Abstract
Transfer RNA (tRNA) is traditionally considered to be an adaptor molecule that helps ribosomes to decode messenger RNA (mRNA) and synthesize protein. Recent studies have demonstrated that tRNAs also serve as a major source of small non-coding RNAs that possess distinct and varied functions. These tRNA fragments are heterogeneous in size, nucleotide composition, biogenesis and function. Here we describe multiple roles that tRNA fragments play in cell physiology and discuss their relevance to human health and disease.
Keywords: Transfer RNA, tRNA-derived fragment, RNA processing, Angiogenin, Stress, Apoptosis
1. Introduction
Cellular RNAs are traditionally divided into three ubiquitous classes: mRNAs possessing protein-coding function, whose sequence is decoded to produce proteins, ribosomal (r) RNAs possessing enzymatic activities required for protein synthesis, and tRNAs that deliver amino acids to the growing polypeptide chain serving non-(protein)-coding functions. Both rRNAs, as an integral part of the ribosome, and tRNAs, as carriers of amino acids, decode nucleotide information on mRNA into the specific amino acid sequence synthesized by the ribosome. The discovery of small non-coding RNAs (ncRNAs) (20–40 nucleotides long) with diverse regulatory functions has revolutionized modern biology and medicine. Among these ncRNAs are siRNAs, miRNAs and piRNAs with well-defined biological functions that rely on the complementarity of their sequences to RNA targets (reviewed in [1]).
With the development of high-throughput sequencing technologies, it became apparent that there are many more classes of small ncRNAs [2]. These novel ncRNAs are produced from other cellular RNA species by specific and regulated RNA processing or cleavage [3]. The sources of such “non-canonical” ncRNAs are diverse cellular RNAs including tRNAs.
The evolutionarily conserved function of tRNA is to help the ribosome synthesize proteins by decoding nucleotide triplets thereby linking nucleotide information on mRNA to amino acid sequence [4,5]. Beyond this canonical function, tRNA is also implicated in a number of other biological processes including cell signaling, cell survival, apoptosis, metabolism of amino acids and porphyrines, and stress response programs [5,6]. Despite their small size (~73–95 nucleotides), tRNAs are characterized by their evolutionarily conserved secondary and tertiary structures, extensive post-transcriptional processing and modifications (up to 100 nucleoside modifications have been described), extreme stability and resistance to nucleases [4,5,7]. Moreover, the human genome contains more than 500 genes encoding over 40 different tRNAs [8–10].
Identification of novel ncRNAs derived from tRNAs, known as tRNA-derived RNA fragments, has recently gained significant attention. Some of these fragments are derived from precursor tRNA molecules, others from mature cytoplasmic tRNAs. Some fragments are constitutive components of all cells, whereas others are only produced in cells exposed to adverse conditions. The identification of tRNA fragments found in different organisms, as well as their biogenesis and possible functions, are described in several recent reviews [5,11–15]. Here, we will only review the biogenesis and functions of tRNA fragments found in human cells and discuss their potential roles in the pathobiology of human diseases.
2. tRNA fragments in human cells
High-throughput RNA sequencing has detected distinct classes of tRNA fragments in mammalian cells. Their abundance varies between different cell types and tissues showing generally no correlation with abundance of parent tRNAs. Moreover, some of the fragments are only produced under specific conditions such as developmental stage, proliferative status, stress or viral infection [11–15]. Although tRNA pieces are heterogeneous in size (10–45 nucleotides), they are not products of random tRNA cleavage or degradation because their ends are precisely defined by RNA cleavage sequence determinants.
Biogenesis of tRNA is a complex process. Initially, tRNA is transcribed in the form of a precursor (pre-tRNA) containing 5′-leader and 3′-trailer sequences and, in some cases, introns in the anticodon loop [5,6,16–18]. During tRNA maturation, the 5′-trailer is processed by RNase P, the 3′-trailer is removed by RNase Z, and following 3′-trailer removal, the 3′-end of all human tRNAs is modified by the addition of the universal 3′-CCA triplet by the CCA-adding enzyme (also known as TRNT1) (Fig. 1). Intron-containing tRNAs (32 in humans, which make up less than 10% of all nuclear-encoded human tRNAs), are processed by a nuclear tRNA splicing endonuclease (TSEN) complex that excises the intron and ligates 5′- and 3′-tRNA exons to form mature tRNA (Fig. 1A). TSEN cleavage leaves a 2′-3′-cyclic phosphate at the 3′end of the 5′ exon and a 5′-hydroxyl (5′-OH) at the 5′-end of 3′-exon. The ligation of 5′- and 3′-exons can go through two different machineries, and it is still a matter of debate which mechanism is more commonly used [19–24] (Fig. 1A). In the first mechanism, RtcB (also known as HSPC17, or C22orf28) works as an RNA 2′,3′-cyclic phosphate and 5′-OH ligase to directly mediate the ligation of both exons. RtcB is a catalytic subunit of a multimeric protein complex consisting at least of five other proteins: ASW (C2orf49), CGI-99 (C14orf166), FAM98B, the DEAD-box helicase DDX1 and archease (ARCH, or ZBTB8OS). The roles of these subunits in the context of RNA ligation remain unclear, although a recent study from Popow et al. suggests that DDX1 and archease facilitate RtcB-mediated ligation of tRNA exons [25]. In the second mechanism, CLP1 (the first RNA kinase ever identified [26]) associates with the TSEN complex to phosphorylate 3′-tRNA exons and facilitate further ligation. Molecular details of CLP1-facilitated ligation of tRNA exons are unknown. While cells can utilize both pathways, their relative contribution, importance for tRNA metabolism or functional redundancy is not known.
Fig. 1.
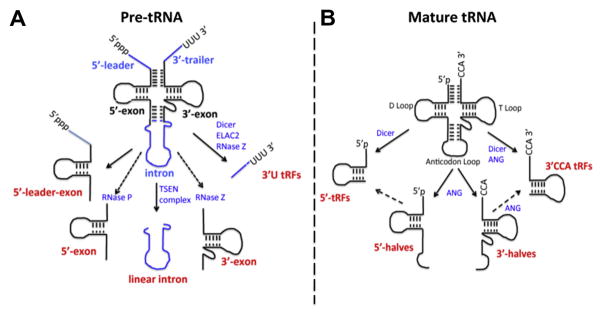
Processing of precursor and mature forms of tRNA in tRNA fragments. (A) Processing of precursor tRNA. Transcribed pre-tRNA contains distinct 5′- and 3′-extensions (5′-leader and 3′-trailer (blue)) that have to be removed by RNase P and RNase Z during tRNA processing. In some cases, the 5′-leader is not removed giving rise to 5′-leader exon fragments. Intron (blue)-containing pre-tRNAs are spliced by the TSEN complex. Usually, such introns are quickly degraded but in some pathological cases linear introns can accumulate. 5′-exons and 3′-exons are naturally present in the nucleus as products of intron-containing tRNA splicing. Upon 3′-trailer removal, the CCA-adding enzyme TRNT1 adds CCA to the 3′-ends of tRNA. In some cases, 3′-trailers accumulate and are exported to the cytoplasm as 3′-U tRFs. (B) Processing of mature cytoplasmic tRNA. Mature tRNA can be cleaved in the anticodon loop by angiogenin (ANG) to produce 5′- and 3′-tRNA halves (or 5′- and 3′-tiRNAs, respectively. Dicer-dependent cleavage of tRNA in the D arm of tRNA results in the production of small tRNA fragments, 5′-tRFs. Similarly, cleavage in the T arm by Dicer or ANG results in the production of tRFs containing CCA at their 3′-ends (so called 3′CCA tRFs). Both 5′-tRFs and 3′CCA tRFs may also be processed from 5′- and 3′-tRNA halves, respectively (dashed arrows).
Importantly, the processing of pre-tRNA and mature tRNA results in the production of distinct classes of tRNA-derived ncRNAs (Fig. 1A and B). tRNA fragments derived from mature tRNA can be divided into two major types: tRNA halves (or tRNA-derived, stress-induced RNAs, known as tiRNAs (see below)) and smaller tRNA fragments (tRFs) (Fig. 1B). tRNA fragments derived from pre-tRNA are distinguished by the presence of sequences derived from 5′-leader or 3′-trailers (Fig. 1A).
2.1. tRNA halves (tiRNAs)
tRNA halves are produced by specific cleavage in the anticodon loop to produce 30–35 nucleotide 5′-tRNA halves and 40–50 nucleotide 3′-tRNA halves. Small quantities of these fragments are found in human cells under normal growth conditions [3,27,28]. Many of these are splicing intermediates derived from intron-containing tRNAs [23,29]. In contrast, tiRNAs are produced by stress-induced (e.g. oxidative stress, heat shock or UV irradiation) cleavage of mature cytoplasmic tRNAs by the ribonuclease angiogenin (ANG) [27,30–33]. Cytoplasmic tRNA halves and tiRNAs are probably the same molecules and these terms can be used interchangeably.
ANG, a member of RNase A superfamily, is a predominantly nuclear protein that is also found in complex with RNH1 (also known as RI, or ANG inhibitor) in the cytoplasm (reviewed in [34]). In response to stress, ANG translocates into cytoplasm from the nucleus and dissociates from RNH1. Under these conditions, ANG cleaves cytoplasmic tRNA into 5′- and 3′-tiRNAs [33]. While intuitively such cleavage should cause abrupt translational arrest, this is not observed because only a minor fraction (<5%) of the tRNA pool is cleaved [27,31,33]. When cells are treated with recombinant ANG, global translation is reduced by 10–15%. A similar reduction in protein synthesis is observed when isolated endogenous tiRNAs are transfected into cells suggesting that it is the tRNA fragments rather then the tRNA cleavage that inhibit protein synthesis. Surprisingly, transfection of 5′-tiRNAs, but not 3′-tiRNAs, inhibits translation in cultured cells [30]. Moreover only 5′-tiRNAs derived from tRNAAla and tRNACys are responsible for the inhibition of translation [32]. Mechanistically, active tiRNAs inhibit translation initiation by interfering with the assembly of the cap binding complex eIF4F (the complex required for canonical translation initiation), and requires cooperation with the translational silencer YB-1 [35–38]. 5′-tiRNAsAla/Cys possess a terminal oligoguanine motif that is required to displace the eIF4F complex and inhibit translation [32]. Inhibition of translation initiation by 5′-tiRNAs also induces the assembly of stress granules (SGs) [30,32], dynamic cytoplasmic RNA foci that possess adaptive and pro-survival functions. SGs contain translationally-stalled mRNAs, associated preinitiation factors, specific RNA-binding proteins and signaling molecules [39–42]. SGs are typically formed upon activation of stress-sensing serine/threonine kinases that phosphorylate serine residue 51 of eIF2α [43], a molecule that regulates the Integrated Stress Response to coordinate cell adaptation and survival under stress conditions (reviewed in [44–46]). However, tiRNAs assemble SGs in a phospho-eIF2α-independent manner [30,32] suggesting the existence of an alternative route of signaling and SG assembly. It is likely that tiRNA-induced translational reprogramming is not limited to translation inhibition of specific transcripts but also involves a number of cellular events initiated by the assembly of SGs.
While our data suggest that ANG directly cleaves tRNA within the anticodon loop, Czech et al. proposed two-step mechanism for ANG-mediated tRNA cleavage [47]. They showed that, in response to oxidative stress, ANG first cleaves the conserved CCA-end of all tRNAs to remove associated amino acids, and only then cleaves in the anticodon loop. Moreover, tRNAs with cleaved CCA ends can serve as substrates for TRNT1 (CCA-adding enzyme) allowing repair of CCA ends when stress is relieved. While this is a very attractive scenario, these data are based on in vitro studies and await in vivo confirmation. Moreover, because ANG cleaves substrates to produce 2′-3′-cyclic phosphate at the 3′-end of cleavage products (e.g. tRNA without CCA-end), TRNT1 must be able to add CCA trinucleotide to 2′-3′-phosphate. While some CCA-adding enzymes (e.g. from Escherichia coli) also have an additional C-terminal domain with phosphohydrolase activity to resolve cyclic phosphate and/or remove 3′-phosphate from the 3′-end (reviewed in [48]), currently it is not known whether Homo sapiens’s TRNT1 alone or in complex with other enzymes can do that in vivo.
ANG-mediated tRNA cleavage is a highly regulated process. Genome-wide analysis of tiRNAs demonstrates that patterns of tRNA cleavage are stress-specific (e.g., oxidative stress vs hypertonic stress), or dependent on stress intensity [27], and phosphorylation of eIF2α, a main trigger of SGs and major player in the Integrated Stress Response, regulates levels of selected tiRNAs [27,33]. Interestingly, higher levels of tRNA cleavage correlate with higher translation rates (e.g., translation inhibitors down-regulate both translation and tRNA cleavage) suggesting that ANG has better access to tRNA when protein synthesis is active [27]. In agreement with this hypothesis, ANG was found on polysomes (pool of actively translating ribosomes), and in SGs when translation is inhibited [49]. Modifications of tRNA anticodon loops also influences ANG-mediated cleavage. In Drosophila melanogaster, methylation of selected tRNAs (ValAAC, GlyGCC, AspGTC) protects these tRNAs from stress- and ANG-induced tRNA cleavage [50].
It must be noted that precise molecular roles for the majority of 5′- and 3′-tiRNAs as well as their localization, relative abundance, and stability are yet to been determined. Our studies identified a number of RNA-binding proteins as partners of 5′- and 3′-tiRNAs, and some of them are both general to tiRNAs and also specific to certain tiRNAs (unpublished data and [32]) suggesting possible specialized tiRNA functions. Moreover, a recent study showed that ANG-induced tiRNAs inhibit hyperosmotic apoptosis in stressed cells [51]. This is achieved by the direct binding of tiRNAs to cytochrome c (Cyt c), which is released from mitochondria during stress to promote apoptosome formation with subsequent caspase activation. Cyt c bound to tiRNAs fails to trigger apoptosome formation and execute cell death. Importantly, RNA sequencing of tiRNAs bound to Cyt c suggests that only a subpopulation of tiRNAs (both 5′- and 3′-, about 20 different species) is highly enriched in these complexes. This study parallels previously reported anti-apoptotic effects of full size tRNAs that are also able to bind Cyt c and inhibit apoptosome formation in vitro [52,53]. However, under stress conditions Cyt c preferentially binds to tiRNAs and not tRNAs in vivo [51]. Further studies are necessary to characterize the composition of Cyt c:tiRNA complexes, sequence/structural determinants of tiRNAs contributing to complex formation and also specificity of stresses triggering formation of these complexes.
2.2. Diverse tRNA fragments (tRFs)
tRFs are fragments of tRNA or pre-tRNA that are typically shorter than tRNA halves (~12–30 nucleotides). There are several subclasses of tRFs, which are often difficult to classify. Here, we will discuss them in terms of their biogenesis and whether they are derived from mature tRNA or pre-tRNA. Mature tRNAs have a 5′-monophosphate group and 3′-CCA charged with cognate amino acid. All nuclear-encoded tRNAs have four base-paired stems (in 5′- 3′-direction: D arm, anticodon arm, T arm, and acceptor stem) bridging the conserved D-loop, tRNA-specific anticodon loop, variable loop and T-loop (Fig. 1B). Compared to mature tRNAs, pre-tRNAs lack the CCA tail but include a 5′-leader starting with triphosphate, a 3′-trailer with an oligouridine stretch and, for intron-containing tRNA, a variable-sized intron (Fig. 1B).
Mature tRNAs can give rise to 5′-tRFs and 3′ CCA-tRFs that are produced by cleavage of tRNA in the D-loop and in/near the T-loop, respectively (Fig. 1B). 5′-tRFs are 19–21 nt fragments that are formed by specific cleavage after conserved nucleotides G18-G19 in the D-loop [54,55]. Interestingly, the abundance of selected 5′-tRF species (e.g. derived from tRNAsGln/Lys/Val/Arg) in HeLa cells is comparable to the levels of abundant microRNAs such as miR-21 or members of let-7 family [54]. Similarly, the abundance of selected 5′-tRFs (e.g. derived from tRNAsGlu/Ser/Leu/Gln) in prostate cancer cell lines is greater than more than 90% of individual microRNAs in these cells [55]. Dicer, an endoribonuclease involved in the biogenesis of mature microRNA from microRNA precursors [1], is responsible for processing of 5′-tRFs from tRNA because siRNA-mediated silencing of Dicer significantly decreases the abundance of 5′-tRFs [54]. However, it is also possible that 5′-tRFs are processed from larger tRNA intermediates, e.g. 5′-tRNA halves, and also in Dicer-independent manner [56]. 5′-tRFs are cytoplasmic and also associate, although poorly, with Argounaute (Ago 1–4) proteins central to RISC (RNA-induced silencing complex) functions [57]. Whether 5′-tRFs play a direct role in RNAi (RNA interference) is unknown but recently their role in translation inhibition was proposed [58]. Sobala and Hutvagner reported that selected 5′-tRFs (3 out of 4 tested) inhibit translation of mRNA reporters in vitro [58]. The molecular details of this inhibition remain to be determined but they are proposed to affect translation elongation [58], which is different from the inhibition of translation initiation caused by 5′-tiRNAs [30,32]. Interestingly, the universally conserved G18–G19 dinucleotide at the 3′-end of 5′-tRF is absolutely required for translation inhibition. The same mutation in 5′-tiRNAAla also affects 5′-tiRNA-mediated repression of translation approximately twofold [32]. An interesting parallel is coming from studies in archaea: a stress-induced 26-nt 5′-tRF derived from tRNAVal inhibits translation by direct binding to the 30S small ribosomal subunit where it inhibits the peptidyl transferase activity of the ribosome [59]. Thus inhibition of translation by 5′-tRNA fragments appears to be an evolutionarily conserved phenomenon.
The 3′ CCA-tRFs are tRNA fragments characterized by the presence of a universal CCA trinucleotide at their 3′-ends and are produced from mature tRNAs by cleavage in the T-loop [55,56,60–63] (Fig. 1B). Lee et al. [55] reported that the abundance of 3′CCA-tRFs is comparable to that of 5′-tRFs and microRNAs. The biogenesis of 3′CCA-tRFs is proposed to be both Dicer-dependent [60–63] and Dicer-independent (for example by ANG-mediated cleavage in the T arm) [56]. Selected 3′CCA-tRFs are found in association with Argonautes and possess RNAi activities [61,62].
Pre-tRNAs can also be processed to produce fragments containing 5′-leader, 3′-trailer or intron-derived sequences (Fig. 1A). The best-studied pre-tRNA-derived fragments belong to so-called 3′U-tRFs (19–25 nt) that include the 3′-trailer from pre-tRNA followed by a 2–6 nt oligouridine stretch produced by terminating RNA polymerase III [55,61,64]. Although biogenesis of 3′U-tRFs is natural and depends on the RNase Z-dependent cleavage of pretRNA [55], one study proposed Dicer-dependent processing [60]. Surprisingly, 3′U-tRFs are relatively stable molecules that are concentrated in the cytoplasm [55,64]. They are also present in human embryonic stem cells and embryoid bodies [65]. Moreover, the biogenesis of 3′U-tRFs is proposed to occur in the cytoplasm from pre-tRNA by a cytoplasmic RNase Z homologue ELAC2 and might be regulated by cell proliferation [55]. It is not clear how pre-tRNAs are exported from the nucleus to the cytoplasm to produce 3′U-tRFs. In the cytoplasm, 3′U-tRFs bind to Argonautes (preferentially Ago3 and Ago4) and compete with cellular microRNAs and siRNAs for incorporation into RISC complexes [61].
Mammalian intron-containing pre-tRNAs also produce a distinct subset of relatively stable tRFs [29,66,67] (Fig. 1A). Some of these tRFs are splicing products of pre-tRNAs that lack leader and trailer sequences. Although these fragments resemble tRNA halves, they are concentrated in the nucleus rather than the cytoplasm. Much larger tRFs containing 5′-leader sequences followed by the entire 5′-exon of pre-tRNA (5′-leader-exon-tRF, e.g. produced from pre-tRNATyr/Arg) are predominantly nuclear and induced by oxidative stress in an ANG-independent manner [29]. In some pathological conditions (discussed below), accumulation of relatively abundant linear introns derived from pre-tRNA is also observed [66]. Additional tRFs (e.g. derived from pre-tRNAIle/Tyr) containing 5′-trailer sequences followed by a partial exon (5′-leader-partial-tRF) or 3′-exon followed by 3′-trailer sequences (3′-partial-exon-trailer) have been reported [66]. However these fragments are less abundant than intron-containing and 5′-leader-exon-tRFs, and their functions are unknown.
3. Possible roles of tRNA fragments in human disease
The production of tRNA-derived ncRNAs has been observed in a number of human diseases. In some cases, these tRNA fragments may serve as useful biomarkers. It remains to be determined whether these tRNA fragments contribute to disease pathogenesis. Below, we discuss examples in which the production of tRNA fragments has been linked to cancer, infection, neurodegeneration and other pathological conditions.
3.1. tRNA fragments and pathological stress injuries
Cellular damage is central to disease pathogenesis. Stress imparted by hypoxia, nutrient deprivation, oxidative conditions and metabolic imbalance can damage cells and promote disease. We and others observed that these stresses stimulate production of tiRNAs [31,33]. In a recent study using animal models of tissue damage (e.g., toxic injury, irradiation and ischemic reperfusion) tiRNAs were found to serve as in vivo biomarkers whose production correlated with the degree of tissue damage [68]. Combining in vitro and in vivo studies Mishima et al. showed that oxidative stress induces a change in tRNA conformation that promotes ANG-mediated production of tiRNAs. This stress-induced conformational change allows 1-methyladenosine nucleoside (m1A), a modification important for stabilizing the L-shaped structure of tRNA, to be recognized by an m1A-specific (Anti-m1a) antibody. This antibody recognizes 3′-tiRNAs (containing m1A) as well as free m1A nucleosides. This antibody was used to show that renal ischemia/reperfusion (I/R) injury and cisplatin-mediated nephrotoxicity (which both induce tissue damage via oxidative stress) generate tiRNAs in damaged kidneys [68]. Similar results were obtained using m1A-based immunohistochemistry to directly visualize damaged areas of kidneys, brain and liver. Importantly, the production of tiRNAs was found to be ANG dependent. Interestingly, the conformational change in tRNA caused by various injuries precedes DNA damage and apoptosis in vivo, which makes detection of tRNA changes useful as an early stress biomarker. Consequently, an m1A-based ELISA that detects tRNA derivatives (unfolded tRNA, tiRNAs, smaller tRFs and free m1A) was used to quantify circulating tRNA derivatives in both humans and animals under different pathological settings (e.g., I/R, γ-irradiation, surgery, chronic kidney disease). The levels of circulating tRNA derivatives were much higher under these conditions than in healthy patients or control animals suggesting that levels of tRNA derivatives can be used as early biomarkers of oxidative stress and tissue damage [68].
Mishima et al. further demonstrated that tRNA-derived fragments avoid degradation in the blood because they are associated with circulating exosomes [68], membranous extracellular vesicles packed with proteins and nucleic acids [69]. Dhabbi et al. similarly quantified tRNA fragments in mouse and human serum [70]. These 30–33 nt fragments were found to be derived exclusively from the 5′-ends of mature tRNAs. These fragments are not included in exosomes but are parts of 100–300 kDa protein–RNA complexes. Interestingly, the source of these circulating 5′-tiRNAs is mainly hematopoietic and lymphoid tissues, and such physiological changes as aging and calorie restriction modulate the appearance of 5′-tiRNAs from specific tRNA isoacceptors. Further studies are required to decipher a casual relationship between circulating 5′-tiRNAs and specific physiological/pathophysiological conditions.
3.2. tRNA fragments and cancer
It has been observed that cancer patients and tumor-bearing animals excrete elevated levels of modified purines and pyrimidines, and these modified nucleosides can originate only from tRNAs [71–73]. Moreover, compared to unmodified RNA, such as mRNAs or microRNAs, the high content of modified nucleotides in tRNA confers ribonuclease resistance making it possible to detect tRNA fragments even after excretion from cells [74]. It was also shown that tumor tissues have a high rate of tRNA turnover [71], and tRNA fragments can be detected in the serum and urine of cancer patients [72,73]. Moreover the levels of tRNA breakdown products correlate with the clinical stage of some cancers.
The levels of tRNAs vary widely among normal (non-diseased) human tissues (the level of variation is as much as tenfold) [75]. As cancer cells demand elevated levels of protein synthesis for their increased proliferation and growth, this also requires a coordinated adjustment of translation machinery components. Cancer cells have higher levels of ribosomal RNAs, tRNAs and ribosomes than non-cancerous cells. Genome-wide analysis of tRNA levels in breast cancer cells versus normal breast tissues revealed that tRNA levels (both nuclear-encoded and mitochondrial) are significantly increased in transformed cells [76]. This increase is selective with over-expression of certain tRNA isoacceptors. Similarly, the levels of tRNAs in multiple myeloma cells are significantly elevated compared to normal bone marrow cells [77]. Moreover, overexpression of initiator tRNA alone leads to global reprograming of gene expression, as well as increased proliferation and metabolism of epithelial cells [78].
Whether the increase in tRNA levels results in an increase in tRNA fragments in cancer cells is unknown. ANG is over-expressed in almost all types of cancer (reviewed in [34,79,80]). It is an angiogenic ribonuclease that promotes cancer cell proliferation and induces tumor angiogenesis by stimulation of rRNA transcription in endothelial and cancer cells [81]. The ribonuclease activity of ANG is critical for its ability to promote angiogenesis [82], and inhibition of the RNase activity by small molecule inhibitors significantly inhibits tumor formation in mouse xenograft tumor models [83]. As ANG produces tiRNAs that re-program translation and promote the assembly of SGs that help cells to survive under adverse conditions, it is possible that ANG-induced tiRNAs directly contribute to ANG-mediated angiogenesis and cancer cell proliferation. Similarly, tiRNAs can help cancer cells to prevent apoptosis by binding to Cyt c [51].
One of the 3′ U-tRFs (namely tRF-1001), derived from pre-tRNASer, is highly expressed in different cancer cell lines, and is required for proliferation of prostate cancer cells [55]. This fragment is produced by cleavage of cytoplasmic pre-tRNA by ELAC2, a prostate cancer susceptibility gene [84]. The levels of tRF-1001 in cancer cells are directly related to levels of cellular proliferation. Knocking down this tRF causes a dramatic loss of cell viability and inhibition of cell proliferation with specific accumulation of cells in G2 phase and inhibition of DNA synthesis [55]. The molecular mechanism by which tRF-1001 affects cell physiology is not known. In addition, analysis of the small RNA transcriptome in prostate cancer revealed enrichment of tRFs in both non-metastatic and metastatic lymph node prostate cancer samples. There may be a differential processing of tRNA in prostate cancer because tRFs in non-metastatic samples were 18-nt long whereas tRFs in metastatic tissues were 27 nt long [85].
Two reports showed the existence of tRFs in human B-cell lymphomas. Li et al. observed accumulation of both 5′-tRFs (predominantly 14–15 nt) and 3′CCA-tRFs (predominantly 17–18 nt, but also shorter abundant species were observed) precisely matching 5′- and 3′-ends of mature tRNAs in the human primary-effusion lymphoma cell line BCP1 [56]. At least two 3′CCA-tRFs (derived from His(GTG) and Leu(CAG) tRNAs) produced in a Dicer-independent manner associate with Ago2 and direct Ago2-mediated cleavage of an mRNA reporter. Intriguingly, various 3′CCA-tRFs are complementary to the replication sites of human endogenous retroviruses (HERV), which exist in the form of long terminal repeats and comprise about 7% of the human genome [86]. Since retroviral elements and retroviruses (such as HIV) use the binding of tRNAs to retroviral primer binding sites (PBS) to initiate retroviral genome replication [87], it is possible that 3′CCA-tRFs can direct Ago2-mediated cleavage of retroviral RNAs and act as inhibitors of HERV replication. In fact, small ncRNA cloned from T cells infected with HIV-1 included an abundant 3′-CCA-tRF corresponding to an 18-nt fragment derived from tRNALys [63]. This tRF is complimentary to the PBS of HIV-1, is enriched only in infected T cells, and its levels correlate with HIV-1 expression.
Interestingly, another study describes a 22-nt 3′CCA-tRF (which they called CU1276, derived from tRNAGly) in mature B cells that has the functional characteristics of a microRNA [62]. CU1276 is produced in a Dicer-dependent manner, associates with all four human Argonautes (Ago 1–4) and functions as a miRNA. Moreover, in B cell lymphomas, the expression of CU1276 is down-regulated and its loss causes derepression of CU1276 endogenous targets including RPA1, a protein involved in DNA replication and repair. Further analysis suggests that CU1276 regulates cell proliferation and DNA damage in an RPA1-dependent manner in lymphoma cell lines. Decreased CU1276 expression in lymphomas may confer a selective growth advantage to malignant cells via increased RPA1 expression [62].
3.3. tRNA fragments and neurodegenerative diseases
Although tRNA metabolism is absolutely essential for all human cells, a number of neurological disorders are caused by defects in tRNA metabolism and tRNA processing enzymes. In 2006, ANG mutants possessing reduced ribonuclease (RNase) activity were implicated in the pathogenesis of Amyotrophic Lateral Sclerosis (ALS), a fatal neurodegenerative disease [88]. In 2012, a subset of ALS-associated ANG mutants was also found in Parkinson’s Disease (PD) patients [89]. Recombinant ANG is neuroprotective for cultured motor neurons [90,91] and administration of ANG to SOD1(G93A) mice, a standard laboratory model for ALS, significantly promotes both life-span and motor function [90]. As most ALS/PD-associated mutations are RNase loss-of-function mutations [92] and ANG-mediated neuroprotection is RNase-dependent, we speculate that they affect production of tiRNAs required for cell survival. Mechanistically, formation of tiRNAs may contribute to motor neuron survival via its ability to inhibit apoptosis [51] or promotion of SGs [30,32]. We are currently examining these possibilities.
Recent work from Blanco et al. further strengthens the link between ANG-induced tiRNAs, cellular stress and neurodevelopmental disorders [93]. Several mutations in the cytosine-5 RNA methyltransferase NSun2 have been identified to cause a syndromic form of intellectual disability (ID) and a Dubowitz-like syndrome in humans [94–96]. Human NSun2 methylates cytosine residues in the anticodon loop (position C34) and at the intersection of the variable loop and the T arm of tRNA (positions 48/49/50). The tRNA targets of NSun2 are limited to a subset of tRNA isotypes (Asp, Glu, Gly, His, Lys and Val). In the absence of NSun2, these tRNAs are non-methylated and prone to accumulate as 5′-tRNA fragments as a result of stress-induced ANG-mediated cleavage. Like classical 5′-tiRNAs, these fragments repress translation, and trigger a stress response and cell death in cortical, hippocampal and striatal neurons. As a consequence, NSun2 knockout mouse brains exhibit reduced neuronal size and impaired formation of synapses. Ultimately, these mechanisms may explain the general growth retardation and impaired intellectual development in patients with mutations of NSun2 gene [93].
tRNA fragments derived from intron-containing tRNAs are also strongly implicated in neurodegeneration [29,66,67]. Mutations in the CLP1 gene (R140A) encoding an RNA kinase involved in tRNA splicing, are found in patients with pontocerebellar hypoplasia (PCH), a heterogeneous group of inherited neurodegenerative disorders characterized by impaired development of various parts of the brain. Animal models of CLP1 deficiency (in both mice and zebrafish) phenocopy developmental and neuromuscular defects observed in PCH patients [29,66,67]. On the cellular level, the R140A mutation leads to the depletion of mature tRNAs, accumulation of unspliced pre-tRNAs in patient-derived neurons [67] and accumulation of linear introns [66]. Transfection of 5′-unphosphorylated tRF corresponding to the 3′-exon of pre-tRNATyr (the natural substrate of CLP1) into patient cells results in reduced neuron survival under oxidative stress compared to the transfection of the 5′-exon, which had no effect on viability [67]. The CLP1 kinase-dead mice accumulate 5′-leader-exon-tRFs that sensitize cells to oxidative stress and promote cell death in a p53-dependent fashion [29]. While the exact mechanistic details on the interplay between CLP1 activity, tRNA splicing and functions of pre-tRNA-derived tRFs are yet to be uncovered, these studies provide a basis for further investigations linking aberrant tRNA metabolism and development of neurodegeneration.
4. Conclusions and perspectives
The human genome encodes hundreds of tRNA genes and pseudogenes. A great number of tRNA genes possess the same anti-codon but different sequences in the tRNA body. The benefit of having a collection of tRNA isodecoders in translation is unclear. Such diversity in the nucleotide composition within the group of isodecoders may have important consequences for tRNA expression and/or allow the regulation of tRNA stability/pattern of nucleotide modifications in tissue-specific and developmental stage-specific manners. We propose here that tRNA diversity may also provide substrates for the production of diverse classes of small ncRNAs that have important effects on cell physiology.
The list of possible functions of tRNA-derived fragments is growing. The recent discovery that the abundance of 5′-tRNA halves found in sperm and oocytes rapidly decreases upon fertilization suggests that this class of molecules can be physiologically regulated [97]. Similarly, 5′-tRNA halves and 5′-tRFs are found in exosomes within semen [98]. The biogenesis and functions of these sperm-born tRNA fragments is an open question. Additionally, tRNAs and tRNA fragments were reported as novel sources of piRNAs (PIWI-interacting RNAs [1]) in both mouse gametes and zygotes [99] as well as in human somatic cells [100]. The functions of these tRNA-derived piRNAs are not known but may be connected to epigenetic inheritance, silencing of retrotransposons and other genetic elements or post-transcriptional regulation of mRNAs. Finally, tRNA fragments are also induced by viral infections such as by Respiratory Syncytial Virus (RSV) [101]. RSV activates ANG to produce tiRNAs, and selected 5′-tiRNAs (such as derived from tRNAGlu) suppress expression of target mRNAs. Significantly, induction of tRNA fragments is required for replication of RSV [101], although the exact molecular mechanisms of their involvement in RSV infection remain unknown. Whether similar mechanisms operate during infection by other viruses or pathogenic microorganisms remains to be determined.
Studies on tRNA-derived fragments are still at a very early stage. It is likely that additional functions for tRNA fragments will be uncovered in the near future. Improved understanding of the functions of tRNA-derived fragments will provide valuable insights into human physiology and pathophysiology.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, Carninci P, Hayashizaki Y. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giege R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- 5.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medha R, Ibba M. TRNAs as regulators of biological processes. Front Genet. 2014:5. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20:306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Abe T, Inokuchi H, Yamada Y, Muto A, Iwasaki Y, Ikemura T. TRNADB-CE: tRNA gene database well-timed in the era of big sequence data. Front Genet. 2014;5:114. doi: 10.3389/fgene.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parisien M, Wang X, Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013;10:1853–1867. doi: 10.4161/rna.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mleczko AM, Celichowski P, Bakowska-Zywicka K. Extranslational function of tRNAs and their fragments in cancer. Acta Biochim Pol. 2014;61:211–216. [PubMed] [Google Scholar]
- 13.Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 14.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194:43–67. doi: 10.1534/genetics.112.147470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA. 2011;2:362–375. doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio MA, Hopper AK. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip Rev RNA. 2011;2:802–817. doi: 10.1002/wrna.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty AK, Subbotin R, Chait BT, Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci USA. 2012;109:6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai KK, Cheng CL, Bingman CA, Phillips GN, Jr, Raines RT. A tRNA splicing operon: archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res. 2014;42:3931–3942. doi: 10.1093/nar/gkt1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mair B, Popow J, Mechtler K, Weitzer S, Martinez J. Intron excision from precursor tRNA molecules in mammalian cells requires ATP hydrolysis and phosphorylation of tRNA-splicing endonuclease components. Biochem Soc Trans. 2013;41:831–837. doi: 10.1042/BST20130025. [DOI] [PubMed] [Google Scholar]
- 22.Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 23.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, Martinez J. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popow J, Jurkin J, Schleiffer A, Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 27.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schutz K, Hesselberth JR, Fields S. Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA. 2010;16:621–631. doi: 10.1261/rna.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, Minis A, Sato F, Mimata H, Yoshimura A, Tamir I, Rainer J, Kofler R, Yaron A, Eggan KC, Woolf CJ, Glatzel M, Herbst R, Martinez J, Penninger JM. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474– 480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol. 2012;227:2822–2826. doi: 10.1002/jcp.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 2001;20:5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 37.Nekrasov MP, Ivshina MP, Chernov KG, Kovrigina EA, Evdokimova VM, Thomas AA, Hershey JW, Ovchinnikov LP. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J Biol Chem. 2003;278:13936–13943. doi: 10.1074/jbc.M209145200. [DOI] [PubMed] [Google Scholar]
- 38.Pisarev AV, Skabkin MA, Thomas AA, Merrick WC, Ovchinnikov LP, Shatsky IN. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40 S ribosomal subunits to the initiation codon of beta-globin mRNA. J Biol Chem. 2002;277:15445–15451. doi: 10.1074/jbc.M111954200. [DOI] [PubMed] [Google Scholar]
- 39.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19:R397–R398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov P, Kedersha N, Anderson P. Stress puts TIA on TOP. Genes Dev. 2011;25:2119–2124. doi: 10.1101/gad.17838411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci: CMLS. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Czech A, Wende S, Morl M, Pan T, Ignatova Z. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 2013;9:e1003767. doi: 10.1371/journal.pgen.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong Y, Steitz TA. A story with a good ending: tRNA 3′-end maturation by CCA-adding enzymes. Curr Opin Struct Biol. 2006;16:12–17. doi: 10.1016/j.sbi.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Pizzo E, Sarcinelli C, Sheng J, Fusco S, Formiggini F, Netti P, Yu W, D’Alessio G, Hu GF. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci. 2013;126:4308–4319. doi: 10.1242/jcs.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, Hu GF, Pusztai-Carey M, Gorla M, Sepuri NB, Pan T, Hatzoglou M. Angiogenin-Cleaved tRNA Halves Interact with Cytochrome c, Protecting Cells from Apoptosis during Osmotic Stress. Mol Cell Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. TRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suryanarayana T, Uppala JK, Garapati UK. Interaction of cytochrome c with tRNA and other polynucleotides. Mol Biol Rep. 2012;39:9187–9191. doi: 10.1007/s11033-012-1791-9. [DOI] [PubMed] [Google Scholar]
- 54.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787–6799. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gebetsberger J, Zywicki M, Kunzi A, Polacek N. TRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. TRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, Erdin S, Isikay S, Franco LM, Gonzaga-Jauregui C, Gambin T, Gelowani V, Hunter JV, Yesil G, Koparir E, Yilmaz S, Brown M, Briskin D, Hafner M, Morozov P, Farazi TA, Bernreuther C, Glatzel M, Trattnig S, Friske J, Kronnerwetter C, Bainbridge MN, Gezdirici A, Seven M, Muzny DM, Boerwinkle E, Ozen M, Clausen T, Tuschl T, Yuksel A, Hess A, Gibbs RA, Martinez J, Penninger JM, Lupski JR Baylor Hopkins Center for Mendelian G. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, Rosti RO, Abdellateef M, Caglar C, Kasher PR, Cazemier JL, Weterman MA, Cantagrel V, Cai N, Zweier C, Altunoglu U, Satkin NB, Aktar F, Tuysuz B, Yalcinkaya C, Caksen F, Bilguvar K, Fu XD, Trotta CR, Gabriel S, Reis A, Gunel M, Baas F, Gleeson JG. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mishima E, Inoue C, Saigusa D, Inoue R, Ito K, Suzuki Y, Jinno D, Tsukui Y, Akamatsu Y, Araki M, Araki K, Shimizu R, Shinke H, Suzuki T, Takeuchi Y, Shima H, Akiyama Y, Toyohara T, Suzuki C, Saiki Y, Tominaga T, Miyagi S, Kawagisihi N, Soga T, Ohkubo T, Yamamura K, Imai Y, Masuda S, Sabbisetti V, Ichimura T, Mount DB, Bonventre JV, Ito S, Tomioka Y, Itoh K, Abe T. Conformational Change in Transfer RNA Is an Early Indicator of Acute Cellular Damage. J Am Soc Nephrol: JASN. 2014 doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–R134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 70.Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Boffelli D, Mote P, Martin DI. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977;37:3362–3366. [PubMed] [Google Scholar]
- 72.Gehrke CW, Kuo KC, Waalkes TP, Borek E. Patterns of urinary excretion of modified nucleosides. Cancer Res. 1979;39:1150–1153. [PubMed] [Google Scholar]
- 73.Lakings DB, Waalkes TP, Borek E, Gehrke CW, Mrochek JE, Longmore J, Adamson RH. Composition, associated tissue methyltransferase activity, and catabolic end products of transfer RNA from carcinogen-induced hepatoma and normal monkey livers. Cancer Res. 1977;37:285–292. [PubMed] [Google Scholar]
- 74.Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. TRNA breakdown products as markers for cancer. Cancer. 1979;44:2120–2123. doi: 10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 75.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. TRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun. 2009;385:160–164. doi: 10.1016/j.bbrc.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao X, Xu Z. Mechanisms of action of angiogenin. Acta Biochim Biophys Sin. 2008;40:619–624. doi: 10.1111/j.1745-7270.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 80.Tello-Montoliu A, Patel JV, Lip GY. Angiogenin: a review of the pathophysiology and potential clinical applications. J Thromb Haemost. 2006;4:1864–1874. doi: 10.1111/j.1538-7836.2006.01995.x. [DOI] [PubMed] [Google Scholar]
- 81.Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2002;294:287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 82.Shapiro R, Vallee BL. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 83.Kao RY, Jenkins JL, Olson KA, Key ME, Fett JW, Shapiro R. A small-molecule inhibitor of the ribonucleolytic activity of human angiogenin that possesses antitumor activity. Proc Natl Acad Sci USA. 2002;99:10066–10071. doi: 10.1073/pnas.152342999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, Desrochers M, Dumont M, Farnham JM, Frank D, Frye C, Ghaffari S, Gupte JS, Hu R, Iliev D, Janecki T, Kort EN, Laity KE, Leavitt A, Leblanc G, McArthur-Morrison J, Pederson A, Penn B, Peterson KT, Reid JE, Richards S, Schroeder M, Smith R, Snyder SC, Swedlund B, Swensen J, Thomas A, Tranchant M, Woodland AM, Labrie F, Skolnick MH, Neuhausen S, Rommens J, Cannon-Albright LA. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 85.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 86.Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr Opin Genet Dev. 2000;10:651–655. doi: 10.1016/s0959-437x(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 87.Das AT, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA(3Lys) J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr, Hardiman O. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 89.van Es MA, Schelhaas HJ, van Vught PW, Ticozzi N, Andersen PM, Groen EJ, Schulte C, Blauw HM, Koppers M, Diekstra FP, Fumoto K, LeClerc AL, Keagle P, Bloem BR, Scheffer H, van Nuenen BF, van Blitterswijk M, van Rheenen W, Wills AM, Lowe PP, Hu GF, Yu W, Kishikawa H, Wu D, Folkerth RD, Mariani C, Goldwurm S, Pezzoli G, Van Damme P, Lemmens R, Dahlberg C, Birve A, Fernandez-Santiago R, Waibel S, Klein C, Weber M, van der Kooi AJ, de Visser M, Verbaan D, van Hilten X, Heutink P, Hennekam EA, Cuppen E, Berg D, Brown RH, Jr, Silani V, Gasser T, Ludolph AC, Robberecht W, Ophoff RA, Veldink JH, Pasterkamp RJ, de Bakker PI, Landers JE, van de Warrenburg BP, van den Berg LH. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol. 2013;70:964–973. doi: 10.1002/ana.22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kieran D, Sebastia J, Greenway MJ, King MA, Connaughton D, Concannon CG, Fenner B, Hardiman O, Prehn JH. Control of motoneuron survival by angiogenin. J Neurosci. 2008;28:14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sebastia J, Kieran D, Breen B, King MA, Netteland DF, Joyce D, Fitzpatrick SF, Taylor CT, Prehn JH. Angiogenin protects motoneurons against hypoxic injury. Cell Death Differ. 2009;16:1238–1247. doi: 10.1038/cdd.2009.52. [DOI] [PubMed] [Google Scholar]
- 92.Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, Xin W, Sims K, Hu GF. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol. 2007;62:609–617. doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Holter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus-Durner V, Hrabe de Angelis M, Karadottir RT, Helm M, Ule J, Gleeson JG, Odom DT, Frye M. Aberrant methylation of tRNAs links cellular stress to neurodevelopmental disorders. EMBO J. 2014 doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K, Weissmann R, Tzschach A, Garshasbi M, Abedini SS, Najmabadi H, Ropers HH, Sigrist SJ, Kuss AW. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, Ishak GE, Doherty D, Weksberg R, Ayub M, Windpassinger C, Ibrahim S, Frye M, Ansar M, Vincent JB. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al-Gazali L, Gleeson JG. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012;49:380–385. doi: 10.1136/jmedgenet-2011-100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, Zhou Q, Chen Q, Duan E. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garcia-Lopez J, deHourcade JD, Alonso L, Cardenas DB, del Mazo J. Global characterization and target identification of piRNAs and endo-siRNAs in mouse gametes and zygotes. Biochim Biophys Acta. 2014;1839:463– 475. doi: 10.1016/j.bbagrm.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 100.Keam SP, Young PE, McCorkindale AL, Dang TH, Clancy JL, Humphreys DT, Preiss T, Hutvagner G, Martin DI, Cropley JE, Suter CM. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther: J Am Soc Gene Ther. 2013;21:368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]


