Abstract
In this study, the effect of plant genotype, soil type and nutrient use efficiency on the composition of different bacterial communities associated with rice roots were investigated. Thus, total bacteria, Alpha- and Beta-proteobacteria, Pseudomonas and Actinobacteria were studied using PCR followed by denaturing gradient gel electrophoresis (PCR-DGGE). Rice genotype determined to a large extent the composition of the different bacterial communities across cultivars. Several cultivars belonging to Oryza sativa subspecies indica tended to select similar bacterial communities, whereas those belonging to subspecies japonica and aromatica selected ones with divergent community structures. An effect of soil type was pronounced for the Actinobacteria communities, while a small effect of ‘improved’ and ‘traditional’ plants was noted for all communities analysed. A few dominant bands in PCR-DGGE, affiliated with Rhizobium radiobacter, Dickeya zeae, Mycobacterium bolletii and with members of the Rhizobiales, Rhodospirillaceae and Paenibacillaceae were spread across cultivars. In contrast, a majority of bands (e.g. affiliated with Enterobacter cloacae or Burkholderia kururiensis) was only present in particular cultivars or was erratically distributed amongst rice replicates. The data suggested that both bacterial adaptation and plant genotype contribute to the shaping of the dynamic bacterial communities associated with roots of rice plants.
Keywords: Rice bacterial community, plant genotype, agricultural management regime, generic and cultivar-specific PCR-DGGE bands
1. Introduction
Half of the Earth’s population depends on rice (Oryza sativa) as the source of essential proteins and calories, and hence rice represents an important source of food for mankind (FAO, 2010). The current availability of the sequence of the rice genome, which is small and reveals the presence of genes representative for other gramineous species, has turned rice into a model plant for studies on monocotyledonous plants (Phillips et al., 2007). In particular, the genomes of two rice subspecies (Oryza sativa subsp. indica and japonica) provide rich sources of information on potential functions and interactions during rice crop growth (Cantrell & Reeves, 2002). Despite the advance in rice genome studies, little is known about the putative interactions between rice plants and their associated bacteria (Mano & Morisaki, 2008; Reinhold-Hurek & Hurek, 1998; Ikeda et al., 2007).
Many plant-associated bacteria are involved in processes that affect the plant life cycle, such as fixation of atmospheric N2, production and modulation of phytohormones and the biocontrol of phytopathogens that frequently affect crops (Rosenblueth & Martínez-Romero, 2006; Compant et al., 2010). These bacteria often interact with their host plants, modulating their physiology and morphology (Feng et al., 2006). For instance, the growth of rice plants has been shown to be promoted by the introduction of specific rhizobia, which enhance growth rates by increasing the water utilization efficiency (Chi et al., 2005). Moreover, the induction of systemic resistance in rice was shown to be triggered by Methylobacterium spp. (Madhaiyan et al., 2004), Pseudomonas fluorescens (Nandakumar et al., 2001) and by the harpin protein HpaG from Xanthomonas oryzae pv. oryzicola (Chen et al,. 2008). Azoarcus sp. strain BH72 has been shown to fix N2 in the aerenchymous tissue of young rice plants (Hurek & Reinhold-Hurek, 2003). Active sulfate-reducing and ammonia-oxidizing bacterial communities have also been found in rice root tissues (Briones et al., 2003; Nicolaisen et al., 2004; Scheid & Stubner, 2001). All of these bacterial activities contribute to, or affect, sustainable rice production.
The make-up of plant-associated bacterial communities is very likely affected by deterministic factors as well as stochastic (neutral) events (Hardoim et al., 2008). Different factors are known to play a role, both qualitatively and quantitatively. The soil, in all of its facets, is one of these factors, because it acts as a major reservoir of bacteria that can colonize the internal tissue of plants (Hallmann et al., 1997). On the other hand, plants offer an environment that is selective to microorganisms (Hallmann & Berg, 2006), “filtering out” specific microbial groups from the diversity found at plant roots. Thus, factors such as plant genotype and physiological status, bacterial colonization traits, abiotic conditions (e.g. temperature, pH) and agricultural management regimes all can affect the diversity of bacterial communities in root tissues (Hardoim et al., 2008; Van Overbeek and Van Elsas, 2008; Andreote et al., 2010). Among these factors, plant genotype may play a key role in the selection of distinct bacterial communities that associate with plants (Hartmann et al., 2009; Andreote et al., 2009). Although this seems like a simple and easy-to-study phenomenon, so far only diazotrophic communities have been investigated in great detail across various rice cultivars (Knauth et al., 2005; Muthukumarasamy et al., 2007).
In this study, we examined the diversity of bacterial communities associated with rice roots across ten cultivars. All plants except those of the cultivar Moroberekan had been cultivated in the same (flooded) soil under the same agricultural management regime. Our hypothesis was that each cultivar, by virtue of its genetic make-up, selects its own bacterial community from the pool of microorganisms present in the soil.
2. Material and methods
Field location and sampling procedure
Ten rice cultivars were selected based on: i) the divergence of the host genotypes, i.e. cultivar Basmati is characterized as Oryza sativa subspecies aromatica, cultivars Azucena and Moroberekan are O. sativa subsp. tropical japonica and cultivars DEE, Peta, APO, IR36, IR64, IR65600 and IR72 are O. sativa subsp. indica; ii) the soil type where they were cultivated: plants of cultivar Moroberekan were harvested from an upland soil, whereas the others nine cultivars were harvested from a homogenized (rotary spading, once yearly) paddy field; and iii) the response to nutrients, cultivars Azucena, Basmati, Moroberekan, DEE and Peta are denoted as ‘traditional’ (i.e. low nutrient use efficiency), whereas cultivars APO, IR36, IR64, IR65600 and IR72 are denoted as ‘improved’ (i.e. high nutrient use efficiency; Peng et al., 2005) (Table 1). Replicate plants of all rice cultivars were sampled from the fields used for rice plant breeding experiments, located at the International Rice Research Institute (IRRI, Los Baños, Philippines). For each of the ten cultivars, three individual plants, at minimal distances of 0.5 m from each other, were harvested at flowering stage (Itoh et al., 2005).
Table 1.
Characteristics of ten rice cultivars used to assess the bacterial community associated with roots.
| Cultivars | O. sativa subspecies | Observations | Sampled | Origina | |
|---|---|---|---|---|---|
| Improved | APO | indica | Good performance under aerobic conditions and responsiveness to nutrients | Wetland | PI |
| IR 36 | indica | High yield crop, resistance to many insect pets and plant diseases | Wetland | PI | |
| IR 64 | indica | Replace cultivar IR 36 as the largest planted cultivar in the 1980s | Wetland | PI | |
| IR 65600 | indica | An elite breeding line of New Plant Type (NPT) developed by crossing subspecies indica with tropical japonica | Wetland | PI | |
| IR 72 | indica | Replace cultivar IR 64 in the 1990s | Wetland | PI | |
|
| |||||
| Traditional | DEE | indica | Parent donor of modern cultivars. Spontaneous mutant, with dwarfing gene | Wetland | TW |
| Peta | indica | Parent donor of modern cultivars, photoperiod insensitivity. Resistance to tungro virus | Wetland | ID | |
|
| |||||
| Basmati | aromatica | Superfine grain qualities, distinct aroma. Tolerant to aluminium | Wetland | IN | |
|
| |||||
| Azucena | tropical | Aromatic. | Wetland | PI | |
| japonica | Tolerant to aluminium | ||||
| Morobere kan | tropical | Resistance to rice blast and tolerant to | Upland | GN | |
| japonica | drought and aluminium | ||||
Countries of origin: PI- Philippines; IN- India; ID- Indonesia; TW- Taiwan; GN- Guinea
Within one hour after sampling, the plants were processed in the laboratory. Therefore, the root mass of each plant was carefully washed under running tap water for removal of adhering soil particles. Root bundles were then separated from the plant aerial tissues using a sterilized scalpel. Selected root parts were further cut into 5-cm pieces and immediately snap-frozen in liquid N2 and stored at −70 °C.
DNA extraction from rice cultivars
The snap-frozen root parts from the rice cultivars were subjected to surface sterilization. Briefly, roots were thawed, immersed in 70% ethanol for 2 min, followed by 2% sodium hypochlorite (NaOCl) solution for 2 min, and three successive washes in sterile demineralised water. Such treatment also helps to remove bacterial cells attached to roots surfaces. The thus surface-sterilized roots (ca 4 g) were transferred to sterile plastic bags containing 2 ml of sterile demineralised water and homogenized by squeezing with a soft-headed hammer in order to release the root-associated/endophytic bacterial cells. Homogenates (400 μl) were directly used for DNA extraction by applying the DNeasy Plant extraction kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions with one modification: the cell disruption step was extended from 10 min to 1 h to optimize bacterial lysis. The DNeasy Plant extraction kit has been successfully employed for microbial community studies with vine plants (Dreo et al., 2007; Gambetta et al., 2007) and potato plants (Andreote et al., 2009; Andreote et al, 2010).
PCR amplification of 16S ribosomal RNA (rRNA) genes for denaturing gradient gel electrophoresis (DGGE) analyses
For amplification of 16S rRNA gene regions at total bacterial, nested PCR approaches were applied. The purpose was to suppress the amplification of plant plastid DNA. Thus, primer 799F in combination with universal bacterial primer 1492R was used in the first PCR (Chelius & Triplett, 2001). Each 25 μl PCR mixture contained 1 μl of DNA template (5 - 20 ng), 1× Stoffel buffer, 3.75 mM MgC12, 200 μM of each dNTP, 400 nM of each primer, 1% formamide, 0.5 mg ml−1 bovine serium albumin (BSA), 0.25 μg T4 gene 32 protein (Roche Diagnostics GmbH, Mannheim, Germany) and 2.5 U AmpliTaq DNA polymerase Stoffel fragment (Applied Biosystems, Foster City, CA). The thermal cycling conditions and cycle number were as described previously (Chelius & Triplett, 2001). The amplicons were electrophoretically separated in agarose gels (1%), and bands of the expected sizes (ca 740 bp) were excised and extracted using the QIAquick gel extraction kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. Purified amplicons were diluted to final DNA template concentration (5-20 ng) and used in the second (nested) PCR’s with primers 968F-GC (carrying a GC clamp at its 5′ end) and 1401R-1a (Brons & Van Elsas, 2008). PCR amplifications (50 μl mixes) were performed as described in Brons & Van Elsas (2008). The resulting PCR products were used for DGGE analysis.
To analyze each of the specific bacterial groups, a first PCR specifically targeted the 16S rRNA gene regions of Alpha- and Betaproteobacteria (Gomes et al., 2001), Actinobacteria (Heuer et al., 1997) and Pseudomonas (Milling et al., 2004). Each 25 μl reaction mixture for the first PCR amplifications contained 1 μl of DNA template (5 - 20 ng) and PCRs were performed according to their respective protocols (Heuer et al., 1997; Gomes et al., 2001; Milling et al. 2004). The thus obtained amplicons were used (1 μl) as the templates in the second (nested) PCR as previously described, with the exception of the Pseudomonas system, in which the reverse primer 1459R was employed (Milling et al., 2004). All PCRs were carried out in a PTC-200 thermal cycler (MJ Research, Inc., Tilburg, NL).
DGGE profiles and statistical analyses
DGGE analysis was performed in a PhorU-2 apparatus, (Ingeny, Goes, The Netherlands) in 0.5 × Tris-acetate-EDTA (TAE) buffer and gels were run at 100 V for 16 h at 60 °C. Gel casting was performed as described by Muyzer et al. (2004), with a gradient consisting of 45-65% denaturant (100% denaturant contained 7 M urea and 40% formamide). The amplicons (150 ng) from three plants per treatment were loaded side-by-side on the gel, while markers (Garbeva et al., 2001) were loaded at both edges and one in the middle of the gel for normalization purposes. After the run, gels were stained with SYBR gold (Molecular Probes, Leiden, The Netherlands) and the DGGE patterns were made visible by illumination with UV. The profiles were digitized using a digital camera and stored as TIFF files.
All PCR-DGGE profiles were analyzed using GelCompar II v 4.06 (Applied Maths, Sint-Martens-Latem, Belgium). After normalization, the position and intensity of individual bands in the profiles (species parameter) were recorded. To assess the complexity of the bacterial communities, bands with similar motility (1% tolerance) were assigned to the same band migration position. The band intensity and position were used for subsequent redundancy analysis (RDA) using CANOCO 4.5 software (Ter Braak and Šmilauer, 2002). The biplot ordinations were generated by scores of samples (plotted from the ordination of the species data) and effects analyzed (environmental variables). In addition, a Monte Carlo permutation test (with 1000 repetitions) was applied to evaluate the correlation of all microbial communities within the assigned environmental variables (Ter Braak, 1994).
Identification of selected PCR-DGGE bands
Dominant bands from universal, as well as Alpha and Betaproteobacteria PCR-DGGE profiles were selected for identification. Following excision, the bands were treated for re-amplification, cloning and subsequent sequencing following the methodology described by Costa et al. (2006). In addition, 16S rRNA gene amplicons of identified rice isolates were used to classify PCR-DGGE bands with identical denaturation motility. The sequences from excised PCR-DGGE bands and co-migrated isolates were deposited were deposited in the GenBank under the accession numbers HQ702192 to HQ702205.
3. Results
Analysis of bacterial communities associated with the roots of different rice cultivars
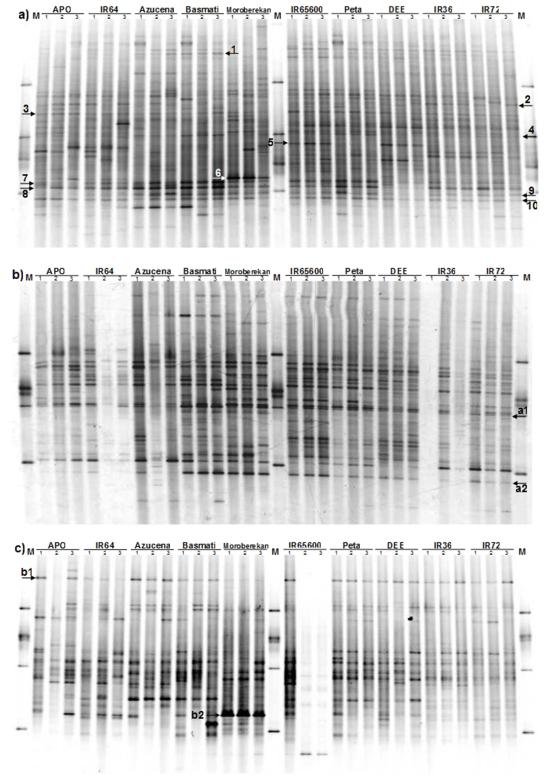
Ten rice cultivars growing at the experimental fields of IRRI were selected at the mature flowering stage (plant height on average 50-70 cm) in accordance with their characteristics (Table 1). Using PCR-DGGE, the compositions of their dominant communities, as related to plant genotype, soil type and nutrient use efficiency, were then investigated. Across the board, total bacterial communities showed the highest complexities with a total of 56 band migration positions; each cultivar (three replicates) containing, on average, 28 bands (Fig. 1a). Communities of Alphaproteobacteria were also highly complex, with 52 band migration positions in total and averages of 24 bands per cultivar (Fig 1b). The communities of Betaproteobacteria were intermediate, with totals of 38 migration positions and averages per cultivar of 16 bands (Fig 1c). Communities of Actinobacteria and Pseudomonas showed the lowest complexities with, respectively, 19 and 25 total band migration positions and 7.5 bands on average per cultivar for each community.
Fig. 1.
PCR-DGGE profiles of bacterial (a), alphaproteobacterial (b) and betaproteobacterial (c) communities associated with root tissues of ten rice cultivars. Arrows indicate identified DGGE bands obtained from excised bands and co-migrated isolates, respectively, right and left ‘head’ directions. Internal arrows are cultivar-specific bands, whereas ‘edge’ arrows are generic bands.
Total bacteria
All bacterial PCR-DGGE profiles contained between 23 and 33 bands within each replicate pattern, suggesting that a considerable bacterial diversity was associated with rice root tissue. Cultivars DEE, Peta, IR65600 and IR72 revealed the highest richness (30 bands), while replicates of cultivars APO, Azucena, Basmati, Moroberekan and IR64 were among the lowest (26 bands). Eight similar dominant bands were present in at least seven of the ten cultivars. From these eight bands, six were identified (Fig 1a; Table 2). Three showed high 16S rRNA gene sequence similarity (respectively 98.5, 100 and 98.4%) to the type species Dickeya zeae CFBP 2052T (band u3, HQ702194), Rhizobium radiobacter NCPPB 2437T (band u4, HQ702195) and Mycobacterium bolletii CIP 108541T (band u9, HQ702200). In contrast, three bands showed low sequence similarity (respectively, 93.5, 92.4 and 93.2%) to the closest type strains Azospirillum lipoferum DSM 1691T (band u7, HQ702198), Paenibacillus terrae AM141T (band u8, HQ702199) and Methylocella silvestris BL2T (band u10). DGGE bands u10 (HQ702201) and a2 (HQ702203) were closely related (99.5% sequence similarity) to Alphaproteobacterium strain CCBAU 45397 isolated from root nodules of peanuts in China. Furthermore, a dominant band (u2, HQ702193) closely related to Enterobacter cloacae ATCC 13047T (98.4% sequence similarity) was found associated with roots of all replicates of cultivars APO, IR36, IR64, IR65600 and IR72, while it was present but erratically distributed among replicates of cultivars Azucena, DEE and Peta (Fig. 1a). Three cultivar-specific DGGE bands were also observed. A dominant band which was closely related to the 16S rRNA sequence of Escherichia coli ATCC 11775T (100% sequence similarity) was found only on replicates of cultivars IR65600 and DEE (band u5, HQ702196), a second band, which was closely related to that of Burkholderia kururiensis KP23T (98.6% sequence similarity) was dominant on the replicates of cultivar Moroberekan, whereas it was faintly present in the replicates of cultivars APO and IR64 (band u6, HQ702197; Fig. 1a). A third faint band (u1, HQ702192), which was present in the replicates of cultivars Basmati and Azucena, was identified as Staphylococcus epidermidis ATCC 14990T (99.7% sequence similarity; Fig. 1a). The remaining bands (12 to 21) were erratically distributed over the replicate PCR-DGGE patterns.
Table 2.
16S rRNA gene identification of generic and cultivar-specific PCR-DGGE bands, retrieved from universal and group-specific PCR-DGGE profiles.
| DGGE band IDa | Closest match | Similarity (%) | Closest type strain | Similarity (%) | Present inb |
|---|---|---|---|---|---|
| u1* | AY741152|Staphylococcus epidermidis|S09 | 99.7 | D83363|Staphylococcus epidermidis|ATCC 14990T | 99.7 | 2 |
| u2* | DQ091238|Enterobacter sp. FL13-2-1 | 99.7 | AJ251469|Enterobacter cloacae|ATCC 13047T | 98.4 | 4 |
| u3 | AF373199|Dickeya chrysanthemi|571 | 98.8 | AF520711|Dickeya zeae|CFBP 2052T | 98.5 | 10 |
| u4* | AY504963|Rhizobium radiobacter|CCBAU 65237 | 100 | D14500|Rhizobium radiobacter| NCPPB 2437T | 100 | 7 |
| u5 | AB272358|Escherichia sp. IF4 | 100 | X80725|Escherichia coli|ATCC 11775T | 100 | 2 |
| u6 | EF397576|Burkholderia sp. ATSB13 | 99.7 | AB024310|Burkholderia kururiensis|KP23T | 98.6 | 3 |
| u7 | AB049112|Azospirillum sp. B518 | 94.0 | GU256441|Azospirillum lipoferum|DSM 1691T | 93.5 | 10 |
| u8 | AB486660|Uncultured bacteria | 95.2 | AF391124|Paenibacillus terrae|AM141T | 92.4 | 10 |
| u9* | AY457082|Mycobacterium chelonae|ATCC 19237 | 98.7 | AY859681|Mycobacterium bolletii|CIP 108541T | 98.4 | 8 |
| u10* | HM107183|Alphaproteobacterium| CCBAU 45397 | 99.5 | AJ491847|Methylocella silvestris|BL2T | 93.2 | 10 |
| a1* | AY504963|Rhizobium radiobacter|CCBAU 65237 | 100 | D14500|Rhizobium radiobacter| NCPPB 2437T | 100 | 10 |
| a2* | HM107183|Alphaproteobacterium| CCBAU 45397 | 99.5 | AJ491847|Methylocella silvestris|BL2T | 93.2 | 6 |
| b1 | AB531409|Collimonas sp. III-9 | 94.0 | DQ665916| Uliginosibacterium gangwonense|5YN10-9T | 95.1 | 10 |
| b2 | EF397576|Burkholderia sp. ATSB13 | 99.4 | AB024310|Burkholderia kururiensis|KP23T | 98.3 | 2 |
PCR-DGGE bands were identified by sequencing the excised bands or by co-migration with known isolate*
Number of ribotypes present in all replicates out of ten cultivars
Ordination of DGGE patterns
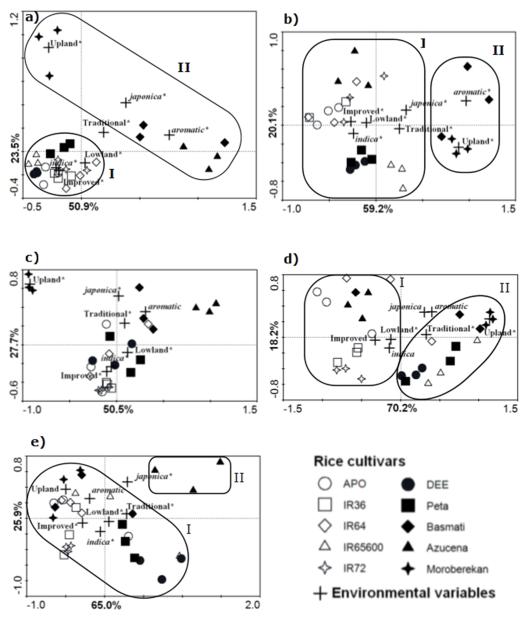
A biplot ordination generated via redundancy analysis (RDA) of the bacterial PCR-DGGE patterns showed a major dichotomy between the total bacterial communities associated with indica plants versus those associated with japonica and aromatica plants (Fig. 2a). Plant genotype explained most of the variability (48.8%) followed by soil type and nutrient use efficiency with 36.6 and 14.6%, respectively. All effects significantly influenced the variation of the total bacterial composition (P < 0.05). Two main clusters, denoted I and II, could thus be distinguished. Cluster I consisted of very similar communities of bacteria exclusively associated with the indica cultivars APO, IR36, IR64, IR65600, IR72, DEE and PETA. Cluster II included scattered bacterial communities from two japonica and one aromatica cultivars, Azucena, Moroberekan and Basmati, respectively. Although the profiles from cluster II were distributed along the second axis, the replicates within cultivars were relatively close to each other, suggesting that, at this level, plant genotype plays a determinative role in the selection of root-associated bacterial communities.
Fig. 2.
Biplot ordination generated by redundancy analysis (RDA) of the PCR-based denaturing gradient gel electrophoresis (PCR-DGGE) profiles from total bacterial (a), alphaproteobacterial (b), betaproteobacterial (c), actinobacterial (d) and pseudomonads (e) communities associated with roots of ten rice cultivars. Each symbol represents the bacterial community composition associated with roots of an individual plant. Open and close symbols represent improved and traditional cultivars, respectively. Crosses represent the centroid position of the nominal environmental variables (effects). Environmental variable with significant influence on the variation of community composition is shown with asterisks (P < 0.05). The variations of each ordination axis are presented in percentage.
Bacterial group-specific PCR-DGGE analyses
Alphaproteobacteria
The alphaproteobacterial PCR-DGGE patterns showed between 16 and 34 bands across all rice cultivars. The highest richness was found within replicates of cultivars DEE and Peta (average of 28.5 bands), while cultivar IR64 showed the lowest richness, with 16 bands. Ten dominant bands were consistently present in at least eight of the ten cultivars, indicating that these members of the Alphaproteobacteria occur in rice roots largely irrespective of plant genotype. Two of these conspicuous bands were identified as Rhizobium radiobacter (band a1, HQ702202) and Methylocella silvestris (band a2, HQ702203; Fig. 1b, Table 2), whereas eight remained unidentified. The remaining bands (6 to 24) were erratically distributed across cultivars. A biplot ordination constructed by RDA revealed two main clusters, denoted I and II (Fig. 2b), which were separated along the first axis. Cluster I consisted of patterns of all indica cultivars APO, IR36, IR64, IR65600, IR72, DEE, PETA coupled to one japonica cultivar (Azucena), while cluster II consisted exclusively of the cultivar Moroberekan and Basmati replicates. The effects plant genotype and soil type significantly influenced the variation in the relative abundance of DGGE bands (P < 0.05), where 68.4 and 21.0% of the total variability were explained, respectively. Nutrient use efficiency explained 10.5% of the total variability.
Betaproteobacteria
The betaproteobacterial-specific PCR-DGGE profiles showed considerable variation amongst, and even within, replicates of the same rice cultivars. Totals of 10 to 20 bands were discernable for each of the cultivar replicates (Fig. 1c). Communities of cultivars Peta, Moroberekan and IR36 showed the highest richness, with 18 bands each, while cultivars Azucena and IR64 had communities with the lowest ones, with 14 bands on average. A conspicuous band (b1, HQ702204) found in eight of the nine cultivars analyzed was most closely related to Uliginosibacterium gangwonense 5YN10-9T, at 95.1% 16S rRNA gene sequence similarity. A dominant band (b2, HQ702205) occurring in patterns from communities associated with roots of Moroberekan was closest related to Burkholderia kururiensis KP23T, at 98.3% sequence similarity. The remaining bands (8 to 18) were distributed scattered amongst the replicates (Fig. 1c). The RDA ordination diagram revealed a main cluster which consisted of all replicate profiles of the indica and aromatica cultivars distributed along the second axis (Fig. 2c). Profiles of the two japonica cultivars (Moroberekan and Azucena) were adjacent to the main cluster, however opposite to each other. This indicated that the community of Betaproteobacteria associated with the roots of japonica cultivar Moroberekan differs greatly from those associated with the other japonica cultivar Azucena. Plant genotype explained most of the variability (50%), followed by soil type and nutrient use efficiency with, respectively, 38 and 12% of the total variability. All variables, but aromatica, significantly influenced the structure composition of betaproteobacterial communities (P < 0.05).
Actinobacteria
The actinobacterial PCR-DGGE profiles revealed low richness, with only 4 to 14 bands (Fig. S1a). Three dominant bands were observed across eight out of ten cultivars, while the remaining bands were erratically distributed across cultivar replicates. Although with high variability, two main clusters emerged following ordination by RDA of the PCR-DGGE profiles (Fig. 2d). Cluster I consisted of all replicates of cultivars APO, IR36, IR72 and Azucena, two replicates of IR64 and one of Basmati. Cluster II consisted of all replicates of cultivars DEE, PETA, Moroberekan and IR65600, two replicates of Basmati and one replicate of IR64. The variables lowland and upland significantly differ (P < 0.05) from each other and explained 45.7% of the total variation in the ordination. Plant genotype and nutrient use efficiency explained 22.8 and 20% of the total variability, respectively.
Pseudomonas
The pseudomonad-specific PCR-DGGE profiles consisted of 3 to 11 bands, which were all erratically distributed over cultivars and replicates (Fig. S1b). The highest richness was encountered in the replicates of cultivars Azucena and IR65600, with averages of 10.5 bands per replicate, while cultivars APO, IR36 and Peta revealed the lowest richness, with 6 bands per lane on average. No conspicuous band was observed to occur across six or more cultivars. However, a single conspicuous band was observed in all replicates of cultivars APO, IR36, IR64, IR72 and Moroberekan. Two dominant bands occurred in all replicates of three cultivars; the first one occurred solely in Azucena, Peta and IR65600, whereas a second band appeared in all replicates of cultivars Azucena, DEE and Peta and was erratically distributed over replicates of cultivars APO and IR65600 (Fig. S1b). RDA ordination revealed two main clusters, denoted I and II, weakly separated regarding plant genotype. Cluster I consisted of pseudomonad communities associated with all indica cultivars plus all replicates of cultivars Basmati and Moroberekan (Fig. 2e), whereas cluster II consisted of all replicates of cultivar Azucena. Plant genotype explained 54.1% of the total variation, while nutrient use efficiency and soil type explained 29 and 18.9%, respectively. All variables, but aromatica and upland, significantly influenced the structure composition of pseudomonads communities (P < 0.05).
Cross comparison over bacterial groups
The roots of indica cultivars IR36 and IR72 revealed consistent PCR-DGGE patterns for all bacterial communities analyzed. Moreover, the indica cultivars APO and IR64 also revealed consistency for the total bacterial, alphaproteobacterial and pseudomonads communities, but divergence in the profiles of betaproteobacterial and actinobacterial communities. The remaining indica cultivars IR65600, DEE and Peta showed large similarity to other indica cultivars for total bacterial communities, whereas they formed distinct communities of Alpha- and Beta-proteobacteria (exception for IR65600) as well as Actinobacteria. The two japonica cultivars Azucena and Moroberekan revealed great dissimilarity for all communities analyzed, while the bacterial community associated with roots of aromatica Basmati resembled both japonica cultivars. Cultivar Basmati selected bacterial communities similar to Moroberekan for Alphaproteobacteria, Pseudomonas and Actinobacteria, whereas total and Betaproteobacteria were highly similar to the same communities associated with cultivar Azucena.
4. Discussion
In this study, we present a survey of ten rice cultivars, in which total and group-specific bacterial communities associated with roots were assessed by PCR-DGGE. We were interested in the effect of plant genotype, soil type and nutrient use efficiency on the composition of the selected communities associated with rice and thus examined, at one point in time, ten rice cultivars, of which nine had been grown simultaneously in the same experimental field. Our results indicated that different rice cultivars select specific fractions of the bacterial communities in sometimes highly different and at other times similar fashions. Thus, since the soil habitat, climatic conditions and agricultural practices were similar across all but one cultivar (Moroberekan), we surmised that the effects seen were for the largest part due to plant cultivar, and, by inference, plant genotype. This observation is corroborated by several other studies in the literature (Sessitsch et al., 2002; Zul et al., 2007; reviewed in Hallmann & Berg, 2006). For instance, in previous studies in rice, the diazotrophic endophytic communities of wild and modern cultivars were shown to be different (i.e. lower diversity in wild cultivars), possibly due to an effect of plant genotype (Elbeltagy et al., 2000; Engelhard et al., 2000). The plant genotype effect most likely results from an effect on rooting and root exudation, thus directly affecting the colonization and nutrient status of bacterial communities associated with roots (Garbeva et al., 2001; Hallmann & Berg, 2006; Bais et al., 2006; van Overbeek & van Elsas, 2008).
The communities of total bacteria and Alphaproteobacteria revealed considerable complexity, with only a few conspicuous bands being widely distributed across cultivars. This suggested that a deterministic effect of plant genotype leading to highly evolved associations may play a role in the selection of bacterial communities. It is known that many different alphaproteobacterial species can form associations with plants, and such associations may be highly evolved. This is certainly the case for rhizobia, which form N2-fixing symbioses with legumes, and agrobacteria, which cause phenomena like crown galls on susceptible plants. Strains of R. radiobacter (formerly Agrobacterium tumefaciens) that do not cause any harm to their hosts are being frequently isolated from root nodules of leguminous plants (Wang et al., 2006). Furthermore, a specific strain of R. radiobacter IRBG74, which was isolated from root nodules of the aquatic legume Sesbania aculeata, was shown to promote rice growth by enhancing the uptake efficiency of N, P, K and Fe and accumulation of indole-3-acetic acid (IAA) on roots of associated plants (Biswas et al., 2000; Tan et al., 2001). The presence of two other conspicuous Alphaproteobacteria bands across all cultivars suggests an almost universal adaptation to rice, and one may assume that rice-beneficial functions might be the driving force for their selection.
Members of Gammaproteobacteria are important rice colonizers (Mano & Morisaki, 2008). Within this group, Enterobacteriaceae species are major players due to their adaptation to broad range of nutritional and physicochemical conditions, as well colonization mechanisms (Holden et al., 2009). The nature of the interactions, being beneficial, deleterious or commensal, may be dictated by the interplay between host and microbe genomes. A conspicuous band (u3), which was present across all rice cultivars, was closely related to the 16S rRNA gene of Dickeya zeae CFBP 2052T (formely Erwinia chrysanthemi), a species that contains members that may cause foot rot across Asian rice (Hussain et al., 2008). Although the sampled rice plants were apparently healthy, the presence of this bacterium may represent a potential threat if conditions lead to disease development. The species E. coli encompasses a range of strains that are either commensalistic or pathogenic to animals, and some of them may form associations with varying plant species (Brandl et al., 2006). We are unfamiliar with the E. coli type that is behind the band found, but it was recently found that the pathogen E. coli O157:H7 can form high population densities on the epidermis and internal plant tissues of alfalfa, reaching up to 108 CFU g−1 fresh weight within 5 days of inoculation (Teplitski et al., 2009). This may indicate its tight association with these cultivars and possibly relates to the physiology of these. In addition, we isolated, from root tissues of cultivar APO plants, a Candidatus novel species denoted Enterobacter oryzaphilus REICA_142T (Hardoim et al., in prep); this organism yielded 16S rRNA amplicons of identical mobility to band u2. The sequence of E. oryzaphilus REICA_142T was the most abundant sequence in an extensive 16S rRNA gene clone library generated from the endophytic community of APO root tissues (Hardoim et al., in prep). In addition, E. oryzaphilus REICA_142T showed several plant growth-promoting properties (e.g. fixation of N2, phosphate solubilisation, ACC deaminase production) as well plant adaptation characteristics (e.g. production of cellulose and growth on methanol as the sole carbon source), which may endow it with the eponymous “competent endophyte” under field conditions.
Three other competent PCR-DGGE bands (bands u8, u9 and b1, respectively), which were spread across most rice cultivars, were assigned to the classes Firmicutes, Actinobacteria and Betaproteobacteria. The members of these classes are often associated with rice plants (Mano & Morisaki, 2008) and can even be isolated from seed tissues (Mano & Morisaki, 2008; Kaga et al., 2009). Moreover, Moroberekan plants distinctly select for a bacterial species closely related to B. kururiensis KP23T, an N2-fixing bacterium that is capable of increasing rice biomass via the production of IAA (Mattos et al., 2008). Interestingly, the addition of nitrogen to Hoagland’s nutrient solution limited the endophytic colonization of rice by B. kururiensis KP23T in a dose-dependent manner (Mattos et al., 2008), suggesting that agricultural regime has a key role in this interaction.
An effect of soil/agricultural regime was observed for total, alpha-, beta-proteobacterial and actinobacterial communities, as the PCR-DGGE patterns of the cultivar Moroberekan-associated communities differed significantly (P < 0.05) from those of the other cultivars. Although Moroberekan plants were sampled from a different soil type and agricultural management regime, the majority of root-associated bacteria were also encounter on other cultivars, suggesting that rice plants select a restricted range of soil bacteria to form interactions. Both soils (i.e. upland and lowland) have a long history in rice cultivation, thus it might favour the enrichment of rice-adapted bacterial communities. Amongst the bacterial communities studied, only in the Actinobacteria PCR-DGGE profile was most of the ordination explained by the soil type. One might interpret these findings to indicate that most of the Actinobacteria are commonly found in soil as typical soil inhabitants. Thus, factors that relate to the soil, such as those that come about as a result of soil management, may be more determinative than plant genotype as controllers of the structure of the rice root-associated organisms (Zul et al., 2007).
Plant breeding strategy targeting high yield crops have an effect on the root-associated bacterial communities. This effect was small for highly complex communities (i.e. total bacteria, Alpha- and Betaproteobacteria), whereas more pronounced in pseudomonads and actinobacterial PCR-DGGE profiles, where ‘traditional’ and ‘improved’ cultivars formed distinct bacterial associations. These results support our observation that each bacterial community responds differently (i.e. in a similar or a dissimilar manner) to a specific effect. Our results are in agreement with those of previous studies on bacterial guilds associated with rice cultivars. ‘Improved’ plants select similar communities of nitrogen-fixing bacteria, which often differ from those of ‘traditional’ plants (Knauth et al., 2005). In another study, the abundance of ammonia-oxidizing bacteria (AOB) on the roots of different rice cultivars did not differ, however a marked contrast in their population structure was observed (Briones et al., 2003), being that ‘improved’ plants clearly selected for Nitrosomonas spp. (Briones et al.,2003).
This study provides new insight in the community structures of bacteria that live in association with rice roots across a range of rice cultivars. We showed that key parts of the bacterial communities respond differently to different plant genotypes. The presence of cultivar-specific phylotypes as well as a few phylotypes that occurred across cultivars emphasized that deterministic factors in accordance with “long-term” interactions contributed to the formation of bacterial communities associated with rice roots. Further studies are needed to understand the mechanisms of the interactions between rice roots and the root-associated bacteria, with a focus on the functioning of these communities in terms of their close interconnections with rice roots. The ultimate aim should be to apply the knowledge in the field, and consequently develop more sustainable agricultural systems.
Supplementary Material
Acknowledgements
We thank Dr. Darshan Brar at IRRI for providing the rice material and Dr. Siegfried Kropf for helping in the statistic analyses with permutation tests. This study was supported by the joint RUG-WUR initiative on rice endophytes in the context of a DOE-JGI project on the rice endophyte metagenome. This work fits within the Ecogenomics program, which is sponsored by the Dutch National Genomics Initiative, and the basic research program on sustainable agriculture (KB4), sponsored by the Dutch Ministry of Agriculture, Nature and Food Safety.
References
- Andreote FD, Nunes da Rocha U, Araújo WL, Azevedo JL, van Overbeek LS. Effect of bacterial inoculation, plant genotype and developmental stage on root-associated and endophytic bacterial communities in potato (Solanum tuberosum) Ant Leeuw. 2010;97:389–399. doi: 10.1007/s10482-010-9421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreote FD, Araújo WL, Azevedo JL, van Elsas JD, Nunes da Rocha U, van Overbeek LS. Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl Environ Microbiol. 2009;75:3396–3406. doi: 10.1128/AEM.00491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plant and other organisms. Annu Rev Plant Biology. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- Biswas JC, Ladha JK, Dazzo FB, Yanni YG, Rolfe BG. Rhizobial inoculation influences seedling vigor and yield of rice. Agron J. 2000;92:880–886. [Google Scholar]
- Brandl MT. Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol. 2006;44:367–392. doi: 10.1146/annurev.phyto.44.070505.143359. [DOI] [PubMed] [Google Scholar]
- Briones AM, Okabe S, Umemiya Y, Ramsing NB, Reichardt W, Okuyama H. Ammonia-oxidizing bacteria on root biofilms and their possible contribution to N use efficiency of different rice cultivars. Plant Soil. 2003;250:335–348. [Google Scholar]
- Brons JK, Van Elsas JD. Analysis of bacterial communities in soil by use of denaturing gradient gel electrophoresis and clone libraries, as influenced by different reverse primers. Appl Environ Microbiol. 2008;74:2717–2727. doi: 10.1128/AEM.02195-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell RP, Reeves TG. The cereal of the world’s poor takes center stage. Science. 2002;296:53. doi: 10.1126/science.1070721. [DOI] [PubMed] [Google Scholar]
- Chelius MK, Triplett EW. The diversity of Archaea and Bacteria in association with the roots of Zea mays L. Microb Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang SJ, Zhang SS, Qu S, Ren X, Long J, et al. A fragment of the Xanthomonas oryzae pv. oryzicola harpin HpaG(Xooc) reduces disease and increases yield of rice. Phytopathol. 2008;98:792–802. doi: 10.1094/PHYTO-98-7-0792. [DOI] [PubMed] [Google Scholar]
- Chi F, Shen S, Cheng H, Jing Y, Yanni Y, Dazzo F. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol. 2005;71:7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42:669–678. [Google Scholar]
- Costa R, Götz M, Mrotzek N, Lottmann J, Gabriele Berg G, Smalla K. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol Ecol. 2006;56:236–249. doi: 10.1111/j.1574-6941.2005.00026.x. [DOI] [PubMed] [Google Scholar]
- Dreo T, Gruden K, Manceau C, Janse JD, Ravnikar M. Development of a real-time PCR-based method for detection of Xylophilus ampelinus. Plant Pathol. 2007;56:9–16. [Google Scholar]
- Elbeltagy A, Nishioka K, Suzuki H, et al. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci Plant Nutr. 2000;46:617–629. [Google Scholar]
- Engelhard M, Hurek T, Reinhold-Hurek B. Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ Microbiol. 2000;2:131–141. doi: 10.1046/j.1462-2920.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- FAO 2010 www.fao.org.
- Feng Y, Shen D, Song W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J Appl Microbiol. 2006;100:938–945. doi: 10.1111/j.1365-2672.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- Gambetta G, Fei J, Rost T, Matthews M. Leaf scorch symptoms are not correlated with bacterial populations during Pierce’s disease. J Exp Bot. 2007;58:4037–4046. doi: 10.1093/jxb/erm260. [DOI] [PubMed] [Google Scholar]
- Garbeva P, van Overbeek LS, van Vuurde JWL, van Elsas JD. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16s rDNA based PCR fragments. Microbial Ecol. 2001;41:369–383. doi: 10.1007/s002480000096. [DOI] [PubMed] [Google Scholar]
- Gomes NCM, Heuer H, Schonfeld J, Costa R, Mendonca-Hagler L, Smalla K. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil. 2001;232:167–180. [Google Scholar]
- Hallmann J, Berg G. Spectrum and population dynamics of bacterial root endophytes. In: Schulz BJE, Boyle CJC, Sieber TN, editors. Microbial Root Endophytes. Springer; Dordercht, NL: 2006. pp. 15–31. [Google Scholar]
- Hallmann J, QuadtHallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43:895–914. [Google Scholar]
- Hardoim P, van Overbeek L, van Elsas J. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, van Tuinen D, Berg G. Plant-driven selection of microbes. Plant Soil. 2009;321:235–257. [Google Scholar]
- Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. Analysis of Actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden N, Pritchard L, Toth I. Colonization outwith the colon: plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol Rev. 2009;33:689–703. doi: 10.1111/j.1574-6976.2008.00153.x. [DOI] [PubMed] [Google Scholar]
- Hurek T, Reinhold-Hurek B. Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes. J Biotechnol. 2003;106:169–178. doi: 10.1016/j.jbiotec.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Hussain M, Zhang HB, Xu JL, Liu QG, Jiang Z, Zhang LH. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J Bacteriol. 2008;190:1045–1053. doi: 10.1128/JB.01472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Fuji S-I, Sato T, Furuya H, Naito H, Ytow N, et al. Microbial diversity in milled rice as revealed by ribosomal intergenic spacer analysis. Microb Environ. 2007;22:165–174. [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Kaga H, Mano H, Tanaka F, Watanabe A, Kaneko S, Morisaki H. Rice seeds as sources of endophytic bacteria. Microb Environ. 2009;24:154–162. doi: 10.1264/jsme2.me09113. [DOI] [PubMed] [Google Scholar]
- Knauth S, Hurek T, Brar D, Reinhold-Hurek B. Influence of different Oryza cultivars on expression of nifH gene pools in roots of rice. Environ Microbiol. 2005;7:1725–1733. doi: 10.1111/j.1462-2920.2005.00841.x. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Senthilkumar M, Seshadri S, Chung H, Yang J, et al. Growth promotion and induction of systemic resistance in rice cultivar Co-47 (Oryza sativa L.) by Methylobacterium spp. Bot Bul Acad Sin. 2004;45:315–324. [Google Scholar]
- Mano H, Morisaki H. Endophytic bacteria in the rice plant. Microb Environ. 2008;23:109–117. doi: 10.1264/jsme2.23.109. [DOI] [PubMed] [Google Scholar]
- Mattos KA, Padua VLM, Romeiro A, Hallack LF, Neves BC, Ulisses TMU, et al. Endophytic colonization of rice (Oryza sativa L.) by the diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. An Acad Bras Cienc. 2008;80:477–493. doi: 10.1590/s0001-37652008000300009. [DOI] [PubMed] [Google Scholar]
- Milling A, Smalla K, Maidl FX, Schloter M, Munch JC. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil. 2004;266:23–39. [Google Scholar]
- Muthukumarasamy R, Kang UG, Park KD, Jeon WT, Park CY, Cho YS, et al. Enumeration, isolation and identification of diazotrophs from Korean wetland rice varieties grown with long-term application of N and compost and their short-term inoculation effect on rice plants. J Appl Microbiol. 2007;102:981–991. doi: 10.1111/j.1365-2672.2006.03157.x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD, editors. Molecular Microbial Ecology Manual. Springer; Dordercht, NL: 2004. pp. 743–770. [Google Scholar]
- Nandakumar R, Babu S, Viswanathan R, Raguchander T, Samiyappan R. Induction of systemic resistance in rice against sheath blight disease by Pseudomonas fluorescens. Soil Biol Biochem. 2001;33:603–612. [Google Scholar]
- Nicolaisen MH, Risgaard-Petersen N, Revsbech NP, Reichardt W, Ramsing NB. Nitrification-denitrification dynamics and community structure of ammonia oxidizing bacteria in a high yield irrigated Philippine rice field. FEMS Microbiol Ecol. 2004;49:359–369. doi: 10.1016/j.femsec.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Peng S, Laza RC, Visperas RM, Khush GS, Virk P. Progress in breeding the new plant type for yield improvement: a physiological view. In: Toriyama K, Heong KL, Hardy B, editors. Rice is life: scientific perspectives for the 21st century. International Rice Research Institute; Manila, Philippines: 2005. pp. 130–132. [Google Scholar]
- Phillips RL, Odland WE, Kahler AL. Rice as a reference genome and more. In: Brar DS, Mackill DJ, Hardy B, editors. Rice Genetics V. World Scientific Publishing & International Rice Research Institute; Manila, Philippines: 2007. pp. 3–16. [Google Scholar]
- Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact. 2006;19:827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- Scheid D, Stubner S. Structure and diversity of Gram-negative sulfate-reducing bacteria on rice roots. FEMS Microbiol Ecol. 2001;36:175–183. doi: 10.1111/j.1574-6941.2001.tb00838.x. [DOI] [PubMed] [Google Scholar]
- Sessitsch A, Reiter B, Pfeifer U, Wilhelm E. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol. 2002;39:23–32. doi: 10.1111/j.1574-6941.2002.tb00903.x. [DOI] [PubMed] [Google Scholar]
- Tan Z, Hurek T, Vinuesa P, Müller P, Ladha JK, Reinhold-Hurek B. Specific detection of Bradyrhizobium and Rhizobium strains colonizing rice (Oryza sativa) roots by 16S-23S ribosomal DNA. Appl Environ Microbiol. 2001;67:3655–3664. doi: 10.1128/AEM.67.8.3655-3664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Braak CJF, Šmilauer P. CANOCO reference manual and Canodraw for windows user’s guide: software for canonical community ordination (version 4.5) Microcomputer Power; Ithaca, NY, USA: 2002. p. 500. [Google Scholar]
- Ter Braak CJF. Canonical community ordination. Part I: Basic theory and linear methods. Ecoscience. 1994;1:127–140. [Google Scholar]
- Van Overbeek L, Van Elsas J. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.) FEMS Microbiol Ecol. 2008;64:283–296. doi: 10.1111/j.1574-6941.2008.00469.x. [DOI] [PubMed] [Google Scholar]
- Wang FQ, Wang ET, Zhang YF, Chen WX. Characterization of rhizobia isolated from Albizia spp. in comparison with microsymbionts of Acacia spp. and Leucaena leucocephala grown in China. Syst Appl Microbiol. 2006;29:502–517. doi: 10.1016/j.syapm.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Zul D, Denzel S, Kotz A, Overmann J. Effects of plant biomass, plant diversity, and water content on bacterial communities in soil lysimeters: implications for the determinants of bacterial diversity. Appl Environ Microbiol. 2007;73:6916–6929. doi: 10.1128/AEM.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.