Abstract
In 2000, the first chemical screen using living zebrafish in a multi-well plate was reported. Since then, more than 60 additional screens have been published describing whole-organism drug and pathway discovery projects in zebrafish. To investigate the scope of the work reported in the last 14 years and to identify trends in the field, we analyzed the discovery strategies of 64 primary research articles from the literature. We found that zebrafish screens have expanded beyond the use of developmental phenotypes to include behavioral, cardiac, metabolic, proliferative and regenerative endpoints. Additionally, many creative strategies have been used to uncover the mechanisms of action of new small molecules including chemical phenocopy, genetic phenocopy, mutant rescue, and spatial localization strategies.
INTRODUCTION
Traditional methods of small molecule drug discovery relied on trial-and-error testing of chemical compounds on phenotypic outcomes in cells or animals. This approach yielded many of the drugs currently used in the clinic today. By contrast, target-driven approaches, which seek to identify novel therapeutics based on a priori knowledge of a single biological target, have received greater emphasis in recent decades but have delivered fewer first-in-class drugs [1].
There are several possible reasons (not mutually exclusive) why phenotype-driven approaches have out-performed target-driven approaches. The first is that target driven approaches depend on selection of the correct, disease-modifying target—an uncertain proposition—whereas phenotype-driven approaches can identify disease-modifying drugs even in the absence of a validated target. Second, the most efficacious drugs may benefit from activity at multiple targets. For example, complex polygenetic disorders may require a ‘magic shotgun’ drug (one exhibiting polypharmacology) rather than a ‘magic bullet’ (one exhibiting specificity for a single target) [2]. Some of the most successful drugs in use today are known to benefit from engagement of multiple targets throughout the body. Third, small molecules derived from phenotypic screens often have been further selected for positive pharmacological properties, such as low toxicity, the ability to make it to the appropriate site(s) of action, and the ability to avoid or exploit endogenous chemical metabolizing enzymes and transporters.
Whole-organism, phenotypic screening holds several advantages over other approaches to small molecule discovery. The approach is target agonistic (therefore not mechanistically biased) and holistic (all possible targets in the organism are available). This includes targets relevant not only to disease intervention but also chemical activation, chemical transport, toxicity and other side effects.
In 2000, it was demonstrated for the first time that a chemical screen could be carried out using live zebrafish in a 96-well plate simply by adding small amounts of compounds directly to the fish water [3]. Though simpler than humans, zebrafish are also complex vertebrates and maintain similarly elaborate mechanisms for activating or mitigating the effects of exogenous chemical substances. Although differences in pharmacological effects between zebrafish and humans certainly do exist, there are now hundreds of examples of small molecules that have conserved biological activities in fish and humans. It is therefore reasonable to expect that many bioactive compounds identified in zebrafish screens will maintain their activity in humans.
In this review, we summarize the work reported in 66 zebrafish chemical screens over the past 15 years. We start by giving a bird’s-eye view of the field to give readers a feel for the scope of what has been accomplished to date. Many of the design details will likely be of interest to those contemplating setting up their own zebrafish screens. We then highlight some of the more interesting examples of the phenotypic endpoints that have been examined and methods of follow-up used to uncover mechanisms of action.
Zebrafish screens by the numbers
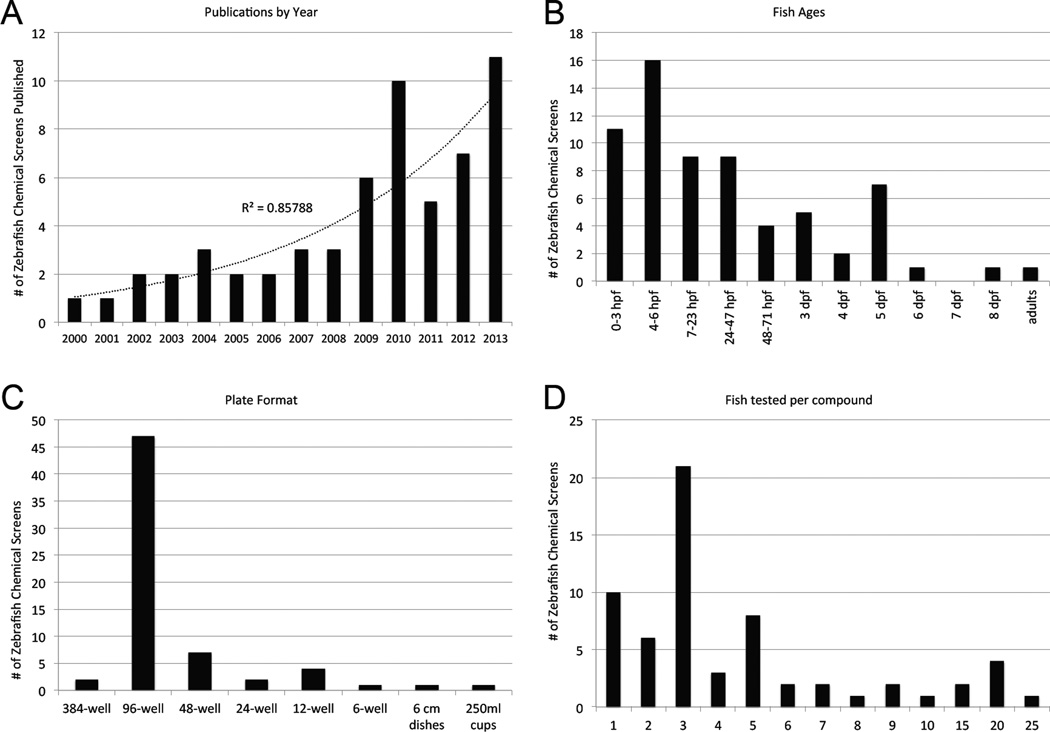
In a survey of the literature, we identified 66 primary research articles each reporting results of a zebrafish chemical screen. These range from the year 2000 to the present time and form the basis for our in-depth analysis. We believe these provide a good representation of the field, but we do not claim this list is exhaustive and apologize for any studies we may have omitted. A simple plot of the number of publications per year demonstrates that zebrafish chemical screens are becoming more widespread, with the number increasing substantially in recent years (Figure 1a). The types of journals publishing these reports ranges in scope from specialized publications, like the journal Zebrafish [4, 5], to journals with very broad appeal, such as Nature [6, 7]. Of the 37 journals represented, only five had published more than two papers on zebrafish chemical screens. On average, the impact factor for papers reporting zebrafish chemical screens has been 9.5.
Figure 1.
a) Publications reporting zebrafish chemical screens by year. b) Ages of zebrafish used in chemical screens. c) Plate formats used in zebrafish chemical screens. d) Numbers of zebrafish tested per chemical.
Of the 66 screens, 49 (74%) were conducted using zebrafish age 48 hours post fertilization (hpf) or younger (Figure 1b). The most frequent treatment age was 4–6 hpf. This bias toward use of zebrafish in the embryonic stage reflects the historical importance of the organism to developmental research. Including the very first screen published [3], 14 screens (22%) examined embryogenesis or gross development as the phenotypic endpoint.
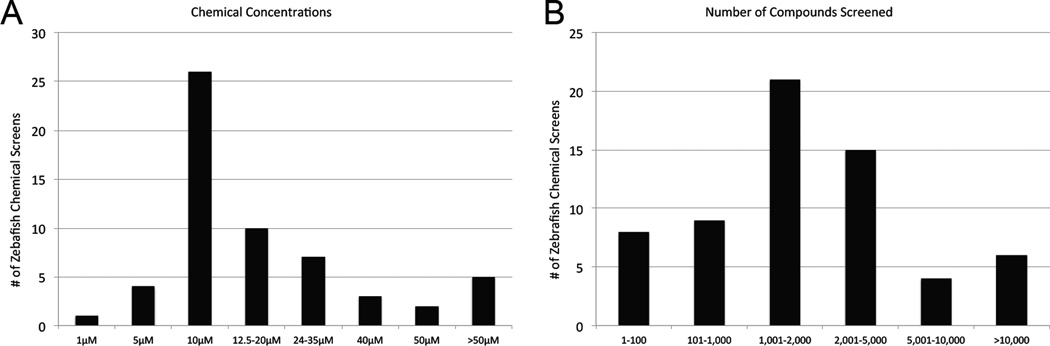
Several different multi-well plates have been used including one screen that was conducted using a 384-well plate [8]. By far the most common format has been the 96-well plate (72%, Figure 1c). Numbers of animals used varies between 1 animal per well per compound to more than 30 animals per chemical treatment. The most common number of zebrafish used per compound has been 3 (33%, Figure 1d). Chemical libraries were obtained from diverse academic, government and commercial sources. The most frequently used were the Chembridge DIVERSetE collection of synthetic compounds (21% of screens), the LOPAC collection of 1280 well-characterized pharmacologically active compounds (15%) and the MicroSource Spectrum collection (17%) which contains 1040 US clinical trial stage drugs, 240 additional drugs marketed internationally and 800 natural products. Chemical concentrations used center around 10 μM, the most commonly used concentration (45%, Figure 2a), but were as low as 1 μM [3].
Figure 2.
a) Chemical concentrations used in zebrafish chemical screens. b) Total number of compounds screened.
The elapsed times between initial chemical treatment and assay readout varied considerably from 15 min to 120 hrs with the average at 42 hrs. The most frequent chemical treatment duration was 48 hours (17%). We also found a wide range in the total number of compounds tested in each screen (Figure 2b). Seven so-called ‘screens’ tested 26 or fewer compounds, while the largest screen examined 26,400 small molecules [9]. Only 6 of 66 (6%) have screened more than 10,000.
Recent advances in phenotyping
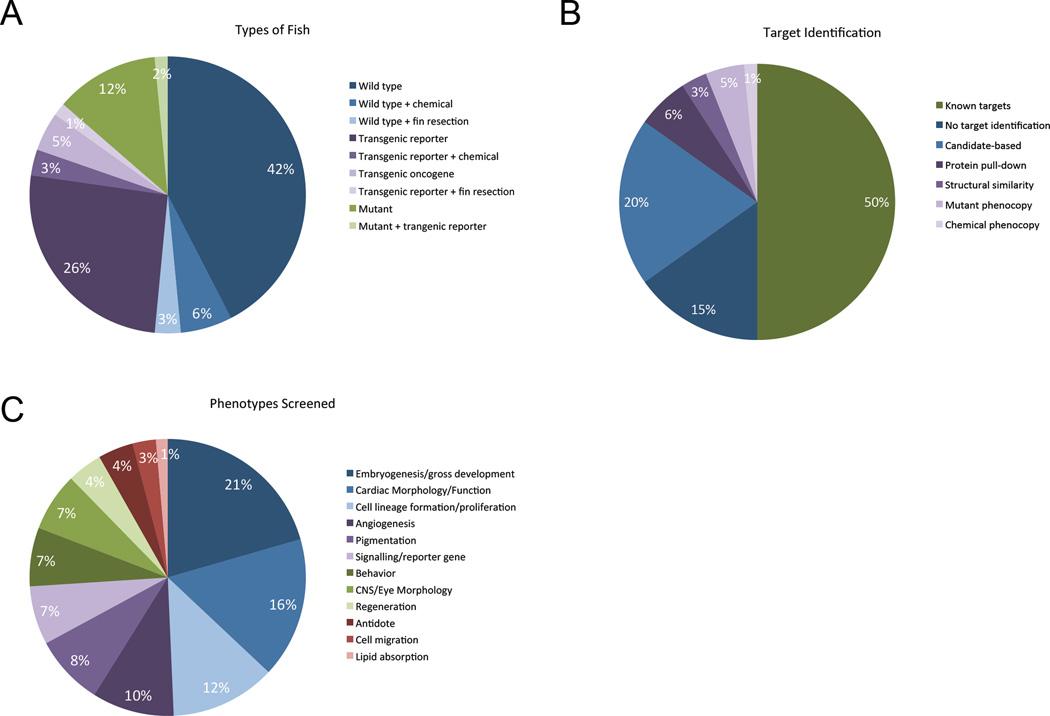
The earliest screens, from 2000 to 2003, were all carried out using wild-type zebrafish, typically with developmental or morphological changes scored by eye as the phenotypic endpoint [3, 10–14]. To date, wild-type fish remain the most commonly used (51% of screens) as compared to transgenic (35%) and mutant (14%) animals (Figure 3a), however the assays have become much more diverse. Screens have now been reported that examine many different phenotypic endpoints including regeneration [4, 15, 16], cell migration [17, 18], cell proliferation [19, 20], lipid absorption [21], heartbeat [13, 22], and animal behavior [23–27].
Figure 3.
a) Types of zebrafish used in chemical screens. b) Follow up work done on small molecules identified in zebrafish chemical screens. c) Phenotypic endpoints used.
This expansion of phenotypes has been made possible by several recent advances. These include the discovery of new, readily assayable behaviors, such as the photomotor response (PMR) [23]. By simply shining light onto a 30 hpf embryo one can elicit an instantaneous bout of motion followed by a brief refractory period where the animal no longer responds. This simple behavior has been exploited for multiple screens and has led to the identification of many new neuroactive small molecules, including structurally novel acetylcholine esterase inhibitors, monoamine oxidase inhibitors, and TRP channel agonists [23, 24].
Advances have also been made in data collection and analysis. High-content imaging, for example, is becoming a powerful tool when combined with translucent zebrafish larvae. Using high-content screening researchers have identified butafenacil as a potent inducer of anemia [28], stimulators of pancreatic beta-cell proliferation [29], enhancers of FGF signaling [30], and the novel compound ‘lenaldekar,’ which selectively kills leukemic cells [9]. On the data analysis side, phenotypic ‘barcoding,’ where a complex endpoint such as a behavioral response is divided into discrete numeric units enabling comparisons of the phenotypes produced by thousands of chemicals to one another [31]. This allows for the generation of signature heat maps and chemical clustering based on phenotypic similarities instead of structure.
The creation of new genetic tools has also advanced the field. For example, the creation of transgenic fish containing inducible oncogenes under the control of the heat shock promoter (hsp) has facilitated the discovery of cancer suppressing pathways, such as a COX/β-catenin-dependent signaling pathway in the case of an acute myelogenous leukemia (AML)-ETO suppressor screen [19] and the MAPK/ERK and AKT/S6K1 pathways in an HRASG12V suppressor screen [32]. Another example of two useful genetic tools is the vasculature marker lines Tg(fli:EGFP) and Tg(flk1:EGPF); their importance is highlighted by their use in no less than six chemical screens for angiogenesis modulators [28, 33–37]. Some screens have become quite sophisticated. The screen conducted by Gutierrez et al. is noteworthy for its elaborate design which required the pigment-sorted progeny of a Tg(rag2:MYC-ER;mitfa)+/− × Tg(rag2:dsRed2)+/+ adult cross where both parents were germline null for the mitfa (pigment gene), treated with 4-hydroxytamoxifen [20].
Current limitations
Despite all the advances, several limitations of zebrafish chemical screening remain. Among the most significant limitations is age. Though there has been at least one chemical screen attempted in adult fish [4], to be able to conduct a high-throughput screen using reasonably small amounts of small molecules, the animals need to fit in a multi-well plate. It is fortunate that even larval fish are developed enough to demonstrate complex behaviors like hunting and possess a blood brain barrier [38, 39], however there are limitations to what can be done in embryos and larvae. Screen for chemicals that reverse natural fish aging or modulate the response of the adaptive immune response to infection, for examples, would likely be very challenging.
A second set of limitations, more likely to be overcome eventually, involve imaging. Classically, much zebrafish work has involved in situ hybridization for RNA detection. This can be made reasonably high-throughput, and in fact in situ screens have been successful (for examples see references [6, 7, 19]), but they are laborious. 17 screens (26%) used fluorescent reporters for which the visualization has been quite good. However imaging of deeper tissues becomes problematic. Another imaging limitation has been the non-uniform orientation of the fish in each well, in three dimensions, which automated imaging systems must cope with. To compare images from different chemical treatments across very large assay datasets, the variability in spatial orientation during image acquisition must be minimized. Live imaging of freely swimming fish must take into account the upright position of the animals, which typically presents a dorsal or ventral view of the fish to a scope objective. Several potential solutions are currently in development. We will discuss these in the ‘Future Directions’ section below.
A third limitation is the time it takes to count and pipet animals into each well. Here again, is a surmountable challenge, which will eventually be worked out. Several platforms are currently in development, including two originally designed for C. elegans screens and at least two designed directly for zebrafish embryos [40, 41]. At least one automated embryo placement system has been used in a zebrafish high-throughput chemical screen [42].
THE IMPACT OF ZEBRAFISH CHEMICAL BIOLOGY
Discovery of new uses for existing bioactive compounds
The majority of small molecule libraries used to date in zebrafish chemical screens have been collections of known bioactive compounds. There are advantages to using libraries of known pharmacological entities. For one, many of these compounds are already in use in humans and therefore could be fast-tracked through any potential clinical trial period, having previously been assessed for safety. Furthermore, small molecule hits with known targets provide instant hypotheses concerning mechanisms of action and the biological pathways involved in generating the unique phenotypes observed. This advantage is not insignificant because, as we will discuss later on, it can sometimes be challenging to identify the mechanism(s) of action of newly discovered small molecules.
33 screens (50%) identified previously known compounds acting on expected targets but producing unexpected phenotypes. These include the compounds known to affect prostaglandin E2 (PGE2) synthesis and signaling identified by North et al. in a screen for compounds that alter hematopoietic stem cell numbers [6]. These authors demonstrated that chemicals enhancing PGE2 synthesis increase hematopoietic stem cell (HSC) numbers, while COX inhibitors have the opposite effect, decreasing HSCs. In an independent screen, our group found that COX-2 inhibitors (such as nimesulide) also block the effect of the leukemic oncogene AML1-ETO on hematopoietic differentiation [19]. Other unexpected tumor growth suppressors identified using zebrafish chemical screens include a commercially available statin, rosuvastatin, which was shown to suppresses the growth of prostate cancer [37], and several phenothiazine antipsychotics, which surprisingly induce apoptosis in T cell acute lymphoblastic leukemia (T-ALL) [20].
Using a screen for chemical modifiers of polycystic kidney disease 2 (pkd2) mutant zebrafish, which present with kidney cyst formation and laterality defects and body curvature, Cao and colleagues identified the class I histone deacetylase (HDAC) inhibitors trichostatin-A (TSA) and valproic acid (VPA) as suppressors of kidney cysts formation [43]. These HDAC inhibitors are structurally unrelated, suggesting that HDAC inhibition is responsible for the rescue phenotype. The effects of the HDAC inhibitors on reduction of kidney cyst formation in zebrafish were also observed in a mouse model of PKD. Both kidney weight and kidney to body weight ratios were significantly reduced in VPA treated mice. There was also a significant decrease in cystic area measured in histological sections, and improved kidney function as measured by blood urea nitrogen.
A chemical modifier screen was also used to identify small molecule suppressors of the breakdance (kcnh2) mutant, which causes a 2:1 atrial to ventricular beating pattern in zebrafish larvae [22]. From this screen three steroid compounds, including the topically used anti-inflammatory drug flurandrenolide, were identified as unexpected suppressors of long QT syndrome. Follow-up studies determined that modulation of the glucocorticoid signaling pathway, specifically, can affect QT length.
A zebrafish chemical screen was also recently used to uncover a surprising effect that known translator protein (TSPO) ligands PK 11195 and Ro5-4864 have on glucose homeostasis [44]. TSPO ligands are typically considered neuroactive compounds. PK 11195, Ro5-4864 and many other neuroactive benzodiazepines bind to TSPO with nM Ki values. For this reason TSPO has also been called the ‘peripheral benzodiazepine receptor.’ Gut and colleagues designed a screen to identify chemical activators of the fasting-inducible gluconeogenic gene pck1. To their surprise they found that although PK 11195 and Ro5-4864 treatments cause a gluconeogenic fasting response, they are ultimately potent glucose-lowering agents. They demonstrate this effect is conserved in mammals showing that PK 11195 improves hepatosteatosis and glucose tolerance in diet-induced obese mice. These two TSPO ligands are structurally diverse, suggests that their glucose lowering effects are likely mediated via a mechanism involving TSPO.
Discovery of new targets for existing bioactive compounds
Zebrafish chemical screening has also identified new targets for old medications. For example, phenothiazine antipsychotics were identified as hit compounds in a small molecule screen for compounds that are toxic to MYC-overexpressing thymocytes [20]. It was shown that phenothiazine compounds slowed T-ALL progression in zebrafish and also induced apoptosis in human T cell acute lymphoblastic leukemia (T-ALL) cell lines.
Antipsychotics are thought to exert their biologic activity through modulation of dopamine receptors. When structurally unrelated antipsychotics including haloperidol, domperidone, and clozapine were tested they did not kill Myc-overexpressing thymocytes, nor did they slow T-ALL progression in zebrafish or affect human T-ALL cell growth. Conversely promethazine, a phenothiazine that does not inhibit dopaminergic signaling, phenocopied the effects of the other hit phenothiazines on T-ALL. This led the authors to search for an alternate target using activity correlation proteomics. They performed affinity purification with a perphenazine analogue bound to steady-state support followed by competition with four different phenothiazines having a range of potency in human cells. They then looked for a human protein with binding activity to the different phenothiazines correlated to the IC50 activity in the human cells. This led to the identification of serine/threonine-protein phosphatase subunit PP2A as a previously unknown target of phenothiazine antipsychotics and mediator of phenothiazine-induced apoptosis.
Discovery of novel classes of compounds
Many novel bioactive small molecules have also now been identified using zebrafish chemical screens. By design, whole-organism phenotypic drug screens are target-agnostic. This is a considerable advantage because these screens are not dependent on (or biased by) assumptions concerning mechanism(s) of action. On the other hand, this presents a challenge if one wants to know the functional targets of hits that come out of the screen. Of the 66 screens we examined, 33 (50%) focused on hit compounds with known mechanisms of action. Of the remaining 33 screen, 23 identified novel compounds as well as their targets, and 10 identified structurally novel compounds but without follow-up target identification (Figure 3b). Many of the reports in this second category, while they do not identify specific targets, do point to pathways affected by the new chemical entities they find.
For example, our group identified several compounds, including ‘GS4012,’ which suppressed the aortic coarctation phenotype in zebrafish caused by the hey2 mutation in gridlock fish [45]. By examining the expression of genes known to be involved in angiogenesis, we found that GS4012 acts by upregulating expression of vascular endothelial growth factor (VEGF). We then demonstrated that the gridlock suppressing effect of GS4012 was mediated by its effect on VEGF expression by phenocopying the GS4012 rescue with injection of vegf cDNA.
The novel compound ‘lenaldekar’ was identified in a chemical screen for small molecules that selective eliminate immature T cells in the developing zebrafish [9]. Lenaldekar exhibits selective toxicity for hematopoietic malignancy lines and primary leukemias and was shown to inhibit growth of human T-ALL xenografts in mice. Importantly it has favorable pharmacokinetics in mice and no observed toxicity other than reduction in the size of thymi and thymocyte numbers. Because the PI3K/AKT/mTOR pathway is known to be important for survival of immature T cells, the effects of lenaldekar on the activation (phosphorylation) status of downstream members of this pathway were examined. Though the precise targets were not elucidated in this initial report, two important clues about lenaldekar’s mechanism of action were discovered. It was found that in leukemic cells, lenaldekar 1) does indirectly inhibit the PI3K/AKT/mTOR pathway and 2) causes a cell cycle delay late in mitosis [9]. Interestingly, these two effects were found to be independent of one another, suggesting that lenaldekar has polypharmacological properties. In other words, it is likely that the effects of lenaldekar are mediated by at least two distinct molecular targets. This discovery nicely illustrates the utility of a zebrafish phenotypic screen for the identification of a multifunctional small molecule.
Zebrafish chemical screens have also been used to identify novel active components in naturally occurring substances. A bioactivity was discovered in an extract from the Jasminum gilgianum plant using a zebrafish screen [46]. This extract was found to induce the formation of ectopic tailbuds in larvae. Various compounds comprising the extract were isolated and then tested using the ectopic tailbud assay. Para-coumaric acid methyl ester (pCAME) was identified as the bioactive component in the extract. Once again, candidate pathways, in this case BMP and non-canonical Wnt signaling, were tested using established knowledge of zebrafish developmental biology. When an inhibitor of JNK, an important kinase in the Wnt noncanonical planar cell polarity (PCP) pathway was tested on ectopic tailbud formation, the inhibitor (SP600125) was found to phenocopy pCAME [46]. In addition, strong synergy was observed when pCAME was combined with BMP inhibitor dorsomorphin (also identified in a zebrafish chemical screen [47]) providing further evidence that the new compound modulates non-canonical Wnt signaling.
DETERMINATION OF MECHANISMS OF ACTION
Mechanism of action studies can be challenging, nevertheless 23 of the screens we examined (35%, Figure 3b) included follow-up work describing the discovery of a new mechanism of action (MOA) for at least one new compound. We found that these studies fall into four distinct categories based on the initial clues provided by the structure, binding, phenotype, or site of action of each compound.
Clues about mechanism based on structure
One of the most straightforward ways to determine a mechanistic target of a newly discovered small molecule is by structural comparison to known bioactives. In a zebrafish chemical screen to identify suppressors of neural crest cell differentiation, White and colleagues identified novel compound ‘NSC210627’ [7]. Using a chemoinformatic algorithm that compares chemicals of interest against a database of known ligands and their targets, it was discovered that NSC210627 shares structural similarity with brequinar a known inhibitor of dihydroorotate dehydrogenase (DHODH). Based on the hypothesis that NSC210627 suppressed neural crest cell differentiation via DHODH inhibition, the authors tested a structurally distinct inhibitor, leflunomide and found that it phenocopied the effects of NSC210627 in zebrafish. DHODH inhibition also reduced the self-renewal effects of mammalian neural crest cells and caused a decrease in human melanocyte growth [7]. This finding may have important clinical applications, as leflunomide, a well-tolerated anti-arthritis drug already in clinical use, may be useful in the treatment of melanoma in humans.
Similarly, the compound ‘PTBA,’ identified based on its ability to induce pericardial edema in a screen for small molecule disrupters of kidney organogenesis, was identified as an HDAC1 inhibitor based on its structural relatedness to known carboxylic acid HDAC1 inhibitors [48]. Further investigation determined that PTBA inhibits fish HDAC activity in vivo, but also human HDAC activity from human (HeLa) cell extracts. Both PTBA and known HDAC1 inhibitors, including the structurally unrelated inhibitor TSA, were shown to expand renal progenitor cells in zebrafish embryos.
Clues about mechanism based on binding
As discussed earlier, a protein affinity purification strategy was used to identify a novel cellular target for phenothiazine antipsychotics [20]. In this case, the target, PP2A, was identified based not only on its ability to bind a perphenazine ‘bait’ molecule, but also the correlation between PP2A binding across four different phenothiazine analogues and IC50 values of those compounds in a cell-based assay. Other zebrafish MOA studies involving binding assays have identified roles for the mitochondrial ATPase in skin pigmentation [49] and mitochondrial malate dehydrogenase (MDH2) in doxorubicin-induced cardiomyopathy [50]. Affinity purification was also used to identify ALDH2 as a bioactivating enzyme responsible for the conversion of 5-nitrofuran antiparasitic medications into cytotoxins, possibly responsible for the side effects seen in patients treated with those drugs [51].
To conduct ‘fishing’ expeditions like these, a chemical of interest must be linked to a magnetic bead or affinity probe, such as a biotin moiety. This requires some amount of structure activity relationship (SAR) work to determine which part(s) of the compound of interest can be modified without losing biological activity. The same assay used to conduct the initial zebrafish chemical screen can also be used to quickly make conclusions about the effects of structural modifications on chemical activity. There are now many examples of SAR studies using zebrafish assays [10–12, 14, 35, 36, 48, 52–54]. In vivo SAR has advantages over in vitro SAR. Modified derivatives must not only maintain their bioactivity at their final targets, but also maintain their ability to get to the target(s), remain unmetabolized by the organism and maintain the ability to be bioactivated if the compound turns out to be a prodrug. Inactive analogues discovered via SAR have also been tools for zebrafish researchers; they have been used, for example, as negative controls in mechanistic studies [51].
Clues about mechanism based on phenocopy
Phenotypic comparisons can be a valuable tool for the discovery of the mechanisms of action of novel small molecules. Our group was able to discover the compound dorsomorphin in a screen based on its dorsalizing effects on developing zebrafish [47]. Dorsomorphin induced changes in the expression of dorsal and ventral markers in the developing embryo similar to what had been observed in zebrafish BMP pathway mutants. Dorsomorphin also counteracted the ventralization phenotype observed in morphant fish depleted of the endogenous BMP antagonist chordin. These observations led to the identification of BMP receptor ALK2 as the functional target.
Using this same principle of chemical-genetic phenocopy, Sandoval et al. looked for chemicals that would induce effects similar to what is observed in well-characterized mutant fish [55]. They found a natural marine product, kalihinol F, that copied the calamity mutant, which is characterized by an undulated notochord and defects in pigmentation formation, hematopoiesis and neural development. Calamity fish contain a mutation in their copper (Cu) transporter gene atp7a. Treatment with exogenous CuCl2 completely prevented the effects of kalihinol F. Sandoval et al. then demonstrated that kalihinol F is a Cu chelator and is able to reverse Cu toxicity in animals.
Clues concerning mechanism of action can also come from chemical-chemical phenocopy. Our group identified a novel compound ‘DTAB’ using a developmental screen for compounds causing anterior-posterior axis defects [56]. The DTAB phenotype resembled the phenotype caused by treatment with retinoic acid. We found that indeed DTAB does have selective agonist activity towards retinoic acid receptors gamma and beta. We also used the principle of chemical-chemical phenocopy and the photomotor behavioral response in 1 dpf fish to identify several new classes of neuroactive compounds, including previously unknown monoamine oxidase and acetylcholinesterase inhibitors [23].
Clues about mechanism based on localization of activity
Some of the most interesting mechanism of action stories reported following zebrafish chemical screens have involved the determination of the loci of chemical activity in vivo. For example, when Molina et al. identified the compound ‘BCI’ as an inducer of dusp6 expression (using transgenic dusp6:EGFP fish), the authors noticed that EGFP expression was localized to places where fibroblast growth factors (FGFs) are known to be expressed in the developing fish [57]. This led the authors to test BCI on Tg(dusp6:EGFP)ace Fgf8 signaling mutants, which they found did not respond. This result convinced them that BCI activity depends on the presence of Fgf ligand and led them to examine targets known to inhibit the pathway downstream of FGFR (feedback attenuators). They then used the zebrafish embryos to screen target candidates in vivo. RNA injection of each candidate led to decreases in Fgf signaling, as expected, measured by expression of downstream sef gene expression. When BCI was added, the only candidate whose effect on sef expression was blocked was Dusp6 itself. Thus anatomical localization led to the identification of Dusp6 as an in vivo molecular target of BCI.
In another example of anatomical localization, this time in larval zebrafish, our group localized the affect of a novel behavior-modifying small molecule named ‘optovin’ to sensory neurons [24]. Optovin was initially discovered in our photomotor response behavioral screen as a compound that caused hyperlocomotion in response to a light stimulus during the refractory period, a time when untreated fish are typically unresponsive to light. To determine whether optovin affected the central or peripheral nervous systems, we tested the compound on spinalized fish. Spinalized fish lacking brain-periphery connections still responded to optovin. This provided the clue needed to solve the mechanistic mystery – that optovin likely functioned at sensory neurons. We tested optovin on murine dorsal root ganglia (DRG) cells and further narrowed down the localization of optovin to sensory neurons responsive to mustard oil, a Trpa1 agonist. Trpa1 was confirmed as the target when we found that DRGs from trpa1 mutant mice were no longer responsive to optovin.
Like anatomical localization, mechanistic clues can also be provided by chronologic localization and genetic localization using live zebrafish assays. A report by Hao et al. nicely demonstrates both of these principles [58]. In this study, a compound called ‘windorphin’ (WD) was identified based on its ability to dorsalize embryos. The researchers were able to chronologically localize the effect window of WD to between 4 and 6 hpf, which corresponds to epiboly. The narrow window of time gave the researchers a clue that the target mediating dorsalizaiton was distinct from the dorsomorphin target (BMP signaling), since dorsomorphin is effective over a much wider timeframe [47]. This led to the examination of a second pathway with known involvement in dorsal-ventral patterning, the canonical Wnt pathway. As predicted, expression of a Wnt-dependent reporter was disrupted by WD during epiboly, WD disrupted Wnt3a-inducible and BIO (GSK3 inhibitor)-inducible reporter assays in dose dependent manner, and WD rescued the masterblind mbl (axin) mutant, which exhibits aberrant activation of Wnt signaling (resulting in no eyes among other things).
Hao and colleagues then used genetic localization to identify the molecular target of WD. Unlike mammals, zebrafish have two beta-catenin genes. They found that while the activity of beta-catenin-1 was blocked by WD, beta-catenin-2 activity was unaffected [58]. The major difference between these two ohnologues resides in the C-term transactivation (TA) domain, which in humans is required for the recruitment of histone deacetylases (HATs). The authors found that when WD is present, the TA domain is no longer able to recruit the HAT p300 to promoters.
SPOTLIGHT ON INTERESTING DESEASE STORIES
As zebrafish chemical screening turns 15 years old, the field is entering a new developmental stage. The discoveries of the past decade and a half are now reaching clinical trial stages. Several compounds discovered from a zebrafish chemical screen have shown preclinical promise and are being developed for clinical indications.
One of these is dorsomorphin, a small molecule inhibitor of bone morphogenetic protein (BMP) signaling. As described earlier, this compound was discovered by our group in a developmental screen for compounds that perturbed normal embryonic development [47]. While important for development, BMP signals also play important roles in adults, and excessive BMP signaling contributes to diseases such as fibrodysplasia ossificans progressiva (FOP) and anemia of inflammation (AI). In the case of FOP, soft tissues (e.g., muscles and tendons) turn to bone, leading to immobility and death, while in AI chronic inflammation causes anemia via BMP signaling in the liver. A derivative of dorsomorphin called LDN-193189 is well tolerated in mice, is orally available and is efficacious in treating FOP and AI in mouse models of these diseases [59, 60]. Development of this compound is now supported by the NIH Center for Advancing Translational Sciences (NCATS) Bridging Interventional Development Gaps (BrIDGs) program.
The discovery that flurandrenolide suppresses long QT interval defects in zebrafish larvae has led to new understanding of the role of glucocorticoid signaling in cardiac function [22]. As a result of these findings, a glucocorticoid steroid similar to flurandrenolide, dexamethasone, is currently being investigated to treat patients with long QT syndrome.
Another ongoing clinical investigation that has resulted directly from a discovery made in a zebrafish chemical screen is the evaluation of ProHema (Fate Therapeutics). It was discovered that dimethyl prostaglandin E2 (dmPGE2) could increase the numbers of hematopoietic stem cell (HSC) in live zebrafish [6]. Based on this discovery, dmPGE was used to treat murine whole bone marrow ex vivo, prior to a bone marrow transplant. The experiment resulted in an increase in repopulation efficiency of the treated cells in recipient animals compared to untreated controls [6]. This has led to the current Phase II clinical trial in which human umbilical cord derived cells are being treated, also ex vivo, with an investigational new drug ProHema to determine if it can enrich the number of umbilical cord HSCs available for transplant into adult patients afflicted with hematologic malignancies.
In addition to these three candidates, there are several more which could follow in short succession. Several of these are directed against cancer including rosuvastatin (an FDA approved cholesterol lowering agent) for suppression of prostate cancer growth [37], phenothiazines (approved as antipsychotics) for human T-ALL [20] and the novel compound ‘lenaldekar’ as an antileukemic agent which demonstrates less in vivo toxicity than AKT inhibitors [9]. Each of these was shown to have efficacy in rodent cancer models using xenografts of human tumors.
FUTURE DIRECTIONS
As zebrafish research develops further, new knowledge and tools are becoming available that will significantly impact the range and quality of in vivo chemical screening. Recent and future advances in zebrafish research sure to have an impact will include improvements in data acquisition (particularly imaging), increased automation capability, advances in genetic manipulation and development of new phenotypic endpoints.
Advances in genetics
Genetically engineered zebrafish lines can be powerful translational tools when used in high-throughput in vivo chemical screens. For example, a transgenic zebrafish model of hereditary arrhythmogenic cardiomyopathy (ACM) was created by driving expression of a pathogenic form of the human plakoglobin gene in zebrafish cardiac myocytes, in vivo [61]. Transgenic fish expressing this mutation develop abnormal cardiac physiology and eventually die as a result. These fish were used in a chemical screen resulting in the discovery that a GSK3beta inhibitor (SB216763) can reverse the electrophysiological defects not only in zebrafish, but also in cardiac myocytes derived from patient iPS cells endogenously expressing the pathogenic plakoglobin gene [61].
Of the 66 screens we examined, all of the genetically engineered zebrafish used were transgenic lines (Figure 3a). Nine screens used non-transgenic mutant fish, but all nine of these mutants were originally derived from forward genetic screens rather than targeted mutation. This is because the reverse genetics technology needed to efficiently create targeted alterations in the zebrafish genome had been unavailable until recently.
During the past few years, however, several new technologies enabling site-directed mutagenesis in live zebrafish embryos have been developed. These involve endonucleases that are recruited to target sites by either zinc-finger domains [62–64], ‘transcription activator-like effector’ (TALE) motifs [65, 66], or ‘clustered regularly interspaced short palindromic repeat’ (CRISPR)-derived guide RNAs [67, 68]. We predict that this new wave of technological advancement will soon be incorporated into high-throughput zebrafish chemical screening efforts and that an even larger portion of zebrafish screens will make use of mutant phenotypes compared to what we have observed to date. We further predict that zebrafish chemical screens will be used to provoke synthetic phenotypes in newly engineered mutant lines which initially exhibit no obvious pathologies.
Advances in imaging & automation
As mentioned earlier, there is a need for improved imagining techniques and automation, particularly solutions to the problem of organism orientation. One solution to this problem has been the invention of a 96-well plate containing built-in prisms adjacent to each well, which refracts both the excitation light and fluorescence emission to and from an objective mounted below the plate [69]. In this way an image could be captured of the lateral view of the larvae while they are upright in the well. Another solution proposed is a plastic mold which can be inserted into an agarose gel thereby creating cavities which properly orient larval fish [70]. These molds can be generated inexpensively with 3D printers and can be used to image living zebrafish over a timespan of at least 24 hrs.
Another orientation tool in development is the ‘Vertebrate Automated Screening Technology’ (VAST) BioImager (Union Biometrica). This elegant device uses a simple fluidic system to capture live fish from a multi-well plate and load them head-first sequentially into a capillary tube which is then able to rotate the organism to precisely the right angle to capture many images of different larvae in identical orientations [71]. Union Biometrica is also developing the COPAS XL and BioSorter platforms for automatic animal plating. We anticipate that advances in animal handling automation will improve the quality of zebrafish chemical screening, increase the number of screens and number of compounds screened, and reduce overall screening costs.
Another noteworthy advance in imaging has been the creation of the roy, nacre (mitfa) double knockout casper fish line, which completely lacks pigmentation outside of the retina [72]. Wild-type zebrafish are already remarkably transparent, especially at younger stages of life, however pigmentation in the skin and scales can often obscure certain tissues. Casper fish are even more transparent, and will likely be used extensively in future chemical screens.
Advances in phenotyping
For historical reasons, the majority of zebrafish chemical screens have been conducted using embryonic fish (1–48 hpf, Figure 1b) and predominately developmental phenotypes (Figure 3c). We expect that within the next 15 years, as zebrafish use in small molecule discovery continues to grow (Figure 1a) there will also be a diversification of the phenotypic repertoire. This will include an expansion of the use of fish at larval and juvenile stages. For this to take place, we must expand our understanding of zebrafish biology at these older stages. For example, we expect there will be increased interest in the normal behaviors of larval fish. There are currently very few robust behavioral assays described in the literature during this stage of life that would be amenable to high-throughput chemical screening.
While zebrafish larvae are advantageous for imaging studies, we also expect there to be advances in non-imaging endpoints as researchers find new creative ways to assess chemical function. Chemically induced changes in metabolomic profiles, for example, could be used as phenotypic endpoints in zebrafish chemical screens [73, 74] in addition to epigenetic, gene expression or proteomic readouts.
Chemically induced phenoptypes are also becoming starting points for antidote or ‘chemical rescue’ screens where an initial compound is added uniformly to all test wells to induce a particular phenotypic state and a screen is conducted in search of a second compound which modulates or counteracts the effects of the first chemical. Our group has recently used antidote screening to identify antidotes for organophosphate poisoning [74], cyanide toxicity [73], and doxorubicin-induced cardiomyopathy [50]. Others have identified compounds able to counteract the hair-damaging effects of ototoxic drugs, such as the aminoglycosidic antibiotic neomycin [75].
CONCLUSION
We have presented here an in-depth summary of more than 60 zebrafish chemical screens conducted over the past 15 years. We conclude that there are several consistent trends across the field. The majority of screens that have taken place were done using 96-well plates, with about three fish per well. The chemical concentration used is usually in the range of 5–25 μM, and all screens (with the exception of one conducted in adults) administer chemical compounds simply by adding the small molecules directly to the fish medium.
There was wide variability in the numbers of compounds screened, with only a few screens testing more than 10,000. Relatively few screens were conducted on fish older than 48 hrs, though it is entirely possible and has been done in several instances. There was also wide variability in the amount of time needed to carry our each screen. The most rapid assays were completed in less than 2 hours, but in many cases the time required between fish plating, compound addition and endpoint measurement exceeded 2 days.
We have summarized the contributions of zebrafish chemical screening to basic research, providing both new knowledge as well as new tools for further study, and also the promise zebrafish screens have for identifying clinically relevant disease pathways and small molecule drug candidates. Many zebrafish screens have produced novel classes of compounds, and we have explored the ways the functional targets of these compound have been identified. Other screens have identified new uses and new mechanisms of action for previously known bioactives.
We predict that the number zebrafish chemical screens conducted will continue to grow each year into the foreseeable future, but that there will be a shift in the field toward the use of older (larval) stage fish, the development of more rapid assays, the increased use of genetically engineered mutant fish, and the use of more diverse phenotypic endpoints.
Time will tell if drug candidates discovered using zebrafish screens fare better in clinical trials than small molecules discovered using target-based approaches. Therapeutic strategies developed as the direct result of zebrafish screens have only just begun to reach patients, and at the moment there are only a few. But given the lag between screening and clinical readiness, there’s reason to believe that the next 15 years will see even more compounds discovered in zebrafish find utility in the laboratory and the clinic.
HIGHLIGHTS.
More than 60 zebrafish chemical screens have been reported since 2000.
We summarize the current practices in design of high-throughput zebrafish screens.
We summarize the types of phenotypic endpoints that have been used.
We describe the impact of zebrafish on chemical biology.
We highlight interesting examples of how mechanisms of action have been discovered.
ACKNOWLEDGEMENTS
This work was supported by US National Institutes of Health grant R01 MH 086867 (RTP), by the Charles and Ann Sanders MGH Research Scholar Award (RTP), and NIH training grant T32 HL 007208 (AJR). We would like to acknowledge the support of our former colleague, mentor and friend the late Dr. Kenneth D. Bloch, who played important roles in the administration of the T32 grant and in the preclinical development of dorsomorphin/LDN-193189, which we have described in this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Swinney DC, Anthony J. How were new medicines discovered? Nature reviews Drug discovery. 2011;10(7):507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 2.Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: Selectively non-selective drugs for mood disorders and schizophrenia. Nature reviews Drug discovery. 2004;3(4):353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 3.Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97(24):12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oppedal D, Goldsmith MI. A chemical screen to identify novel inhibitors of fin regeneration in zebrafish. Zebrafish. 2010;7(1):53–60. doi: 10.1089/zeb.2009.0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik EJ, de Jong JL, Pugach E, Opara P, Zon LI. A chemical genetic screen in zebrafish for pathways interacting with cdx4 in primitive hematopoiesis. Zebrafish. 2010;7(1):61–68. doi: 10.1089/zeb.2009.0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, et al. Prostaglandin e2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, Chen F, et al. Dhodh modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471(7339):518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran TC, Sneed B, Haider J, Blavo D, White A, Aiyejorun T, Baranowski TC, Rubinstein AL, Doan TN, Dingledine R, Sandberg EM. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer research. 2007;67(23):11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- 9. Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L, Choudhry P, Manos EJ, Sofla H, Sanati A, Welborn S, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119(24):5621–5631. doi: 10.1182/blood-2011-12-398818.. ** This is one of the largest chemical screens conducted in zebrafish. A small molecule with anti leukemic properties in mice and activity against human malignant lymphoblasts was discovered based on its ability to selectively eliminate immature T cells in the developing zebrafish. This paper also provides a nice example of polypharmacological discovery.
- 10.Spring DR, Krishnan S, Blackwell HE, Schreiber SL. Diversity-oriented synthesis of biaryl-containing medium rings using a one bead/one stock solution platform. Journal of the American Chemical Society. 2002;124(7):1354–1363. doi: 10.1021/ja017248o. [DOI] [PubMed] [Google Scholar]
- 11.Sternson SM, Louca JB, Wong JC, Schreiber SL. Split--pool synthesis of 1,3-dioxanes leading to arrayed stock solutions of single compounds sufficient for multiple phenotypic and protein-binding assays. Journal of the American Chemical Society. 2001;123(8):1740–1747. doi: 10.1021/ja0036108. [DOI] [PubMed] [Google Scholar]
- 12.Moon HS, Jacobson EM, Khersonsky SM, Luzung MR, Walsh DP, Xiong W, Lee JW, Parikh PB, Lam JC, Kang TW, Rosania GR, et al. A novel microtubule destabilizing entity from orthogonal synthesis of triazine library and zebrafish embryo screening. Journal of the American Chemical Society. 2002;124(39):11608–11609. doi: 10.1021/ja026720i. [DOI] [PubMed] [Google Scholar]
- 13.Milan DJ, Peterson TA, Ruskin JN, Peterson RT, MacRae CA. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107(10):1355–1358. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- 14.Khersonsky SM, Jung DW, Kang TW, Walsh DP, Moon HS, Jo H, Jacobson EM, Shetty V, Neubert TA, Chang YT. Facilitated forward chemical genetics using a tagged triazine library and zebrafish embryo screening. Journal of the American Chemical Society. 2003;125(39):11804–11805. doi: 10.1021/ja035334d. [DOI] [PubMed] [Google Scholar]
- 15.Mathew LK, Sengupta S, Kawakami A, Andreasen EA, Lohr CV, Loynes CA, Renshaw SA, Peterson RT, Tanguay RL. Unraveling tissue regeneration pathways using chemical genetics. The Journal of biological chemistry. 2007;282(48):35202–35210. doi: 10.1074/jbc.M706640200. [DOI] [PubMed] [Google Scholar]
- 16.Namdaran P, Reinhart KE, Owens KN, Raible DW, Rubel EW. Identification of modulators of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2012;32(10):3516–3528. doi: 10.1523/JNEUROSCI.3905-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YJ, Fan HB, Jin Y, Ren CG, Jia XE, Wang L, Chen Y, Dong M, Zhu KY, Dong ZW, Ye BX, et al. Cannabinoid receptor 2 suppresses leukocyte inflammatory migration by modulating the jnk/c-jun/alox5 pathway. The Journal of biological chemistry. 2013;288(19):13551–13562. doi: 10.1074/jbc.M113.453811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson AL, Holmes GR, Bojarczuk AN, Burgon J, Loynes CA, Chimen M, Sawtell AK, Hamza B, Willson J, Walmsley SR, Anderson SR, et al. A zebrafish compound screen reveals modulation of neutrophil reverse migration as an anti-inflammatory mechanism. Science translational medicine. 2014;6(225):225ra229. doi: 10.1126/scitranslmed.3007672.. * This work illustrates the advantage of using zebrafish for transparent in vivo imaging of cell migration. A zebrafish wound model is used to identify a natually-occuring small molecule capable of accelerating resolution of inflamation after injury.
- 19.Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5(4):236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gutierrez A, Pan L, Groen RW, Baleydier F, Kentsis A, Marineau J, Grebliunaite R, Kozakewich E, Reed C, Pflumio F, Poglio S, et al. Phenothiazines induce pp2a-mediated apoptosis in t cell acute lymphoblastic leukemia. The Journal of clinical investigation. 2014;124(2):644–655. doi: 10.1172/JCI65093.. ** This article details one of the most elaborately designed chemical screening strategies used in zebrafish, requiring mutants, flourescent transgenics and chemically controlled expression of an oncogene. The authors discovered that phenothiazine antipsychotics are able to slow cancer progression in fish and induce apoptosis in human T-ALL cell lines. This paper nicely illustrates how zebrafish screens can be exploited to identify new uses for old drugs and to uncover previously unknown targets of those drugs.
- 21.Clifton JD, Lucumi E, Myers MC, Napper A, Hama K, Farber SA, Smith AB, 3rd, Huryn DM, Diamond SL, Pack M. Identification of novel inhibitors of dietary lipid absorption using zebrafish. PloS one. 2010;5(8):e12386. doi: 10.1371/journal.pone.0012386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peal DS, Mills RW, Lynch SN, Mosley JM, Lim E, Ellinor PT, January CT, Peterson RT, Milan DJ. Novel chemical suppressors of long qt syndrome identified by an in vivo functional screen. Circulation. 2011;123(1):23–30. doi: 10.1161/CIRCULATIONAHA.110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokel D, Bryan J, Laggner C, White R, Cheung CY, Mateus R, Healey D, Kim S, Werdich AA, Haggarty SJ, Macrae CA, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kokel D, Cheung CY, Mills R, Coutinho-Budd J, Huang L, Setola V, Sprague J, Jin S, Jin YN, Huang XP, Bruni G, et al. Photochemical activation of trpa1 channels in neurons and animals. Nat Chem Biol. 2013;9(4):257–263. doi: 10.1038/nchembio.1183.. ** This report describes the use of a zebrafish chemical screen for the discovery of a novel compound that sesitizes neurons to light. Using the compound 'optovin' the authors are able to stimulate motor neurons with a laser, causing spinalized fish to swim. This paper also provides a nice example of how a mechanism of action can be determined using a 'localization of activity' strategy.
- 25.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, Schier AF. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327(5963):348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolman MA, Jain RA, Liss L, Granato M. Chemical modulation of memory formation in larval zebrafish. Proc Natl Acad Sci U S A. 2011;108(37):15468–15473. doi: 10.1073/pnas.1107156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baraban SC, Dinday MT, Hortopan GA. Drug screening in scn1a zebrafish mutant identifies clemizole as a potential dravet syndrome treatment. Nature communications. 2013;4:2410. doi: 10.1038/ncomms3410.. * Here a zebrafish model for dravet syndrome is used to identify a small molecule specifically able to overcome both the convulsive behaviours and electrographic seizures characterisitc of the disease.
- 28.Leet JK, Lindberg CD, Bassett LA, Isales GM, Yozzo KL, Raftery TD, Volz DC. High-content screening in zebrafish embryos identifies butafenacil as a potent inducer of anemia. PloS one. 2014;9(8):e104190. doi: 10.1371/journal.pone.0104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuji N, Ninov N, Delawary M, Osman S, Roh AS, Gut P, Stainier DY. Whole organism high content screening identifies stimulators of pancreatic beta-cell proliferation. PloS one. 2014;9(8):e104112. doi: 10.1371/journal.pone.0104112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saydmohammed M, Vollmer LL, Onuoha EO, Vogt A, Tsang M. A high-content screening assay in transgenic zebrafish identifies two novel activators of fgf signaling. Birth defects research Part C, Embryo today : reviews. 2011;93(3):281–287. doi: 10.1002/bdrc.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokel D, Rennekamp AJ, Shah AH, Liebel U, Peterson RT. Behavioral barcoding in the cloud: Embracing data-intensive digital phenotyping in neuropharmacology. Trends in biotechnology. 2012;30(8):421–425. doi: 10.1016/j.tibtech.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le X, Pugach EK, Hettmer S, Storer NY, Liu J, Wills AA, DiBiase A, Chen EY, Ignatius MS, Poss KD, Wagers AJ, et al. A novel chemical screening strategy in zebrafish identifies common pathways in embryogenesis and rhabdomyosarcoma development. Development. 2013;140(11):2354–2364. doi: 10.1242/dev.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitambi SS, McCulloch KJ, Peterson RT, Malicki JJ. Small molecule screen for compounds that affect vascular development in the zebrafish retina. Mechanisms of development. 2009;126(5–6):464–477. doi: 10.1016/j.mod.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez Y, Astudillo O, Jensen L, Reynolds AL, Waghorne N, Brazil DP, Cao Y, O'Connor JJ, Kennedy BN. Selective inhibition of retinal angiogenesis by targeting pi3 kinase. PloS one. 2009;4(11):e7867. doi: 10.1371/journal.pone.0007867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z, Li Y, Xiang Q, Pei Z, Liu X, Lu B, Chen L, Wang G, Pang J, Lin Y. Design and synthesis of novel xyloketal derivatives and their vasorelaxing activities in rat thoracic aorta and angiogenic activities in zebrafish angiogenesis screen. Journal of medicinal chemistry. 2010;53(12):4642–4653. doi: 10.1021/jm1001502. [DOI] [PubMed] [Google Scholar]
- 36.Zhang ZR, Li JH, Li S, Liu AL, Hoi PM, Tian HY, Ye WC, Lee SM, Jiang RW. In vivo angiogenesis screening and mechanism of action of novel tanshinone derivatives produced by one-pot combinatorial modification of natural tanshinone mixture from salvia miltiorrhiza. PloS one. 2014;9(7):e100416. doi: 10.1371/journal.pone.0100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Tao W, Wang Y, Bikow J, Lu B, Keating A, Verma S, Parker TG, Han R, Wen XY. Rosuvastatin, identified from a zebrafish chemical genetic screen for antiangiogenic compounds, suppresses the growth of prostate cancer. European urology. 2010;58(3):418–426. doi: 10.1016/j.eururo.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Eliceiri BP, Gonzalez AM, Baird A. Zebrafish model of the blood-brain barrier: Morphological and permeability studies. Methods in molecular biology (Clifton, NJ) 2011;686:371–378. doi: 10.1007/978-1-60761-938-3_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umans RA, Taylor MR. Zebrafish as a model to study drug transporters at the blood-brain barrier. Clinical pharmacology and therapeutics. 2012;92(5):567–570. doi: 10.1038/clpt.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graf SF, Hotzel S, Liebel U, Stemmer A, Knapp HF. Image-based fluidic sorting system for automated zebrafish egg sorting into multiwell plates. Journal of laboratory automation. 2011;16(2):105–111. doi: 10.1016/j.jala.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Mandrell D, Truong L, Jephson C, Sarker MR, Moore A, Lang C, Simonich MT, Tanguay RL. Automated zebrafish chorion removal and single embryo placement: Optimizing throughput of zebrafish developmental toxicity screens. Journal of laboratory automation. 2012;17(1):66–74. doi: 10.1177/2211068211432197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truong L, Reif DM, St Mary L, Geier MC, Truong HD, Tanguay RL. Multidimensional in vivo hazard assessment using zebrafish. Toxicol Sci. 2014;137(1):212–233. doi: 10.1093/toxsci/kft235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Y, Semanchik N, Lee SH, Somlo S, Barbano PE, Coifman R, Sun Z. Chemical modifier screen identifies hdac inhibitors as suppressors of pkd models. Proc Natl Acad Sci U S A. 2009;106(51):21819–21824. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gut P, Baeza-Raja B, Andersson O, Hasenkamp L, Hsiao J, Hesselson D, Akassoglou K, Verdin E, Hirschey MD, Stainier DY. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol. 2013;9(2):97–104. doi: 10.1038/nchembio.1136.. * This paper illustrates how a finding from a zebrafish chemical screen can translate to mammalian biology. In a screen to identify chemical modulators of glucose metabolism, the authors discover a previously unknown role for the TSPO protein in the induction of fasting metabolism. They go on to demonstrate that TSPO ligands produce reduced blood glucose, reduced weight gain and improved glucose tolerance in a mouse model of diabetes and obesity.
- 45.Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nature biotechnology. 2004;22(5):595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 46.Gebruers E, Cordero-Maldonado ML, Gray AI, Clements C, Harvey AL, Edrada-Ebel R, de Witte PA, Crawford AD, Esguerra CV. A phenotypic screen in zebrafish identifies a novel small-molecule inducer of ectopic tail formation suggestive of alterations in non-canonical wnt/pcp signaling. PloS one. 2013;8(12):e83293. doi: 10.1371/journal.pone.0083293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits bmp signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4(1):33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Groh ED, Swanhart LM, Cosentino CC, Jackson RL, Dai W, Kitchens CA, Day BW, Smithgall TE, Hukriede NA. Inhibition of histone deacetylase expands the renal progenitor cell population. Journal of the American Society of Nephrology : JASN. 2010;21(5):794–802. doi: 10.1681/ASN.2009080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung DW, Williams D, Khersonsky SM, Kang TW, Heidary N, Chang YT, Orlow SJ. Identification of the f1f0 mitochondrial atpase as a target for modulating skin pigmentation by screening a tagged triazine library in zebrafish. Molecular bioSystems. 2005;1(1):85–92. doi: 10.1039/b417765g. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Asnani A, Zou L, Bentley VL, Yu M, Wang Y, Dellaire G, Sarkar KS, Dai M, Chen HH, Sosnovik DE, et al. Visnagin protects against doxorubicin-induced cardiomyopathy through modulation of mitochondrial malate dehydrogenase. Science translational medicine. doi: 10.1126/scitranslmed.3010189. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou L, Ishizaki H, Spitzer M, Taylor KL, Temperley ND, Johnson SL, Brear P, Gautier P, Zeng Z, Mitchell A, Narayan V, et al. Aldh2 mediates 5-nitrofuran activity in multiple species. Chemistry & biology. 2012;19(7):883–892. doi: 10.1016/j.chembiol.2012.05.017.. * The authors start with a chemical screen to identify small molecule modulators of zebrafish melanocyte development and unexpectedly uncover a mechanism of action that might explain some of the benefits and side effects of a class of drugs frequently used to treat trypanosome infection.
- 52.Wong JC, Sternson SM, Louca JB, Hong R, Schreiber SL. Modular synthesis and preliminary biological evaluation of stereochemically diverse 1,3-dioxanes. Chemistry & biology. 2004;11(9):1279–1291. doi: 10.1016/j.chembiol.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Torregroza I, Evans T, Das BC. A forward chemical screen using zebrafish embryos with novel 2-substituted 2h-chromene derivatives. Chemical biology & drug design. 2009;73(3):339–345. doi: 10.1111/j.1747-0285.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das BC, McCartin K, Liu TC, Peterson RT, Evans T. A forward chemical screen in zebrafish identifies a retinoic acid derivative with receptor specificity. PloS one. 2010;5(4):e10004. doi: 10.1371/journal.pone.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandoval IT, Manos EJ, Van Wagoner RM, Delacruz RG, Edes K, Winge DR, Ireland CM, Jones DA. Juxtaposition of chemical and mutation-induced developmental defects in zebrafish reveal a copper-chelating activity for kalihinol f. Chemistry & biology. 2013;20(6):753–763. doi: 10.1016/j.chembiol.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sachidanandan C, Yeh JR, Peterson QP, Peterson RT. Identification of a novel retinoid by small molecule screening with zebrafish embryos. PloS one. 2008;3(4):e1947. doi: 10.1371/journal.pone.0001947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M. Zebrafish chemical screening reveals an inhibitor of dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5(9):680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hao J, Ao A, Zhou L, Murphy CK, Frist AY, Keel JJ, Thorne CA, Kim K, Lee E, Hong CC. Selective small molecule targeting beta-catenin function discovered by in vivo chemical genetic screen. Cell reports. 2013;4(5):898–904. doi: 10.1016/j.celrep.2013.07.047.. ** Here the authors use the genetic duplication in zebrafish to their advantage to identify the mechanism of action of their newly discovered compound 'windorphin.' They find that windorphin blocks the activity of zebrafish beta-catenin-1 but not beta-catenin-2. Mammals have only the first homolog, however the second, unique to zebrafish, provided an important clue which ultimately led to elucidation of the chemical mechanism of action.
- 59.Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, Kamiya N, et al. Bmp type i receptor inhibition reduces heterotopic [corrected] ossification. Nat Med. 2008;14(12):1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinbicker AU, Sachidanandan C, Vonner AJ, Yusuf RZ, Deng DY, Lai CS, Rauwerdink KM, Winn JC, Saez B, Cook CM, Szekely BA, et al. Inhibition of bone morphogenetic protein signaling attenuates anemia associated with inflammation. Blood. 2011;117(18):4915–4923. doi: 10.1182/blood-2010-10-313064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Asimaki A, Kapoor S, Plovie E, Karin Arndt A, Adams E, Liu Z, James CA, Judge DP, Calkins H, Churko J, Wu JC, et al. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Science translational medicine. 2014;6(240):240ra274. doi: 10.1126/scitranslmed.3008008.. ** This paper describes a zebrafish chemical screen used to identify a small molecule that can suppress cardiac defects caused by a pathogenic form of a human gene. To demonstrate the utility of the compound in humans, the authors show this compound reverses the electrophysiological defects apparent in the cardiac myocytes derived from patient iPS cells.
- 62.Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nature protocols. 2009;4(12):1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by oligomerized pool engineering (open) PloS one. 2009;4(2):e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sander JD, Yeh JR, Peterson RT, Joung JK. Engineering zinc finger nucleases for targeted mutagenesis of zebrafish. Methods Cell Biol. 2011;104:51–58. doi: 10.1016/B978-0-12-374814-0.00003-3. [DOI] [PubMed] [Google Scholar]
- 65.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric talens. Nucleic acids research. 2012;40(16):8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered talens. Nature biotechnology. 2011;29(8):697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a crispr-cas system. PloS one. 2013;8(7):e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a crispr-cas system. Nature biotechnology. 2013;31(3):227–229. doi: 10.1038/nbt.2501.. ** First demonstration that the CRISPR/Cas9 system can be used in vivo to create site-directed genetic modifications in zebrafish.
- 69.Rovira M, Huang W, Yusuff S, Shim JS, Ferrante AA, Liu JO, Parsons MJ. Chemical screen identifies fda-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc Natl Acad Sci U S A. 2011;108(48):19264–19269. doi: 10.1073/pnas.1113081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wittbrodt JN, Liebel U, Gehrig J. Generation of orientation tools for automated zebrafish screening assays using desktop 3d printing. BMC biotechnology. 2014;14:36. doi: 10.1186/1472-6750-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pardo-Martin C, Allalou A, Medina J, Eimon PM, Wahlby C, Fatih Yanik M. High-throughput hyperdimensional vertebrate phenotyping. Nature communications. 2013;4:1467. doi: 10.1038/ncomms2475.. * Description of the 'Vertebrate Automated Screening Technology' (VAST) system which is able to capture and orient live zebrafish larvae for high throughput comparative imaging.
- 72.White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, Zon LI. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2(2):183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nath AK, Roberts LD, Liu Y, Mahon SB, Kim S, Ryu JH, Werdich A, Januzzi JL, Boss GR, Rockwood GA, MacRae CA, et al. Chemical and metabolomic screens identify novel biomarkers and antidotes for cyanide exposure. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27(5):1928–1938. doi: 10.1096/fj.12-225037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin S, Sarkar KS, Jin YN, Liu Y, Kokel D, Van Ham TJ, Roberts LD, Gerszten RE, Macrae CA, Peterson RT. An in vivo zebrafish screen identifies organophosphate antidotes with diverse mechanisms of action. Journal of biomolecular screening. 2013;18(1):108–115. doi: 10.1177/1087057112458153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Owens KN, Santos F, Roberts B, Linbo T, Coffin AB, Knisely AJ, Simon JA, Rubel EW, Raible DW. Identification of genetic and chemical modulators of zebrafish mechanosensory hair cell death. PLoS genetics. 2008;4(2):e1000020. doi: 10.1371/journal.pgen.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]