Abstract
Proteins carry out important functions as they fold themselves. Protein misfolding occurs during different biochemical processes and may lead to the development of diseases such as cancer, which is characterized by genetic instability. The cancer microenvironment exposes malignant cells to a variety of stressful conditions that may further promote protein misfolding. Tumor development and progression often arises from mutations that interfere with the appropriate function of tumor-suppressor proteins and oncogenes. These may be due to alteration of catalytic activity of the protein, loss of binding sites for effector proteins or alterations of the native folded protein conformation. Src family kinases, p53, mTOR and C-terminus of HSC70 interacting protein (CHIPs) are some examples associated with protein misfolding and tumorigenesis. Molecular chaperones, such as heat-shock protein (HSP)70 and HSP90, assist protein folding and recognize target misfolded proteins for degradation. It is likely that this misfolding in cancer is linked by common principles, and may, therefore, present an exciting possibility to identify common targets for therapeutic intervention. Here we aim to review a number of examples that show how alterations in the folding of tumor-suppressor proteins or oncogenes lead to tumorigenesis. The possibility of targeting the targets to repair or degrade protein misfolding in cancer therapy is discussed.
Keywords: cancer, cancer targets, chaperones, heat-shock proteins, protein misfolding, proteomics
Protein folding is the process by which the newly synthesized protein molecule folds into its unique, 3D structure in order to acquire its functionally active, native state. The folding of a polypeptide chain to a unique, functionally competent structure is a key step in cellular protein synthesis. In vivo, protein folding can be: initiated before the completion of protein synthesis, where the nascent chain is still attached to the ribosome [1]; undergo the major part of their folding in the cytoplasm after release from the ribosome; or fold in specific compartments, such as mitochondria or the endoplasmic reticulum (ER), after trafficking and translocation through membranes (Figure 1) [2–4]. The misfolding and mis-assembly that occur within the ER are largely controlled by the interplay of chaperones and proteasome machinery and occur later in the secretory pathway, making it a challenge to correct [5].
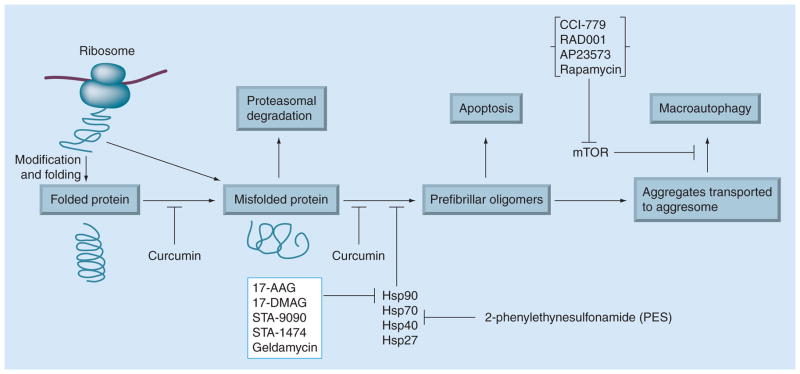
Figure 1. Model of protein misfolding and therapeutic targets in cancer.
Proteins are synthesized on ribosomes from the genetic information encoded in cellular DNA and undergo the major part of their folding in the cytoplasm after release from the ribosome. Newly synthesized proteins are translocated into the endoplasmic reticulum, where they fold into their 3D structures. Correctly folded proteins are transported to the Golgi complex and then delivered to the extracellular environment. Under certain circumstances, a folded protein is converted to a misfolded protein by various stresses, gene mutations or proteolysis. During normal conditions, misfolded proteins are detected by a quality-control mechanism and are degraded by the ubiquitin–proteosome pathway. A misfolded protein can be induced to form prefibrillar oligomers at higher concentrations and transported to form aggresomes. These aggregated proteins at the aggresome are targeted for degradation via macroautophagy. Accumulation of prefibrillar oligomers at the aggresome pathway may induce apoptosis. Curcumin, a potential antioxidant or anticancer compound may inhibit protein misfolding and/or aggregation. Protein aggregation can also be blocked by upregulation of molecular chaperones with heat shock protein (HSP)90 inhibitors (17-allylamino-demethoxygeldanamycin (AAG), 17-[dimethylaminoethylamino]-17-demethoxygeldanamycin (DMAG), STA-9090, STA-1474 and geldamycin) or HSP70 inhibitors, such as 2-phenylethyneusulfonamide. Aggregate clearance can be upregulated via inhibition of mTOR by rapamycin that inhibits macroautophagy. Analogues of rapamycin, such as CCI-779 (temsirolimus), RAD001 (everolimus) and AP23573, are likely to be the first mTOR-perturbing molecules to be approved for anticancer use in humans. Redrawn with permission of Cambridge University Press from [106].
Numerous factors can induce the accumulation of misfolded or unfolded proteins including cytotoxic stresses, such as hypoxia, nutrient deprivation, redox, cellular environment due to aging or temperature fluctuation, genetic mutation, or exposure to amino acid analogues and Ca2+ misregulation [6]. Protein misfolding or unfolding is normally counterbalanced by quality control machinery, including chaperones, activated due to stress or heat-shock responses. The folding process requires extremely high fidelity to ensure that misfolded proteins do not increase in concentration and mediate undesirable effects. The cellular folding machinery is adept at correctly folding large quantities of protein, however, when this system is overwhelmed with protein in its non-native conformation, a diverse range of protein-folding diseases, such as cancer, may occur [7].
Understanding carcinogenesis, tumor progression and metastasis, requires a careful analysis of effector molecules, such as key proteins, which have critical functions in the complex network of signaling pathways in cancer [8,9]. Proteomic techniques can play a considerable role in global analysis of cellular response to determine protein expression which assists in understanding gene functions [10]. Two key technologies underpinning these studies in cancer tissues are 2D polyacrylamide gel electrophoresis (2DE) and mass spectrometry (MS). Utilizing these techniques, evaluation of protein expression levels, regardless of the pattern, may potentially diagnose and/or identify molecular therapeutic targets that can lead to drug development. One particular area that has clinical implications for proteomics studies in this regard includes identifying a series of modification of heat-shock proteins (HSPs) in cancer. HSPs seem to have fundamental tumor promoting activities and are among the most repeatedly identified proteins utilizing proteomic approaches [11]. Here we discuss the major events and targets of misfolded proteins in cancer. Tumor-suppressor proteins and oncogenes targeting protein misfolding will be explored and highlighted as potential pharmacological targets in cancer therapy. We will also shed light on recent proteomic strategies that are likely to be most effective for cancer prevention and treatment.
Proteomic techniques
Proteomic technologies identify changes in protein expression, PTMs, subcellular distribution, and decipher protein–protein interactions. In cancer, proteomic approaches aim to isolate, identify and recognize the pattern of expression of diverse proteins involved in tumor progression, development and therapeutic targeting and have been used by cancer researchers to look at proteins that are differentially displayed in a given tissue, body fluid or serum to identify biomarkers that could be important for early detection, diagnosis and treatment. The study of proteomics also allows for a greater understanding of protein interactions within cellular signaling pathways and responses to internal and external signals. Proteomic techniques are broadly divided into gel-based and non-gel-based techniques, each with its own advantages and disadvantages.
Gel-based proteomics
Although 2DE is limited by low sensitivity and low solubility of proteins, it is still the mainstay of proteomic techniques. This technique allows for exploration of hundreds to thousands of proteins at once using an extensive separation technique based on the charge, according to their different electrofocusing points in the first dimension and SDS-PAGE, according to their different molecular weights in the second dimension. If a protein of interest is found, it can be broken down further into peptides by digestion with site-specific proteases. This technique can be labor intensive, suffers from poor resolution at high pI values, and lacks reproducibility. The development of 2D difference in-gel electrophoresis (DIGE) by Unlu et al. in 1997 significantly improved the accuracy of protein identification and led to more precise quantification [12]. This advance introduced pre-labeling of proteins with positively charged, amine reactive and molecular weight-matched fluorescent cyanine dyes (Cy2, Cy3 and Cy5), followed by simultaneous electrophoresis on the same 2D gel [13]. This resolved many of the above described problems, including reduction in inter-gel variability, the number of gels required, accurate spot matching and spot identification using MS.
Non-gel-based proteomics
Non-gel-based proteomic approaches involve isotope-coded affinity tagging (ICAT), isobaric tags for relative and absolute quantitation (iTRAQ) and electron spray ionization tandem MS (ESI MS/MS), which rely on liquid chromatography (LC) for protein separation interfaced with high-end mass spectrometers for protein identification [14]. The advantages of these techniques include automation and reduced sample requirement, but lack universal availability and have higher costs [15].
Surface-enhanced laser desorption/ionization (SELDI) time of flight (TOF) MS enables high-throughput analysis of individual clinical samples, such as serum, urine and other biofluids, using protein chips with various surface characteristics, but it usually does not provide the identity of differentially expressed proteins [16]. Methods for quantitative comparison of protein abundance between two biological samples using label-free shotgun proteomics are well established based on spectral counting techniques [17].
Recent progress in non-gel-based proteomics has included development of better surface chemistry, capture molecule attachment, and protein labeling [14]. Non-gel-based proteomics approaches or protein chips include chemical (e.g., ionic, hydrophobic or hydrophilic) or biochemical (e.g., antibody, receptor or DNA) surfaces to capture proteins of interest. The chemically modified surfaces are used to retain a group of proteins on the basis of a specific physical property, such as hydrophobicity or charge. Biologically modified surfaces are typically used to isolate a specific protein or functional class of proteins.
Major targets of protein misfolding in cancer
A failure to adequately respond to increases in the requirement for cellular folding may lead to an accumulation of misfolded proteins and development of cancer (Table 1), as summarized in Figure 1. Misfolded tumor suppressors are simply inactive and result in a loss-of function phenotype (VHL and NF2) or the mutated protein may adopt an aberrant conformation that is regulated differently than the wild-type protein (p53 and Src family kinases [SFKs]) leading to tumorigenesis [18]. The unambiguous mediators of protein folding are the cellular chaperones, which include the heat-shock family proteins. HSPs constitute an evolutionarily conserved family that is ubiquitous in nature and exerts prominent functions in protein synthesis, transport, maintenance and degradation. The molecular chaperones of the HSP family can be classified into two groups – stress-repressible HSPs and stress-inducible HSPs – which actively correct folding and refolding mechanism upon denaturation [19]. HSP70 and HSP90 play key roles in assisting protein folding and in recognizing and targeting misfolded proteins for degradation [20]. The C-terminus of HSP70-interacting protein (CHIP) suppresses tumorigenesis and metastatic cellular phenotypes in cancer cells. The mTOR, integrates diverse signals to regulate fundamental cellular processes, such as translation, cell growth, autophagy and stress response [21–23].
Table 1.
Proteins involved in misfolding cancer.
Overexpression of HSP has been reported in many solid tumors during cancer progression [24] and may in part account for the ability of cancer cells to maintain protein homoeostasis [25]. Therefore, targeting HSPs with chemical inhibitors may disrupt multiple cancer promoting processes. Therefore, there has been widespread interest in the development of small-molecule inhibitors that target individual HSPs to alter their function and cause the proteasomal degradation of oncogenic proteins [26]. Upregulation of HSP90 and HSP70 is relatively common in human tumors and is also often associated with increased chemotherapy resistance and poor patient prognosis [19]. Both of these chaperones vary in response to the change in folding requirements of normal and cancer cell types.
Major tumor-suppressor proteins & oncogenes associated with protein misfolding & cancer p53
The tumor-suppressor protein p53 is a sequence-specific transcription factor whose function is to maintain genome integrity. Owing to its ability to initiate cell cycle arrest, apoptosis and senescence, it is a key protein involved in the cell’s defense against tumorigenesis [27]. p53 is a modular protein containing an N-terminal transactivation domain, followed by a proline-rich region, a central DNA binding domain, a tetramerization domain, and a C terminus [28]. The central or core domain of p53, comprising residues 94-312, is responsible for specific DNA interactions, and 97% of the point mutations in p53 are in this domain [28].
Based on these functions, p53 represents the classic example of protein misfolding-mediated tumor development. Inactivation of p53 by mutation is a key molecular event, and is seen in more than 50% of human cancers, making it a likable target for cancer therapies [29]. Due to this frequent mutation-induced misfolding of p53 in human cancers, an efficient treatment strategy would be to reverse these folding defects that can restore wild-type functions to mutant p53 in tumors. However, structural studies of p53 show that the extent of misfolding differs among mutants. Therefore, it appears unlikely that different mutants possess a defined alternative fold [30].
p53 has been observed to interact with several HSPs, such as HSP40, HSP70 and HSP90 [31]. Interestingly, the unfolded mutant p53 binds to HSP70 with higher affinity than the wild-type protein [32], suggesting that binding of HSPs stabilizes p53 in an unfolded conformation. Therefore, disruption of HSP binding to mutant p53 should rescue p53 unfolded conformation.
Src family kinases
The proto-oncogene c-Src (Src) encodes a non-receptor tyrosine kinase whose expression and activity are correlated with cancer progression, advanced malignancy and poor prognosis in a variety of human cancers [33–36]. The SFKs are involved in regulating important mechanisms of receptor tyrosine kinases, G-protein-coupled receptors, and focal adhesion kinase (FAK), thereby influencing many aspects of tumor cell behavior, including proliferation, survival, angiogenesis, adhesion, invasion and metastasis [33–37]. Destabilization of Src in relation to oncogenic mutations supports a possible relationship between tumor development and protein misfolding [38]. Proper folding by molecular chaperones, especially HSP90, is an important part of the regulation of SFKs, including c-Src [39]. In addition to its role in folding, HSP90 appears to protect constitutively activated SFK proteins from degradation by the ubiquitin–proteasome system. In doing so, HSP90 allows the accumulation of mutant activated SFK associated with tumor development [39]. Src requires HSP90 as a substrate for the regulatory kinase Csk and for the maturation of its catalytic activity [40,41]. The site of interaction of HSP90 with SFKs has been narrowed down to the catalytic domain [42]. This has been demonstrated by the ability of geldanamycin to inhibit folding and induce misfolding of the catalytic domain of the SFK Lck [43].
CHIP
CHIP is a cytoplasmic protein with highly conserved amino acid sequences across species. CHIP interacts with the molecular chaperone complex HSC70–HSP70–HSP90 through a tetratricopeptide repeat (TPR) domain [44,45]. The interaction with this chaperone complex results in client substrate ubiquitylation and degradation by the proteasome. Thus, CHIP acts to tilt the folding–refolding machinery toward the degradative pathway. CHIP’s ability to degrade proteins that are signatures of disease, for example, ErbB2 in breast and ovarian cancers, could prove to be a point of therapeutic intervention [46]. CHIP has dual activities as a co-chaperone (through its interactions with the cytoplasmic chaperones HSP70 and HSP90) as well as a quality control ubiquitin ligase, binding to molecular chaperones and ubiquitylating misfolded proteins to target them for proteasome-dependent degradation [46,47].
HSP90
The molecular chaperone HSP90 was initially identified as one of the highly conserved HSPs involved in the stress response. Evidence suggests that expression of HSP90 and other HSPs is mediated by the transcription factor heat-shock factor 1 (HSF1). Under normal conditions, HSP90 binds to HSF1, preventing HSF1 release and transcriptional activation of the heat-shock response [48]. However, it has been postulated that, under stress, the HSF1/HSP90 complex is disassembled and HSF1 trimerizes and ultimately translocates to the nucleus, where it binds to the heat-shock binding elements and initiates transcription of the heat-shock genes that encode for multiple HSPs (HSP27, HSP40, HSP70 and HSP90) [49]. Accumulation of misfolded substrates diverts the chaperone and triggers nuclear localization of HSF1 resulting the upregulation of targets involved in protection from cellular stress. The HSP90 family stabilizes the structure and function of different nuclear receptors, protein kinases (AKT, c-Src and Raf-1) and transcription factors (MyoD, NF-κB, nuclear steroid receptors) [11,50,51]. In addition, the HSP90 protein-folding machinery assists in the solubilization and refolding of aggregated and denatured proteins [52]. As a result, small-molecule HSP90 inhibitors can exert differing activities.
Owing to an overwhelming need to fold overexpressed and mutated proteins, HSP90 and associated chaperones have been shown to be elevated in human tumors [53,54]. HSP90 can facilitate genetic variation in cancer by permitting the function of mutated and less stable proteins with tumor promotion or progression activities [11]. The environmental conditions found in tumors, such as hypoxia, low pH and poor nutritional status, can further destabilize proteins, making them even more dependent on HSP90 activity [26]. Therefore, targeting HSP90 is warranted and may be a new and powerful therapeutic approach in the treatment of cancer [55,56]. Interestingly, it has been demonstrated that HSP90 in cancer cells exhibits a higher affinity for selective inhibitors than in normal cells, although the mechanisms underlying the tumor selectivity of HSP90 inhibitors are not fully understood [57,58]. HSP90 is expressed on tumor cell surfaces and is present in advanced, malignant melanomas, suggesting that extracellular HSP90 may also be a viable anticancer target [59]. This provides an opportunity for drug development that exhibits higher differential selectivity.
HSP70
HSP70 is involved in the folding of nonnative proteins and can prevent aggregation, promote folding to the native state, and solubilize and refold aggregated proteins (Figure 1) [42]. The interaction of HSP70 with exposed hydrophobic amino acids in polypeptides [20,42] assist in the folding of newly synthesized proteins and minimizes their aggregation [60]. Levels of HSP70 protein are elevated in several cancer cell lines, are abundantly expressed in a variety of malignant tumors and often correlate with tumor metastasis that result into poor outcomes in cancer patients [61–63]. Downregulation of HSP70 in cancer cells induce differentiation and cell death [64]. The cancer microenvironment exposes cancer cells to a variety of stressful conditions that promote protein misfolding. As opposed to normal cells, HSP70 helps cancer cells to survive under stress and interact with numerous substrates and co-chaperones. The secreted and/or membrane-anchored forms of HSP70 can chaperone tumor antigens to various antigen-presenting cells, thereby inducing an immune response to neoplasia that could have diagnostic, prognostic and therapeutic value [65,66]. The interaction of HSP70 with regulatory proteins continues in activation cycles that involves HSP90 and a number of co-chaperones [42]. HSP90 and HSP70 share common substrates in anti-apoptotic pathway and both are over-expressed in tumor cells. Therefore, the neutralization of the function of HSP70 or inhibition of its expression may inhibit tumor growth by chemotherapeutic agents and, therefore, is a potentially interesting molecular target for cancer.
mTOR
The mTOR is a serine/threonine protein kinase, with a large molecular size near 300 kDa, belongs to the phosphatidylinositol kinase-related kinase (PIKK) family. Recent studies have revealed the existence of two different mTOR complexes [67] (mTORC1 and mTORC2), which differ in molecular composition, cellular function, upstream regulation and sensitivity to rapamycin (Figure 1). The complex of rapamycin with its intracellular receptor FKB12 binds directly to mTORC1 and, at least in vitro, suppresses mTORC1-mediated phosphorylation of the substrates S6K1 and 4EBP1 [68]. As a target of rapamycin, mTORC1 has been the main focus in most mTOR-related studies. Furthermore, rapamycin and its analogues inhibit several processes that are relevant to the anticancer properties [69]. Biomarkers indicate that the mTOR pathway is hyperactive in certain cancers, suggesting mTOR as an attractive target for cancer therapy [70].
Role of proteomics in cancer relates to misfolded proteins
Proteomic approaches play a major role in understanding the diagnosis, prognosis and potential treatment of human cancer. Profiling of differentially expressed proteins is the most important and useful approach in the study of misfolding proteins and cancer. Key technologies underpinning these studies in cancer tissue are 2DE and MS. Although MALDI-TOF and SELDI-TOF-MS is the mainstay for serum or plasma analysis, other methods, including isotope-coded affinity tag technology, shotgun proteomics, reverse-phase protein arrays and antibody microarrays, are emerging as alternative proteomic technologies [17,71,72].
A number of proteins associated with misfolding-associated cancer development have been identified using proteomic approaches. Numerous studies have correlated changes in HSP70 and HSP90 expression in different cancer types [56,62,66,73]. Different proteomic approaches have confirmed the correlation between HSP70 overexpression and the aggressiveness of various types of cancer [74,75]. Grossmann et al. employed peptide elution followed by MS analysis in a novel fashion to identify endogenous HSP70-binding peptides [76]. A number of peptides that were identified led to the determination of a HSP70 binding motif for endogenously bound peptides. This study also identified a number of endogenously bound peptides, providing the opportunities to further characterize the role of HSP70 in the processing, presentation, and degradation of peptides, leading to a better understanding of the interactions between HSP70, co-activators, the proteosome and MHC in fundamental areas of tumor cells.
Recently, Fujita et al. identified novel tumor antigens in the sera of patients with esophageal squamous cell carcinoma (ESCC) using a proteomic-based approach [77]. They showed that concentrations of the serum HSP70 autoantibody were significantly higher in patients with ESCC than for patients with gastric or colon cancer or healthy individuals, suggesting that proteomic approaches have the potential to define the precise role of the cancer-related immune response.
Similar to HSP70, several proteomic-based studies have shown HSP90 overexpression in multiple tumor cell lines [56,66,78–80]. In a study by Hayashi et al., fibrosarcoma cells were characterized by differential expression of nine proteins, including two HSP90 isoforms HSP90-α and HSP90-β [78]. Seven of these proteins (calreticulin precursor, tropomyosin 1 α-chain, annexin A5, PEBP and Prx 1) were overexpressed and two (Anp32e and HDGF) were downregulated. Numerous studies have also studied the HSP90 interactome, and its dynamic interactions influenced by HSP90 ligands [81–85].
A recent study revealed comprehensive information on the influence of p53 in the modulation of cell-to-cell and cell-to-stroma interactions by the differential release of proteins [86]. The release of p53-modulated proteins in the H358/TetOn/p53 cell line was analyzed using liquid chromatography-matrix assisted laser desorption ionization-MS/MS (LC-MALDI-MS/MS) for differential analysis with iTRAQ labeling. Moreover, our studies using 2-DE analysis reveals the influence of p53 expression on the modulation of PTMs of secretary proteins [Singh O et al., Unpublished Data]. Differential display analysis was performed for the expression of proteins in human breast carcinoma cell lines with mutated p53 after treatment with PRIMA-1 using a 2DE coupled with MS analyses and revealed an important complementary role of both functional and expression proteomics in the identification of molecular targets involved in the restoration of p53 signaling with small-molecule inhibitors [87]. Recently, Shor et al. performed quantitative phospho-proteomics for cancer cells treated with the mTOR active-site inhibitor WYE-125132 (WYE-132), a highly potent and specific inhibitor of mTORC1 and mTORC2 [88]. They mechanistically identified Ser-75 of Maf1, a known repressor of RNA polymerase III (Pol III) transcription, further supporting the role of mTOR signaling to fine tune the rates of Pol III transcription in cancer cells to provide growth stimulatory signals.
Targets for protein repair & cancer therapy
Chemical chaperones are small molecules that bind to a protein, stabilize the folded state and thereby reduce protein misfolding. Extensive studies on protein misfolding have described the inhibition of protein aggregation by small compounds such as curcumin (Figure 1), a biologically active phytochemical isolated from turmeric root [89]. Emerging data suggest that it may be possible to upregulate molecular chaperones using several different pharmacological strategies. Notably, the HSP90 inhibitor geldanamycin (GA) and its derivatives (17-allylamino-demethoxygeldanamycin) 17-AAG and (17-[dimethylaminoethylamino]-17-demethoxygeldanamycin) 17-DMAG induce the expression of HSP70 and HSP27 in cancer models [90,91], raising the possibility that these agents may be useful for treating protein-misfolding diseases (Figure 1) [92]. These inhibitors could hamper various client proteins that act as key players in different aberrant signaling pathways [56]. Data from earlier studies supports a model in which GA inhibits the formation of toxic protein aggregates by regulating HSP70 [92,93]. The 17-AAG derivative is less toxic than GA in rats and causes growth inhibition in breast, melanoma, and ovarian mouse xenograft models [94]. 17-AAG or tanespimycin and 17-DMAG are currently being tested in Phase I and II clinical trials as potential anticancer agents (Table 2) [91], suggesting that they may be suitable for administration to patients with protein-misfolding disorders. Toxicity of 17-AAG is of concern; however, new formulations and structural analogues have improved tolerability [94–96]. It has been shown that simultaneous suppression of two cytosolic HSP70s, HSC70 and HSP72, sensitize cancer cells to 17-AAG [62,63,73,97].
Table 2.
Pharmacological inhibitors and protein-based cancer therapeutics.
| Inhibitors | Protein | Clinical trial | Cancer type | Ref. |
|---|---|---|---|---|
| 17-AAG | HSP90 | Phase I | Breast, solid tumors, lymphoma | [108,119,201] |
| 17-DMAG | HSP90 | Phase I | Lung, solid tumors | [120,121,201] |
| STA-9090 | HSP90 | Phase II | Non-small-cell lung cancer, gastrointestinal stroma tumor | [99,201] |
| STA-1474 | HSP90 | Preclinical | Osteosarcoma | [122] |
| Geldamycin | HSP90 | Phase I | Breast | [123] |
| CCI-779 | mTOR | Phase III, Phase II, Phase I | Breast, endometrial, myeloma, glioma | [124–127] |
| RAD001 | mTOR | Phase III, Phase II | Breast, carcinoid, pancreatic neuroendocrine | [128,129] |
| AP23573 | mTOR | Phase II | Breast | [130] |
Heat-shock protein 90 inhibition offers additional promise in the treatment of a wide variety of solid tumors. Various HSP90 inhibitors have entered clinical trials and are reviewed and summarized in Table 2 [26]. These inhibitors represent multiple drug classes in adult and pediatric oncology clinical trials [94]. Recent observations suggest that the efficacy of HSP90 inhibitors may be improved by co-administration with HSP70 inhibitors. Consistent with this approach, siRNA was used against HSP70 to increase the efficacy of 17-AAG in cancer cells [98]. STA-9090 is a potent and synthetic small molecule that acts as a HSP90 inhibitor and shows 100-times greater potential than the first-generation HSP90 inhibitors, as well as activity against a wider range of kinases (Table 2). Clinical trials of STA-9090 on multiple cancer types [99] highlight the inter-connectivity of chaperone networks and the ability of this basic biological approach to assist in designing therapeutic strategies [201].
Multiple lines of evidence suggest that the accumulation of cytosolic protein aggregates can be suppressed dramatically via pharmacological upregulation of macroautophagy (Figure 1). This approach may have greater therapeutic potential than the enhancement of proteasome activity, given that upregulation of the ubiquitin–proteasome pathway (in contrast to autophagy) is likely to increase the risk of cancer by causing depletion of short-lived proteins with anti-tumorigenic activity (e.g., p53) [100]. Potential approaches to stabilize the structure of mutant p53 include the use of several peptides and small-molecule compounds that act to restore specific DNA-binding, transcription and apoptosis functions to mutant p53. These synthetic peptides derived from the C-terminus of p53, as well as peptides such as CDB3, and compounds isolated from chemical library screening, such as CP-31398, a small synthetic molecule, reactivate p53 and induce massive apoptosis (PRIMA-1 [101]. CDB3 stabilizes the structure of mutant p53 proteins [81] by binding to mutant p53 and efficiently inducing the refolding of two hot spot p53 mutants, His273 and His175, in cancer cells [82]. PRIMA-1 selectively inhibits the growth of tumor cells by provoking apoptosis in a transcription-dependent fashion through conformational manipulation of p53 mutants to restore sequence-specific DNA binding [83]. CP-31398 [84] has the capacity to restore wild-type p53 function from its mutant form [85]. However, the exact molecular mechanism by which these molecules act upon the misfolded mutant p53 to restore its activity is yet unclear.
2-phenylethynesulfonamide (PES) is a lead compound that specifically inhibits HSP70 (Figure 1) [86], interfering with its cancer cell survival activity [102]. Studies show that the interaction of PES with HSP70 blocks its stress-relieving functions and also induces HSP70-dependent cell death by disrupting the cell’s ability to remove damaged components. Paradoxically, for a compound first identified for blocking apoptosis, PES does kill cells, however, through a different mechanism. Given the heterogeneity of cancer cells, simultaneously disabling networks of signaling pathways may be important.
The dysregulation of mTOR signaling is implicated in a number of human diseases, including cancer. Despite the substantial pre-clinical data indicating that rapamycin and its analogues have antitumor effects and that mTOR participates in many cancer-related pathways, these molecules have not shown universal antitumor activity in early clinical trials. Analogues of rapamycin, such as CCI-779 (also known as temsirolimus; Wyeth), RAD001 (also known as everolimus; Novartis) and AP23573 (Ariad Pharmaceuticals), are likely to be the first mTOR-perturbing molecules to be approved for anticancer use in humans (Table 2) (reviewed in [69]). The US FDA has approved the rapamycin analogs CCI-779 and RAD001 for the treatment of advanced-stage renal cell carcinoma and sarcoma, respectively [103–105].
Expert commentary
There has been tremendous progress over the past few years in the development of novel therapeutic approaches targeting protein misfolding in cancer. In the case of proteins that normally have a compact, globular structure, misfolding and aggregation have been inhibited successfully by compounds that stabilize the native conformation [106]. Continued high-throughput screening efforts will lead to the identification of additional compounds that interfere with protein self-assembly. Emerging data also suggest that the depletion of aggregating proteins using RNAi methods in vivo may be a powerful approach to prevent misfolding diseases [106,107].
Mutant p53 reactivation by small-molecule inhibitors is a rapidly evolving approach with great potential for the development of novel anticancer therapeutics [30]. The inactivation of the p53 pathway is most likely a universal feature of tumor cells. The successful identification of p53-targeting therapeutics in recent years has encouraged investigators to initiate screening of chemical libraries with the aim of identifying novel molecular therapeutics targeting the p53 pathway in human cancer.
Considering the role of HSPs in the induction of cancer cell resistance to different therapeutic approaches, targeting HSP inhibition may be a useful therapeutic strategy supplementing conventional anticancer therapy. HSP90 inhibitors have already entered preclinical and clinical evaluation and have revealed promising results as single agents or in combination with chemotherapy. Targets to the HSP90 in the treatment of cancer will focus on progress in developing agents that exhibit reduced toxicity compared with ansamycin-based molecules, such as 17-AAG. Based on preclinical results, HSP90 inhibitors for anti-cancer therapeutics have a potentially bright future. Moreover, HSP70 proteins are now seen as attractive targets in cancer. Among challenges, the relatively high levels of HSP70 expression, the large number of family members, the complex series of functions and the mechanistic details of multiple co-chaperone interactions remain to be analyzed [62]. The preclinical and clinical results of HSP90 inhibitors [108,109] have provided proof-of-concept for targeting at least one mediator of the HSF1 stress response pathway; however, it will be necessary to accurately define the toxicologic profile of HSP-modulating drugs, especially in terms of global cell proteome modification.
Proteomics of HSPs have revealed a significant role of drug-induced overexpression and highlighted multiple potential clinical applications. For example, numerous proteomic studies clearly correlate the level of HSP70 overexpression with therapeutic resistance [110–112] and this finding has fundamental implications in drug therapy or targeting. Moreover, proteomics studies have also revealed the modulation of HSP induction and release through the presentation of tumor-specific peptides to antigen-presenting cells, which could ameliorate the immune response towards cancer. Therefore, proteomic approaches could be useful to obtain a better understanding of some of the biological activities and mechanisms related to the induction of HSPs.
There are a number of outstanding challenges that still remain to be addressed. However, the use of -omics-based technologies and genome-wide RNAi [113–115] will improve our understanding by dissecting the mechanism of tumorigenesis. The insights highlighted above will make it possible to devise therapeutic strategies that modulate cellular signaling pathways disrupted by protein misfolding and aggregation, including apoptosis, tumorigenicity, signal transduction and transcription.
Five-year view
The continued elucidation of protein-folding pathways of oncogenes and tumor-suppressor proteins will provide new mechanisms to correct the defects of misfolded variants. The mechanistic views of aggregated oligomers and protofibrils/fibrils for cell death in cancer cells are still unclear. Understanding the regulatory pathways of cellular chaperones has extended targets for drug intervention. Induction in signaling pathway-based approaches may provide a better therapeutic benefit in the near future.
Drug combinations targeting protein aggregation as well as the resulting perturbation of cellular pathways will provide the most effective treatment strategies to protein misfolding in cancer. Approaches targeting misfolded proteins in cancer therapy are already under Phase I and Phase II clinical trials. Additionally, anti-apoptotic approaches for HSPs are being tested in clinical trials and in ongoing combination drug trials. The results from current clinical trials will significantly impact future cancer treatment strategies. The identification of protein folding and rescue in cancer opens new-insight into cancer-causing pathways and therapies.
Key issues.
Proteomic approaches have identified a series of modification of heat-shock protein (HSP) expression patterns in cancer.
Proteomic and RNAi methods have resulted in the designing of several novel protein-aggregation inhibitors.
Proteomics technology in protein misfolding research has improved our understanding of signaling pathways involved in tumor progression and enhanced our abilities of molecular targeting.
Antiapoptotic approaches for HSPs are now being tested in clinical trials.
Drug candidates preventing the formation of oligomers, protofibrils or fibrils, and reducing toxicity in animal models of misfolding diseases, are a high priority.
Drug combinations targeting protein aggregation provide the most effective treatment in cancer associated with protein misfolding.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Prog Nucleic Acid Res Mol Biol. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125(3):443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 4.Luheshi LM, Dobson CM. Bridging the gap: from protein misfolding to protein misfolding diseases. FEBS Lett. 2009;583(16):2581–2586. doi: 10.1016/j.febslet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 5.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 6•.So AY, de la Fuente E, Walter P, Shuman M, Bernales S. The unfolded protein response during prostate cancer development. Cancer Metastasis Rev. 2009;28(1–2):219–223. doi: 10.1007/s10555-008-9180-5. Comprehensive and good review on the overview over unfolded protein response (UPR) pharmacological mediators and important biomarkers for selection of targeted therapy. [DOI] [PubMed] [Google Scholar]
- 7.Blagosklonny MV. Loss of function and p53 protein stabilization. Oncogene. 1997;15(16):1889–1893. doi: 10.1038/sj.onc.1201374. [DOI] [PubMed] [Google Scholar]
- 8.Hoehn GT, Suffredini AF. Proteomics. Crit Care Med. 2005;33(12 Suppl):S444–S448. doi: 10.1097/01.ccm.0000187001.44171.5e. [DOI] [PubMed] [Google Scholar]
- 9.Alessandro R, Fontana S, Kohn E, De Leo G. Proteomic strategies and their application in cancer research. Tumori. 2005;91(6):447–455. doi: 10.1177/030089160509100601. [DOI] [PubMed] [Google Scholar]
- 10.Yarmush ML, Jayaraman A. Advances in proteomic technologies. Annu Rev Biomed Eng. 2002;4:349–373. doi: 10.1146/annurev.bioeng.4.020702.153443. [DOI] [PubMed] [Google Scholar]
- 11.Bottoni P, Giardina B, Scatena R. Proteomic profiling of heat shock proteins: an emerging molecular approach with direct pathophysiological and clinical implications. Proteom Clin Appl. 2009;3(6):636–653. doi: 10.1002/prca.200800195. [DOI] [PubMed] [Google Scholar]
- 12.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 13.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382(3):669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 14.DeSouza L, Diehl G, Rodrigues MJ, et al. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2005;4(2):377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 15.Pastwa E, Somiari SB, Czyz M, Somiari RI. Proteomics in human cancer research. Proteom Clin Appl. 2007;1(1):4–17. doi: 10.1002/prca.200600369. [DOI] [PubMed] [Google Scholar]
- 16.Hariharan D, Weeks ME, Crnogorac-Jurcevic T. Application of proteomics in cancer gene profiling: two-dimensional difference in gel electrophoresis (2D-DIGE) Methods Mol Biol. 2010;576:197–211. doi: 10.1007/978-1-59745-545-9_11. [DOI] [PubMed] [Google Scholar]
- 17.Old WM, Meyer-Arendt K, Aveline-Wolf L, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Scott MD, Frydman J. Aberrant protein folding as the molecular basis of cancer. Methods Mol Biol. 2003;232:67–76. doi: 10.1385/1-59259-394-1:67. [DOI] [PubMed] [Google Scholar]
- 19•.Morano KA. New tricks for an old dog: the evolving world of Hsp70. Ann NY Acad Sci. 2007;1113:1–14. doi: 10.1196/annals.1391.018. Discusses in detail the discovery and application of heat shock protein (HSP)70, from molecule to man, including small-molecule modulation and clinical impact. [DOI] [PubMed] [Google Scholar]
- 20.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 21••.Kajiro M, Hirota R, Nakajima Y, et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol. 2009;11(3):312–319. doi: 10.1038/ncb1839. C-terminus of HSC70 interacting protein (CHIP) suppresses the expression of oncogenic proteins that enhance anchorage-independent tumor growth. [DOI] [PubMed] [Google Scholar]
- 22.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25(48):6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- 24.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31(3):164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 25••.Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ. Targeting HSP90 for cancer therapy. Br J Cancer. 2009;100(10):1523–1529. doi: 10.1038/sj.bjc.6605066. Combinations of HSP90 inhibitors with proteosomal inhibitors, histone deacetylase inhibitors (HDACis), small-molecule RTK inhibitors or TRAIL may lead to greater efficacy and improved clinical outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solit DB, Chiosis G. Development and application of Hsp90 inhibitors. Drug Discov Today. 2008;13(1–2):38–43. doi: 10.1016/j.drudis.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 28.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 29.Nikolova PV, Wong KB, DeDecker B, Henckel J, Fersht AR. Mechanism of rescue of common p53 cancer mutations by second-site suppressor mutations. EMBO J. 2000;19(3):370–378. doi: 10.1093/emboj/19.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selivanova G, Wiman KG. Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene. 2007;26(15):2243–2254. doi: 10.1038/sj.onc.1210295. [DOI] [PubMed] [Google Scholar]
- 31.Walerych D, Kudla G, Gutkowska M, et al. Hsp90 chaperones wild-type p53 tumor suppressor protein. J Biol Chem. 2004;279(47):48836–48845. doi: 10.1074/jbc.M407601200. [DOI] [PubMed] [Google Scholar]
- 32.Rudiger S, Freund SM, Veprintsev DB, Fersht AR. CRINEPT-TROSY NMR reveals p53 core domain bound in an unfolded form to the chaperone Hsp90. Proc Natl Acad Sci USA. 2002;99(17):11085–11090. doi: 10.1073/pnas.132393699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irby RB, Yeatman TJ. Role of Src expression and activation in human cancer. Oncogene. 2000;19(49):5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- 34.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4(6):470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 35.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22(4):337–358. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 36.Wheeler DL, Iida M, Dunn EF. The role of Src in solid tumors. Oncologist. 2009;14(7):667–678. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 38.Falsone SF, Leptihn S, Osterauer A, Haslbeck M, Buchner J. Oncogenic mutations reduce the stability of SRC kinase. J Mol Biol. 2004;344(1):281–291. doi: 10.1016/j.jmb.2004.08.091. [DOI] [PubMed] [Google Scholar]
- 39.Bijlmakers MJ, Marsh M. Hsp90 is essential for the synthesis and subsequent membrane association, but not the maintenance, of the Src-kinase p56(lck) Mol Biol Cell. 2000;11(5):1585–1595. doi: 10.1091/mbc.11.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci USA. 1993;90(15):7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Singer MA, Lindquist S. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc Natl Acad Sci USA. 1999;96(1):109–114. doi: 10.1073/pnas.96.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartson SD, Ottinger EA, Huang W, Barany G, Burn P, Matts RL. Modular folding and evidence for phosphorylation-induced stabilization of an hsp90-dependent kinase. J Biol Chem. 1998;273(14):8475–8482. doi: 10.1074/jbc.273.14.8475. [DOI] [PubMed] [Google Scholar]
- 44.Demand J, Luders J, Hohfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18(4):2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballinger CA, Connell P, Wu Y, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Connell P, Ballinger CA, Jiang J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 48.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9(2):122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson LB, Blagg BS. To fold or not to fold: modulation and consequences of Hsp90 inhibition. Future Med Chem. 2009;1(2):267–283. doi: 10.4155/fmc.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296(5576):2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 51.Arthur JC, Lich JD, Aziz RK, Kotb M, Ting JP. Heat shock protein 90 associates with monarch-1 and regulates its ability to promote degradation of NF-κB-inducing kinase. J Immunol. 2007;179(9):6291–6296. doi: 10.4049/jimmunol.179.9.6291. [DOI] [PubMed] [Google Scholar]
- 52.Weis F, Moullintraffort L, Heichette C, Chretien D, Garnier C. The 90-kDa heat shock protein Hsp90 protects tubulin against thermal denaturation. J Biol Chem. 2010;285(13):9525–9534. doi: 10.1074/jbc.M109.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jameel A, Skilton RA, Campbell TA, Chander SK, Coombes RC, Luqmani YA. Clinical and biological significance of HSP89 α in human breast cancer. Int J Cancer. 1992;50(3):409–415. doi: 10.1002/ijc.2910500315. [DOI] [PubMed] [Google Scholar]
- 54.Yufu Y, Nishimura J, Nawata H. High constitutive expression of heat shock protein 90 α in human acute leukemia cells. Leuk Res. 1992;16(6–7):597–605. doi: 10.1016/0145-2126(92)90008-u. [DOI] [PubMed] [Google Scholar]
- 55.Soti C, Nagy E, Giricz Z, Vigh L, Csermely P, Ferdinandy P. Heat shock proteins as emerging therapeutic targets. Br J Pharmacol. 2005;146(6):769–780. doi: 10.1038/sj.bjp.0706396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 57.Kamal A, Thao L, Sensintaffar J, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425(6956):407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 58.Maroney AC, Marugan JJ, Mezzasalma TM, et al. Dihydroquinone ansamycins: toward resolving the conflict between low in vitro affinity and high cellular potency of geldanamycin derivatives. Biochemistry. 2006;45(17):5678–5685. doi: 10.1021/bi0524969. [DOI] [PubMed] [Google Scholar]
- 59.Didelot C, Lanneau D, Brunet M, et al. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr Med Chem. 2007;14(27):2839–2847. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- 60•.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16(6):589–597. doi: 10.1038/nsmb.1614. Recent and comprehensive review on the mechanism of early co-translational events involving the ribosome that guide cytosolic proteins to their native state. [DOI] [PubMed] [Google Scholar]
- 61.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10(2):86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powers MV, Jones K, Barillari C, Westwood I, van Montfort RL, Workman P. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010;9(8) doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 63.Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6(11):1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 64.Nylandsted J, Brand K, Jaattela M. Heat shock protein 70 is required for the survival of cancer cells. Ann NY Acad Sci. 2000;926:122–125. doi: 10.1111/j.1749-6632.2000.tb05605.x. [DOI] [PubMed] [Google Scholar]
- 65.Parmiani G, Testori A, Maio M, et al. Heat shock proteins and their use as anticancer vaccines. Clin Cancer Res. 2004;10(24):8142–8146. doi: 10.1158/1078-0432.CCR-04-1194. [DOI] [PubMed] [Google Scholar]
- 66.Schmitt E, Gehrmann M, Brunet M, Multhoff G, Garrido C. Intracellular and extracellular functions of heat shock proteins: repercussions in cancer therapy. J Leukoc Biol. 2007;81(1):15–27. doi: 10.1189/jlb.0306167. [DOI] [PubMed] [Google Scholar]
- 67.Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 68.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6(9):729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 69.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4(5):335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 70•.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. Another recent review on the use of rapamycin in oncology and discussion on the future of mTOR-targeted therapy. [DOI] [PubMed] [Google Scholar]
- 71.Reymond MA, Schlegel W. Proteomics in cancer. Adv Clin Chem. 2007;44:103–142. doi: 10.1016/s0065-2423(07)44004-5. [DOI] [PubMed] [Google Scholar]
- 72.Sun Y, Zang Z, Xu X, et al. Differential proteomics identification of HSP90 as potential serum biomarker in hepatocellular carcinoma by two-dimensional electrophoresis and mass spectrometry. Int J Mol Sci. 2010;11(4):1423–1433. doi: 10.3390/ijms11041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73••.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell. 2008;14(3):250–262. doi: 10.1016/j.ccr.2008.08.002. Demonstrated that the simultaneous reduction of expression of HSC70 and HSP72 induces proteasome-dependent degradation of HSP90 client proteins, G1 cell cycle arrest and apoptosis in tumorigenic cell lines but not in nontumorigenic lines. [DOI] [PubMed] [Google Scholar]
- 74.Bottoni P, Giardina B, Martorana GE, et al. A two-dimensional electrophoresis preliminary approach to human hepatocarcinoma differentiation induced by PPAR-agonists. J Cell Mol Med. 2005;9(2):462–467. doi: 10.1111/j.1582-4934.2005.tb00371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He QY, Cheung YH, Leung SY, Yuen ST, Chu KM, Chiu JF. Diverse proteomic alterations in gastric adenocarcinoma. Proteomics. 2004;4(10):3276–3287. doi: 10.1002/pmic.200300916. [DOI] [PubMed] [Google Scholar]
- 76.Grossmann ME, Madden BJ, Gao F, et al. Proteomics shows Hsp70 does not bind peptide sequences indiscriminately in vivo. Exp Cell Res. 2004;297(1):108–117. doi: 10.1016/j.yexcr.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 77.Fujita Y, Nakanishi T, Miyamoto Y, et al. Proteomics-based identification of autoantibody against heat shock protein 70 as a diagnostic marker in esophageal squamous cell carcinoma. Cancer Lett. 2008;263(2):280–290. doi: 10.1016/j.canlet.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 78.Hayashi E, Kuramitsu Y, Okada F, et al. Proteomic profiling for cancer progression: differential display analysis for the expression of intracellular proteins between regressive and progressive cancer cell lines. Proteomics. 2005;5(4):1024–1032. doi: 10.1002/pmic.200401132. [DOI] [PubMed] [Google Scholar]
- 79.Schumacher JA, Crockett DK, Elenitoba-Johnson KS, Lim MS. Proteome-wide changes induced by the Hsp90 inhibitor, geldanamycin in anaplastic large cell lymphoma cells. Proteomics. 2007;7(15):2603–2616. doi: 10.1002/pmic.200700108. [DOI] [PubMed] [Google Scholar]
- 80.Maloney A, Clarke PA, Naaby-Hansen S, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67(7):3239–3253. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 81.Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res. 2007;6(5):1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- 82.Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579(28):6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 83.Falsone SF, Gesslbauer B, Rek A, Kungl AJ. A proteomic approach towards the Hsp90-dependent ubiquitinylated proteome. Proteomics. 2007;7(14):2375–2383. doi: 10.1002/pmic.200600996. [DOI] [PubMed] [Google Scholar]
- 84.Tsaytler PA, Krijgsveld J, Goerdayal SS, Rudiger S, Egmond MR. Novel Hsp90 partners discovered using complementary proteomic approaches. Cell Stress Chaperones. 2009;14(6):629–638. doi: 10.1007/s12192-009-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gano JJ, Simon JA. A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol Cell Proteomics. 2010;9(2):255–270. doi: 10.1074/mcp.M900261-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chenau J, Michelland S, de Fraipont F, et al. The cell line secretome, a suitable tool for investigating proteins released in vivo by tumors: application to the study of p53-modulated proteins secreted in lung cancer cells. J Proteome Res. 2009;8(10):4579–4591. doi: 10.1021/pr900383g. [DOI] [PubMed] [Google Scholar]
- 87.Lee K, Wang T, Paszczynski AJ, Daoud SS. Expression proteomics to p53 mutation reactivation with PRIMA-1 in breast cancer cells. Biochem Biophys Res Commun. 2006;349(3):1117–1124. doi: 10.1016/j.bbrc.2006.08.152. [DOI] [PubMed] [Google Scholar]
- 88.Shor B, Wu J, Shakey Q, et al. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem. 2010;285(20):15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J Biol Chem. 2007;282(14):10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 90.Drysdale MJ, Brough PA, Massey A, Jensen MR, Schoepfer J. Targeting Hsp90 for the treatment of cancer. Curr Opin Drug Discov Devel. 2006;9(4):483–495. [PubMed] [Google Scholar]
- 91.McCollum AK, Teneyck CJ, Sauer BM, Toft DO, Erlichman C. Up-regulation of heat shock protein 27 induces resistance to 17-allylamino-demethoxygeldanamycin through a glutathione-mediated mechanism. Cancer Res. 2006;66(22):10967–10975. doi: 10.1158/0008-5472.CAN-06-1629. [DOI] [PubMed] [Google Scholar]
- 92.McLean PJ, Klucken J, Shin Y, Hyman BT. Geldanamycin induces Hsp70 and prevents α-synuclein aggregation and toxicity in vitro. Biochem Biophys Res Commun. 2004;321(3):665–669. doi: 10.1016/j.bbrc.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 93.Hay DG, Sathasivam K, Tobaben S, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13(13):1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 94.Kim YS, Alarcon SV, Lee S, et al. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem. 2009;9(15):1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410(3):439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 96.Taldone T, Sun W, Chiosis G. Discovery and development of heat shock protein 90 inhibitors. Bioorg Med Chem. 2009;17(6):2225–2235. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patury S, Miyata Y, Gestwicki JE. Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem. 2009;9(15):1337–1351. doi: 10.2174/156802609789895674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo F, Rocha K, Bali P, et al. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 2005;65(22):10536–10544. doi: 10.1158/0008-5472.CAN-05-1799. [DOI] [PubMed] [Google Scholar]
- 99.Lin TY, Bear M, Du Z, et al. The novel HSP90 inhibitor STA-9090 exhibits activity against Kit-dependent and -independent malignant mast cell tumors. Exp Hematol. 2008;36(10):1266–1277. doi: 10.1016/j.exphem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 101.Selivanova G, Iotsova V, Okan I, et al. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat Med. 1997;3(6):632–638. doi: 10.1038/nm0697-632. [DOI] [PubMed] [Google Scholar]
- 102• •.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. Demonstrated that phenylethynesulfonamide, a small molecule, disrupts actions of HSP70 in multiple signaling pathways and also aids in the development of new cancer therapies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 104.Kapoor A. Inhibition of mTOR in kidney cancer. Curr Oncol. 2009;16(Suppl 1):S33–S39. doi: 10.3747/co.v16i0.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mita MM, Mita AC, Chu QS, et al. Phase I trial of the novel mammalian target of rapamycin inhibitor deforolimus (AP23573; MK-8669) administered intravenously daily for 5 days every 2 weeks to patients with advanced malignancies. J Clin Oncol. 2008;26(3):361–367. doi: 10.1200/JCO.2007.12.0345. [DOI] [PubMed] [Google Scholar]
- 106.Rochet JC. Novel therapeutic strategies for the treatment of protein-misfolding diseases. Expert Rev Mol Med. 2007;9(17):1–34. doi: 10.1017/S1462399407000385. [DOI] [PubMed] [Google Scholar]
- 107.Saito Y, Yokota T, Mitani T, et al. Transgenic small interfering RNA halts amyotrophic lateral sclerosis in a mouse model. J Biol Chem. 2005;280(52):42826–42830. doi: 10.1074/jbc.M507685200. [DOI] [PubMed] [Google Scholar]
- 108.Banerji U, O’Donnell A, Scurr M, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23(18):4152–4161. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 109.Modi S, Stopeck AT, Gordon MS, et al. Combination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2 overexpressing breast cancer: a Phase I dose-escalation study. J Clin Oncol. 2007;25(34):5410–5417. doi: 10.1200/JCO.2007.11.7960. [DOI] [PubMed] [Google Scholar]
- 110.Castagna A, Antonioli P, Astner H, et al. A proteomic approach to cisplatin resistance in the cervix squamous cell carcinoma cell line A431. Proteomics. 2004;4(10):3246–3267. doi: 10.1002/pmic.200400835. [DOI] [PubMed] [Google Scholar]
- 111.Short DM, Heron ID, Birse-Archbold JL, Kerr LE, Sharkey J, McCulloch J. Apoptosis induced by staurosporine alters chaperone and endoplasmic reticulum proteins: identification by quantitative proteomics. Proteomics. 2007;7(17):3085–3096. doi: 10.1002/pmic.200600964. [DOI] [PubMed] [Google Scholar]
- 112.Hu W, Wu W, Verschraegen CF, et al. Proteomic identification of heat shock protein 70 as a candidate target for enhancing apoptosis induced by farnesyl transferase inhibitor. Proteomics. 2003;3(10):1904–1911. doi: 10.1002/pmic.200300547. [DOI] [PubMed] [Google Scholar]
- 113.Nollen EA, Garcia SM, van Haaften G, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2004;101(17):6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114•.Nagaraj NS. Evolving ‘omics’ technologies for diagnostics of head and neck cancer. Brief Funct Genomic Proteomic. 2009;8(1):49–59. doi: 10.1093/bfgp/elp004. Well reviewed on ‘omics’ technologies towards biomarker discovery, identification of signaling molecules associated with cell growth, cell death, cellular metabolism and early detection of cancer. [DOI] [PubMed] [Google Scholar]
- 115.Nagaraj NS, Singh OV. Integrating genomics and proteomics-oriented biomarkers to comprehend lung cancer. Expert Opin Med Diagn. 2009;3(2):167–180. doi: 10.1517/17530050902725125. [DOI] [PubMed] [Google Scholar]
- 116.Dome JS, Coppes MJ. Recent advances in Wilms tumor genetics. Curr Opin Pediatr. 2002;14(1):5–11. doi: 10.1097/00008480-200202000-00002. [DOI] [PubMed] [Google Scholar]
- 117.Ivan M, Kaelin WG., Jr The von Hippel–Lindau tumor suppressor protein. Curr Opin Genet Dev. 2001;11(1):27–34. doi: 10.1016/s0959-437x(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 118.Bashour AM, Meng JJ, Ip W, MacCollin M, Ratner N. The neurofibromatosis type 2 gene product, merlin, reverses the F-actin cytoskeletal defects in primary human Schwannoma cells. Mol Cell Biol. 2002;22(4):1150–1157. doi: 10.1128/MCB.22.4.1150-1157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Usmani SZ, Bona R, Li Z. 17 AAG for HSP90 inhibition in cancer – from bench to bedside. Curr Mol Med. 2009;9(5):654–664. doi: 10.2174/156652409788488757. [DOI] [PubMed] [Google Scholar]
- 120.Kubo T, Kobayashi N, Jida M, et al. The antitumor effect of orally active heat shock protein 90 inhibitor, 17-DMAG, on the growth of gefitinib-resistant non-small cell lung cancer. J Clin Oncol (Meeting Abstracts) 2009;27(15 Suppl):Abstract e22064. [Google Scholar]
- 121.Pacey SC, Wilson R, Walton M, et al. A Phase I trial of the HSP90 inhibitor, alvespimycin (17-DMAG) administered weekly, intravenously, to patients with advanced, solid tumours. J Clin Oncol (Meeting Abstracts) 2009;27(15 Suppl):Abstract 3534. [Google Scholar]
- 122.McCleese JK, Bear MD, Fossey SL, et al. The novel HSP90 inhibitor STA-1474 exhibits biologic activity against osteosarcoma cell lines. Int J Cancer. 2009;125(12):2792–2801. doi: 10.1002/ijc.24660. [DOI] [PubMed] [Google Scholar]
- 123.Zheng FF, Kuduk SD, Chiosis G, et al. Identification of a geldanamycin dimer that induces the selective degradation of HER-family tyrosine kinases. Cancer Res. 2000;60(8):2090–2094. [PubMed] [Google Scholar]
- 124.Oza AM, Elit L, Biagi J, et al. A Phase II study of temsirolimus (CCI-779) in patients with metastatic and/or locally recurrent endometrial cancer – NCICCTG IND 160. Clinical Cancer Res. 2005;11(24):9099s–9099s. [Google Scholar]
- 125.Roccaro AM, Leduc R, Nelson M, et al. Phase I trial of CCI-779 and weekly bortezomib in relapsed and/or refractory multiple myeloma. Clin Lymphoma Myelom. 2009;9:S86–S86. [Google Scholar]
- 126.Wen P, Chang S, Kuhn J, et al. Phase I/II study of erlotinib and CCI-779 (temsirolimus) for patients with recurrent malignant gliomas (NABTC 04-02) Neuro Oncol. 2009;11(2):232–232. doi: 10.1093/neuonc/not247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chan S, Scheulen ME, Johnston S, et al. Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. J Clin Oncol. 2005;23(23):5314–5322. doi: 10.1200/JCO.2005.66.130. [DOI] [PubMed] [Google Scholar]
- 128.Baselga J, Semiglazov V, Van Dam P, et al. Phase II double-blind randomized trial of daily oral RAD001 (everolimus) plus letrozole (LET) or placebo (P) plus LET as neoadjuvant therapy for ER plus breast cancer. Breast Cancer Res. 2007;106:S107–S107. [Google Scholar]
- 129.Yao JC, Phan AT, Chang DZ, et al. Efficacy of RAD001 (everolimus) and octreotide LAR in advanced low- to intermediate-grade neuroendocrine tumors: results of a Phase II study. J Clin Oncol. 2008;26(26):4311–4318. doi: 10.1200/JCO.2008.16.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yardley DA, Seiler M, Ray-Coquard I, et al. Ridaforolimus (AP23573; MK-8669) in combination with trastuzumab for patients with HER2-positive trastuzumab-refractory metastatic breast cancer: a multicenter Phase 2 clinical trial. Cancer Res. 2009;69(24):676s–676s. [Google Scholar]
Website
- 201.Clinical Trials website. www.clinicaltrials.gov.