Abstract
Working memory, a theoretical construct from the field of cognitive psychology, is crucial to everyday life. It refers to the ability to temporarily store and manipulate task-relevant information. The identification of genes for working memory might shed light on the molecular mechanisms of this important cognitive ability and—given the genetic overlap between, for example, schizophrenia risk and working-memory ability—might also reveal important candidate genes for psychiatric illness. A number of genome-wide searches for genes that influence working memory have been conducted in recent years. Interestingly, the results of those searches converge on the mediating role of neuronal excitability in working-memory performance, such that the role of each gene highlighted by genome-wide methods plays a part in ion channel formation and/or dopaminergic signaling in the brain, with either direct or indirect influence on dopamine levels in the prefrontal cortex. This result dovetails with animal models of working memory that highlight the role of dynamic network connectivity, as mediated by dopaminergic signaling, in the dorsolateral prefrontal cortex. Future work, which aims to characterize functional variants influencing working-memory ability, might choose to focus on those genes highlighted in the present review and also those networks in which the genes fall. Confirming gene associations and highlighting functional characterization of those associations might have implications for the understanding of normal variation in working-memory ability and also for the development of drugs for mental illness.
Keywords: Working memory, Genomics, Cognition, GWA, Dynamic network connectivity
Introduction
Working memory is crucial to everyday life; it plays a key role in everyday tasks—following spoken directions, reading a magazine article, calculating a tip in a restaurant—that require information to be temporarily stored and manipulated [1, 2]. It has been described as being core to reasoning and judgment in humans; in other words, working memory is crucial to other important aspects of cognitive performance, such as attention and executive functioning [3, 4], and is a determinant of an individual’s level of intelligence [5, 6]. As a consequence, working memory is one of the most studied concepts in cognitive neuroscience [7]. In addition, working memory is impaired in psychiatric and neurodegenerative illnesses such as schizophrenia and Alzheimer’s disease [8, 9]. Moreover, there is thought to be a substantial genetic overlap between those genes that mediate illness risk and those that influence working-memory ability [10, 11]. The importance of working memory to cognition in general, combined with the key role that working memory plays in the symptomatology of certain illnesses, behooves the research community to provide insights into the molecular underpinnings of working-memory ability. The field of behavior genetics is ideally suited to fulfill this task and, to date, several genome-wide searches for genes influencing working memory have been conducted. The present manuscript provides a qualitative review of this literature. The work reviewed here converges on the role of neuronal excitability in working-memory ability, with a focus on dopaminergic signaling, which is in line with the work of Goldman-Rakic and colleagues and, more recently, Arnsten and colleagues [12–14, 15••, 16].
A Definition of Working Memory
Working memory is a theoretical construct from the field of cognitive psychology. The term was coined by Miller et al. [17] to refer to the ability to temporarily store and manipulate task-relevant information. As a concrete example, imagine that someone asks you to solve a difficult multiplication problem without using a pen and paper (e.g., “What is 8 times 35?”). It is unlikely that you could simply recall the answer from your long-term memory—instead, you would probably need to split the problem into several easier multiplications (e.g., 8×5 and 8×3×10) and then sum together those answers. Working memory is needed to choose which smaller multiplications to perform, to remember the answers before summing them (40 and 240), and to remember the final answer before saying it out loud (280). A critical feature of this definition of working memory is that it includes both storage and manipulation. Working memory is not just a synonym for short-term memory; rather, it subsumes many functions that allow the retrieval, integration, transformation, and disposal of stored information (for a review, see Baddeley [7]).
It is easy to see how working memory would play a role in most everyday tasks, and empirical work supports this intuition: working memory is believed to constrain other aspects of cognition, such that the better an individual’s working memory, the better their attentional control and executive functioning [3, 4, 18, 19]. Moreover, measures of working memory usually correlate with general intellectual ability better than almost all other cognitive measures [5, 6]. Thus, working memory ubiquitously influences cognition.
Heritability of Working Memory
Heritability (h2) is a measure of effect size that describes the amount of phenotypic variance that is due to genetic differences between individuals. It is bounded between 0 (indicating no genetic influence) and 1 (complete genetic influence), and is estimated using the correlations between relatives in the phenotype of interest [20]. Estimates of h2 for working memory range between 0.32 and 0.66—indicating moderate to high heritability—in both healthy individuals [21–26, 27•] and clinical samples [28–31].
Multivariate analyses have demonstrated that the strong correlations between measures of working memory and general intelligence can be almost entirely attributed to shared genetic influences. In other words, the majority (~95 %) of the genes that influence working memory also influence general intelligence [26, 27•]. Although there is this substantial genetic overlap, there is utility in focusing on working memory rather than on general intelligence when attempting to isolate genetic effects on cognitive ability. Compared with broad abilities, specific cognitive measures are associated with relatively distinct brain circuits [32]. For example, the neural systems that support different aspects of working memory are consistently linked to portions of the dorsolateral and ventrolateral prefrontal cortex, the medial posterior parietal cortex, and regions of the medial temporal cortex, depending on the particular task used [33–40]. While it is unlikely that brain regions are rigidly specialized for domain-specific cognitive processing, there is clear evidence for differential engagement of particular brain networks for classes of information processing [41]. To the extent that genes are uniquely expressed in anatomic regions of the adult brain [42], it is possible that specific genes or gene networks may have relatively more control on domain-specific cognitive processing associated with those regions. This line of thought suggests that identifying genes that influence distinct cognitive domains may be more tractable than finding genes for general intelligence [43]. Some might argue that because heritability estimates for intelligence are higher than those for working memory (between 0.50 and 0.80 [44]), gene-finding efforts should focus on intelligence. However, heritability estimates reflect the overall cumulative genetic effect on a trait but do not reveal the subtle complexities therein or the specific composition, architecture, and number of the underlying causal genes [45]; therefore, the strength of a heritability estimate is not necessarily predictive of the ease with which genes will be detected.
The Molecular Basis of Working Memory
Because working memory is a collection of largely transient processes, the underlying neural circuitry and molecular mechanisms must be well suited to moment-to-moment processing [16, 46]. By contrast, it is known that long-term memory is governed by long-lasting architectural changes induced by long-term potentiation in brain regions, including the hippocampus [47, 48]. Previous work has shown that layer III of the dorsolateral prefrontal cortex (DLPFC) is particularly important in working memory, where synaptic connections are found on long, thin dendritic spines ideally suited to dynamic processing [12, 49]. Research in primates has revealed that networks of DLPFC neurons are persistently active during the execution of a visual delayed response task (an archetypal measure of working memory) and become inactive upon task completion [50, 51]. The same type of neuron has also been found in other brain areas associated with working memory, including the inferior temporal cortex, hippocampus, posterior parietal cortex, and entorhinal cortex [52–56].
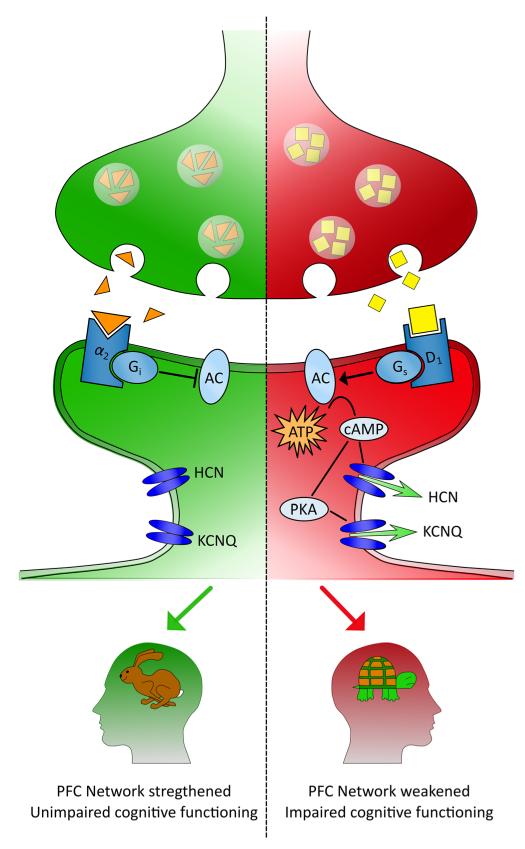
It has been consistently shown that dopamine levels in the DLPFC influence performance on working-memory tasks [57–60]. Modulation of neurons in the DLPFC is thought to take place, broadly speaking, by dopaminergic signaling, which acts in concert with excitatory glutamatergic and inhibitory GABAergic systems [61]. In more detail, the binding of dopamine to its receptor (D1) allows stimulatory G-protein (Gs) to increase adenylyl cyclase (AC) activity. AC enhances intracellular cyclic adenosine monophosphate (cAMP) concentrations, and cAMP in turn activates protein kinase A (PKA), resulting in the opening of ion channels HCN (hyperpolarization-activated channels) and KCNQ [61]. The opening of potassium (K+) KCNQ channels decreases firing and results in rapid weakening of the network of neurons that are otherwise active during a working-memory task. Conversely, the binding of norepinephrine to its receptor (α2A) strengthens the network by inhibiting the release of cAMP [16]. Arnsten and colleagues coined the term “dynamic network connectivity” to describe the mediating role of these neurotransmitters on the neural activity associated with working memory [15••]. A schematic of this process is shown in Fig. 1 where, on the left (green) side, the inhibition of cAMP results in strengthened network connections and fast cognitive operations and, on the right (red) side, the increase in cAMP results in the slowing down of cognition. Thus, the strength of the network underlying working-memory ability is reliant on cAMP mediation of ion channels and, specifically, the transfer of positive ions (or cations) across voltage-gated channels, both independently and also via activation of PKA [16, 62, 63].
Fig. 1.
Dynamic network connectivity in the dorsolateral prefrontal cortex (PFC) [adapted from Arnsten and colleagues [15••]]. The neuromodulators norepinephrine and dopamine mediate the strength of the network, residing in the dorsolateral PFC, that underlies working-memory ability. On the left (green) side of the figure, the binding of norepinephrine (represented by the orange triangles) to its receptor (α2A) strengthens the network by inhibiting the release of cyclic adenosine monophosphate (cAMP), which results in uninterrupted cognitive function. Conversely, on the right (red) side of the figure, the binding of dopamine to its receptor (D1) results in increased adenylyl cyclase (AC) activity via the activation of stimulatory G-protein (Gs). Increased AC activity results in enhanced cAMP concentration, and this results in the opening of sodium and potassium ion channels (HCN and KCNQ, the latter via the activation of protein kinase A [PKA]). The opening of ion channels results in depolarization of the neuron and, consequently, weakening of the network and impaired cognitive function. ATP adenosine triphosphate, Gi inhibitory G-protein
Genes Contributing to Healthy Variation in Working Memory
Identifying genes for working memory in humans might further elucidate the underlying molecular mechanisms by confirming and possibly extending those that have already been discovered via animal research [16]. Many genetic associations have been identified for cognition and, more specifically, for working memory, under the candidate-gene approach [64]. However, attempts to replicate those associations have failed [65]. Even associations that were considered to be robust—for example, with COMT [66]—have been revealed to be false positives when tested in large samples of healthy individuals [67]. Consequently, the present review focuses on genome-wide approaches. The majority of candidate-gene documented variants for working memory are common, and consequently those same associations should be amenable to detection by genome-wide association (GWA), an approach that focuses on common variation [68]. Upon reviewing the genome-wide searches of working memory carried out in recent years, it becomes evident that the results of most studies appear to converge on the role of neuronal excitability via voltage-gated ion channels—a key feature of the experimental work carried out in animals by Arnsten and colleagues [16].
Genome-Wide Association Studies of Working Memory
Cirulli and colleagues conducted a GWA study principally focused on executive function in 1,086 healthy individuals of mixed ethnicity. Additional cognitive tests, including some that indexed working memory, were conducted in a subsample of 514 individuals [69•]. No individual test yielded any genome-wide significant results. The top result was for digit-span backward (p=6.3×10−8; another classic measure of working memory), but this result was intergenic. The second-best hit for the same test was located in the gene KIAA1217 (p=3.9×10−7). KIAA genes are a group of novel human genes identified by the cDNA project, and the functional characterization of many of these genes remains unknown [70–72].
Cirulli and colleagues also found suggestively significant hits for animal fluency and the trail-making test, both of which measure aspects of working-memory ability. The animal fluency task requires participants to name as many animals as possible in 60 seconds and necessarily relies on working memory to retain and inhibit already uttered responses and to implement memory retrieval strategies [73]. The top hit for animal fluency (p=6.41×10−7) was located in an intron of the gene KCNB2, which encodes a type of voltage-gated potassium channel called Kv2.2 [74]. KCNB2 contributes to the maintenance of overall excitability of neurons, such that neuronal networks can be disrupted if there is disruption in the placement or functionality of voltage-gated ion channels [75, 76]. Kv2.2 channels are preferentially expressed in the basal forebrain and particularly in GABAergic neurons [77]. Research in rats suggests that these neurons contribute to working-memory performance, possibly via modulation of ascending dopaminergic projections to the frontal cortex [78, 79].
The other suggestively significant result from Cirulli and colleagues was for the trail-making test part A (TMT-A); this test is primarily considered a measure of visuomotor processing, cognitive flexibility, and attention, but it also includes a strong working-memory component [80, 81]. Participants must consecutively connect numbered circles on a work sheet, without lifting the pen, as quickly as possible [82]. The top hit for TMT-A (p=2.55×10−7) was located in an intron of the gene ATL1 (or SPG3A), which encodes the alastin-1 protein [83]. This gene is associated with hereditary spastic paraplegia, a group of disorders characterized by movement disorder and cognitive impairment, and which, on the basis of evidence from case studies, may be related to Parkinson’s disease; the two illness types are thought to have shared etiologies, possibly within nigrostriatal dopamine projections [84–87]. Research using Drosophila with a disrupted ATL1-homolog exhibits age-related degeneration of dopaminergic neurons— degeneration that can be restored by the administration of a D1 receptor agonist [88]. Lee and colleagues maintain that this finding might have treatment implications for spastic paraplegia sufferers, although this has not been found to be the case in the small number of patients tested to date [89]. The full story is not yet clear, but the ATL1 gene may indirectly mediate prefrontal dopamine levels, which underlie working-memory performance.
Need and colleagues conducted a GWA study of the Cambridge Neuropsychological Test Automated Battery (CANTAB). They calculated heritability estimates of each test by comparing correlations in 100 monozygotic and 100 dizygotic twin pairs, and then conducted association analysis in a sample comprising one member of each twin pair plus additional unrelated subjects, totaling ~700 individuals [90•]. No genome-wide significant variants were identified. However, the top-ranked variant (p=6.03×10−6) for working-memory strategy, a score that indexes the efficiency of the search strategy employed in a spatial working-memory task (h2= 0.53), was located 1.5 kb upstream of the gene FXYD2, which encodes a membrane protein (Na+, K+-ATPase gamma subunit) that regulates sodium and potassium ion transportation; this protein is phosphorylated by PKA [91, 92]. Dopamine inhibits this particular membrane protein by increasing cAMP via binding to D1 receptors [93–97]. Thus, the gene FXYD2 plays a part in maintaining the excitability of neurons that might be crucial to the completion of a working-memory task.
Papassotiropoulos and colleagues conducted a multiphase GWA study and found that a variant within the gene SCN1A, which mediates the construction of a type of sodium (Na+) channel (Nav1.1), was associated with working-memory performance at a genome-wide significant level [98•]. The study was conducted in multiple phases, from discovery through to replication in several additional samples, totaling 2,032 individuals. The neuropsychological task was immediate recall performance, a type of short-term memory storage that is essential to working memory. Moreover, the authors also showed an association between the same variant and brain activation associated with the performance of an n-back task, a classic measure of working memory [98•]. Sodium channels for which SCN1A is responsible control the flow of sodium into a cell and therefore play a crucial role in neuronal excitability [99, 100]. Indeed, mutations in SCN1A are thought to contribute to the neuronal excitability that leads to epilepsy [101, 102]. GABAergic neurotransmission is impaired in SCN1A mutant mice, for whom there is a 50 % reduction in the proportion of Nav1.1 sodium channels in the prefrontal cortex [103]. This is a particularly interesting result, because the role of GABAergic neurons in the prefrontal cortex is thought to involve fine-tuning of activated neuronal networks, such that said activity can be focused onto task-relevant items [104]. GABAergic hypofunction in the prefrontal cortex makes up one theory of cognitive impairment in schizophrenia, of which one of the primary deficits is in working memory [8, 105, 106].
Seshadri and colleagues conducted a GWA study of a cognitive test battery in a sample of 694 individuals taken from families that make up the Framingham extended-pedigree study [107•]. The top hit for any cognitive test (p= 3.2×10−6) was for an index of abstract reasoning. This is a relevant finding because working memory and reasoning ability are similar constructs [5]. The variant was located within an intron of SORL1, which encodes a low-density lipoprotein receptor (LDLR). LDLR is thought to be involved in endocytosis of amyloid precursor protein (APP) [108]. Specifically, SORL1 activity modulates APP processing, such that down-regulation of the gene results in increased APP sorting into amyloid-β (Aβ) [109–111]. Alzheimer’s disease is associated with an accumulation of Aβ protein in the brain [112]. Indeed, SORL1 has been found to mediate Alzheimer’s disease risk [113–116], and levels of SOR1 are decreased in the frontal cortex and lymphoblasts of patients with Alzheimer’s disease [117]. An association between cognitive impairment and SPOR1 has also been shown [118]. Moreover, an association has been shown between hippocampal volume and SPOR1 in a sample of healthy adults [119]. It is thought that elevated Aβ destabilizes those neuronal networks that underlie cognition by suppressing excitatory activity at the postsynaptic level [112]. Specifically, increased levels of Aβ decrease the number of N-methyl-D-aspartate (NMDA) receptors, a subclass of glutamate-gated ion channels [60, 89].
Knowles and colleagues conducted a genome-wide linkage analysis of factor models of genetically clustered cognitive traits, followed by quantitative trait locus (QTL) region-specific association analyses in a sample of 1,269 Mexican American individuals from extended pedigrees [27•]. This study is distinct from the other studies discussed so far because rather than relying on an individual neuropsychological measure to index working memory, detailed phenotypic models were built. More specifically, the authors built a three-tier hierarchical model of cognition, which included a “g” (general intelligence) factor that subsumed several correlated cognitive domains onto which loaded the individual neuropsychological tasks. In this way, the working-memory phenotype in this study might be said to more reliably reflect the underlying construct than those from previous studies. By definition, a latent variable reflects the shared variance between the multiple tasks that load upon it and thus, rather than reflecting the version of working memory indexed by a particular neuropsychological task, the domain reflects the overlapping working-memory portion of each task in a single score [120]. Two genome-wide significant QTLs were revealed for working memory on chromosome 8 (8q21.11–13 and 8q24.22), and each exhibited pleiotropy with the other cognitive domains in the model. Post hoc association analysis in the region beneath the linkage peak at 8q21.13 revealed two common variants that were associated with working memory near the HEY1 gene. HEY1 is a transcription target of Notch and makes up part of the hairy and enhancer of split (HES) and hairy/enhancer-of-split-related with YRPW-like motif (HEY, also named HERP) gene families. Notch is associated with the formation of long-term memories [121]. Furthermore, HEY1 binds to DAT1, the dopamine transporter gene, such that deficits in the HEY1 gene result in enhanced expression of DAT1 and in turn increase expression of D1 receptor genes [122]. HEY1 knockout mice exhibit impaired performance on the Y-maze test, a measure of working memory in rodents [123, 124]. The same type of knockout mice also exhibit enhanced prepulse inhibition due to altered dopamine sensitivity [125]. The full story is not clear, but it seems likely that HEY1 has a mediating role in dopamine activity related to successful working-memory performance. Two genome-wide significant QTLs were also observed for the spatial-memory domain, for which a contributing measure was a spatial working-memory task, on chromosome 17 (17q22– 24.2 and 17q25.1–3). For the first QTL, post hoc association analysis highlighted two variants as being peak-wide significant. Several genes were highlighted in post hoc analysis as being implicated in spatial-memory ability, including BCAS3, MGAT5B, and also APPBP2. APPBP2, which is functionally associated with the production of Aβ, is particularly interesting given previous research highlighting the importance of Aβ in Alzheimer’s disease, and also given the results from the GWA study by Seshadri and colleagues outlined above [107•].
Taken together, each GWA study of working memory has a top-ranked gene that is implicated in neuronal excitability via ion-gated channels, and/or prefrontal dopa-mine expression (Table 1). Cirulli and colleagues found a role for KCNB2, which encodes a type of voltage-gated potassium channel [69•]. Need and colleagues highlighted the involvement of FXDY2, which encodes a membrane protein that regulates sodium and potassium ion transportation [90•]. Papassotiropoulos and colleagues emphasized the gene SCN1A, which is responsible for control of the flow of sodium into a cell [98•]. Seshadri and colleagues noted the importance of SORL1, which increases Aβ and consequently destabilizes neuronal networks by suppressing excitatory activity [107•], and Knowles and colleagues further underscored the role of APPBP2, which is functionally associated with the production of Aβ [27•]. Knowles and colleagues also highlighted the gene HEY1, which results in enhanced expression of the DAT1 and D1 receptor genes [27•]. Neuronal excitability appears to be key to working-memory performance across samples and neuropsycho-logical tasks. It is difficult, upon reviewing this literature, not to be struck by the way in which the majority of the results highlight genes with a moderating role in neuronal excitability, which fits in neatly with the dynamic network connectivity model of working memory [15••].
Table 1.
Summary of the top-ranked variants, taken from genome-wide searches, for working-memory ability
| Authors | N | Measure | Variant | p value | Gene; gene function |
|---|---|---|---|---|---|
| Seshadri et al. (2007) [107•] | 694 | Abstract reasoning | rs1131497 | 3.2×0-6 |
SORL1; encodes an LDLR that modulates APP processing into Aβ, which can suppress excitatory neuronal activity |
| Need et al. (2009) [90•] | 721 | Working-memory strategy |
rs4472969 | 6.03×10-6 |
FXYD2; encodes a membrane protein (Na+, K+-ATPase gamma subunit) that regulates sodium and potassium ion transportation |
| Cirulli et al. (2010) [69•] | 514 | Animal fluency | rs2247572 | 6.41×10-7 |
KCNB2; encodes a type of voltage-gated potassium channel (Kv2.2) that mediates potassium ion transportation |
| TMT-A | rs17122693 | 2.55×10-7 |
ATL1; encodes alastin-1 protein. Disrupted ATL1 results in age-related degeneration of dopaminergic neurons in Drosophila |
||
| Papassotiropoulos et al. (2011) [98•] | 333discovery 1,699replication |
Immediate recall | rs10930201 | 3.0×10-4a |
SCN1A; mediates the construction of a type of sodium (Na+) channel (Nav1.1) |
| Knowles et al. (2014) [27•] | 1,269 | Factor score | rs2467774 | 1.1×10-5b |
HEY1; encodes a protein belonging to the HES and HEY gene families |
Aβ amyloid-β, APP amyloid precursor protein, FDR false discovery rate, HES hairy and enhancer of split, HEYhairy/enhancer-of-split-related with YRPW-like motif, LDLR low-density lipoprotein receptor, TMT-A trail-making test part A
FDR corrected
Linkage peak-wide corrected
Enrichment Studies of Working Memory
Heck and colleagues recently published cohesive evidence for a link between neuronal excitability and working memory, using enrichment analysis [126••]. While GWA studies have been successful in isolating genes for working memory, many of the results discussed in the present review failed to meet genome-wide significance. This might be because GWA, which focuses on single variants, has limited power to identify genetic influences on a complex trait because of the vast number of tests conducted and the associated necessity for multiple-testing correction [127]. Enrichment analysis, on the other hand, uses a more powerful approach whereby the association of biologically related gene sets is tested with a trait [128].
Heck and colleagues conducted gene-set enrichment of analysis of working memory using an n-back task, first in a discovery sample and then in two replication samples, totaling 2,824 individuals [126••]. They found that the voltage-gated cation channel activity gene set was significantly associated with working memory in the discovery sample and in one of the replication samples, and was the top-ranked gene set in the remaining sample. The authors extended this finding to show that alleles from the gene set correlated with working-memory–associated brain activation in brain regions previously shown to be important for working-memory performance. Voltage-gated cation channel activity refers to the transfer of a positively charged ion (for example, calcium, sodium, or potassium) across a cell membrane via ion channels, the permeability of which is mediated by the membrane potential of the cell. Thus, neuronal excitability, which is underlain by the proper functioning of ion channels, was again shown to be crucial for working-memory performance in this study.
Conclusion
Genetic studies have the potential to provide key insights into the molecular mechanisms that underlie working memory. The work of Arnsten and colleagues highlights the role of dynamic networks of neurons, the activity of which is mediated by dopaminergic signaling, such that non-optimal levels of dopamine result in rapid weakening of the network via cAMP-mediated depolarization of neurons [15••]. Genome-wide studies of working memory confirm and extend this work. The associations documented to date converge on the role of neuronal excitability, which fits in neatly with the dynamic network connectivity model of working memory [15••]. Each result highlights the role of genes with a functional role in voltage-gated ion channels and/or prefrontal dopamine expression. These findings represent QTLs, not true functional gene localizations, but nonetheless they provide aninteresting starting point. Future work that aims to characterize functional variants influencing working-memory ability might choose to focus on those genes highlighted in the present review and also those networks in which the genes fall. Those networks should relate to voltage-gated ion channel activity and neuronal excitability. This will expand upon the work discussed here and, in so doing, will shed light on working memory, a construct thought to be extremely important for normal variation in general cognitive ability, as well as in psychiatric and neurodegenerative illness. Confirming gene associations and highlighting functional characterization of those associations might have implications for the development of treatments for mental illness.
Acknowledgments
Financial support for this study was provided by National Institute of Mental Health grants MH078143 (principal investigator: D. C. Glahn), MH078111 (principal investigator: J. Blangero), MH083824 (principal investigator: D. C. Glahn). John Blangero as PI on grant MH083824.
Footnotes
Conflict of Interest Emma Knowles, Laura Almasy, Samuel Mathias, and David McKay have no conflicts of interest. Emma Sprooten is employed at Yale University via the National Institute of Mental Health (NIMH) and has received standard reimbursement for travel to conferences from Yale University/NIMH. David Glahn received a grant from the National Institutes of Health (NIH).
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by the authors.
Contributor Information
E. E. M. Knowles, Department of Psychiatry, Yale University School of Medicine, New Haven, CT, USA; Olin Neuropsychiatric Research Center, Institute of Living, Hartford, Hospital, Hartford, CT, USA
S. R. Mathias, Department of Psychiatry, Yale University School of Medicine, New Haven, CT, USA; Olin Neuropsychiatric Research Center, Institute of Living, Hartford, Hospital, Hartford, CT, USA
D. R. McKay, Department of Psychiatry, Yale University School of Medicine, New Haven, CT, USA; Olin Neuropsychiatric Research Center, Institute of Living, Hartford, Hospital, Hartford, CT, USA
E. Sprooten, Department of Psychiatry, Yale University School of Medicine, New Haven, CT, USA; Olin Neuropsychiatric Research Center, Institute of Living, Hartford, Hospital, Hartford, CT, USA
John Blangero, Department of Genetics, Texas Biomedical Research Institute, San Antonio, TX, USA.
Laura Almasy, Department of Genetics, Texas Biomedical Research Institute, San Antonio, TX, USA.
D. C. Glahn, Department of Psychiatry, Yale University School of Medicine, New Haven, CT, USA; Olin Neuropsychiatric Research Center, Institute of Living, Hartford, Hospital, Hartford, CT, USA
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Logie RH. Working memory in everyday cognition. In: Davies G, Logie RH, editors. Memory in everyday life. Elsevier; Amsterdam: 1993. pp. 173–218. [Google Scholar]
- 2.Shah P, Miyake A. Models of working memory: an introduction. In: Miyake A, Shah P, editors. Models of working memory: mechanisms of active maintenance and executive control. Cambridge University Press; Cambridge: 1999. pp. 1–27. [Google Scholar]
- 3.Fukuda K, Vogel EK. Human variation in overriding attentional capture. J Neurosci. 2009;29(27):8726–33. doi: 10.1523/JNEUROSCI.2145-09.2009. doi:10.1523/JNEUROSCI.2145-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCabe DP, Roediger HL, McDaniel MA, Balota DA, Hambrick DZ. The relationship between working memory capacity and executive functioning: evidence for a common executive attention construct. Neuropsychology. 2010;24(2):222–43. doi: 10.1037/a0017619. doi:10.1037/a0017619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyllonen PC, Christal RE. Reasoning ability is (little more than) working-memory capacity?! Intelligence. 1990;14:389–433. [Google Scholar]
- 6.Süß HM, Oberauer K, Wittmann WW, Wilhelm O, Schulze R. Working-memory capacity explains reasoning ability—and a little bit more. Intelligence. 2002;30:261–88. [Google Scholar]
- 7.Baddeley A. Working memory: theories, models, and controversies. Annu Rev Psychol. 2012;63:1–29. doi: 10.1146/annurev-psych-120710-100422. doi:10.1146/annurev-psych-120710-100422. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Park S. Working memory impairments in schizophrenia: a meta-analysis. J Abnorm Psychol. 2005;114(4):599–611. doi: 10.1037/0021-843X.114.4.599. doi:10. 1037/0021-843X.114.4.599. [DOI] [PubMed] [Google Scholar]
- 9.Buckner RL. Memory and executive function in aging and AD: multiple factors that cause decline and reserve factors that compensate. Neuron. 2004;44(1):195–208. doi: 10.1016/j.neuron.2004.09.006. doi:10.1016/j.neuron. 2004.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Pirkola T, Tuulio-Henriksson A, Glahn D, Kieseppa T, Haukka J, Kaprio J, et al. Spatial working memory function in twins with schizophrenia and bipolar disorder. Biol Psychiatry. 2005;58(12):930–6. doi: 10.1016/j.biopsych.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 11.Owens SF, Picchioni MM, Rijsdijk FV, Stahl D, Vassos E, Rodger AK, et al. Genetic overlap between episodic memory deficits and schizophrenia: results from the Maudsley Twin Study. Psychol Med. 2011;41(3):521–32. doi: 10.1017/S0033291710000942. [DOI] [PubMed] [Google Scholar]
- 12.Goldman-Rakic PS. Cellular basis of working memory. Neuron. 1995;14(3):477–85. doi: 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- 13.Krimer LS, Goldman-Rakic PS. Prefrontal microcircuits: membrane properties and excitatory input of local, medium, and wide arbor interneurons. J Neurosci. 2001;21(11):3788–96. doi: 10.1523/JNEUROSCI.21-11-03788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muly EC, 3rd, Szigeti K, Goldman-Rakic PS. D1 receptor in interneurons of macaque prefrontal cortex: distribution and sub-cellular localization. J Neurosci. 1998;18(24):10553–65. doi: 10.1523/JNEUROSCI.18-24-10553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Arnsten AF, Paspalas CD, Gamo NJ, Yang Y, Wang M. Dynamic network connectivity: a new form of neuroplasticity. Trends Cogn Sci. 2010;14(8):365–75. doi: 10.1016/j.tics.2010.05.003. doi:10.1016/j.tics.2010.05.003. Coined the term “Dynamic network connectivity” to describe the mediating role of dopamine and norepinephrine on dorsolateral prefrontal cortex within the context of working memory performance.
- 16.Arnsten AF, Jin LE. Molecular influences on working memory circuits in dorsolateral prefrontal cortex. Prog Mol Biol Transl Sci. 2014;122:211–31. doi: 10.1016/B978-0-12-420170-5.00008-8. doi:10.1016/B978-0-12-420170-5.00008-8. [DOI] [PubMed] [Google Scholar]
- 17.Miller GA, Galanter E, Pribram KH. Plans and the structure of behavior. Holt, Rinehart and Winston, Inc; New York: 1960. [Google Scholar]
- 18.Awh E, Jonides J. Overlapping mechanisms of attention and spatial working memory. Trends Cogn Sci. 2001;5(3):119–26. doi: 10.1016/s1364-6613(00)01593-x. [DOI] [PubMed] [Google Scholar]
- 19.Oberauer K, Süß HM, Schulze R, Wittmann WW. Working memory capacity—facets of a cognitive ability construct. Personal Individ Differ. 2006;29:1017–45. [Google Scholar]
- 20.Plomin R, DeFries JC, McClearn GE, McGuffin P. Nature, nurture and behavior. In: Plomin R, et al., editors. Behavioral genetics. 5th ed Worth; New York: 2008. pp. 59–91. [Google Scholar]
- 21.Rijsdijk FV, Vernon PA, Boomsma DI. The genetic basis of the relation between speed-of-information-processing and IQ. Behav Brain Res. 1998;95(1):77–84. doi: 10.1016/s0166-4328(97)00212-x. [DOI] [PubMed] [Google Scholar]
- 22.Ando J, Ono Y, Wright MJ. Genetic structure of spatial and verbal working memory. Behav Genet. 2001;31(6):615–24. doi: 10.1023/a:1013353613591. [DOI] [PubMed] [Google Scholar]
- 23.Luciano M, Wright MJ, Geffen GM, Geffen LB, Smith GA, Martin NG. Multivariate genetic analysis of cognitive abilities in an adolescent twin sample. Aust J Psychol. 2004;56:79–88. [Google Scholar]
- 24.Hansell NK, Wright MJ, Luciano M, Geffen GM, Geffen LB, Martin NG. Genetic covariation between event-related potential (ERP) and behavioral non-ERP measures of working-memory, processing speed, and IQ. Behav Genet. 2005;35(6):695–706. doi: 10.1007/s10519-005-6188-2. doi:10.1007/s10519-005-6188-2. [DOI] [PubMed] [Google Scholar]
- 25.Karlsgodt KH, Kochunov P, Winkler AM, Laird AR, Almasy L, Duggirala R, et al. A multimodal assessment of the genetic control over working memory. J Neurosci. 2010;30(24):8197–202. doi: 10.1523/JNEUROSCI.0359-10.2010. doi:10.1523/JNEUROSCI.0359-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg X, Alemany S, Rosa A, Picchioni M, Nenadic I, Owens SF, et al. Substantial genetic link between IQ and working memory: implications for molecular genetic studies on schizophrenia. the European Twin Study of Schizophrenia (EUTwinsS) Am J Med Genet B Neuropsychiatr Genet. 2013;162B(4):413–8. doi: 10.1002/ajmg.b.32158. doi:10.1002/ajmg.b.32158. [DOI] [PubMed] [Google Scholar]
- 27•.Knowles EE, Carless MA, de Almeida MA, Curran JE, McKay DR, Sprooten E, et al. Genome-wide significant localization for working and spatial memory: identifying genes for psychosis using models of cognition. Am J Med Genet B Neuropsychiatr Genet. 2014;165(1):84–95. doi: 10.1002/ajmg.b.32211. doi:10.1002/ajmg.b.32211. Used linkage in extended pedigrees to isolate the gene HEY1 as being influential on a factor-score of workingmemory performance.
- 28.Tuulio-Henriksson A, Haukka J, Partonen T, Varilo T, Paunio T, Ekelund J, et al. Heritability and number of quantitative trait loci of neurocognitive functions in families with schizophrenia. Am J Med Genet. 2002;114(5):483–90. doi: 10.1002/ajmg.10480. doi:10.1002/ajmg. 10480. [DOI] [PubMed] [Google Scholar]
- 29.Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the Consortium on the Genetics of Schizophrenia. Arch Gen Psychiatry. 2007;64(11):1242–50. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Husted JA, Lim S, Chow EW, Greenwood C, Bassett AS. Heritability of neurocognitive traits in familial schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(6):845–53. doi: 10.1002/ajmg.b.30907. doi:10.1002/ajmg.b.30907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calkins ME, Tepper P, Gur RC, Ragland JD, Klei L, Wiener HW, et al. Project Among African-Americans to Explore Risks for Schizophrenia (PAARTNERS): evidence for impairment and heritability of neurocognitive functioning in families of schizophrenia patients. Am J Psychiatry. 2010;167(4):459–72. doi: 10.1176/appi.ajp.2009.08091351. doi:10.1176/appi.ajp.2009.08091351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenartowicz A, Kalar DJ, Congdon E, Poldrack RA. Towards an ontology of cognitive control. Top Cogn Sci. 2010;2:678–92. doi: 10.1111/j.1756-8765.2010.01100.x. [DOI] [PubMed] [Google Scholar]
- 33.Jonides J, Smith EE, Koeppe RA, Awh E, Minoshima S, Mintun MA. Spatial working memory in humans as revealed by PET. Nature. 1993;363(6430):623–5. doi: 10.1038/363623a0. doi:10.1038/363623a0. [DOI] [PubMed] [Google Scholar]
- 34.Petrides M, Alivisatos B, Meyer E, Evans AC. Functional activation of the human frontal cortex during the performance of verbal working memory tasks. Proc Natl Acad Sci U S A. 1993;90(3):878–82. doi: 10.1073/pnas.90.3.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Esposito M, Detre JA, Alsop DC, Shin RK, Atlas S, Grossman M. The neural basis of the central executive system of working memory. Nature. 1995;378(6554):279–81. doi: 10.1038/378279a0. doi:10.1038/378279a0. [DOI] [PubMed] [Google Scholar]
- 36.Glahn DC, Kim J, Cohen MS, Poutanen VP, Therman S, Bava S, et al. Maintenance and manipulation in spatial working memory: dissociations in the prefrontal cortex. Neuroimage. 2002;17(1):201–13. doi: 10.1006/nimg.2002.1161. [DOI] [PubMed] [Google Scholar]
- 37.Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci. 2003;3(4):255–74. doi: 10.3758/cabn.3.4.255. [DOI] [PubMed] [Google Scholar]
- 38.Owen AM, McMillan KM, Laird AR, Bullmore E. N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Map. 2005;25(1):46–59. doi: 10.1002/hbm.20131. doi: 10.1002/hbm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller NG, Knight RT. The functional neuroanatomy of working memory: contributions of human brain lesion studies. Neuroscience. 2006;139(1):51–8. doi: 10.1016/j.neuroscience.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Niendam TA, Laird AR, Ray KL, Dean YM, Glahn DC, Carter CS. Meta-analytic evidence for a superordinate cognitive control network subserving diverse executive functions. Cogn Affect Behav Neurosci. 2012;12:241–68. doi: 10.3758/s13415-011-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Reilly RC, Rudy JW. Conjunctive representations in learning and memory: principles of cortical and hippocampal function. Psychol Rev. 2001;108(2):311–45. doi: 10.1037/0033-295x.108.2.311. [DOI] [PubMed] [Google Scholar]
- 42.Rasmus F. Genes, brain, and cognition: a roadmap for the cognitive scientist. Cognition. 2006;101:247–69. doi: 10.1016/j.cognition.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 43.van der Sluis S, Verhage M, Posthuma D, Dolan CV. Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS One. 2010;5(11):e13929. doi: 10.1371/journal.pone.0013929. doi:10.1371/journal.pone.0013929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deary IJ. Intelligence. Annu Rev Psychol. 2012;63:453–82. doi: 10.1146/annurev-psych-120710-100353. doi:10.1146/annurev-psych-120710-100353. [DOI] [PubMed] [Google Scholar]
- 45.Almasy L. Quantitative risk factors as indices of alcoholism susceptibility. Ann Med. 2003;35(5):337–43. doi: 10.1080/07853890310004903. [DOI] [PubMed] [Google Scholar]
- 46.Fuster JM. The prefrontal cortex: anatomy, physiology, and neuropsychology of the frontal lobe. 3rd edition Lippincott-Raven; Philadelphia: 1997. [Google Scholar]
- 47.Eichenbaum H. Declarative memory: insights from cognitive neurobiology. Annu Rev Psychol. 1997;48:547–72. doi: 10.1146/annurev.psych.48.1.547. doi:10.1146/annurev.psych.48.1.547. [DOI] [PubMed] [Google Scholar]
- 48.Bearden CE, Karlsgodt KH, Bachman P, van Erp TG, Winkler AM, Glahn DC. Genetic architecture of declarative memory: implications for complex illnesses. Neuroscientist. 2012;18(5):516–32. doi: 10.1177/1073858411415113. doi:10.1177/1073858411415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dumitriu D, Hao J, Hara Y, Kaufmann J, Janssen WG, Lou W, et al. Selective changes in thin spine density and morphology in monkey prefrontal cortex correlate with aging-related cognitive impairment. J Neurosci. 2010;30(22):7507–15. doi: 10.1523/JNEUROSCI.6410-09.2010. doi:10.1523/JNEUROSCI.6410-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuster JM, Alexander GE. Neuron activity related to short-term memory. Science. 1971;173(3997):652–4. doi: 10.1126/science.173.3997.652. [DOI] [PubMed] [Google Scholar]
- 51.Kojima S, Goldman-Rakic PS. Functional analysis of spatially discriminative neurons in prefrontal cortex of rhesus monkey. Brain Res. 1984;291(2):229–40. doi: 10.1016/0006-8993(84)91255-1. [DOI] [PubMed] [Google Scholar]
- 52.Miller EK, Li L, Desimone R. Activity of neurons in anterior inferior temporal cortex during a short-term memory task. J Neurosci. 1993;13(4):1460–78. doi: 10.1523/JNEUROSCI.13-04-01460.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T, Niki H. Hippocampal unit activity and delayed response in the monkey. Brain Res. 1985;325(1-2):241–54. doi: 10.1016/0006-8993(85)90320-8. [DOI] [PubMed] [Google Scholar]
- 54.Koch KW, Fuster JM. Unit activity in monkey parietal cortex related to haptic perception and temporary memory. Exp Brain Res. 1989;76(2):292–306. doi: 10.1007/BF00247889. [DOI] [PubMed] [Google Scholar]
- 55.Zhou YD, Fuster JM. Neuronal activity of somatosensory cortex in a cross-modal (visuo-haptic) memory task. Exp Brain Res. 1997;116(3):551–5. doi: 10.1007/pl00005783. [DOI] [PubMed] [Google Scholar]
- 56.Egorov AV, Hamam BN, Fransen E, Hasselmo ME, Alonso AA. Graded persistent activity in entorhinal cortex neurons. Nature. 2002;420(6912):173–8. doi: 10.1038/nature01171. doi:10.1038/nature01171. [DOI] [PubMed] [Google Scholar]
- 57.Simon H, Scatton B, Le Moal M. Definitive disruption of spatial delayed alternation in rats after lesions in the ventral mesencephalic tegmentum. Neurosci Lett. 1979;15(2-3):319–24. doi: 10.1016/0304-3940(79)96133-0. [DOI] [PubMed] [Google Scholar]
- 58.Sawaguchi T, Goldman-Rakic PS. D1 dopamine receptors in prefrontal cortex: involvement in working memory. Science. 1991;251(4996):947–50. doi: 10.1126/science.1825731. [DOI] [PubMed] [Google Scholar]
- 59.Phillips AG, Ahn S, Floresco SB. Magnitude of dopamine release in medial prefrontal cortex predicts accuracy of memory on a delayed response task. J Neurosci. 2004;24(2):547–53. doi: 10.1523/JNEUROSCI.4653-03.2004. doi:10.1523/JNEUROSCI.4653-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aalto S, Bruck A, Laine M, Nagren K, Rinne JO. Frontal and temporal dopamine release during working memory and attention tasks in healthy humans: a positron emission tomography study using the high-affinity dopamine D2 receptor ligand [11C]FLB 457. J Neurosci. 2005;25(10):2471–7. doi: 10.1523/JNEUROSCI.2097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dash PK, Moore AN, Kobori N, Runyan JD. Molecular activity underlying working memory. Learn Mem. 2007;14(8):554–63. doi: 10.1101/lm.558707. [DOI] [PubMed] [Google Scholar]
- 62.Llinas RR. The intrinsic electrophysiological properties of mammalian neurons: insights into central nervous system function. Science. 1988;242(4886):1654–64. doi: 10.1126/science.3059497. [DOI] [PubMed] [Google Scholar]
- 63.Goldman-Rakic PS. Regional and cellular fractionation of working memory. Proc Natl Acad Sci U S A. 1996;93(24):13473–80. doi: 10.1073/pnas.93.24.13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Payton A. The impact of genetic research on our understanding of normal cognitive ageing: 1995 to 2009. Neuropsychol Rev. 2009;19(4):451–77. doi: 10.1007/s11065-009-9116-z. doi:10.1007/s11065-009-9116-z. [DOI] [PubMed] [Google Scholar]
- 65.Houlihan LM, Harris SE, Luciano M, Gow AJ, Starr JM, Visscher PM, et al. Replication study of candidate genes for cognitive abilities: the Lothian birth cohort 1936. Genes Brain Behav. 2009;8(2):238–47. doi: 10.1111/j.1601-183X.2008.00470.x. doi:10.1111/j.1601-183X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- 66.Barnett JH, Scoriels L, Munafo MR. Meta-analysis of the cognitive effects of the catechol-O-methyltransferase gene Val158/108Met polymorphism. Biol Psychiatry. 2008;64(2):137–44. doi: 10.1016/j.biopsych.2008.01.005. doi:10.1016/j.biopsych.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 67.Chabris CF, Hebert BM, Benjamin DJ, Beauchamp J, Cesarini D, van der Loos M, et al. Most reported genetic associations with general intelligence are probably false positives. Psychol Sci. 2012;23(11):1314–23. doi: 10.1177/0956797611435528. doi:10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014;81(3):484–503. doi: 10.1016/j.neuron.2014.01.027. doi:10.1016/j.neuron.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18(7):815–20. doi: 10.1038/ejhg.2010.2. doi:10.1038/ejhg.2010.2. Used association analysis of the animal fluency task to highlight the influence of the gene KCNB2 as being influential on aspects of working memory associated with fluency tasks.
- 70.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, et al. Prediction of the coding sequences of unidentified human genes: I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1(1):27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 71.Nagase T, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes: XXII. The complete sequences of 50 new cDNA clones which code for large proteins. DNA Res. 2001;8(6):319–27. doi: 10.1093/dnares/8.6.319. [DOI] [PubMed] [Google Scholar]
- 72.Nagase T, Koga H, Ohara O. Kazusa mammalian cDNA resources: towards functional characterization of KIAA gene products. Brief Funct Genom Proteomic. 2006;5(1):4–7. doi: 10.1093/bfgp/ell005. [DOI] [PubMed] [Google Scholar]
- 73.Azuma T. Working memory and perseveration in verbal fluency. Neuropsychology. 2004;18(1):69–77. doi: 10.1037/0894-4105.18.1.69. doi:10.1037/0894-4105.18.1.69. [DOI] [PubMed] [Google Scholar]
- 74.Fink M, Duprat F, Lesage F, Heurteaux C, Romey G, Barhanin J, et al. A new K+ channel beta subunit to specifically enhance Kv2.2 (CDRK) expression. J Biol Chem. 1996;271(42):26341–8. doi: 10.1074/jbc.271.42.26341. [DOI] [PubMed] [Google Scholar]
- 75.Lai HC, Jan LY. The distribution and targeting of neuronal voltage-gated ion channels. Nat Rev Neurosci. 2006;7(7):548–62. doi: 10.1038/nrn1938. [DOI] [PubMed] [Google Scholar]
- 76.Misonou H. Homeostatic regulation of neuronal excitability by K(+) channels in normal and diseased brains. Neuroscientist. 2010;16(1):51–64. doi: 10.1177/1073858409341085. doi:10.1177/1073858409341085. [DOI] [PubMed] [Google Scholar]
- 77.Hermanstyne TO, Kihira Y, Misono K, Deitchler A, Yanagawa Y, Misonou H. Immunolocalization of the voltage-gated potassium channel Kv2.2 in GABAergic neurons in the basal forebrain of rats and mice. J Comp Neurol. 2010;518(21):4298–310. doi: 10.1002/cne.22457. doi:10.1002/cne.22457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeSousa NJ, Beninger RJ, Jhamandas K, Boegman RJ. Stimulation of GABAB receptors in the basal forebrain selectively impairs working memory of rats in the double Y-maze. Brain Res. 1994;641(1):29–38. doi: 10.1016/0006-8993(94)91811-2. [DOI] [PubMed] [Google Scholar]
- 79.Carr DB, Sesack SR. GABA-containing neurons in the rat ventral tegmental area project to the prefrontal cortex. Synapse. 2000;38(2):114–23. doi: 10.1002/1098-2396(200011)38:2<114::AID-SYN2>3.0.CO;2-R. doi:10.1002/1098-2396(200011)38:2<114::AID-SYN2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez-Cubillo I, Perianez JA, Adrover-Roig D, Rodriguez-Sanchez JM, Rios-Lago M, Tirapu J, et al. Construct validity of the trail making test: role of task-switching, working memory, inhibition/interference control, and visuomotor abilities. J Int Neuropsychol Soc. 2009;15(3):438–50. doi: 10.1017/S1355617709090626. doi:10.1017/S1355617709090626. [DOI] [PubMed] [Google Scholar]
- 81.Hobert MA, Niebler R, Meyer SI, Brockmann K, Becker C, Huber H, et al. Poor trail making test performance is directly associated with altered dual task prioritization in the elderly—baseline results from the TREND study. PLoS One. 2011;6(11):e27831. doi: 10.1371/journal.pone.0027831. doi:10.1371/journal.pone.0027831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lezak MD. Orientation and attention. In: Lezak MD, Howieson DB, Loring DW, editors. Neuropsychological assessment. 4th ed. Oxford University Press; New York: 2004. pp. 365–7. [Google Scholar]
- 83.Hazan J, Lamy C, Melki J, Munnich A, de Recondo J, Weissenbach J. Autosomal dominant familial spastic paraplegia is genetically heterogeneous and one locus maps to chromosome 14q. Nat Genet. 1993;5(2):163–7. doi: 10.1038/ng1093-163. doi:10.1038/ng1093-163. [DOI] [PubMed] [Google Scholar]
- 84.Kang SY, Lee MH, Lee SK, Sohn YH. Levodopa-responsive parkinsonism in hereditary spastic paraplegia with thin corpus callosum. Parkinsonism Relat Disord. 2004;10(7):425–7. doi: 10.1016/j.parkreldis.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 85.Micheli F, Cersosimo MG, Zuniga Ramirez C. Hereditary spastic paraplegia associated with dopa-responsive parkinsonism. Mov Disord. 2006;21(5):716–7. doi: 10.1002/mds.20800. doi:10.1002/mds.20800. [DOI] [PubMed] [Google Scholar]
- 86.Guidubaldi A, Piano C, Santorelli FM, Silvestri G, Petracca M, Tessa A, et al. Novel mutations in SPG11 cause hereditary spastic paraplegia associated with early-onset levodopa-responsive parkinsonism. Mov Disord. 2011;26(3):553–6. doi: 10.1002/mds.23552. doi:10.1002/mds.23552. [DOI] [PubMed] [Google Scholar]
- 87.Kim JS, Kim JM, Kim YK, Kim SE, Yun JY, Jeon BS. Striatal dopaminergic functioning in patients with sporadic and hereditary spastic paraplegias with parkinsonism. J Korean Med Sci. 2013;28(11):1661–6. doi: 10.3346/jkms.2013.28.11.1661. doi:10.3346/jkms.2013.28.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee Y, Paik D, Bang S, Kang J, Chun B, Lee S, et al. Loss of spastic paraplegia gene atlastin induces age-dependent death of dopaminergic neurons in drosophila. Neurobiol Aging. 2008;29(1):84–94. doi: 10.1016/j.neurobiolaging.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 89.Albin RL, Koeppe RA, Rainier S, Fink JK. Normal dopaminergic nigrostriatal innervation in SPG3A hereditary spastic paraplegia. J Neurogenet. 2008;22(4):289–94. doi: 10.1080/01677060802337307. doi:10.1080/01677060802337307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90•.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18(23):4650–61. doi: 10.1093/hmg/ddp413. doi:10.1093/hmg/ddp413. Used association analysis of the CANTAB, and specifically the efficiency index of the search strategy employed in the spatial working-memory task, to highlight the role of the gene FXYD2 in working memory.
- 91.Kim JW, Lee Y, Lee IA, Kang HB, Choe YK, Choe IS. Cloning and expression of human cDNA encoding Na+, K(+)-ATPase gamma-subunit. Biochim Biophys Acta. 1997;1350(2):133–5. doi: 10.1016/s0167-4781(96)00219-9. [DOI] [PubMed] [Google Scholar]
- 92.Silva EC, Masui DC, Furriel RP, McNamara JC, Barrabin H, Scofano HM, et al. Identification of a crab gill FXYD2 protein and regulation of crab microsomal Na, K-ATPase activity by mammalian FXYD2 peptide. Biochim Biophys Acta. 2012;1818(11):2588–97. doi: 10.1016/j.bbamem.2012.05.009. doi:10.1016/j.bbamem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 93.Bertorello AM, Hopfield JF, Aperia A, Greengard P. Inhibition by dopamine of (Na(+)+K+)ATPase activity in neostriatal neurons through D1 and D2 dopamine receptor synergism. Nature. 1990;347(6291):386–8. doi: 10.1038/347386a0. doi:10.1038/347386a0. [DOI] [PubMed] [Google Scholar]
- 94.Rajanna B, Hobson M, Harris L, Ware L, Chetty CS. Effects of cadmium and mercury on Na(+)-K+, ATPase and uptake of 3H-dopamine in rat brain synaptosomes. Arch Int Physiol Biochim. 1990;98(5):291–6. doi: 10.3109/13813459009113989. [DOI] [PubMed] [Google Scholar]
- 95.Chen C, Lokhandwala MF. Inhibition of Na+, K(+)-ATPase in rat renal proximal tubules by dopamine involved DA-1 receptor activation. Naunyn Schmiedeberg’s Arch Pharmacol. 1993;347(3):289–95. doi: 10.1007/BF00167447. [DOI] [PubMed] [Google Scholar]
- 96.Shahedi M, Laborde K, Azimi S, Hamdani S, Sachs C. Mechanisms of dopamine effects on Na-K-ATPase activity in Madin-Darby canine kidney (MDCK) epithelial cells. Pflugers Arch. 1995;429(6):832–40. doi: 10.1007/BF00374808. [DOI] [PubMed] [Google Scholar]
- 97.Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev. 1998;78(1):189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 98•.Papassotiropoulos A, Henke K, Stefanova E, Aerni A, Muller A, Demougin P, et al. A genome-wide survey of human short-term memory. Mol Psychiatry. 2011;16(2):184–92. doi: 10.1038/mp.2009.133. doi:10.1038/mp.2009.133. Used association analysis of short-term memory storage to highlight the role of SCN1A in working memory.
- 99.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26(1):13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 100.Gulledge AT, Jaffe DB. Multiple effects of dopamine on layer V pyramidal cell excitability in rat prefrontal cortex. J Neurophysiol. 2001;86(2):586–95. doi: 10.1152/jn.2001.86.2.586. [DOI] [PubMed] [Google Scholar]
- 101.Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24(4):343–5. doi: 10.1038/74159. doi:10.1038/74159. [DOI] [PubMed] [Google Scholar]
- 102.Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68(6):1327–32. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han S, Tai C, Westenbroek RE, Yu FH, Cheah CS, Potter GB, et al. Autistic-like behaviour in Scn1a+/- mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489(7416):385–90. doi: 10.1038/nature11356. doi:10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seamans JK, Gorelova N, Durstewitz D, Yang CR. Bidirectional dopamine modulation of GABAergic inhibition in prefrontal cortical pyramidal neurons. J Neurosci. 2001;21(10):3628–38. doi: 10.1523/JNEUROSCI.21-10-03628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Silver H, Feldman P, Bilker W, Gur RC. Working memory deficit as a core neuropsychological dysfunction in schizophrenia. Am J Psychiatry. 2003;160(10):1809–16. doi: 10.1176/appi.ajp.160.10.1809. [DOI] [PubMed] [Google Scholar]
- 106.Enomoto T, Tse MT, Floresco SB. Reducing prefrontal gamma-aminobutyric acid activity induces cognitive, behavioral, and dopaminergic abnormalities that resemble schizophrenia. Biol Psychiatry. 2011;69(5):432–41. doi: 10.1016/j.biopsych.2010.09.038. doi:10.1016/j.biopsych.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 107•.Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. Used association analysis of an abstract reasoning task to highlight the role of SORL1 in working memory.
- 108.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102(38):13461–6. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carey RM, Balcz BA, Lopez-Coviella I, Slack BE. Inhibition of dynamin-dependent endocytosis increases shedding of the amyloid precursor protein ectodomain and reduces generation of amyloid beta protein. BMC Cell Biol. 2005;6:30. doi: 10.1186/1471-2121-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26(5):1596–603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choy RW, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid beta (abeta) production in the trans-golgi network. Proc Natl Acad Sci U S A. 2012;109(30):E2077–82. doi: 10.1073/pnas.1208635109. doi:10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Palop JJ, Mucke L. Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci. 2010;13(7):812–8. doi: 10.1038/nn.2583. doi:10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun J, Jia P, Fanous AH, Oord E v d, Chen X, Riley BP, et al. Schizophrenia gene networks and pathways and their applications for novel candidate gene selection. PLoS One. 2010;5(6):e11351. doi: 10.1371/journal.pone.0011351. doi:10.1371/journal.pone.0011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wen Y, Miyashita A, Kitamura N, Tsukie T, Saito Y, Hatsuta H, et al. SORL1 is genetically associated with neuropathologically characterized late-onset Alzheimer’s disease. J Alzheimers Dis. 2013;35(2):387–94. doi: 10.3233/JAD-122395. doi:10.3233/JAD-122395. [DOI] [PubMed] [Google Scholar]
- 116.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.05.006. doi:10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61(8):1200–5. doi: 10.1001/archneur.61.8.1200. doi:10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 118.Sager KL, Wuu J, Leurgans SE, Rees HD, Gearing M, Mufson EJ, et al. Neuronal LR11/sorLA expression is reduced in mild cognitive impairment. Ann Neurol. 2007;62(6):640–7. doi: 10.1002/ana.21190. doi:10.1002/ana.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bralten J, Arias-Vasquez A, Makkinje R, Veltman JA, Brunner HG, Fernandez G, et al. Association of the Alzheimer’s gene SORL1 with hippocampal volume in young, healthy adults. Am J Psychiatry. 2011;168(10):1083–9. doi: 10.1176/appi.ajp.2011.10101509. doi:10.1176/appi.ajp.2011.10101509. [DOI] [PubMed] [Google Scholar]
- 120.Kline RB. Principles and practice of structural equation modeling. 3rd ed Guilford; New York: 2011. [Google Scholar]
- 121.Costa RM, Drew C, Silva AJ. Notch to remember. Trends Neurosci. 2005;28(8):429–35. doi: 10.1016/j.tins.2005.05.003. doi:10.1016/j.tins.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 122.Fuke S, Sasagawa N, Ishiura S. Identification and characterization of the Hesr1/Hey1 as a candidate trans-acting factor on gene expression through the 3′ non-coding polymorphic region of the human dopa-mine transporter (DAT1) gene. J Biochem. 2005;137(2):205–16. doi: 10.1093/jb/mvi020. [DOI] [PubMed] [Google Scholar]
- 123.Fuke S, Minami N, Kokubo H, Yoshikawa A, Yasumatsu H, Sasagawa N, et al. HESR1 knockout mice exhibit behavioral alterations through the dopaminergic nervous system. J Neurosci Res. 2006;84(7):1555–63. doi: 10.1002/jnr.21062. doi:10.1002/jnr.21062. [DOI] [PubMed] [Google Scholar]
- 124.Dudchenko PA. An overview of the tasks used to test working memory in rodents. Neurosci Biobehav Rev. 2004;28(7):699–709. doi: 10.1016/j.neubiorev.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 125.Kanno K, Kokubo H, Takahashi A, Koide T, Ishiura S. Enhanced prepulse inhibition and low sensitivity to a dopamine agonist in HESR1 knockout mice. J Neurosci Res. 2014;92(3):287–97. doi: 10.1002/jnr.23291. doi:10.1002/jnr.23291. [DOI] [PubMed] [Google Scholar]
- 126••.Heck A, Fastenrath M, Ackermann S, Auschra B, Bickel H, Coynel D, et al. Converging genetic and functional brain imaging evidence links neuronal excitability to working memory, psychiatric disease, and brain activity. Neuron. 2014;81(5):1203–13. doi: 10.1016/j.neuron.2014.01.010. doi:10.1016/j.neuron.2014.01.010. Used enrichment analysis to highlight the role of neuronal excitability, and more specifically the voltage-gated cation channel activity gene set, in n-back task performance, a classic measure of working memory, and brain activity associated with task performance.
- 127.Ohashi J, Tokunaga K. The power of genome-wide association studies of complex disease genes: statistical limitations of indirect approaches using SNP markers. J Hum Genet. 2001;46(8):478–82. doi: 10.1007/s100380170048. doi:10.1007/s100380170048. [DOI] [PubMed] [Google Scholar]
- 128.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11(12):843–54. doi: 10.1038/nrg2884. doi:10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]