronaridine
- RPMI
Roswell park memorial institute
- SD
Standard deviation
- WANECAM
West african network for clinical trials of antimalarial drugs
- WHO
World health organization
- WWARN
Worldwide antimalarial resistance network
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SP, FG, KT designed the study and experiments, KT, AL, GB, MC, FS, GCB performed data collection; SP, OKD, FG and KT performed data analysis and wrote the manuscript. All the authors have read and approved the content of this manuscript.
Contributor Information
Karim Traore, Email: karim@icermali.org.
Adeline Lavoignat, Email: adeline.lavoignat@univ-lyon1.fr.
Guillaume Bonnot, Email: guillaume.bonnot@univ-lyon1.fr.
Fatimata Sow, Email: fatimata.sow@etu.univ-lyon1.fr.
Giuliana C Bess, Email: giuliana.constantino-bess@etu.univ-lyon1.fr.
Marjorie Chavant, Email: chavant.marjorie@gmail.com.
Frederick Gay, Email: frederick.gay@upmc.fr.
Ogobara Doumbo, Email: okd@icermali.org.
Stephane Picot, Email: stephane.picot@univ-lyon1.fr.
References
- 1.WHO . World Malaria Report 2011. Geneva: World Health Organization; 2011. [Google Scholar]
- 2.Verma R, Khanna P, Chawla S. Malaria vaccine can prevent millions of deaths in the world. Hum Vaccines Immunother. 2013;9:1268–71. doi: 10.4161/hv.23950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridley RG. Medical need, scientific opportunity and the drive for antimalarial drugs. Nature. 2002;415:686–93. doi: 10.1038/415686a. [DOI] [PubMed] [Google Scholar]
- 4.Abdul-Ghani R, Al-Maktari MT, Al-Shibani LA, Allam AF. A better resolution for integrating methods for monitoring Plasmodium falciparum resistance to antimalarial drugs. Acta Trop. 2014;137:44–57. doi: 10.1016/j.actatropica.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 5.Vestergaard LS, Ringwald P. Responding to the challenge of antimalarial drug resistance by routine monitoring to update national malaria treatment policies. Am J Trop Med Hyg. 2007;77:153–9. [PubMed] [Google Scholar]
- 6.Laufer MK. Monitoring antimalarial drug efficacy: current challenges. Curr Infect Dis Rep. 2009;11:59–65. doi: 10.1007/s11908-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedt BL, Laufer MK, Cohen T. Drug resistance surveillance in resource-poor settings: current methods and considerations for TB, HIV, and malaria. Am J Trop Med Hyg. 2011;84:192–9. doi: 10.4269/ajtmh.2011.10-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basco LK. Field application of in vitro assays for the sensitivity of human malaria parasites to antimalarial drugs. Geneva: World Health Organization; 2007. [Google Scholar]
- 9.Wendler JP, Okombo J, Amato R, Miotto O, Kiara SM, Mwai L, et al. A genome wide association study of Plasmodium falciparum susceptibility to 22 antimalarial drugs in Kenya. PLoS One. 2014;9:e96486. doi: 10.1371/journal.pone.0096486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacon DJ, Latour C, Lucas C, Colina O, Ringwald P, Picot S. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob Agents Chemother. 2007;51:1172–8. doi: 10.1128/AAC.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vossen MG, Pferschy S, Chiba P, Noedl H. The SYBR green I malaria drug sensitivity assay: performance in low parasitemia samples. Am J Trop Med Hyg. 2010;82:398–401. doi: 10.4269/ajtmh.2010.09-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51:1926–33. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacon DJ, Jambou R, Fandeur T, Le Bras J, Wongsrichanalai C, Fukuda MM, et al. World Antimalarial Resistance Network (WARN) II: in vitro antimalarial drug susceptibility. Malar J. 2007;6:120. doi: 10.1186/1475-2875-6-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price RN, Dorsey G, Ashley EA, Barnes KI, Baird JK, D’ Alessandro U, et al. World Antimalarial Resistance Network I: clinical efficacy of antimalarial drugs. Malar J. 2007;6:119. doi: 10.1186/1475-2875-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodrow CJ, Dahlstrom S, Cooksey R, Flegg JA, Le Nagard H, Mentre F, et al. High-throughput analysis of antimalarial susceptibility data by the WorldWide Antimalarial Resistance Network (WWARN) in vitro analysis and reporting tool. Antimicrob Agents Chemother. 2013;57:3121–30. doi: 10.1128/AAC.02350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West African Network for Clinical Trials of Antimalarial Drugs: WANECAM. 2013. http://www.wanecam.org/fr.
- 17.WorldWide Antimalarial Resistance Network: WWARN. 2015. http://www.wwarn.org/fr.
- 18.Tinto H, Bonkian LN, Nana LA, Yerbanga I, Lingani M, Kazienga A, et al. Ex vivo anti-malarial drugs sensitivity profile of Plasmodium falciparum field isolates from Burkina Faso five years after the national policy change. Malar J. 2014;13:207. doi: 10.1186/1475-2875-13-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gharbi M, Flegg JA, Hubert V, Kendjo E, Metcalf JE, Bertaux L, et al. Longitudinal study assessing the return of chloroquine susceptibility of Plasmodium falciparum in isolates from travellers returning from West and Central Africa, 2000–2011. Malar J. 2013;12:35. doi: 10.1186/1475-2875-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Issaka M, Salissou A, Arzika I, Guillebaud J, Maazou A, Specht S, et al. Ex vivo responses of Plasmodium falciparum clinical isolates to conventional and new antimalarial drugs in Niger. Antimicrob Agents Chemother. 2013;57:3415–9. doi: 10.1128/AAC.02383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringwald P, Ekobo A, Keundjian A, Mangamba D, Basco LK. Chimiorésistance de P. falciparum en milieu urbain à Yaoundé, Cameroun. Part 1: Surveillance in vitro et in vivo de la résistance de Plasmodium falciparum à la chloroquine entre 1994 et 1999 à Yaoundé, Cameroun. Trop Med Int Health. 2000;5:612–9. doi: 10.1046/j.1365-3156.2000.00613.x. [DOI] [PubMed] [Google Scholar]


