Abstract
Context
Pseudohypoparathyroidism type 1b (PHP-Ib) is characterized by renal resistance to PTH (and, sometimes, a mild resistance to TSH) and absence of any features of Albright's hereditary osteodystrophy. Patients with PHP-Ib suffer of defects in the methylation pattern of the complex GNAS locus. PHP-Ib can be either sporadic or inherited in an autosomal dominant pattern. Whereas familial PHP-Ib is well characterized at the molecular level, the genetic cause of sporadic PHP-Ib cases remains elusive, although some molecular mechanisms have been associated with this subtype.
Objective
The aim of the study was to investigate the molecular and imprinting defects in the GNAS locus in two unrelated patients with PHP-Ib.
Design
We have analyzed the GNAS locus by direct sequencing, Methylation-Specific Multiplex Ligation-dependent Probe Amplification, microsatellites, Quantitative Multiplex PCR of Short Fluorescent fragments and array-Comparative Genomic Hybridization studies in order to characterize two unrelated families with clinical features of PHP-Ib.
Results
We identified two duplications in the GNAS region in two patients with PHP-Ib: one of them, comprising ∼320 kb, occurred ‘de novo’ in the patient, whereas the other one, of ∼179 kb in length, was inherited from the maternal allele. In both cases, no other known genetic cause was observed.
Conclusion
In this article, we describe the to-our-knowledge biggest duplications reported so far in the GNAS region. Both are associated to PHP-Ib, one of them occurring ‘de novo’ and the other one being maternally inherited.
Introduction
Pseudohypoparathyroidism (PHP) is a group of rare endocrine diseases characterized by hypocalcemia, hyperphosphatemia and an elevation of PTH values due to a variable resistance to this hormone in its target organs, mainly the proximal renal tubule [1]. Its exact prevalence in diverse populations is currently unknown, although it has been estimated to be 0.79/100.000 (according to Orphanet Report Series (http://www.orpha.net/consor/cgi-bin/index.php) and [2]).
Depending on the associated phenotype and biochemical features, PHP is divided in different subtypes: PHP-Ia (OMIM#103580), PHP-Ib (OMIM#603233), PHP-Ic (OMIM#612462), PHP-II (OMIM%203330) [2]. PHP type I is caused by defects at the GNAS locus (20q13.2–13.3), a complex cluster which generates five transcripts using alternative first exons. Between them, the alpha subunit of the heterotrimeric stimulatory G protein (Gsα) [3], which is a signaling protein essential for the actions of PTH and many other hormones (TSH, glucagon, gonadotropins,…) through the adenylyl cyclase pathway [4].
As mentioned above, GNAS is a highly complex imprinted locus that generates multiple gene products through the use of multiple promoters and first exons that splice onto a common set of downstream exons. Moreover, these transcripts can be expressed by the maternal, the paternal or both alleles as the locus is regulated by imprinting mechanisms [5] (Fig. 1A).
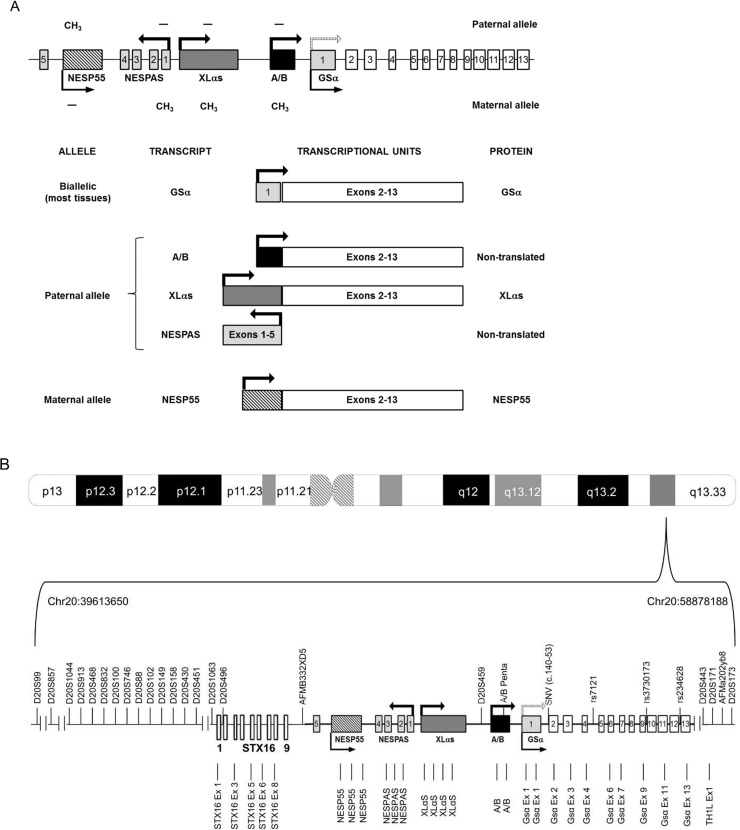
Fig 1. Graphic representation of the GNAS complex locus.
(A) Alternative exons and transcripts in the region. Each first alternative exon is depicted in a different format (the scratched rectangle corresponds to NESP55, the dark grey rectangle to XLαs, the black rectangle to exon A/B and the dotted rectangle to GSα exon 1). NESPAS exons 1–5 are shown as soft grey rectangles. White rectangles represent common exons 2–13 and the continuous line represents introns. Arrows indicate the origin and sense of the transcription (dotted arrow indicates that paternal GSα is not expressed in all tissues). Methylated regions are marked with a methyl group. (B) Probes and markers used for the analysis. Polymorphic markers are depicted above, MS-MLPA probes below. See text for details. Drawings not to scale.
Patients with PHP-Ia present a characteristic phenotype called Albright’s hereditary osteodystrophy (AHO), which comprises short stature, rounded face, brachydactyly, ectopic ossifications and/or mental retardation. There is a minor response in cAMP production after exogenous PTH administration and a variable resistance to multiple hormones that act through the G protein (TSH, glucagon, gonadotropins, ACTH, GHRH…). At a molecular level, most of PHP-Ia cases are caused by haploinsufficiency because of maternally-inherited heterozygous inactivating mutations in Gsα [6]. On the contrary, when the germline mutation is inherited from the paternal allele, patients suffer of pseudo-pseudohypoparathyrodism (P-PHP, OMIM#612463). These patients present with AHO features but do not have biochemical alterations. This occurs because Gsα is predominantly transcribed from the maternal allele in the tissues where PTH, TSH, LH, FSH act (proximal renal tubule, thyroid, ovary, pituitary), so they exert their actions normally if the mutation is transmitted by the paternal allele [7].
PHP-Ib is characterized by renal resistance to PTH and absence of AHO phenotype and resistance to other hormones (although there is sometimes a mild resistance to TSH) [8]. There is also a minor cAMP production after PTH administration. PHP-Ib is due to changes in the methylation pattern of the GNAS locus [9]. There is a loss of methylation in the maternal exon A/B, sometimes combined with other epigenetic defects in other regions in the locus. It can be inherited in an autosomal dominant pattern (AD-PHP-Ib), although most of the cases are sporadic [2]. AD-PHP-Ib is typically characterized by an isolated loss of methylation at exon A/B, associated with microdeletions in the STX16 gene, encoding syntaxin 16, 220 kb upstream of GNAS [10–12]. Less frequently, AD-PHP-Ib is caused by other microdeletions affecting NESP55 and NESPAS differentially methylated regions (DMRs) [13–15], causing broad loss of imprinting at the GNAS locus. Sporadic cases also show broad imprinting abnormalities at the NESPAS, XLαs and A/B DMRs, with no other changes in cis- or trans-acting elements explaining these phenomena up to now. Some PHP-Ib cases present paternal 20q disomies as the cause of the disease [16]). Moreover, an autosomal recessive form has also been hypothesized to explain the molecular mechanism of sporadic PHP-Ib in some families [17].
Recent articles have described methylation defects in some patients with PHP and AHO phenotype [18–21], an overlapping that has also been referred about the activity of the Gsα protein [22], indicating a possible molecular and clinical superposition between PHP-Ia and PHP-Ib [23].
Few genomic rearrangements have been described at the GNAS locus, such as terminal deletions [24] or an inversion [25], both of them related to PHP-Ia patients. To our knowledge, there is only a report describing an intralocus duplication in GNAS in an AD-PHP-Ib kindred [13]. In this article, we report the biggest duplications detected so far in the GNAS region, both associated to a PHP-Ib phenotype and arising in the maternal allele. One of them (which occurred ‘de novo’ in the patient) was associated with a complete loss of methylation at the NESPAS, XLαs and A/B DMRs, whereas the other one (present both in the index case and in her mother) was concomitant with an isolated loss of methylation in exon A/B. We did not detect any other change in the locus in either patient that could explain the differences in the methylation status, although the existence of a still unknown agent co-responsible for the (epi)phenotype cannot be discarded.
Patients
Case 1
The first case (GS0131) is a 16-year-old male, the only child of healthy unrelated parents, born after a normal pregnancy and a normal delivery (weight 3200 g, length: 51 cm). The mother was 26 and the father 27 years of age at the time of the pregnancy (mother’s height 157cm, father’s height 175cm). No familial history of PHP was reported. He has been properly vaccinated, with a good psychomotor development, with weight and height in the range of standard values. He has passed through an ordinary and complete puberty, and is phenotypically normal, with no symptoms of Albright’s hereditary osteodystrophy.
At age 14 years, he presented with generalized convulsions. After an altered electrocardiography was observed in the Emergency Department, he was referred to the Pediatric Cardiology Unit. All analyses were normal except for a long Q-T segment. Referred to Neuropediatrics, he was treated with different anticonvulsants, presenting a normal Magnetic Resonance Imaging (MRI). The electroencephalography showed normal but he still suffered of convulsive crisis. A scanner revealed calcifications and a biochemical evaluation showed the following values: Ca, 6.5mg/dl; P, 7.1mg/dl; PTH, 621.9pg/dl. One month later, hypocalcemia, hyperphosphatemia and PTH high values persisted (Ca, 6.1mg/dl; P, 7.8mg/dl; PTH, 884pg/dl), the patient being diagnosed of pseudohypoparathyroidism. He started oral treatment with 500 mg of calcium carbonate and 0.5 μg of calcitriol each 24 hours. Calcium and phosphate values as well as electrocardiographic alterations normalized in the first month of treatment. He is properly following the treatment and all biochemical controls have turned out to be normal, with no need of changes in the doses. He has not presented any intercurrent disease. The thyroid function analysis showed a slight hyperthyrotropinemia in the beginning, which settled down later.
The last physical evaluation (16y 5m) showed a height of 169.9cm (p25–50) and a weight of 61kg (p25-p50) (BMI: 21.1).
Case 2
The second case (GS0155) is a 27-year-old female. She had had some episodes of right hemiparesis and a bowel volvulus in pediatric age. She was studied in the Neurology Unit after an episode of right hemiparesis, paresthesia and a visual field deficit. Imaging studies did not show any significant alteration. Biochemical analysis showed calcium values in the lower limit of normality (8.5–9.3mg/dl) and elevated PTH (200–300pg/ml), so she was diagnosed of pseudohypoparathyroidism. She did not present any symptom of Albright’s hereditary osteodystrophy (height 1.65m; weight 68.5kg; BMI: 25.1). No other hormone deficit was detected (TSH, 3μU/ml). She referred regular periods. Calcium and vitamin D supplements (both cholecalciferol and calcifediol) were prescribed, although she is not properly following the treatment.
In the last visit, she referred paresthesia in upper extremities with no calcium supplementation for a month. She brought a report with analytical levels: Ca, 8.7mg/dl; P, 3.9mg/dl; Mg, 2.4mg/dl, TSH, 4.15μU/ml; PTH, 349pg/dl; 25(OH)D, 37.2 ng/ml; urine Ca, 76mg/day; CLcr, 177ml/min; Dual-energy X-ray absorptiometry (DXA) with a lumbar T score of -0.7 SD and a femoral T score of -0.3 SD. She is clinically stable, with no symptoms of latent tetany. Biochemical analysis were performed: Ca, 8.9mg/dl; P, 3.7mg/dl; HbA1c 5.4%, PTH, 417pg/dl; 25(OH)D, 44 ng/ml.
Her mother was diagnosed at age 48 years with a multinodular toxic goitre. She was treated with I131 that caused iatrogenic hypothyroidism, so she was prescribed levothyroxine 125μg/day. Her past clinical history showed symptoms of generalized anxiety, being followed in the Mental Health Unit. She does not present any characteristic phenotype, with a normal physic exploration (height 1.64m; weight 70kg; BMI: 26).
Methods
Ethics Statement
The study was approved by the Comité Ético de Investigación Clínica (CEIC) from the Hospital Universitario Cruces. The patients and their relatives provided their written informed consent to participate in this study. In case of the minor included in the cohort, we obtained written informed consent from his guardians prior to enrolment in the study.
Structural analysis of the GNAS locus
Peripheral blood samples from the probands and their parents were collected on EDTA tubes. DNA was extracted from whole blood using QIAamp DNA Blood Minikit (Qiagen) according to the manufacturer’s specifications. The structural analysis of the 13 Gsα exons and GNAS exon A/B was performed by polymerase chain reaction (PCR) amplification, ExoSap (USB) purification and sequencing of both strands on an ABI3130xl Genetic Analyzer (Life Technologies). Sequences were analyzed with Sequencing Analysis v.5.2 software (Life Technologies) and compared with the reference sequence (Ensembl identifier: GNAS locus ENSG00000087460) using SeqScape v.2.5 software (Life Technologies). Primer sequences and PCR conditions are available on request.
Methylation analysis of the GNAS cluster by MS-MLPA
Dosage and methylation analyses of the GNAS locus were studied by methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) using the ME031-A2 kit (MRC-Holland) [26]. This SALSA MS-MLPA probemix is intended to provide information on deletions, duplications and abnormal methylation of sequences in the 20q13.32 GNAS region. The ME031-A2 GNAS probemix contains 27 probes specific for the complex GNAS locus and one for the TH1L gene (located 90 kb q-telomeric of GNAS) (Table 1, Fig. 1B). Of these, 17 are MS-MLPA probes containing a HhaI recognition site specific to the differentially methylated regions of the locus GNAS: XLαs, NESP55, NESPAS and GNAS exon A/B and one in STX16 (located 160 kb p-telomeric of the GNAS locus) and in TH1L. Furthermore, 6 regular MLPA probes (no HhaI site) specific for GNAS and 4 probes for the STX16 gene are included.
Table 1. Genomic location of the probes used for the MS-MLPA analysis and the 27 polymorphic markers in chromosome 20q mentioned in the text.
| Probe number (MS-MLPA) | Marker/Probe | Genomic location GRCh37/hg19 coordinates |
|---|---|---|
| D20S99 | Chr20:39613650–39613812 | |
| D20S857 | Chr20:50108271–50108486 | |
| D20S1044 | Chr20:52452369–52452578 | |
| D20S913 | Chr20:52501053–52501308 | |
| D20S468 | Chr20:53459303–53459503 | |
| D20S832 | Chr20:53895474–53895673 | |
| D20S100 | Chr20:54314065–54314276 | |
| D20S746 | Chr20:54401983–54402234 | |
| D20S88 | Chr20:54903235–54903696 | |
| D20S102 | Chr20:54991350–54991700 | |
| D20S149 | Chr20:55514140–55514427 | |
| D20S158 | Chr20:55589222–55589604 | |
| D20S430 | Chr20:56150061–56150259 | |
| D20S451 | Chr20:56664529–56664842 | |
| D20S1063 | Chr20:57201940–57202140 | |
| P1 | STX16 Ex1 | Chr20:57226476–57226533 |
| D20S496 | Chr20:57236177–57236381 | |
| P2 | STX16 Ex3 | Chr20:57242563–57242626 |
| P3 | STX16 Ex5 | Chr20:57244321–57244372 |
| P4 | STX16 Ex6 | Chr20:57245568–57245646 |
| P5 | STX16 Ex8 | Chr20:57252459–57252516 |
| AFMB332XD5 | Chr20:57257444–57257525 | |
| P6 | NESP55 | Chr20:57414748–57414799 |
| P7 | NESP55 | Chr20:57414927–57415009 |
| P8 | NESP55 | Chr20:57415142–57415234 |
| P9 | NESPAS Ex1 | Chr20:57425810–57425873 |
| P10 | NESPAS Ex1 | Chr20:57425907–57426001 |
| P11 | NESPAS Int1 | Chr20:57426013–57426065 |
| P12 | XLαs | Chr20:57429229–57429286 |
| P13 | XLαs | Chr20:57429288–57429336 |
| P14 | XLαs | Chr20:57430117–57430171 |
| P15 | XLαs | Chr20:57430201–57430258 |
| D20S459 | Chr20:57439859–57440088 | |
| P16 | A/B | Chr20:57464100–57464159 |
| A/B Pentan | Chr20:57464278–57464282 | |
| P17 | A/B | Chr20:57464367–57464424 |
| P18 | GSα Ex1 | Chr20:57466769–57466826 |
| P19 | GSα Ex1 | Chr20:57466829–57466892 |
| SNV (c.140–53) | Chr20: 57470614 | |
| P20 | GSα Ex2 | Chr20:57470713–57470779 |
| P21 | GSα Ex3 | Chr20:57474005–57474058 |
| P22 | GSα Ex4 | Chr20:57478577–57478646 |
| rs7121 | Chr20:57478807 | |
| P23 | GSα Ex6 | Chr20:57480450–57480516 |
| P24 | GSα Ex7 | Chr20:57484181–57484241 |
| P25 | GSα Ex9 | Chr20:57484583–57484643 |
| rs3730173 | Chr20:57484934 | |
| P26 | GSα Ex11 | Chr20:57485057–57485131 |
| rs234628 | Chr20:57485611 | |
| P27 | GSα Ex13 | Chr20:57485737–57485800 |
| D20S443 | Chr20:57495457–57495600 | |
| P28 | TH1L Ex1 | Chr20:57556345–57556400 |
| D20S171 | Chr20:57808030–57808168 | |
| AFMa202yb9 | Chr20:57975178–57975420 | |
| D20S173 | Chr20:58878008–58878188 |
Genomic coordinates (according to GRCh37/hg19 build) of the 28 probes used for the MS-MLPA analysis and the 27 polymorphic markers in chromosome 20q. They are situated in the table according to their chromosomal location. In the case of the MS-MLPA probes, “Probe numbers” indicate the reference used in Fig. 2. Probes highlighted in bold (P6 to P17) are used to identify methylation aberrations in the locus.
The protocol was implemented following the manufacturer's recommendations. Analysis of the MS-MLPA PCR products was performed on an ABI3130xl genetic analyzer using GeneMapper v4.0 software (Life Technologies). Each MS-MLPA reaction generates two samples. For copy number analysis, data generated by the undigested sample was intra-normalized by dividing the peak area of each amplification product by the total area of the reference probes only. The ratios were then obtained by dividing the intra-normalized probe ratio in a sample by the average intra-normalized probe ratio of all reference runs. For methylation analysis (digested sample), the intra-normalized peak area of each MS-MLPA probe from the digested sample was divided by the value obtained for the undigested sample.
Analysis of polymorphic markers
To characterize chromosome 20q and rule out any uniparental disomy (UPD) at this locus, twenty three polymorphic markers (D20S99, D20S857, D20S1044, D20S913, D20S468, D20S832, D20S100, D20S746, D20S88, D20S102, D20S149, D20S158, D20S430, D20S451, D20S1063, D20S496, AFMB332XD5, D20S459, D20S443, D20S171, AFMa202yb9, D20S173 and exon A/B pentanucleotide repeat) were typed by fluorescent PCR and electrophoresis on an ABI3130xl Genetic Analyzer using GeneMapper v.4.0 software (Life Technologies) (primers and conditions are available on request). Further, four single nucleotide variations (SNV) were also typed and used as markers during the structural analysis of Gsα (c.140–53, rs7121, rs3730173 and rs234628). The chromosomal location of all these markers is indicated in Table 1 and Fig. 1B.
Whole genome and 20q13-ter Comparative Genomic Hybridization (CGH) arrays
Detection of gene copy number was performed by array-Comparative Genomic Hybridization (a-CGH) experiments. Whole genome analysis was conducted using a human whole genome 400k array (SurePrint G3 Human CGH microarray 2x400, AMADID 021850, Agilent Technologies, Santa Clara, CA), following standard and manufacturer’s protocol [27]. For the analysis of the region chr20:47897125–62435965, the array was designed using Agilent’s e-array tool (Agilent Technologies) following the manufacturer’s instruction (https://earray.chem.agilent.com/). In both cases, healthy male and female donor samples were used as hybridization controls.
Microarray data were extracted and visualized using Feature Extraction software v10.7 and Agilent Genomic Workbench v5.0 (Agilent Technologies). Copy number altered regions were analyzed using ADAM-2 (set as 6) statistic provided by DNA Analytics, with a minimum number of 5 consecutive probes (the analysis resolution, thus, is approximately 25 kb for most of the regions for the whole genome array and 400 bp for the custom array). Genomic build hg19 was used for the experiments.
STX16 deletions
All samples from both families were screened for the 3-kb and 4.4-kb STX16 deletions by PCR and agarose gel electrophoresis followed by ethidium bromide staining, as previously described [10, 11], with slight modifications in the PCR conditions.
Determination of the parental origin of the duplication in family GS0131
D20S443, D20S459 and D20S1063 microsatellites, located inside the duplicated region (chr20:57381447–57701869), were genotyped by fluorescent PCR and capillary electrophoresis in order to study the inheritance of the alleles. Only D20S459 turned out to be informative (see Table 2). Quantitative multiplex PCR of short fluorescent fragments (QMPSF) was subsequently carried out for this marker together with amplicons from AIRE (21q22), HNF1A (12q24) and/or HNF1B (17q12) genes in a FAM-labeled multiplex PCR under semiquantitative conditions. Resulting PCR products were analyzed on an ABI3130xl Genetic Analyzer using GeneMapper v.4.0 software (Life Technologies). Data analysis was performed in Excel (Microsoft Office 2007).
Table 2. Genotyping results of the 27 polymorphic markers at 20q in both families.
| GS0131 | GS0155 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker/Probe | Index | Mother | Father | Result | Index | Mother | Father | Result | ||||||
| D20S99 | 161 | 159 | 159 | 161 | 161 | 161 | Excludes isodisomy | 161 | 161 | 161 | 161 | 161 | 165 | Excludes paternal heterodisomy |
| D20S857 | 208 | 208 | 208 | 216 | 208 | 214 | Excludes heterodisomy | 198 | 216 | 216 | 216 | 198 | 212 | Informative |
| D20S1044 | 212 | 212 | 212 | 212 | 212 | 212 | Uninformative | 212 | 212 | 212 | 212 | 212 | 212 | Uninformative |
| D20S913 | 255 | 257 | 257 | 257 | 255 | 255 | Informative | 255 | 255 | 255 | 257 | 255 | 265 | Excludes heterodisomy |
| D20S468 | 207 | 207 | 207 | 207 | 207 | 201 | Excludes paternal heterodisomy | 208 | 192 | 192 | 208 | 208 | 202 | Excludes isodisomy |
| D20S832 | 210 | 210 | 210 | 210 | 210 | 212 | Excludes paternal heterodisomy | 212 | 210 | 210 | 210 | 212 | 210 | Excludes isodisomy |
| D20S100 | 203 | 207 | 207 | 203 | 203 | 203 | Excludes isodisomy | 197 | 205 | 205 | 207 | 197 | 207 | Informative |
| D20S746 | 253 | 253 | 253 | 253 | 253 | 253 | Uninformative | 253 | 253 | 253 | 253 | 253 | 253 | Uninformative |
| D20S88 | 445 | 431 | 431 | 443 | 445 | 441 | Informative | 441 | 437 | 437 | 429 | 441 | 433 | Informative |
| D20S102 | 180 | 182 | 182 | 182 | 180 | 180 | Informative | 180 | 180 | 180 | 180 | 180 | 182 | Excludes paternal heterodisomy |
| D20S149 | 296 | 288 | 288 | 308 | 296 | 280 | Informative | 288 | 296 | 296 | 284 | 288 | 288 | Informative |
| D20S158 | 388 | 392 | 392 | 392 | 388 | 382 | Informative | 378 | 382 | 382 | 386 | 378 | 382 | Excludes isodisomy |
| D20S430 | 204 | 224 | 224 | 230 | 204 | 204 | Informative | 204 | 204 | 204 | 216 | 204 | 208 | Excludes heterodisomy |
| D20S451 | 314 | 304 | 304 | 322 | 314 | 318 | Informative | 304 | 314 | 314 | 296 | 304 | 304 | Informative |
| D20S1063 | 202 | 202 | 202 | 202 | 202 | 203 | Excludes paternal heterodisomy | 202 | 202 | 202 | 202 | 202 | 202 | Uninformative |
| D20S496 | 205 | 205 | 205 | 205 | 205 | 205 | Uninformative | 205 | 205 | 205 | 205 | 205 | 205 | Uninformative |
| AFMB332XD5 | 75 | 75 | 75 | 75 | 75 | 75 | Uninformative | 75 | 75 | 75 | 75 | 75 | 75 | Uninformative |
| D20S459 | 237 | 231 | 231 | 235 | 237 | 231 | Excludes isodisomy | 231 | 231 | 231 | 231 | 231 | 231 | Uninformative |
| A/B Pentan | 3 | 3 | 3 | 2 | 3 | 2 | Excludes heterodisomy | 2 | 3 | 3 | 3 | 2 | 3 | Excludes isodisomy |
| SNV (c.140–53) | C | T | C | T | C | T | Excludes isodisomy | C | T | T | C | C | C | Excludes isodisomy |
| rs7121 | C | C | C | T | C | C | Excludes maternal heterodisomy | T | T | T | C | T | C | Excludes heterodisomy |
| rs3730173 | C | C | C | C | C | C | Uninformative | T | C | C | C | T | C | Excludes isodisomy |
| rs234628 | T | T | T | C | T | C | Excludes heterodisomy | C | T | T | C | C | C | Excludes isodisomy |
| D20S443 | 139 | 139 | 139 | 135 | 139 | 139 | Excludes maternal heterodisomy | 139 | 139 | 139 | 135 | 139 | 135 | Excludes heterodisomy |
| D20S171 | 132 | 128 | 128 | 134 | 132 | 132 | Informative | 132 | 138 | 132 | 138 | 132 | 138 | Excludes isodisomy |
| AFMa202yb9 | 243 | 243 | 243 | 243 | 243 | 243 | Uninformative | 243 | 243 | 243 | 243 | 243 | 251 | Excludes paternal heterodisomy |
| D20S173 | 182 | 184 | 184 | 182 | 182 | 182 | Excludes isodisomy | 184 | 182 | 182 | 182 | 184 | 182 | Excludes isodisomy |
Fluorescent PCR and genotyping results of the 27 polymorphic markers in chromosome 20q, situated in the table according to their chromosomal location. Figures indicate the obtained size of the amplicons, or the number of pentanucleotide repeats, in the case of the A/B pentanucleotide marker. In all cases, first columns indicate paternally inherited alleles; the second columns indicate maternally inherited alleles (in the cases in which alleles are not directly informative, this inheritance has been assigned assuming both paternal alleles are present; see text for details). It was not possible to determine the inheritance of the single nucleotide variation (SNV) c.140–53. Bold figures indicate informative markers.
Results
Structural analysis of the GNAS locus
As expected, neither patient GS0131 nor GS0155 present any point mutation along the GNAS gene.
MS-MLPA
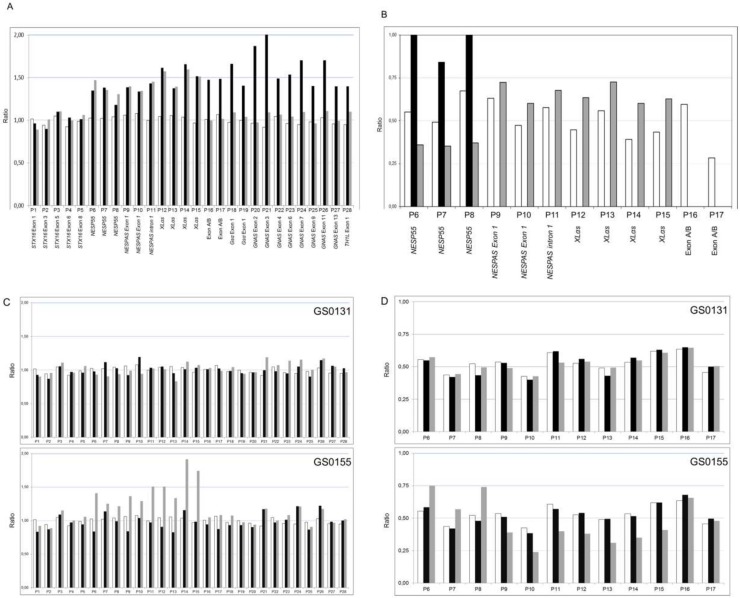
Dosage analysis of the GNAS locus by MS-MLPA revealed that GS0131 carried a duplication which included the 23 probes corresponding to the region from NESP55 to TH1L (Fig. 2A, black bars). Methylation analysis with the same technique revealed a complete alteration of the maternal methylation pattern (loss of methylation at exon A/B, XLαs and NESPAS and a gain of methylation at exon NESP55) (Fig. 2B, black bars).
Fig 2. Dosage and methylation analysis of the GNAS locus.
In all cases, on the X-axis, MS-MLPA probes for copy number determination (P1 to P28) or methylation analysis (P6 to P17) are shown according to their chromosome location (detailed in Table 1). On the Y-axis, the final probe ratio is shown. (A) Dosage analysis of the GNAS locus in GS0131 (black bars) and GS0155 (grey bars) compared against the average of four controls (white bars). (B) Methylation analysis of the GNAS locus in GS0131 (black bars) and GS0155 (grey bars) compared against the average of four controls (white bars). GS0131 shows complete loss of methylation at NESPAS, XLαs and exon A/B, and gain of methylation at NESP55, whereas GS0155 shows complete loss of methylation at A/B, partial gain at XLαs and NESPAS and partial loss at NESP55. (C) Dosage analysis of the GNAS locus in GS0131 parents (upper panel) and GS0155 parents (lower panel) compared against the average of four controls. In all cases, black bars represent the father, grey bars represent the mother and white bars represent the controls. Mother of GS0155 presents the same duplication as her daughter, whereas absence of alterations in GS0131 parents demonstrates its ‘de novo’ origin. (D) Methylation analysis of the GNAS locus in GS0131 parents (upper panel) and GS0155 parents (lower panel) compared against the average of four controls. In all cases, black bars represent the father, grey bars represent the mother and white bars represent the controls. There is not any methylation alteration in the parents of GS0131. Mother of GS0155 presents a partial loss of methylation in XLαs (P12-P15) and NESPAS (P9-P11), and a partial gain of methylation in NESP55 (P6-P8), exon A/B not being affected. See text for details.
The MS-MLPA study of his parents revealed that the duplication occurs ‘de novo’ in GS0131, as we could not see any alteration occurring in either parent. Both their gene dosage and their methylation pattern were normal (Fig. 2C and 2D, upper panels).
In the second family, dosage analysis of the GNAS locus by MS-MLPA showed that GS0155 presented with a duplication affecting 10 probes from NESP55 to XLαs, and not affecting exon A/B or the GSα promoter (Fig. 2A, grey bars). Regarding methylation, she suffered a complete loss of methylation at exon A/B, a partial gain of methylation at exons XLαs and NESPAS and a partial loss of methylation at NESP55 (Fig. 2B, grey bars).
In this case, the study of her parents revealed that GS0155 inherits the duplication from her mother, who also had 10 affected probes (Fig. 2C, lower panel, grey bars). GS0155’s mother also presents an altered methylation pattern (Fig. 2D, lower panel, grey bars), with a partial loss of methylation at exons XLαs and NESPAS (probes P9 to P15), and a partial gain at NESP55 (probes P6-P8). In this case, however, she does not have any epigenetic alteration in exon A/B (probes P16 and P17). GS0155’s father has normal dosage and methylation patterns (Fig. 2C and 2D, lower panels, black bars).
As suggested by Mantovani et al [2], methylation status was validated by methylation-specific PCR (data not shown). MS-PCR confirmed the observed results in GS0131, and the complete loss of methylation at exon A/B in GS0155, while partial methylation results in the family, although apparent, were a little bit more difficult to interpret. We intend to perform pyrosequencing analysis of the methylation status in both families in the future, in order to triple check the obtained results.
Analysis of polymorphic markers
We then characterized twenty seven polymorphic markers in chromosome 20q to rule out the possibility of a uniparental disomy occurring simultaneously with any of both duplications, as it has been described for PHP-Ib [15]. Although several markers are not directly informative about their inheritance, some can individually exclude either parental isodisomies or heterodisomies, whereas the combination of all the results let us assume both paternal alleles where present in the index cases (Table 2).
Whole genome and 20q13-ter Comparative Genomic Hybridization (CGH) arrays
In order to check whether there was any big rearrangement in the genome of these families, we performed a whole genome aCGH analysis, which showed there was no other genomic alteration in either patient or their parents, apart from the 20q duplication. Besides, analysis of the parents confirmed that the duplication occurred ‘de novo’ in GS0131 and was inherited from the mother in GS0155.
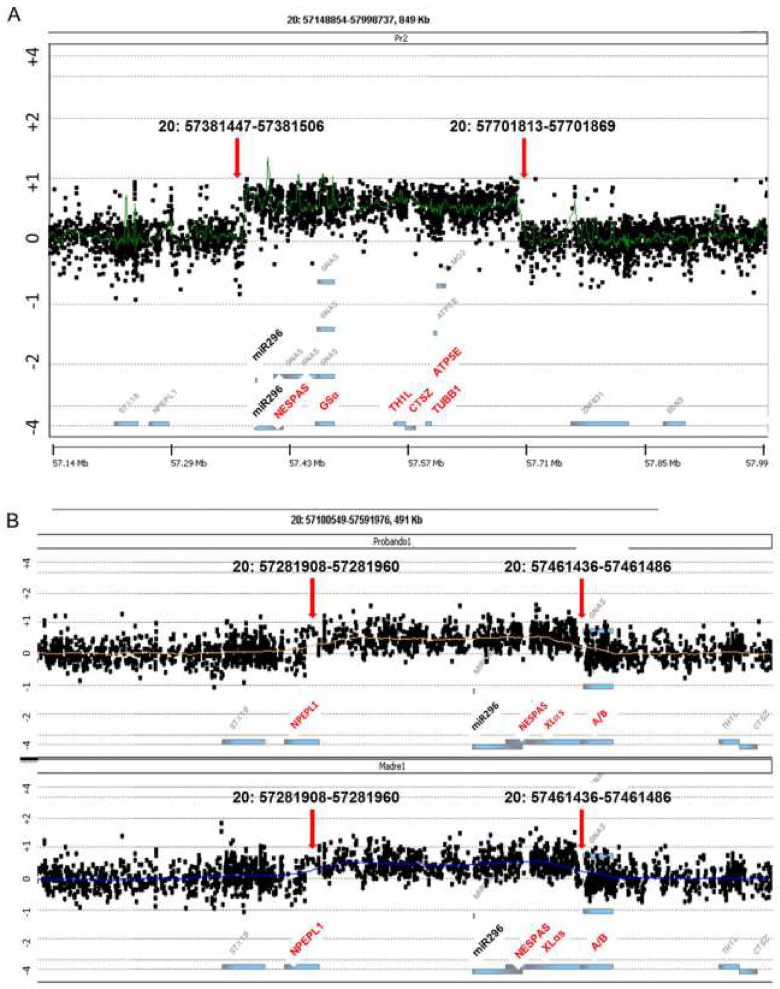
We then focused on the chromosomal region 20q13, using a high-density array specific only for the chromosomal coordinates chr20:47897125–62435965. Using this customized array, we were able to determine the limits of both duplications: according to hg19, GS0131 presented a ∼320 kb duplication, affecting chr20:57381447–57701869 (Fig. 3A), whereas GS0155 presented a ∼179 kb duplication, affecting chr20:57281908–57461486 (Fig. 3B, upper panel). The presence of the same duplication was confirmed in GS0155’s mother (Fig. 3B, lower panel).
Fig 3. High density aCGH assay results for the 20q13 region of both families.
In both cases, affected genes and regions mentioned in the text are highlighted in red on the X-axis. On the Y-axis, binary logarithm of the sample/control ratio. miR296 location is indicated in bold (corresponds to chr20:57392669–57392748). Red arrows indicate the location of the probes at each breakpoint. (A) Results for GS0131 index case. Won probes correspond to coordinates chr20:57381447–57701869. (B) Results of family GS0155 (upper panel, index case; lower panel, mother). Won probes correspond to coordinates chr20:57281908–57461486.
STX16 deletions
We discarded the presence in both families of the 3-kb and 4.4-kb STX16 deletions associated with A/B isolated loss of methylation [10, 11] (data not shown).
Determination of the parental origin of the duplication in family GS0131
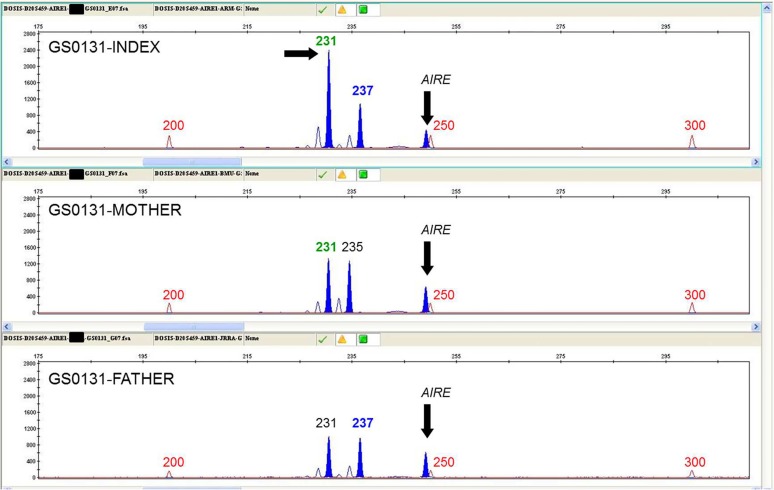
In order to study the parental origin of the allele in which the duplication occurred in the GS0131 family, we carried out a QMPSF analysis of microsatellites inside the duplicated region. Only D20S459 turned out to be informative, revealing the duplication arose in the maternal allele (Fig. 4).
Fig 4. Determination of the parental origin of the duplication in family GS0131.
Electropherogram showing the quantitative multiplex PCR of short fluorescent fragments (QMPSF) of microsatellite D20S459 from family GS0131 members against exon 6 of AIRE (21q22), used as a control amplicon. From up to down, results for the index case, mother and father. Vertical arrows show the control fragment. The horizontal arrow indicates the maternally-inherited duplicated allele in the index case. Figures show the size of the fragments in base pairs (red: weight marker; green: maternally-inherited allele; blue: paternally-inherited allele). Similar data were obtained using other control amplicons: exon 2 of HNF1A (12q24) and exon 3 of HNF1B (17q12). Data of these experiments are not shown.
Search of specific sequences
Trying to clarify the origin of both duplications, we searched within the sequences at breakpoints for different elements susceptible to being responsible for hypothetical non-allelic homologous recombination (NAHR) or non-homologous end joining (NHEJ), such as LINE or SINE elements, using RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker); homologous sequences at the two ends of each duplication, using BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or DNA palindromes, using DNA-palindrome Finder (http://www.alagu-molbio.net/palin.html) [28]. We were not able to localize any significant element in any of the patients.
Discussion
In this article we report the biggest duplications detected so far in the GNAS region, both present in two PHP-Ib patients and arising in the maternal allele.
We did not find any significant common element susceptible to be responsible for both rearrangements (i.e. specific sequences at the breakpoints, such as palindromes or SINE elements), so we cannot conclude whether this is a specific region prone to suffer these recombinations or both duplications are due to stochastic mechanisms.
The duplication in family GS0131 occurred ‘de novo’ in the patient. It was associated with a complete loss of methylation at the NESPAS, XLαs and A/B promoters. No other change was detected in the locus, either in the patient or in his parents. Paternal 20q disomies were also ruled out by polymorphic markers’ analysis.
This 320 kb duplication (chr20:57381447–57701869) also affected four genes located 3’ to the GNAS locus: TH1L (encoding NELFC and NELFD negative elongation factors), CTSZ (cathepsin Z), TUBB1 (tubulin beta-1 chain), and ATP5E (the epsilon subunit of the mitochondrial ATPase F1 complex). Mutations in these genes have been associated with macrothrombocytopenia (TUBB1, [29]), mitochondrial complex V (ATP synthase) deficiency nuclear type (ATP5E, [30]) or even tumour metastasis (CTSZ, [31]). As far as we know, there is not any report in the literature of any duplication in this region. Furthermore, at this point, the patient does not show any sign of these disorders, so the duplication he presents only seems to clinically affect the GNAS locus and its phenotypic manifestations, and the clinical significance of the overdose of the rest of the genes still needs to be elucidated.
In the same way, the duplication also affects miR-296 and miR-298, two microRNAs located in the GNAS region (Fig. 3A). These microRNAs have been described to target IKBKE and other genes regulating signaling pathways [32]. Moreover, they have been shown to have an imprinted expression, as they are paternally expressed and maternally repressed. In the case of GS0131, only the “silenced” maternal allele is duplicated, so the function of both miRNAs should not be affected, although further research should be performed to check this issue.
The second duplication, present in family GS0155, appeared both in the index case and her mother. It comprised 179 kilobases (chr20:57281908–57461486) and did not affect any other gene downstream GNAS, although both miR-296 and miR-298 and the NPEPL1 gene (located outside the GNAS locus on chr20:57264187–57294294) were also included in the affected region. In the case of the miRNAs, again, the duplicated “silenced” maternal allele is predicted not to affect their function, but this should be tested. On the other hand, NPEPL1 codifies for the protein known as “probable aminopeptidase NPEPL1”, whose exact function has not been reported so far, but it probably catalyzes the removal of unsubstituted N-terminal amino acids from various peptides. NPEPL1 proteins might function as tumor suppressors in tumorigenesis under the control of the miR-19a microRNA, although this has also not been consistently proved [33], so the effect of the overdose of this gene also remains elusive.
Duplications and deletions affecting other imprinted loci have also been reported. For instance, ICR1 and ICR2 in 11p15, associated with Silver-Russell and Beckwith-Wiedemann syndromes [34]. The interpretation of the copy number variation in this region is complicated, because the clinical features in carriers of these alterations depend on the size, breakpoint positions and the parental inheritance of the aberrations. Some interstitial microduplications have also been reported in the Prader-Willi and Angelman critical region, in 15q11-q13, also subject to imprinting. These duplications are the most common alteration in autism. In this case, maternal duplications are associated with autism, while paternal duplications usually have a normal phenotype [35].
Regarding the GNAS locus, as mentioned, few genomic rearrangements have been reported, and most of them are related to PHP-Ia. In 2005, Bastepe et al described the up-to-date only mentioned duplication in the GNAS locus, present in an AD-PHP-Ib kindred [13]. In that case, the duplicated region encompassed 15kb from the region between XLαs and A/B. Furthermore, the duplication was located in place of a 4kb deletion removing NESP55 DMR and NESPAS exons 3 and 4. The authors stated that, when inherited maternally, this deletion caused the loss of methylations of the DMRs normally methylated in the maternal allele. Inherited paternally, no epigenetic alteration was observed. The phenotype observed in this patient was thus due to the disappearance of the NESP55 DMR rather than to the simultaneous duplication.
In our study, there is a maternal inheritance of the duplication in the index case in family GS0155, whereas methylation status of NESPAS, NESP55 and XLαs invites to hypothesize that there is a paternal inheritance of the duplication in her mother (the partial loss of methylation at exons XLαs and NESPAS, and partial gain at NESP55 agrees with a doubled paternal allele together with a unique maternal allele). Besides, the duplicated region should have undergone proper methylation. If this is the case, it could be speculated that only the maternal inheritance of the duplication would lead to the phenotypic manifestation of PHP-Ib, as the index’s mother is asymptomatic. Unfortunately, we did not have access to the grandparents’ samples in order to confirm this latter point. Besides, GS0155 index case also suffered a complete loss of methylation at the A/B promoter, which was not present in her mother (Fig. 2B and 2D). Therefore, we hypothesize that two different mutational events could have coincided in the index case: the first one, a duplication of a region in the locus that provoked the methylation level change, inherited from her mother. The presence of two maternal alleles would lead to the observed proportional increase in methylation levels at NESPAS and XLαs and the decrease in NESP55. The second mutation, if present, would be responsible for the isolated total loss of methylation of the A/B promoter in the patient. For this reason, we tested for the presence of the already described 3-kb and 4.4-kb STX16 deletions associated with A/B isolated loss of methylation [10, 11] in this region, not affected by the duplication, but the analysis turned out to be negative (data not shown). We then performed a high-density cGH array analysis in the 20q region in order to be able to find any small deletion inside the duplication capable of causing an isolated loss of A/B methylation, as previously described [15] but, again, we were unable to detect any second alteration. Thus, this apparent isolated loss of A/B methylation remains unexplained. It is possible that the structure of the locus itself, together with the duplication of the maternal allele make it difficult to detect this deletion, if it exists. Or, maybe, there is another cis-acting element (or elements), still not described, controlling the methylation status of the different promoters, located in the region comprising chr20:57381447–57461486 (the common elements to both duplications) whose duplication in the maternal allele may cause a change in the methylation pattern of the region, either in a broad way, as it occurs in GS0131, or just in the A/B promoter, as it happens in GS0155.
Anyway, further studies are required in order to characterize the GNAS locus and the 20q region, as its imprinting pattern can be modified either by small deletions, small duplications or, as in our cases, by big duplications, which indicates that maybe there are several still unknown modifier agents which take part in the establishment or maintenance of the methylation of the different promoters in the region.
Acknowledgments
We kindly thank all family members for their participation.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was partially supported by Grants IT-795-13 and IT-472-07 from the Basque Department of Education (http://www.hezkuntza.ejgv.euskadi.net/r43-2591/es). TV is supported by the FPI Program of the University of Basque Country (UPV-EHU, http://www.ehu.es/p200-home/es). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Albright F, Burnett CH, Smith PH, Parson W (1942) Pseudohypoparathyroidsm- an example of ‘Seabright syndrome’. Endocrinology 30: 922–932. [Google Scholar]
- 2.Mantovani G, Linglart A, Garin I, Silve C, Elli FM, et al. (2012) Clinical utility gene card for: Pseudohypoparathyroidism. Eur J Hum Genet [Epub 2012 Sep 12]. 10.1038/ejhg.2012.211 [DOI] [PMC free article] [PubMed]
- 3. Kozasa T, Itoh H, Tsukamoto T, Kaziro Y (1988) Isolation and characterization of the human Gs alpha gene. Proc Natl Acad Sci USA. 85 (7): 2081–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spiegel AM (1999) Hormone resistance caused by mutations in G proteins and G protein-coupled receptors. J Pediatr Endocrinol Metab 12 Suppl 1: 303–309. [PubMed] [Google Scholar]
- 5. Weinstein LS, Yu S, Warner DR, Liu J (2001) Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev 22 (5):675–705. [DOI] [PubMed] [Google Scholar]
- 6. Mantovani G (2011) Clinical review: Pseudohypoparathyroidism: diagnosis and treatment. J Clin Endocrinol Metab 96 (10): 3020–3030. 10.1210/jc.2011-1048 [DOI] [PubMed] [Google Scholar]
- 7. Wilson LC, Oude Luttikhuis ME, Clayton PT, Fraser WD, Trembath RC (1994) Parental origin of Gs alpha gene mutations in Albright's hereditary osteodystrophy. J Med Genet 31 (11): 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winter JS, Hughes IA (1980) Familial pseudohypoparathyroidism without somatic anomalies. Can Med Assoc J 123 (1): 26–31. [PMC free article] [PubMed] [Google Scholar]
- 9. Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, et al. (2000) A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest 106 (9): 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bastepe M, Fröhlich LF, Hendy GN, Indridason OS, Josse RG, et al. (2003) Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS . J Clin Invest 112 (8): 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linglart A, Gensure RC, Olney RC, Jüppner H, Bastepe M (2005) A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS . Am J Hum Genet 76 (5): 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elli FM, de Sanctis L, Peverelli E, Bordogna P, Pivetta B, et al. (2014). Autosomal Dominant Pseudohypoparathyroidism type Ib: a novel inherited deletion ablating STX16 causes Loss of Imprinting at the A/B DMR. J Clin Endocrinol Metab 99(4):E724–8. Epub 2014 Jan 17 10.1210/jc.2013-3704 [DOI] [PubMed] [Google Scholar]
- 13. Bastepe M, Fröhlich LF, Linglart A, Abu-Zahra HS, Tojo K, et al. (2005) Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet 37 (1): 25–27. [DOI] [PubMed] [Google Scholar]
- 14. Chillambhi S, Turan S, Hwang DY, Chen HC, Jüppner H, et al. (2010) Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type Ib and biparental defects of GNAS methylation in cis. J Clin Endocrinol Metab 95 (8): 3993–4002. 10.1210/jc.2009-2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richard N, Abeguilé G, Coudray N, Mittre H, Gruchy N, et al. (2012) A new deletion ablating NESP55 causes loss of maternal imprint of A/B GNAS and autosomal dominant pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab 97 (5): E863–E867. 10.1210/jc.2011-2804 [DOI] [PubMed] [Google Scholar]
- 16. Bastepe M, Altug-Teber O, Agarwal C, Oberfield SE, Bonin M, et al. (2011) Paternal uniparental isodisomy of the entire chromosome 20 as a molecular cause of pseudohypoparathyroidism type Ib (PHP-Ib). Bone. 48 (3): 659–662. 10.1016/j.bone.2010.10.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernández-Rebollo E, Pérez de Nanclares G, Lecumberri B, Turan S, Anda E, et al. (2011) Exclusion of the GNAS locus in PHP-Ib patients with broad GNAS methylation changes: evidence for an autosomal recessive form of PHP-Ib? J Bone Miner Res 26 (8): 1854–1863. 10.1002/jbmr.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Nanclares GP, Fernández-Rebollo E, Santin I, García-Cuartero B, Gaztambide S, et al. (2007) Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab 92 (6): 2370–2373. [DOI] [PubMed] [Google Scholar]
- 19. Mariot V, Maupetit-Méhouas S, Sinding C, Kottler ML, Linglart A (2008) A maternal epimutation of GNAS leads to Albright osteodystrophy and parathyroid hormone resistance. J Clin Endocrinol Metab 93 (3): 661–665. 10.1210/jc.2007-0927 [DOI] [PubMed] [Google Scholar]
- 20. Mantovani G, de Sanctis L, Barbieri AM, Elli FM, Bollati V, et al. (2010) Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab 95 (2): 651–658. 10.1210/jc.2009-0176 [DOI] [PubMed] [Google Scholar]
- 21. Fernandez-Rebollo E, García-Cuartero B, Garin I, Largo C, Martínez F, et al. (2010) Intragenic GNAS deletion involving exon A/B in pseudohypoparathyroidism type 1A resulting in an apparent loss of exon A/B methylation: potential for misdiagnosis of pseudohypoparathyroidism type 1B. J Clin Endocrinol Metab 95 (2): 765–771. 10.1210/jc.2009-1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zazo C, Thiele S, Martín C, Fernandez-Rebollo E, Martinez-Indart L, et al. (2011) Gsα activity is reduced in erythrocyte membranes of patients with psedohypoparathyroidism due to epigenetic alterations at the GNAS locus. J Bone Miner Res 26 (8): 1864–1870. 10.1002/jbmr.369 [DOI] [PubMed] [Google Scholar]
- 23. Mantovani G, Elli FM, Spada A (2012) GNAS epigenetic defects and pseudohypoparathyroidism: time for a new classification? Horm Metab Res 44 (10): 716–723. 10.1055/s-0032-1314842 [DOI] [PubMed] [Google Scholar]
- 24. Aldred MA, Aftimos S, Hall C, Waters KS, Thakker RV, et al. (2002) Constitutional deletion of chromosome 20q in two patients affected with albright hereditary osteodystrophy. Am J Med Genet 113 (2): 167–172. [DOI] [PubMed] [Google Scholar]
- 25. Fernandez-Rebollo E, Barrio R, Pérez-Nanclares G, Carcavilla A, Garin I, et al. (2008) New mutation type in pseudohypoparathyroidism type Ia. Clin Endocrinol (Oxf) 69 (5): 705–712. 10.1111/j.1365-2265.2008.03255.x [DOI] [PubMed] [Google Scholar]
- 26. Alsum Z, Abu Safieh L, Nygren AO, Al-Hamed MA, Alkuraya FS (2010) Methylation-specific multiplex-ligation-dependent probe amplification as a rapid molecular diagnostic tool for pseudohypoparathyroidism type 1b. Genet Test Mol Biomarkers 14 (1): 135–139. 10.1089/gtmb.2009.0092 [DOI] [PubMed] [Google Scholar]
- 27. Barrett MT, Scheffer A, Ben-Dor A, Sampas N, Lipson D, et al. (2004) Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc Natl Acad Sci USA 101 (51): 17765–17770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13: R57–R64. [DOI] [PubMed] [Google Scholar]
- 29. Kunishima S, Kobayashi R, Itoh TJ, Hamaguchi M, Saito H (2009) Mutation of the beta1-tubulin gene associated with congenital macrothrombocytopenia affecting microtubule assembly. Blood 113 (2): 458–461. 10.1182/blood-2008-06-162610 [DOI] [PubMed] [Google Scholar]
- 30. Mayr JA, Havlícková V, Zimmermann F, Magler I, Kaplanová V, et al. (2010) Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 epsilon subunit. Hum Mol Genet 19 (17): 3430–3439. 10.1093/hmg/ddq254 [DOI] [PubMed] [Google Scholar]
- 31. Wang J, Chen L, Li Y, Guan XY (2011) Overexpression of cathepsin Z contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. PLoS One 6 (9): e24967 10.1371/journal.pone.0024967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robson JE, Eaton SA, Underhill P, Williams D, Peters J (2012) MicroRNAs 296 and 298 are imprinted and part of the GNAS/Gnas cluster and miR-296 targets IKBKE and Tmed9 . RNA 18: 135–144 10.1261/rna.029561.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ouchida M, Kanzaki H, Ito S, Hanafusa H, Jitsumori Y, et al. (2012) Novel direct targets of miR-19a identified in breast cancer cells by a quantitative proteomic approach. PLoS One 7(8):e44095 10.1371/journal.pone.0044095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Begemann M, Spengler S, Gogiel M, Grasshoff U, Bonin M, et al. (2012) Clinical significance of copy number variations in the 11p15.5 imprinting control regions: new cases and review of the literature. J Med Genet 49: 547–553. 10.1136/jmedgenet-2012-100967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cai G, Edelmann L, Goldsmith JE, Cohen N, Nakamine A, et al. (2008) Multiplex ligation-dependent probe amplification for genetic screening in autism spectrum disorders: Efficient identification of known microduplications and identification of a novel microduplication in ASMT . BMC Med Genomics 1:50 10.1186/1755-8794-1-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.