Summary
The regeneration potential of mesenchymal stem cells (MSCs) diminishes with advanced age and this diminished potential is associated with changes in cellular functions. This study compared MSCs isolated from the bone marrow of rhesus monkeys (rBMSCs) in three age groups: young (< 5 years), middle (8–10 years), and old (> 12 years). The effects of aging on stem cell properties and indicators of stem cell fitness such as proliferation, differentiation, circadian rhythms, stress response proteins, miRNA expression, and global histone modifications in rBMSCs were analyzed. rBMSCs demonstrated decreased capacities for proliferation and differentiation as a function of age. The production of heat shock protein 70 (HSP70) and heat shock factor 1 (HSF1) were also reduced with increasing age. The level of a core circadian protein, Rev-erb α, was significantly increased in rBMSCs from old animals. Furthermore, analysis of miRNA expression profiles revealed an up-regulation of mir-766 and mir-558 and a down-regulation of mir-let-7f, mir-125b, mir-222, mir-199-3p, mir-23a, and mir-221 in old rBMSCs compare to young rBMSCs. However, there were no significant age-related changes in the global histone modification profiles of the four histone core proteins: H2A, H2B, H3, and H4 on rBMSCs. These changes represent novel insights into the aging process and could have implications regarding the potential for autologous stem cells therapy in older patients.
Keywords: aging, cell cycle, differentiation, mesenchymal stem cells, miRNA, non-human primate
Introduction
Mesenchymal stem cells (MSCs) are a heterogeneous population of cells that can be isolated from the connective tissue of almost all organs including: adipose, periosteum, synovial fluid, muscle, hair follicles, roots of deciduous teeth, articular cartilage, placenta, dermis, umbilical cord, Wharton's jelly, lung, liver, and spleen and have been described in a variety of species, including humans, nonhuman primates, and rodents (Prockop, 1997; Majumdar et al., 1998; Pittenger et al., 1999; Woodbury et al., 2000; Izadpanah et al., 2005). MSCs have the capacity for self-renewal and the potential to differentiate into multiple lineages, such as osteocytes (Jaiswal et al., 1997), adipocytes (Purpura et al., 2004), chondrocytes (Johnstone et al., 1998) and myo-blast (Wakitani et al., 1995). While it is obvious that MSCs retain their capacity for self-renewal and differentiation, it has become increasingly clear that the therapeutic efficacy mediated by MSCs is through the production of bioactive levels of soluble factors (growth factors and cytokines) that regulate diverse disease-associated processes, including activation of tissue-resident stem ⁄ progenitor cells, apoptosis, stimulation of vasculo-genesis and inhibition of inflammation (Giordano et al., 2007; Kolf et al., 2007).
Biological aging is associated with a progressive loss of regulation of cellular, tissue and organ interaction, ultimately resulting in senescence. Biological aging can influence the decline in regenerative potential of tissue and cellular functions in a variety of organs. Clinical trials as well as animal studies have shown that the regeneration potential of bone and other tissues declines with age due to a decline in the number or frequency of stem cells present in adult organs; these factors may contribute to human aging and age-related disease (Meyer, 2001; Stenderup et al., 2004; Conboy & Rando, 2005; Rando, 2006). Therefore, understanding the age-related functional and biological changes that occur in MSCs will be critical to the success of any therapeutic application of MSCs in regenerative medicine.
Only recently have people begun to collect data on the effects of natural aging on mesenchymal lineage stem cells. Several reports indicate that aging is accompanied by numerous changes in biological processes in MSCs. The number of cells obtained by bone marrow aspiration (Sethe et al., 2006) and their potential to proliferate and differentiate declined with age in both humans and mice (Bellows et al., 2003; Shi et al., 2005; Mareschi et al., 2006; Tokalov et al., 2007). BMSCs isolated from older human donors lack the characteristic spindle-like morphology observed in BMSCs from younger donors (Baxter et al., 2004). Several groups have demonstrated that the frequency of CFU decreased in aged donors among multiple species (Baxter et al., 2004; Stolzing & Scutt, 2006; Zhou et al., 2008). A study performed using MSCs from a broad age range of human donors (17–90 years old), revealed a four-fold increase in the frequency of senescent cells and a doubling rate that was almost twice as long in MSCs from older donors (Zhou et al., 2008). Essential intrinsic cell processes such as telomere shortening (Armanios et al., 2009), DNA damage accumulation (Beausejour, 2007), and oxidative stress (Stolzing & Scutt, 2006; Kasper et al., 2009) are also affected by age in MSCs. It has also been determined that BMSCs from aged human subjects have increased levels of p21 and p53, as well as apoptotic cells (Stolzing et al., 2008; Zhou et al., 2008). Moreover, the cells from aged donors had a marked decrease in the overall expansion rate and multilineage differentiation potential (D'Ippolito et al., 2006; Stolzing & Scutt, 2006; Stolzing et al., 2008; Zhou et al., 2008). Recent studies that compared gene expression profiles from MSCs derived from young and old humans, monkeys, and mice showed down-regulation of genes involved in the cell cycle, DNA replication, and DNA repair with age (Auricchio et al., 2002; Hacia et al., 2008; Wagner et al., 2008). Furthermore, several studies have shown that miRNAs are regulated in various human cells as a result of aging (Hackl et al., 2010) and that a number of miRNAs are differentially expressed in aged MSCs due to long-term culture (Wagner et al., 2008). These changes are controlled by epigenetic alterations such as DNA methylation and histone modifications and could play a role in the changes associated with aging observed in cells (Young et al., 2004; Bork et al., 2009).
In this study, the affects of biological aging on the properties of rBMSCs at both the cellular and molecular levels were analyzed. Assessments of growth and differentiation, as well as cellular and molecular markers of aging, were investigated. The data presented here demonstrate an age-dependent loss of cellular proliferation and differentiation potential. The functional changes correlate with changes in cellular indices of aging including: age-related alterations in the expression of both CD44 and CD90, an increase in the frequency of cells expressing senescence-associated β-galactosidase, and increased levels of reactive oxygen species (ROS) as well as both p53 and p21. As part of this analysis, age-related changes in the miRNA expression patterns and levels were characterized. The affects of aging on epigenetic alteration of histone proteins demonstrated that histone modification is not markedly altered as a function of age. These data provide novel insights into the molecular and biological alterations occurring in rBMSCs that are associated with biologic aging. The data suggest that aging results in the accumulation of altered stem cell-associated properties that directly result in a marked decrease in the functionality and quality of BMSCs. Ultimately, these age-associated alterations may impact the efficacy of BMSCs autologously transplanted in aged populations.
Results
Morphologic appearance of rBMSCs from three age groups
The rBMSCs were characterized by morphology, differentiation capacity, and immunophenotype as previously described (Izadpanah et al., 2005). For this study, cells were divided into three different age groups; young rBMSCs (< 5 years), middle rBMSCs (8–10 years), and old rBMSCs (> 12 years) groups. Young rBMSCs showed a fibroblast-like appearance with few cytoplasmic expansions, while middle and old rBMSCs showed diverse shapes that were widely spread out; this appeared as early as passage 1 (Fig. S1A). The size of the cells isolated from the young, middle, and old rBMSCs was also analyzed. As demonstrated by forward light-scatter characteristics, rBMSCs from animals > 12 years old were found to be significantly larger than rBMSCs from animals < 5 years old (Fig S1B,C).
Immunophenotype analysis in vitro
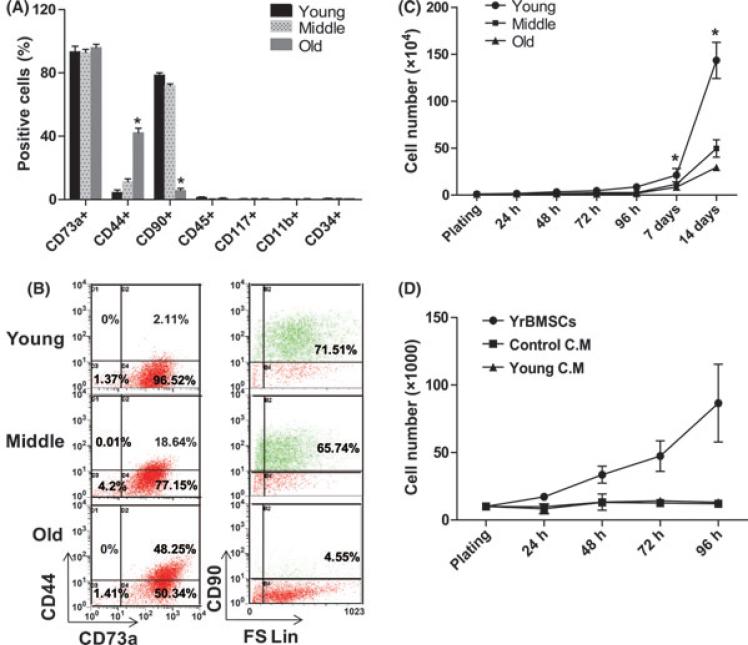
The effects of biological aging on the immunophenotype of rBMSC were analyzed by flow cytometry for cell surface antigens. A panel of seven surface markers composed of antibodies to both hematopoietic and non-hematopoietic lineage proteins typically used for MSC characterization was analyzed (Fig. 1A). All groups of rBMSCs at passage 1 or 2 were negative for hematopoietic markers, CD34, CD45, CD11b, and CD117 and positive for MSC markers, including CD90, CD44, and CD73. Interestingly, the percentage of CD90+ cells in young and middle rBMSCs was much higher than that of old rBMSCs. The percentage of CD90+ cells was 71.51% in young rBMSCs, 65.74% in middle rBMSCs, and only 4.55% in old rBMSCs. However, the percentage of CD44+ cells in old rBMSCs was higher than that from young rBMSCs. The frequency of CD44+ cells was 2.1% in young rBMSCs, 18.64% in middle rBMSCs and 48.25% in old rBMSCs (Fig. 1B).
Fig. 1.
Age-related changes in immunophenotype and growth curves. (A) Age-related changes in the proportion of rBMSCs expressing specific surface marker (*P < 0.05, significant difference from young rBMSCs). (B) Immunophenotypic characterization of rBMSCs in three age groups. Dot plot graphs are representative of three samples per/ each group. (C) Growth curves of rBMSCs derived from young, middle, and old age groups. The young rBMSCs expanded more rapidly than old rBMSCs. The difference in growth rate among the three groups is statistically significant (*P < 0.05, n = 4/ group). (D) Effect of CM obtained from young rBMSCs on cell growth of aged rBMSCs. Old rBMSCs were plated on 12 well plates and cultured in CM obtained from young rBMSCs. Old rBMSCs were counted every 24 h for 4 days. Data are expressed as the mean ± SED, n = 3.
Age-related decreases in proliferation capacity
To assess age dependent changes of rBMSCs, a cellular proliferation assay was performed over a period of 14 days. Cells were harvested by trypisinzation and counted every 24 h for the first 96 h and then again on days 7 and 14. After 72 h, rBMSCs from young animals exhibit more rapid proliferation rates compared to rBMSCs from the other groups (Fig. 1C). The doubling time in rBMSCs from the older animals increased gradually until the cells ultimately underwent senescence at passage 2 (data not shown).
Several studies have reported that conditioned medium (CM) obtained from MSCs contains putative growth factors that accelerate cell division, delay senescence, and enhance cell differentiation (Hakkinen et al., 2001; Chivu et al., 2002; Elder, 2002; Torres et al., 2002). The ability of CM collected from rBMSCs from young rhesus macaques to stimulate the growth of rBMSCs from the old group of animals was investigated. rBMSCs from the old group of animals were plated in 12 well plates, cultured for 24 h and the media was replaced with 100% CM obtained from rBMSCs from young animals. The cells were then counted every 24 h for 4 days. The CM from the young rBMSCs failed to enhance the proliferation of the rBMSCs from old animals, compared to the control samples (Fig. 1D). These results suggest that the presence of factors secreted by the rBMSCs from young animals alone were not sufficient to influence proliferation of rBMSCs from older donors.
Differentiation potential is diminished as a function of biologic age
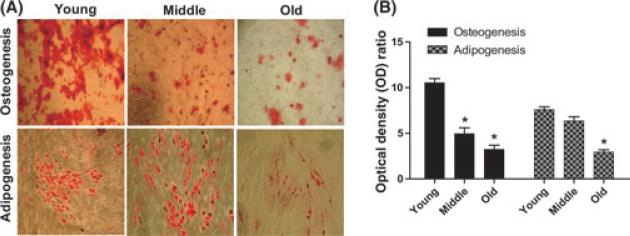
To assess the osteogenic differentiation potential of rBMSCs among the three age groups, cells were cultured in osteogenic induction medium. The cells aggregated, formed nodules, and accumulated calcium deposits over the 2-week period. An Alizarin Red stain was used to detect precipitated calcium salt, which indicates differentiation. The data from a visual assessment demonstrated that rBMSCs from the old and middle age groups showed lower amounts of Alizarin Red staining compared to cells from young animals (Fig. 2A). The extraction and quantification of the levels of calcium accumulation was used as an indication of the extent of differentiation. The results indicated that young rBMSCs [Optical Density (OD) ratio = 10.1] appeared to differentiate into mineralizing cells to a greater degree than the middle rBMSCs (OD ratio = 5.6) and the old rBMSCs (OD ratio = 3.7) (Fig. 2B).
Fig. 2.
Age-related decline in differentiation potential of rBMSCs. (A) Representative image showing Alizarin Red stained mineral deposits (top) and Oil Red O stained lipid inclusions (bottom) on cultured rBMSC for each age group. Young rBMSCs show a markedly increased level of differentiation potential at passage 2. 10× magnification. (B) Graphs represent the ratio of normalized OD of differentiated cells.
The adipogenic differentiation potential of rBMSCs in the three age groups was also monitored. Oil Red O staining was used to quantify cell differentiation following 3 weeks of culture in standard adipogenic medium. Based on the visual assessment of the extent of staining of Oil Red O-positive lipid inclusions in the cells, the adipogenic differentiation potential also decreased with biological age (Fig. 2A). The quantification of the extracted Oil Red O stain showed a statistically significant decrease in adipogenic differentiation of rBMSCs in the middle (OD ratio = 6.4) and old age groups (OD ratio = 3) when compared to the young age group (OD ratio = 7.6) (Fig. 2B).
Cell cycle related perturbations associated with age
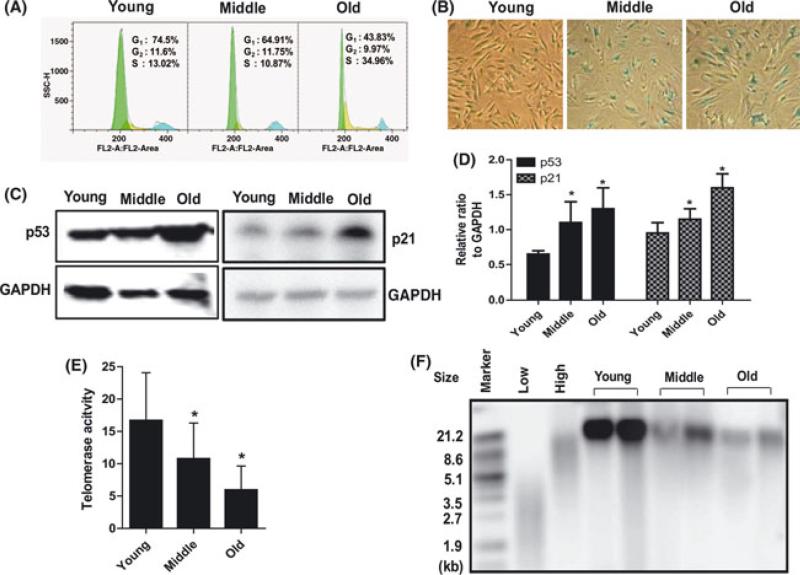
The cell cycle distribution of rBMSCs among the three age groups was determined using flow cytometric analysis, after staining the cellular DNA with propidium iodide (PI). Single-variable histograms of DNA provided data describing the percentages of cells in G0-G1, S, and G2-M phase of the cell cycle. Populations of rBMSCs from old rhesus macaques possess a significantly higher percentage of cells in S phase (34.96 ± 3.3%) than cells from young animals (13.5 ± 2.8%). These data demonstrated an increasing fraction of cells arrested in or with a protracted duration of S phase in the rBMSCs from old and middle age groups (Fig. 3A).
Fig. 3.
Increased cellular senescence in aged rBMSCs. (A) The cell cycle distribution of rBMSCs was determined by flow cytometry analysis after the cellular DNA was stained with propidium iodide (PI). The histograms of DNA content indicate the percentages of cells in G0-G1, S, and G2-M phases of the cell cycle. A representative result is shown. (B) Images of β-galactosidase staining (blue) of rBMSCs in three age groups. (C) Age-related increase in p53 and p21 protein expression was detected by Western blotting. (D) Relative ratio to GAPDH (*P < 0.05, significant difference from young rBMSCs). (E) Telomerase enzyme activity. Telomerase activity was detected by the TRAP assay. Analysis of telomerase activity showed reduced activity in old rBMSCs (*P < 0.01, significant difference from young rBMSCs). Data represent mean ± SEM of three different experiments. (F) Telomere length was determined as a mean terminal restriction fragment (TRF) length of genomic DNA based on Southern hybridization with a telomeric probe. Terminal restriction fragments were visualized, using a labeled (TTAGGG) probe.
Cellular senescence in the three age groups
Senescence is a phenomenon of cellular aging, which can be detected by monitoring senescence associated β-galactosidase activity. The percentage of β-gal positive, early passage cells in old rBMSCs was significantly higher than in young rBMSCs at the same passage (Fig. 3B). To further validate that there is an age-related alteration in cell senescence in cells from the older animals, markers associated with the senescent phenotype of somatic cells such as the tumor suppressor gene p53 and the cell-cycle regulation protein p21 were analyzed by Western blot. The expression of both p53 and p21 were significantly up-regulated in old rBMSCs compared to young rBMSCs (Fig. 3C). The relative quantification by densitometry revealed approximately two-fold up-regulation of p53 protein and a 1.7-fold up-regulation of p21 protein in old rBMSCs compared to young rBMSCs (Fig. 3D).
Telomere maintenance in aged rBMSCs
Telomerase enzyme activity was evaluated in the three age groups of rBMSCs using the amplified telomeric repeat (TRAP) method. The levels of telomerase activity were reduced in old rBMSCs. The telomerase activity was decreased by 11.5% in middle-aged rBMSCs and by 65% in cells from old animals, when compared to cells from young animals at identical passage (Fig. 3E). Telomere length was monitored using a mean terminal restriction fragment (TRF) length of genomic DNA based on Southern hybridization with a telomeric probe. Telomere length was analyzed in the three age groups. Although the signal intensity of telomere length in middle and old rBMSCs was decreased compared to young rBMSCs, shorter telomeres were not detected (Fig. 3F).
Stress response related changes with age
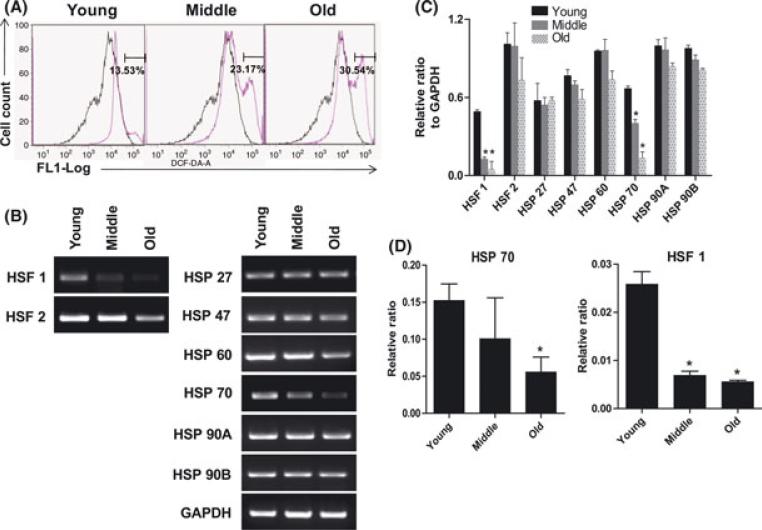
Oxidative damage of many types accumulates with age (Stolzing et al., 2006; Sauvaigo et al., 2007; Satoh et al., 2008). ROS production has been determined to be a major cause of DNA damage and cell senescence in the aged. The accumulation of ROS in the rBMSCs was determined using DCFH-DA fluorescence as an indicator. Flow cytometric analysis showed that the frequency of ROS positive cells increased in middle (27.17%) and old (30.54%) rBMSCs compared to young (13.53%) rBMSCs (Fig. 4A).
Fig. 4.
Age related changes of stress responses genes. (A) ROS accumulation in rBMSCs of three age groups. DCFH-DA fluorescence of cells was determined by FACS cytometry as an indicator of ROS accumulation within the cells. A representative result is shown. Median of fluorescence intensity of unstained rBMSCs (black) and DCFH-DA stained (pink) rBMSCs of three age groups. The X axis represents log F1 fluorescence intensity; the Y axis represents cell number. (B) Alteration of stress response genes with age. Total RNA was isolated from three age groups and analyzed by RT-PCR for the indicated factor (HSF-1, HSF-2, HSP27, HSP 47, HSP 60, HSP 70, HSP 90A, and HSP 90B). A representative result is shown. (C) The quantification of stress response genes expression level via RTPCR was determined using an image analyzer. Values are mean ± SEM. *p < 0.05, significant difference compare to young rBMSCs. (D) To confirm expression of HSF 70 and HSF 1, RNA was quantified using qPCR. The graph is shown a quantitative analysis of relative expression of mRNAs at three age groups. The values were normalized using β-actin as a control. *p < 0.05, significantly difference compare to young rBMSCs.
The effects of age on the expression of stress related genes were also analyzed. The expression of heat shock and stress proteins (HSF-1, HSF-2, HSP27, HSP 47, HSP 60, HSP 70, HSP 90A, and HSP 90B) was examined by RT-PCR in the three age groups (Fig. 4B). While no alterations were observed in the expression of the majority of the genes analyzed, there was a two fold decrease in the expression levels of mRNAs encoding the transcriptional regulator HSF-1 and a four fold decrease in the expression of HSP 70 in middle and old rBMSCs compared to young rBMSCs (P < 0.05) (Fig. 4C).
Circadian rhythm element gene expression
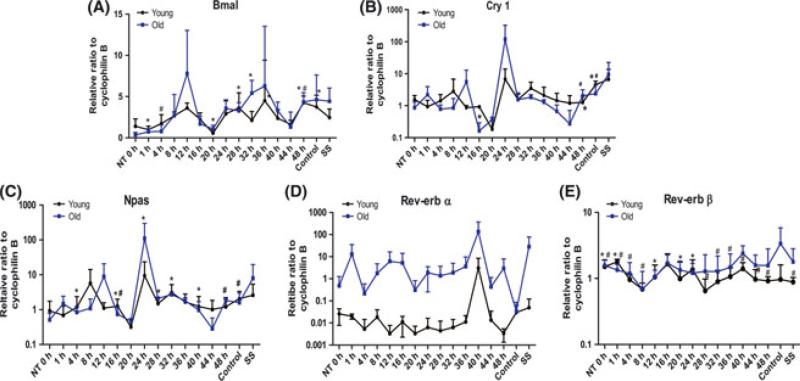
The circadian system and circadian proteins play direct roles in the aging process (Gibson et al., 2009). With advancing age, these daily fluctuations deteriorate, leading to disrupted cycles with reduced amplitude in both experimental organisms and in humans. Such disruptions in the circadian system appear to accelerate the aging process and contribute to senescence with age (Wagner & Mittag, 2009). To investigate the relevance of oscillation of circadian rhythm genes with age in rBMSCs, confluent and quiescent rBMSCs were exposed to 1 μm dexamethasone for 2 h and then cultured in regular medium. For RNA isolation, individual plates were harvested at 4-h intervals over a total period of 48 h. The expression of the core circa-dian transcription factors, Bmal 1, Cry 1, Npas 2, Rev-erb α and Rev-erb β were examined in the three age groups (Fig. 5). Young and old rBMSCs showed similar rhythm patterns of their clock genes and there was no significant difference between the two groups. Of interest is the fact that, the peak level of expression for Rev-erb α in young rBMSCs was much lower than that in old rBMSCs (Fig. 5D). The peak time point for Rev-erb β appeared to be broader in duration and reached a lower zenith in young rBMSCs compared to old rBMSCs (Fig. 5E). Rev-erb α/ β is an essential component of the core circadian clockwork and the data demonstrate that age may have an impact on both the appropriate timing and the level of Rev-erb expression in rBMSCs in aged rhesus.
Fig. 5.
The variations in relative mRNA expression for core circadian oscillator proteins: (A) Bmal1, (B) Cry1, (C) Npas, and (D) Rev-erb α, and (E) Rev-erb β in rBMSCs isolated from young and old group animals. Samples were collected every 4 h for a total of 48 h in young (black) and old (blue) rBMSCs. Cyclophilin B is used as a control. Values are mean ± SEM. (# P < 0.05 in young age group, *P < 0.05 in old age group, n = 3/ group).
Altered microRNA expression with advanced age
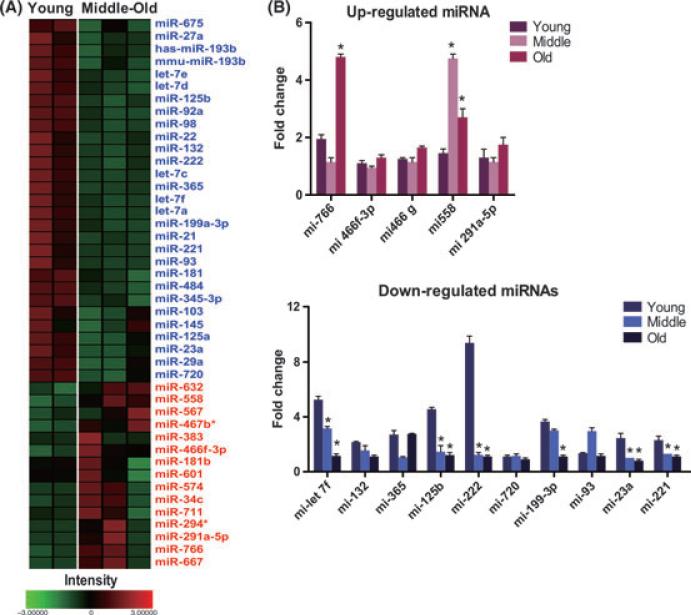
To investigate the role biological aging plays in miRNA expression patterns in rBMSCs, miRNA array analysis was performed on cells from the three age groups using array profiling of global miRNA expression. Our results indicate that multiple miRNAs are differentially expressed as a function of biological aging. A total of 138 miRNAs were up-regulated in middle and old rBMSCs, compared to young rBMSCs, which represents 25% of the total miRNA probes on the array. A total of 206 miRNAs were down-regulated in comparison to young rBMSCs, which represents 38% of total miRNA probes on the array. The analysis permitted the identification of 15 miRNAs that were significantly up-regulated (> 1.5 fold) and 29 that were down-regulated (> 1.5 fold) in middle and old rBMSCs (Fig. 6A). Table 1 highlights the miRNA with the greatest fold variations in middle and old rBMSCs compared to young rBMSCs. The differential expression of miRNAs was validated using quantitative RT-PCR; we confirmed the down-regulation of mir-let-7f, mir-132, mir-365, mir-125b, mir-222, mir-720, mir-199-3p, mir-23a, and mir-221 and the up-regulation of mir-766, mir-466-3p, mir-466, mir-558, and mir-291a-5p (Fig. 6B).
Fig. 6.
MicroRNA expression in rBMSCs. (A) Heat map visualization and clustering of miRNA expression data. Hierarchical clustering of age dependent expression profiles among 556 miRNA in young, middle, and old rBMSCs. Relative expression levels at three age groups visualized in using a heatmap in which red indicates up-regulation in each group, while blue corresponds to down-regulation. See scale bar at bottom. (B) The fold changes in expression of the miRNAs were performed by qPCR. *P < 0.05, significant differences compare to young rBMSCs.
Table 1.
Regulation of miRNAs relative to the young age group
| Sequence code | Middle (8-10 years) fold change | Old (> 12 years) fold change | Sequence code | Middle (8-10 years) fold change | Old (> 12 years) fold change |
|---|---|---|---|---|---|
| miR-720 | –1.9 | –1.61 | miR-667 | 2.03 | 1.61 |
| miR-93 | –1.55 | –1.6 | miR-467b* | 1.75 | 1.58 |
| miR-365 | –1.75 | –1.7 | miR-294* | 1.65 | 1.43 |
| miR-23a | –2.44 | –1.96 | miR-291a-5p | 2.21 | 2.22 |
| miR-222 | –1.51 | –1.52 | miR-512-3p | 2.28 | 1.29 |
| miR-221 | –1.69 | –1.76 | miR-466f-3p | 1.97 | 1.38 |
| miR-199a-3p | –1.64 | –1.96 | miR-466g | 1.68 | 1.31 |
| miR-132 | –1.81 | –1.91 | miR-766 | 1.66 | 1.28 |
| miR-125b | –1.63 | –1.69 | miR-294* | 1.65 | 1.43 |
| miR-let-7f | –1.57 | –1.74 | miR-558 | 1.21 | 1.36 |
| miR-let-7e | –1.57 | –1.8 |
Reverse-Phase Liquid Chromatography-Mass Spectrometry (RPLC-MS)
Histone modifications contribute to epigenetic mechanisms, which have been associated with biological function in stem cell lineages (Wu & Sun, 2006; Chambers et al., 2007; Rugg- Gunn et al., 2010). Global histone profiling (Zhang et al., 2004; Su et al., 2007a, 2009) was performed on cells from cohorts of rBMSCs from three different rBMSC samples from both the young and old animals. Reversed phase LC-MS analysis of the histones extracted from these cells revealed distinct profiles of H4 isoforms for each animal (Fig. S2). However, there was no apparent correlation between the profiles and age.
Discussion
Many studies have described biological age-related changes in human and murine MSCs. In this study, we investigated age-related alterations of the molecular and biological properties of rBMSCs. The data demonstrated that cells from aged rhesus macaques (> 12 years old) have diminished capacities for proliferation and differentiation compared to cells isolated from young animals. rBMSCs from the young animals expressed the typical MSC markers (CD90 and CD73) at high levels and were negative for hematopoietic markers (CD11b, CD117, and CD45). In the rBMSCs from aged macaques, the percentage of CD90+ cells was decreased as previously reported in other species (Campioni et al., 2006; Tokalov et al., 2007). A previous study demonstrated that CD90) human BMSCs have decreased immunosuppressive properties and do not increase the secretion of HLA-G or IL-10 levels as the CD90+ BMSCs (Campioni et al., 2008). The percentage of CD44+ cells was also markedly increased in samples from aged animals, which has not been described for BMSCs in other species. According to a recent report, the expression of CD44 was shown to be increased on T cells in aged mice and correlated with increasing age (Han et al., 2009), which supports our observation.
Cell senescence as a consequence of cellular aging was observed by SA-β-galactosidase assay and up-regulation of p53 and p21 protein in middle and old rBMSCs. Telomerase activity in old rBMSCs was decreased significantly compared to young rBMSCs, whereas there was no detectable shortening of the telomeres in middle or old rBMSCs. Taken together, the data suggest that biological aging affects cell morphology as evidenced by enlargement of the rBMSCs, increases cell senescence, and alters the cell surface antigen profile. These changes may ultimately stop proliferation and decrease the maximal life span of BMSCs.
As illustrated in this study, rBMSCs show a progressive accumulation of senescent cells with age. This is associated with various stress responses such as heat shock and oxidative stress (Hall et al., 2000). One of the most widely studied groups of stress response proteins is a family of molecular chaperones known as heat shock proteins (HSPs). HSPs play a critical role in protecting against cellular damage (Swanlund et al., 2008). Many other studies have shown age-related declines in stress-induced synthesis of HSPs. The observed decrease in HSP 60 indicates an age-related loss of apoptotic flexibility, which has been associated with senescence (Campisi, 2002). Wagner & Margolis (1995) found that HSP90 could be a direct component of the proteasome complex in young, but not in aged bovine tissues. Recently, Chung & Ng (2006) reported that HSP27, )60 and )70 are all increased in senescent rat muscle tissue, when comparing adult and senescent donors. In contrast to these results, Stolzing et al. (2006) reported that HSP 27, HSP70, and HSP90 were up-regulated in young rat MSCs cultured at a reduced temperature. In this study, we found significantly increased levels of ROS generation and decreased expression of HSP70 in old compared to young rBMSCs. There was also a decrease in heat shock factor 1 (HSF1) with increasing age in rBMSCs. HSF1 is essential for protecting cells from protein-damaging stress associated with misfolded proteins, and influences aging process. Studies showed that reducing HSF1 activity accelerated tissue aging and shortens life-span (Hsu et al., 2003; Westerheide et al., 2009). HSF1 also is responsible for modulating the transcriptional expression of HSP70 in response to whole body stress in rodents, highlighting the central role for HSF1 even during physiological stress (Pirkkala et al., 2001). Therefore, further investigation is required to determine whether HSF1 mediates expression of HSP 70 in aging process of rBMSCs. Our results provide that changes in the expression of the stress response genes could be an important factor in advanced donor age.
Circadian rhythms are genetically determined biological rhythms that are considered an important adaptive mechanism to the cyclical light/ dark alterations in the environment. Age-related changes in the circadian time-keeping mechanism are well known, and seemingly contribute to various pathologies associated with biological aging (Kondratov, 2007). Recent studies indicate that the circadian system and circadian clock proteins may be involved in DNA repair and in regulating accumulation of cellular ROS, and that they play a role in aging processes (Geyfman & Andersen, 2010). Multiple lines of evidence demonstrate that core components of the circadian regulatory system control a variety of cellular processes, including: cell division control, DNA damage response pathways, and chromatin remodeling. In animal models, there is a strong link between the expression of circadian regulatory proteins and regulated progression through the cell cycle (Fu et al., 2002; Matsuo et al., 2003; Gery et al., 2005). In fact, deregulated expression of either Per1 or Per2 alters the expression of several cell cycle genes, including c-myc, p21, wee1, cyclin B1, and cdc2, that are key regulators of arrest of the cell cycle and apoptosis (Gery et al., 2005, 2006, 2007a,b; Hua et al., 2006; Gery & Koeffler, 2007a,b). Moreover, dysregulated expression of circadian genes has been observed in a wide variety of human tumors, including hematologic, breast, pancreas, and endometrial cancers (Shih et al., 2006; Yang et al., 2006; Gery & Koeffler, 2007a). Circa-dian pathways also have a direct link to pathways involved in chromatin remodeling and ultimately, transcription regulation (Crosio et al., 2000; Curtis et al., 2004; Naruse et al., 2004; Doi et al., 2006). A previous study reported that exposure to dexamethasone or serum shock initiated the oscillatory expression of mRNA associated with the core circadian transcriptional apparatus (Wu et al., 2008). The rBMSCs in this study were exposed to dexamethasone for 2 h and the expression of circa-dian core genes was measured. Our data demonstrate that circadian core genes (bmal1, cry1, rev-erb α, and npas) are rhythmically expressed in young and old rBMSCs. A significant age-related change in the levels of mRNA expression for one of the core circadian genes was detected. Specifically, the basal expression of rev-erb α was suppressed in young rBMSCs compared to old rBMSCs. Robust oscillations in rev-erb α were observed in young and old rBMSCs at different time point throughout the 48-h period. Rev-erb α, known as nuclear receptor subfamily 1, group D member 1 (Nr1d1), belongs to the family of ‘orphan receptors’ and functions as a member of the clock gene family. Apart from its circadian regulatory functions, it has been reported that Rev-erb α also regulates cell proliferation and differentiation (Chawla & Lazar, 1993; Downes et al., 1995). Until now, there has been no experimental evidence describing the expression of oscillating clock genes in aged rBMSCs. Therefore, this observation is noteworthy as Reverb a, a ‘clock gene’, has also been reported to act as regulator of aging process in rhesus pituitary gland (Sitzmann et al., 2010).
The potential of miRNAs to be central regulators of aging in humans has recently attracted intense interest. In this study, global age-related alterations in miRNA expression in rBMSCs were examined, with a view to identifying pathways that may play key roles in the age-associated declines in MSC function. The data demonstrate for the first time that miRNAs are differentially regulated with increasing age in rBMSCs. Surprisingly, many of the described miRNA functions did not have a direct association with cell growth and cell cycle; however, the full activity of most miRNAs is not well-defined. Among the up-regulated miRNAs, miR-558 is noted for targeting genes involved in cell cycle checkpoint and apoptosis (Maes et al., 2008). Among the down-regulated miRNAs, miR-221, and miR-222 are involved in tumorigenesis and regulate the expression of cell cycle regulatory proteins (Lambeth et al., 2009; Lorimer, 2009). Both miR-365 and miR-720 have functions specifically related to growth (Wenguang et al., 2007; Maes et al., 2009). To date, few studies have analyzed global miRNA expression patterns in aging stem cells and most of the studies were related to the effects of protracted culture on cells. A more recent study identified four miRNAs that were commonly down-regulated as a result of human aging (Hackl et al., 2010). This group analyzed these down-regulated miRNAs from four replicative cell types including human MSCs by miRNA microarray. There was no overlap between these miRNA and those identified in our studies, which could be the result of species differences or differences in analytical techniques. Therefore, the miRNA expression patterns described in the current manuscript may provide novel markers of the aging process on MSCs.
Furthermore, the contribution of epigenetic alterations related to biologic aging was investigated. The epigenetic profile of rBMSCs isolated from young and old animals were compared using LC-MS analysis. There was no apparent correlation between the histone epigenetic profiles and age. While these data suggest that the global pattern of histone isoforms is not significantly altered during aging, there is no way to determine the specific patterns of histone modifications from this LC-MS experiment. More detailed experiments are planned to examine the patterns of histone modification using quantitative middle-down and bottom-up mass spectrometry (Ren et al., 2005; Su et al., 2007b, Knapp, 2007). In summary, these results present insights into age-related alterations in cellular processes and comprehensive information on understanding the biological functions in rBMSCs. The outcomes have implications relating to the potential utility of autologous BMSCs isolated from older donors for regenerative medical therapies.
Experimental procedures
Isolation and expansion of rBMSCs
rBMSCs were isolated and cultured from groups of select ages of rhesus macaques (Macaca mulatta) as previously described (Izadpanah et al., 2005). Healthy rhesus macaques ranging in age from one to 20 years old and of both genders were used for these studies. The bone marrow samples were taken from 22 healthy rhesus macaques from three different age groups; young (< 5 years, n = 9), middle (8–10 years, n = 4), and old (> 12 years, n = 9) group. All animal procedures in this study conformed to the requirements of the Animal Welfare Act and were approved by the Institutional Animal Care and Use Committee (IACUC) of the Tulane National Primate Research Center prior to implementation. The animals were housed under conditions approved by the Association for the Assessment and Accreditation of Laboratory Animal Care International. The rBMSCs were cultured in MEM-alpha (Invitrogen, Carlsbad, CA, USA) supplemented with 20% fetal bovine serum (FBS) (Atlanta Biological, Lawrenceville, GA, USA), 1% l-glutamine, and 1% penicillin/ streptomycin (Invitrogen) at 37 °C in 5% CO2. Cells were routinely passaged when they reached no more than 75–80% confluence, using 0.25% trypsin plus 0.2% EDTA.
Flow cytometry
The immunophenotype of rBMSCs at passage 1 or 2 was analyzed by flow cytometry. Cells were trypsinized, collected, washed twice with PBS, and incubated with 0.1% BSA in PBS for 1 h at 4 °C with PeCy5, PeCy7, FITC or phycoerythrin (PE)-conjugated antibodies against CD34, CD11b, CD45, CD117 (all from Beckman-Coulter, Miami, FL, USA), CD 44, CD73a, and CD106 (Becton Dickinson, San Jose, CA, USA). Excess antibody was removed by washing cells with PBS, and cells were analyzed on Becton Dickinson FACS Calibur (Becton Dickinson) using Cell Quest software.
Proliferation assay
To compare the cell proliferation capacity among the three age groups, rBMSCs at passage 1 or 2 were plated in triplicate at 104 cells per well in six well plates. The medium was replaced every 48 h. Cells were counted every 24 h for 14 days using a hemocytometer, in order to determine their growth rate by cell number.
Collection of conditioned media
rBMSCs obtained from the young animals were cultured in 10 cm2 dishes. Cells were incubated for 24 h with complete medium to allow the cells to permit their adherence to the growth surface. The next day, the complete medium was removed and replaced with serum free medium. After 48 h, the medium was collected and used as CM for in vitro studies.
Differentiation assays
Osteogenic and adipogenic differentiation were induced according to a standard protocol (Izadpanah et al., 2005). For osteogenesis, the cultures were then incubated in complete expansion media (CEM) supplemented with 20 mm glycerol phosphate, 50 ng/ mL thyroxine, 1 nm dexamethasone, and 50 μm ascorbate 2-phosphate (all from Sigma, St Louis, MO, USA). The media was changed two times per week for 2 weeks. The cells were fixed with 10% formalin for 20 min at room temperature (RT) and stained with Alizarin Red, pH 4.1 (Sigma) for 20 min at RT. To quantify the amount of Alizarin Red, the deposition was extracted by 10% (w/ v) cetylpyridinium chloride in 10 mm sodium phosphate (pH 7.0) at RT for 1 h and the Alizarin Red stain in extraction buffer was determined by measuring the optical density (OD) of the solution at 560 nm.
For adipogenesis, the cultures were incubated in CEM supplemented with 5 μg/ mL insulin, 50 μm indomethacin, 1 μm dexamethasone, and 0.5 μm 3-isobutyl-1-methylxanthine (IBMX) (all from Sigma). The medium was changed two times per week for 2 weeks. The cells were fixed with 10% formalin for 20 min at RT and stained with 0.5% Oil Red O (Sigma) in methanol (Sigma) for 20 min at RT. To quantify the amount of Oil Red O, the stained oil droplets were extracted by isopropyl alcohol and Oil Red O stain in extraction buffer was determined by measuring the optical density (OD) of the solution at 520 nm. The results were then normalized to the protein contents of the samples.
Analysis of cell cycle status
Single cell suspensions of each age group were obtained from cultures at passages 1–2. For DNA content analysis, cells were fixed in 70% ethanol, rehydrated in PBS, treated for 30 min with RNase A (1 mg/ mL), and stained with 1 μg/ mL of propidium iodide (PI) for 5 min. The intensity of fluorescence was determined by analysis on a fluorescent-activated cell sorter (Becton Dickinson FACscan, San Jose, CA, USA) equipped with a 488-nm argon laser. Data acquisition was performed with Cell Quest (Becton Dickinson) software, and the percentages of cells in G1, S, and G2 phases of the cell cycle were calculated with Flowjo software (Tree Star, Inc, Ashland, OR, USA).
ROS detection
The intracellular generation of ROS was measured using DCFH-DA. DCFH-DA is cleaved intracellularly by nonspecific esterases to form DCFH, which is further oxidized by ROS to form the fluorescent compound DCF. DCFH-DA working solution was added directly to the medium to reach 10 μm, and then incubated at 37 °C for 15 min. Cells were then washed once, resuspended in PBS and kept on ice for an immediate detection by FACScan (Becton Dickinson). Data was analyzed with Flowjo software (Tree Star, Inc, Ashland, OR, USA).
Senescence-associated β-galactosidase staining
For staining, the cells were washed in PBS, fixed for 5 min at RT in 2% formaldehyde/0.2% glutaraldehyde, washed, and incubated at 37 °C in the absence of CO2 with freshly prepared senescence associated [β-Gal (SA-β-Gal)] stain solution: 1 mg of 5-bromo-4-chloro-3-indolyl P3-D-galactoside (X-gal) per mL (stock = 20 mg of dimethylformamide per mL)/ 40 mm citric acid/ sodium phosphate, pH 6.0/ 5 mm potassium ferrocyanide/ 5 mm potassium ferricyanide/ 150 mm NaCl/ 2 mm MgCl2. Staining was evident within 2–4 h and maximal in 12–16 h.
Telomerase activity assay
Telomerase activity was measured with the telomere repeat amplification protocol (TRAP) by using the Telo TAGGG telomerase PCR ELISA kit according to manufacturer's instructions (Roche Applied Science, Indianapolis, IN, USA). Briefly, telomerase added telomeric repeats (T2AG3) to the end of biotin-labeled primers. The extension products of telomerase were amplified using polymerase chain reaction (PCR). The telomerase-mediated elongated product was detected by hybridization to digoxigenin-labeled probes. The levels of enzyme activity were evaluated and determined by photometric enzyme immunoassay.
Telomere length assay
Telomere length was analyzed using the Telo TAGGG Telomere Length Assay kit according to manufacturer's instructions (Roche Applied Science). Genomic DNA (gDNA) was extracted from rBMSCs using a DNA extraction kit (Qiagen, Valencia, CA, USA). One microgram of gDNA was digested with the mixture of HinfI and RsaI restriction endonucleases, electrophoresed through a 0.8% agarose gel, and transferred to a positively charged nylon membrane. The membrane was then hybridized to a digoxigenin (DIG)-labeled telomeric oligonucleotide (TTAGGG)3. The DNA/ oligonucleotide hybridization products were visualized after reaction with a chemiluminescent substrate, using Versa Doc imaging system (Bio-Rad Laboratories, Hercules, CA, USA).
Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
Total cellular RNA was isolated from rBMSCs and reverse transcribed using conventional protocols. The primer sequences used in the experiment were as follows: HSF-1: 5′-GAA AGT GAC CAG TGT GTC CA-3′, 5′-AGA ACT GCC GGC TAT ACT TG-3′, HSF-2: 5′-GCC CTC TCA TGT CTA GTG CT-3′, 5′-GCT GCT TAT CTG GTT TGG AT-3′, HSP27: 5′-CTG GAT GTC AAC CAC TTC G-3′, 5′-AGT CTC ATC GGA TTT TGC AG-3′, HSP47: 5′-CTG CTC GTC AAC GCC ATG T-3′, 5′-CCA TCC AGG TCT TCA GCT GC-3′, HSP60: 5′-GCC TTA ATG CTT CAA GGT GT-3′, 5′-GTG ATG ACA CCC TTT CTT CC-3′, HSP70: 5′-TTC TAC ACG TCC ATC ACC AG-3′, 5′-TCG GAG TAG GTG GTG AAG AT-3′, HSP90A: 5′-TCC CAG TCT GGA GAT GAG AT-3′, 5′-CAG CAA GGT GAA GAC ACA AG-3′, HSP90B: 5′-AGA AGC AGC CAA AGA AGA GA -3′, 5′-AAA CAT TCA AGG GGA GAT CA-3′. All primer sequences were generated from established GenBank sequences. The PCR products were visualized and analyzed by 1.5% agarose gel electrophoresis.
Analysis of circadian rhythm-associated genes
The medium was removed from confluent cultures of undifferentiated rBMSCs in six well plates and replaced with MEM-alpha medium supplemented with 2% FBS and 1% penicillin/ streptomycin alone or further supplemented with 1 μm dexamethasone. The MSCs were exposed to the dexamethasone for 2 h, after that the medium was replaced with MEM-alpha medium and 1% penicillin/ streptomycin. The cells did not receive any further medium changes from this point onward until the time of harvest. Individual plates were harvested for total RNA at 4-hour intervals for a total of 48 h following the initial exposure (Wu et al., 2008).
Semi-Quantitative Real-time RT-PCR
Total RNA was isolated from rBMSCs of each group after dexamethasone exposure according to the manufacturer's specifications. Approximately 2 μg of total RNA was reverse transcribed using moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI, USA), with Oligo-dT at 42 °C for 1 h. Real time RT-PCR was performed on 10 fold diluted cDNA samples in water with SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) using the 7900HT Real Time PCR system (Applied Biosystems) under universal cycling conditions (95 °C for 10 min; 40 cycles of 95 °C for 15 s; then 60 °C for 1 min). The RT-PCR using gene specific primers for Bmal 1, Cry 1, Npas 2, Rev-erb α, and Rev-erb β had been validated and determined to display single peaks in their dissociation curves. All results were normalized relative to a cyclophilin B expression control.
Western blot analysis
rBMSC cultures were washed twice with ice cold phosphate-buffered saline (PBS) and then lysed in 40 μL of lysis buffer (Sigma) and 1 μL of cocktail proteinase inhibitor (Sigma). Total protein concentration was measured using a Bradford assay containing Coomassie Plus protein reagent (Bio-Rad Laboratories) according to the manufacturer's specifications. Equivalent amounts of total cell lysate were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using 10% polyacrylamide gels. Proteins were electroblotted to PVDF membrane (Millipore, Billerica, MA, USA). The membranes were then blocked and incubated in anti-p53, anti-p21 and anti-GAPDH antibody (all at 1:2000; Abcam, Cambridge, UK) overnight at 4 °C. Alkaline phosphatase-conjugated anti-rabbit IgGs (1:2000) were used as secondary antibodies (Bio-Rad Laboratories) for detection. The membranes were incubated with Western Blotting Detection Reagents (Invitrogen) according to the manufacturer's instructions and detected using the Kodak multimodal imaging system (Carestream Molecular Imaging, Rochester, NY, USA).
Data pre-processing for analysis and validation of miRNA expression
The microRNA expression analysis was performed by employing Thermo Scientific Dharmacon RNAi Discovery and Therapeutic Services (http://www.dharmacon.com). All probes on the array were used in the subsequent analysis. Relative intensity data for three age groups were subjected to statistical filtering to identify miRNA probes associated changes with P-value < 0.05. This resulted in 536 miRNA probes passing statistical filters. The remaining data were inter-array scaled and transformed to log (2). Error-weighted anova was used in the analysis of differential expression, where within-array error term is used as a surrogate for between-replicate error. For comparisons of the age groups, data from each age were compared with the other two ages. Holm multiple test correction was applied in each analysis, which reports P-value.
Quantitative real-time PCR analysis for miRNA array
For quantification of expression for candidate miRNA, RNA was reverse-transcribed with an Ncode miRNA first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer-specified guidelines. Forward sequences used in this study were referenced from the Sanger miRNA database (http://microrna.sanger.ac.uk), and U6 was used as a normalizing control. qRT-PCR reactions were performed with the power SYBR green PCR master mix in a MicroAmp optical 96-well reaction plate with ABI PRISM® 7900HT sequence detector (Applied Biosystems) according to the manufacturer's instructions.
Histone preparation
Samples from rBMSCs were procured and prepared as follows: 1 × 107 rBMSCs at passages 2 were obtained from young and old age groups. Cells were washed twice with PBS. Histones were prepared using a standard acid extraction procedure as previously described (Zhang et al., 2003). Bovine calf thymus histones were used as a single source of histone standards throughout all experiments. The bovine calf thymus tissue was purchased from Worthington Biochemical Co. (Lakewood, NJ, USA).
Histone Protein Liquid Chromatography Mass Spectrometry Profiling (LC-MS)
The acid extracted protein mixture was separated by reverse-phase HPLC (Discovery Bio wide pore C18 column, 1.0 mm i.d., 5 mm, 300Å; Supelco, Bellefonte, Pennsylvania, USA) and detected by an ESI-TOF mass spectrometer (Su et al., 2007a). HPLC separation was carried out using a flow rate of 50 μL/ min with a gradient of mobile phase A (0.1% TFA in water) and mobile phase B (0.1% TFA in acetonitrile) where B increased from 30% to 45% in 2 min, 60% in 20 min and was held at 60% for 4 min. Between each run, the column was washed at 100% B for 2 min and equilibrated at 30% B for 30 min. The eluted histones were infused into the ESI LC-TOF MS or ESI LC-QTOF MS coupled with an auto-sampling Waters HPLC instrument (Waters 2690; Waters, Milford, MA, USA). ESI was performed at the optimal conditions of 3 kV capillary voltage, 100 °C source temperature and 50 V cone voltage. Data were acquired in continuum mode at the rate of 1 spectrum sec-1. All spectra were obtained in the positive ion mode. Sodium iodide (NaI) was used for external mass calibration over the m/z range 500–2500.
Statistical analysis
All experiments were performed at least in triplicate for individual sample. All values are provided as the mean ± SEM. Comparisons among groups were analyzed via two sided t-tests or anova for experiments with more than two groups. P value of < 0.05 was considered to indicate significant difference.
Supplementary Material
Fig. S1 Morphological appearance of rBMSCs in three age groups (A) shows cell morphology at the first passage from young rBMSCs (< 5 years), middle rBMSCs (8–10 years), and old rBMSCs (> 12 years), respectively. Original magnification, 10·. (B) rBMSCs obtained from three age groups were analyzed for cell size. Shown is a representative single-parameter histogram. (C) A representative flow cytometry plot of rBMSCs in three age groups according to light scatter characteristics reflecting cytoplasmic granularity and cell size.
Fig. S2 HPLC separation of histones from rBMSCs cells in young and old group. (A) All core histones are observed. (B) Several variants of histone H2B, H2A, H3 and H4 were separated and their retention times were labeled on the top of their corresponding peaks.
Acknowledgments
The authors would like to acknowledge the expertise, advice, and resources rendered by the Division of Veterinary Medicine and the flow cytometry core at the TNPRC. The research was supported by grant number RR00164 from the National Center for Research Resources, National Institutes of Health, Louisiana Gene Therapy Research Consortium, Pennington Biomedical Research Center, and Tulane University.
Footnotes
Authors contributions
J.M. Yu and B.A. Bunnell conceived, designed, and executed the experiments. X. Wu and M Freitas performed some of the additional experiments. B.A. Bunnell, J.M. Gimble, M. Freitas, and J.M. Yu analyzed the data. J.M. Yu and B.A. Bunnell wrote the paper. J.M. Gimble and M. Freitas edited the paper.
Supporting Information
Additional supporting information may be found in the online version of this article:
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Armanios M, Alder JK, Parry EM, Karim B, Strong MA, Greider CW. Short telomeres are sufficient to cause the degenerative defects associated with aging. Am. J. Hum. Genet. 2009;85:823–832. doi: 10.1016/j.ajhg.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auricchio A, Rivera VM, Clackson T, O'Connor EE, Maguire AM, Tolentino MJ, Bennett J, Wilson JM. Pharmacological regulation of protein expression from adeno-associated viral vectors in the eye. Mol. Ther. 2002;6:238–242. doi: 10.1006/mthe.2002.0660. [DOI] [PubMed] [Google Scholar]
- Baxter MA, Wynn RF, Jowitt SN, Wraith JE, Fairbairn LJ, Bellantuono I. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells. 2004;22:675–682. doi: 10.1634/stemcells.22-5-675. [DOI] [PubMed] [Google Scholar]
- Beausejour C. Bone marrow-derived cells: the influence of aging and cellular senescence. Handb. Exp. Pharmacol. 2007;180:67–88. doi: 10.1007/978-3-540-68976-8_4. [DOI] [PubMed] [Google Scholar]
- Bellows CG, Pei W, Jia Y, Heersche JN. Proliferation, differentiation and self-renewal of osteoprogenitors in vertebral cell populations from aged and young female rats. Mech. Ageing Dev. 2003;124:747–757. doi: 10.1016/s0047-6374(03)00088-5. [DOI] [PubMed] [Google Scholar]
- Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2009;9:54–63. doi: 10.1111/j.1474-9726.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campioni D, Moretti S, Ferrari L, Punturieri M, Castoldi GL, Lanza F. Immunophenotypic heterogeneity of bone marrow-derived mesenchymal stromal cells from patients with hematologic disorders: correlation with bone marrow microenvironment. Haematologica. 2006;91:364–368. [PubMed] [Google Scholar]
- Campioni D, Lanza F, Moretti S, Ferrari L, Cuneo A. Loss of Thy-1 (CD90) antigen expression on mesenchymal stromal cells from hematologic malignancies is induced by in vitro angiogenic stimuli and is associated with peculiar functional and phenotypic characteristics. Cytotherapy. 2008;10:69–82. doi: 10.1080/14653240701762364. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cancer and aging: yin, yang, and p53. Sci. Aging Knowledge Environ. 2002:pe1. doi: 10.1126/sageke.2002.1.pe1. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J. Biol. Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- Chivu M, Diaconu CC, Brasoveanu L, Alexiu I, Bleotu C, Banceanu G, Miscalencu D, Cernescu C. Ex vivo differentiation of umbilical cord blood progenitor cells in the presence of placental conditioned medium. J. Cell Mol. Med. 2002;6:609–620. doi: 10.1111/j.1582-4934.2002.tb00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Ng YC. Age-related alterations in expression of apoptosis regulatory proteins and heat shock proteins in rat skeletal muscle. Biochim. Biophys. Acta. 2006;1762:103–109. doi: 10.1016/j.bbadis.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. Aging, stem cells and tissue regeneration: lessons from muscle. Cell Cycle. 2005;4:407–410. doi: 10.4161/cc.4.3.1518. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Curtis DJ, Hall MA, Van Stekelenburg LJ, Robb L, Jane SM, Begley CG. SCL is required for normal function of short-term repopulating hematopoietic stem cells. Blood. 2004;103:3342–3348. doi: 10.1182/blood-2003-09-3202. [DOI] [PubMed] [Google Scholar]
- D'Ippolito G, Howard GA, Roos BA, Schiller PC. Sustained stromal stem cell self-renewal and osteoblastic differentiation during aging. Rejuvenation Res. 2006;9:10–19. doi: 10.1089/rej.2006.9.10. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Downes M, Carozzi AJ, Muscat GE. Constitutive expression of the orphan receptor, Rev-erbA alpha, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Mol. Endocrinol. 1995;9:1666–1678. doi: 10.1210/mend.9.12.8614403. [DOI] [PubMed] [Google Scholar]
- Elder SH. Conditioned medium of mechanically compressed chick limb bud cells promotes chondrocyte differentiation. J. Orthop. Sci. 2002;7:538–543. doi: 10.1007/s007760200096. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Gery S, Koeffler HP. The role of circadian regulation in cancer. Cold Spring Harb. Symp. Quant. Biol. 2007a;72:459–464. doi: 10.1101/sqb.2007.72.004. [DOI] [PubMed] [Google Scholar]
- Gery S, Koeffler HP. Transcription factors in hematopoietic malignancies. Curr. Opin. Genet. Dev. 2007b;17:78–83. doi: 10.1016/j.gde.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler HP. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–28. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, Marchevsky A, McKenna R, Taguchi H, Koeffler HP. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin. Cancer Res. 2007a;13:1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Onco-gene. 2007b;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- Geyfman M, Andersen B. Clock genes, hair growth and aging. Aging. 2010;2:122–128. doi: 10.18632/aging.100130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EM, Williams WP, Kriegsfeld LJ. Aging in the circadian system: considerations for health, disease prevention and longevity. Exp. Gerontol. 2009;44:51–56. doi: 10.1016/j.exger.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano A, Galderisi U, Marino IR. From the laboratory bench to the patient’s bedside: an update on clinical trials with mesenchymal stem cells. J. Cell. Physiol. 2007;211:27–35. doi: 10.1002/jcp.20959. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Lee CC, Jimenez DF, Karaman MW, Ho VV, Siegmund KD, Tarantal AF. Age-related gene expression profiles of rhesus monkey bone marrow-derived mesenchymal stem cells. J. Cell. Biochem. 2008;103:1198–1210. doi: 10.1002/jcb.21498. [DOI] [PubMed] [Google Scholar]
- Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C, Laschober GT, Lepperdinger G, Sampson N, Berger P, Herndler-Brandstetter D, Wieser M, Kuhnel H, Strasser A, Rinnerthaler M, Breitenbach M, Mildner M, Eckhart L, Tschachler E, Trost A, Bauer JW, Papak C, Trajanoski Z, Scheideler M, Grillari-Voglauer R, Gru-beck-Loebenstein B, Jansen-Durr P, Grillari J. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9:291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkinen L, Koivisto L, Larjava H. An improved method for culture of epidermal keratinocytes from newborn mouse skin. Methods Cell Sci. 2001;23:189–196. doi: 10.1023/a:1016385109922. [DOI] [PubMed] [Google Scholar]
- Hall DM, Xu L, Drake VJ, Oberley LW, Oberley TD, Moseley PL, Kregel KC. Aging reduces adaptive capacity and stress protein expression in the liver after heat stress. J. Appl. Physiol. 2000;89:749–759. doi: 10.1152/jappl.2000.89.2.749. [DOI] [PubMed] [Google Scholar]
- Han GM, Zhao B, Jeyaseelan S, Feng JM. Age-associated parallel increase of Foxp3(+)CD4(+) regulatory and CD44(+)CD4(+) memory T cells in SJL/J mice. Cell. Immunol. 2009;258:188–196. doi: 10.1016/j.cellimm.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, Wang X, Wang Z, Cornelissen-Guillaume G, Halberg F. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97:589–596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izadpanah R, Joswig T, Tsien F, Dufour J, Kirijan JC, Bunnell BA. Characterization of multipotent mesenchymal stem cells from the bone marrow of rhesus macaques. Stem Cells Dev. 2005;14:440–451. doi: 10.1089/scd.2005.14.440. [DOI] [PubMed] [Google Scholar]
- Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J. Cell. Biochem. 1997;64:295–312. [PubMed] [Google Scholar]
- Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp. Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- Kasper G, Mao L, Geissler S, Draycheva A, Trippens J, Kuhnisch J, Tschirschmann M, Kaspar K, Perka C, Duda GN, Klose J. Insights into mesenchymal stem cell aging: involvement of antioxidant defense and actin cytoskeleton. Stem Cells. 2009;27:1288–1297. doi: 10.1002/stem.49. [DOI] [PubMed] [Google Scholar]
- Knapp AR, Ren C, Su X, Lucas DM, Byrd JC, Freitas MA, Parthun MR. Quantitative profiling of histone post-translational modifications by stable isotope labeling. Methods. 2007;41:312–319. doi: 10.1016/j.ymeth.2006.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res. Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing Res. Rev. 2007;6:12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Lambeth LS, Yao Y, Smith LP, Zhao Y, Nair V. MicroRNAs 221 and 222 target p27Kip1 in Marek’s disease virus-transformed tumour cell line MSB-1. J. Gen. Virol. 2009;90:1164–1171. doi: 10.1099/vir.0.007831-0. [DOI] [PubMed] [Google Scholar]
- Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2009;8:2685. doi: 10.4161/cc.8.17.9489. [DOI] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in MicroRNA expression patterns in human fibroblasts after low-LET radiation. J. Cell. Biochem. 2008;105:824–834. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- Maes OC, Sarojini H, Wang E. Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J. Cell. Physiol. 2009;221:109–119. doi: 10.1002/jcp.21834. [DOI] [PubMed] [Google Scholar]
- Majumdar MK, Thiede MA, Mosca JD, Moorman M, Gerson SL. Phenotypic and functional comparison of cultures of marrow-derived mesenchymal stem cells (MSCs) and stromal cells. J. Cell. Physiol. 1998;176:57–66. doi: 10.1002/(SICI)1097-4652(199807)176:1<57::AID-JCP7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Mareschi K, Ferrero I, Rustichelli D, Aschero S, Gammaitoni L, Aglietta M, Madon E, Fagioli F. Expansion of mesenchymal stem cells isolated from pediatric and adult donor bone marrow. J. Cell. Biochem. 2006;97:744–754. doi: 10.1002/jcb.20681. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Meyer KC. The role of immunity in susceptibility to respiratory infection in the aging lung. Respir. Physiol. 2001;128:23–31. doi: 10.1016/S0034-5687(01)00261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilin eage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- Purpura KA, Aubin JE, Zandstra PW. Sustained in vitro expansion of bone progenitors is cell density dependent. Stem Cells. 2004;22:39–50. doi: 10.1634/stemcells.22-1-39. [DOI] [PubMed] [Google Scholar]
- Rando TA. Stem cells, aging and the quest for immortality. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- Ren C, Zhang L, Freitas MA, Ghoshal K, Parthun MR, Jacob ST. Peptide mass mapping of acetylated isoforms of histone H4 from mouse lymphosarcoma cells treated with histone deacetylase (HDACs) inhibitors. J. Am. Soc. Mass Spectrom. 2005;16:1641–1653. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl. Acad. Sci. USA. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Ishikawa Y, Takahashi Y, Itoh T, Minami Y, Nakamura M. Association between oxidative DNA damage and telomere shortening in circulating endothelial progenitor cells obtained from metabolic syndrome patients with coronary artery disease. Atherosclerosis. 2008;198:347–353. doi: 10.1016/j.atherosclerosis.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Sauvaigo S, Bonnet-Duquennoy M, Odin F, Hazane-Puch F, Lachmann N, Bonte F, Kurfurst R, Favier A. DNA repair capacities of cutaneous fibroblasts: effect of sun exposure, age and smoking on response to an acute oxidative stress. Br. J. Dermatol. 2007;157:26–32. doi: 10.1111/j.1365-2133.2007.07890.x. [DOI] [PubMed] [Google Scholar]
- Sethe S, Scutt A, Stolzing A. Aging of mesenchymal stem cells. Ageing Res. Rev. 2006;5:91–116.. doi: 10.1016/j.arr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Shi YY, Nacamuli RP, Salim A, Longaker MT. The osteogenic potential of adipose-derived mesenchymal cells is maintained with aging. Plast. Reconstr. Surg. 2005;116:1686–1696. doi: 10.1097/01.prs.0000185606.03222.a9. [DOI] [PubMed] [Google Scholar]
- Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol. Carcinog. 2006;45:732–740. doi: 10.1002/mc.20198. [DOI] [PubMed] [Google Scholar]
- Sitzmann BD, Lemos DR, Ottinger MA, Urbanski HF. Effects of age on clock gene expression in the rhesus macaque pituitary gland. Neurobiol. Aging. 2010;31:696–705. doi: 10.1016/j.neurobiolaging.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenderup K, Rosada C, Justesen J, Al-Soubky T, Dagnaes-Hansen F, Kassem M. Aged human bone marrow stromal cells maintaining bone forming capacity in vivo evaluated using an improved method of visualization. Biogerontology. 2004;5:107–118. doi: 10.1023/B:BGEN.0000025074.88476.e2. [DOI] [PubMed] [Google Scholar]
- Stolzing A, Scutt A. Age-related impairment of mesenchymal progenitor cell function. Aging Cell. 2006;5:213–224. doi: 10.1111/j.1474-9726.2006.00213.x. [DOI] [PubMed] [Google Scholar]
- Stolzing A, Sethe S, Scutt AM. Stressed stem cells: temperature response in aged mesenchymal stem cells. Stem Cells Dev. 2006;15:478–487. doi: 10.1089/scd.2006.15.478. [DOI] [PubMed] [Google Scholar]
- Stolzing A, Jones E, McGonagle D, Scutt A. Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech. Ageing Dev. 2008;129:163–173. doi: 10.1016/j.mad.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Su X, Jacob NK, Amunugama R, Lucas DM, Knapp AR, Ren C, Davis ME, Marcucci G, Parthun MR, Byrd JC, Fishel R, Freitas MA. Liquid chromatography mass spectrometry profiling of histones. J. Chromatogr. 2007a;850:440–454. doi: 10.1016/j.jchromb.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Zhang L, Lucas DM, Davis ME, Knapp AR, Green- Church KB, Marcucci G, Parthun MR, Byrd JC, Freitas MA. Histone H4 acetylation dynamics determined by stable isotope labeling with amino acids in cell culture and mass spectrometry. Anal. Biochem. 2007b;363:22–34. doi: 10.1016/j.ab.2006.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Lucas DM, Zhang L, Xu H, Zabrouskov V, Davis ME, Knapp AR, Young DC, Payne PR, Parthun MR, Marcucci G, Grever MR, Byrd JC, Freitas MA. Validation of an LC-MS based approach for profiling histones in chronic lymphocytic leukemia. Proteomics. 2009;9:1197–1206. doi: 10.1002/pmic.200800333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanlund JM, Kregel KC, Oberley TD. Autophagy following heat stress: the role of aging and protein nitration. Autophagy. 2008;4:936–939. doi: 10.4161/auto.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokalov SV, Gruner S, Schindler S, Wolf G, Baumann M, Abolmaali N. Age-related changes in the frequency of mesenchymal stem cells in the bone marrow of rats. Stem Cells Dev. 2007;16:439–446. doi: 10.1089/scd.2006.0078. [DOI] [PubMed] [Google Scholar]
- Torres PM, Guilarducci CV, Franco AS, de Araujo EG. Sciatic conditioned medium increases survival, proliferation and differentiation of retinal cells in culture. Int. J. Dev. Neurosci. 2002;20:11–20. doi: 10.1016/s0736-5748(02)00005-9. [DOI] [PubMed] [Google Scholar]
- Wagner BJ, Margolis JW. Age-dependent association of isolated bovine lens multicatalytic proteinase complex (proteasome) with heat-shock protein 90, an endogenous inhibitor. Arch. Biochem. Biophys. 1995;323:455–462. doi: 10.1006/abbi.1995.0067. [DOI] [PubMed] [Google Scholar]
- Wagner V, Mittag M. Probing circadian rhythms in Chlamydomonas rheinhardtii by functional proteomics. Methods Mol. Biol. 2009;479:173–188. doi: 10.1007/978-1-59745-289-2_11. [DOI] [PubMed] [Google Scholar]
- Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, Ho AD. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS ONE. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakitani S, Saito T, Caplan AI. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve. 1995;18:1417–1426. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
- Wenguang Z, Jianghong W, Jinquan L, Yashizawa M. A subset of skin-expressed microRNAs with possible roles in goat and sheep hair growth based on expression profiling of mammalian microRNAs. Omics. 2007;11:385–396. doi: 10.1089/omi.2006.0031. [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury D, Schwarz EJ, Prockop DJ, Black IB. Adult rat and human bone marrow stromal cells differentiate into neurons. J. Neurosci. Res. 2000;61:364–370. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun YE. Epigenetic regulation of stem cell differentiation. Pediatr. Res. 2006;59:21R–25R. doi: 10.1203/01.pdr.0000203565.76028.2a. [DOI] [PubMed] [Google Scholar]
- Wu X, Yu G, Parks H, Hebert T, Goh BC, Dietrich MA, Pelled G, Izadpanah R, Gazit D, Bunnell BA, Gimble JM. Circadian mechanisms in murine and human bone marrow mesenchymal stem cells following dexamethasone exposure. Bone. 2008;42:861–870. doi: 10.1016/j.bone.2007.12.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Chang JG, Lin PM, Tang KP, Chen YH, Lin HY, Liu TC, Hsiao HH, Liu YC, Lin SF. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–1307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Wu S, Hansteen G, Du C, Sambucetti L, Remiszewski S, O'Farrell AM, Hill B, Lavau C, Murray LJ. Inhibitors of histone deacetylases promote hematopoietic stem cell self-renewal. Cytotherapy. 2004;6:328–336. doi: 10.1080/14653240410004899. [DOI] [PubMed] [Google Scholar]
- Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- Zhang L, Freitas MA, Wickham J, Parthun MR, Klisovic MI, Marcucci G, Byrd JC. Differential expression of histone post-translational modifications in acute myeloid and chronic lymphocytic leukemia determined by high-pressure liquid chromatography and mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:77–86. doi: 10.1016/j.jasms.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS, Glowacki J. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7:335–343. doi: 10.1111/j.1474-9726.2008.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Morphological appearance of rBMSCs in three age groups (A) shows cell morphology at the first passage from young rBMSCs (< 5 years), middle rBMSCs (8–10 years), and old rBMSCs (> 12 years), respectively. Original magnification, 10·. (B) rBMSCs obtained from three age groups were analyzed for cell size. Shown is a representative single-parameter histogram. (C) A representative flow cytometry plot of rBMSCs in three age groups according to light scatter characteristics reflecting cytoplasmic granularity and cell size.
Fig. S2 HPLC separation of histones from rBMSCs cells in young and old group. (A) All core histones are observed. (B) Several variants of histone H2B, H2A, H3 and H4 were separated and their retention times were labeled on the top of their corresponding peaks.