Summary
Mycobacterium tuberculosis (Mtb), the primary causative agent of human tuberculosis, has killed more people than any other bacterial pathogen in human history and remains one of the most important transmissible diseases worldwide. Because of the longstanding interaction of Mtb with humans, it is no surprise that human mucosal and innate immune cells have evolved multiple mechanisms to detect Mtb during initial contact. To that end, the cell surface of human cells is decorated with numerous pattern recognition receptors for a variety of mycobacterial ligands. Further, once Mtb is ingested into professional phagocytes, other host molecules are engaged to report on the presence of an intracellular pathogen. In this review, we discuss the role of specific mycobacterial products in modulating the host’s ability to detect Mtb. In addition, we describe the specific host receptors that mediate the detection of mycobacterial infection and the role of individual receptors in mycobacterial pathogenesis in humans and model organisms.
Keywords: Mycobacterium tuberculosis, pattern recognition receptors, innate immunity, microbial pathogenesis
Introduction
Mycobacterium tuberculosis (Mtb) is spread through the inhalation of aerosolized droplets that are released into the air when an infected individual coughs or sneezes. The bacteria are ingested by resident alveolar macrophages that recognize specific pathogen-associated molecular patterns (PAMPs). Following phagocytosis of mycobacteria, infected macrophages release pro-inflammatory cytokines that recruit additional immune cells to the lung, including neutrophils, naive monocytes, and dendritic cells (DCs). DCs that have ingested Mtb leave the lung and migrate to regional lymph nodes, where they drive induction of the adaptive immune response.
Initiation of an adaptive response leads to the formation of a granuloma surrounding the bacteria. The granuloma is composed of a central region of infected macrophages, multinuclear giant cells, epithelioid cells, and foamy macrophages as well as neutrophils and monocytes. An outer layer of B cells and T cells as well as fibroblasts that drive the development of a fibrotic capsule comprise the outer layer of the granuloma.
This entire complex cascade of events would be impossible without the primary step, namely, recognition of Mtb by mucosal and innate immune cells. Here, we review the various mycobacterial PAMPs and their role in triggering the immune response to Mtb via cell surface and cytoplasmic receptors. Whenever possible, we incorporate both a host- and bacterial-centered view, as engagement of host receptors leads to both beneficial and detrimental consequences to each partner in this interaction.
Mycobacterium tuberculosis pathogen-associated molecular patterns
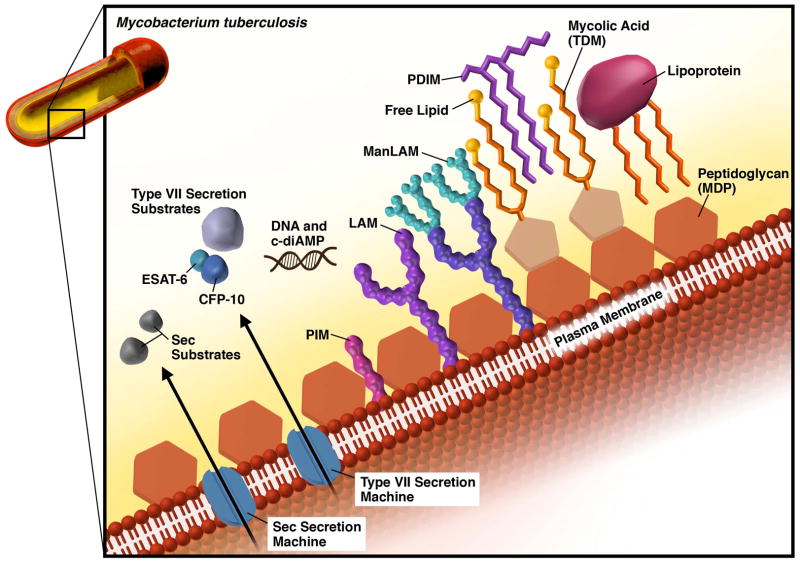
Mtb produces and releases antigens common to all bacteria including components of the peptidoglycan cell wall and nucleic acids. However, the unique makeup of the Mtb cell wall generates unique antigens specific to mycobacterial species (Fig. 1). These include lipomannan (LM), lipoarabinomannan (LAM) and its mannosylated form (ManLAM), lipoproteins, phthiocerol dimycocerosate (PDIM), and mycolic acids. Mtb also secretes effector proteins either via the generalized Sec secretion system or the specialized ESAT-6 (ESX) secretion system and some Mtb secreted proteins can be recognized by pattern recognition receptors (PRRs). As discussed below, many of the cell wall components can be viewed not only as PAMPs that stimulate the immune system but also as bacterial effectors that modulate the host response.
Fig. 1. Schematic representation of the Mycobacterium tuberculosis (Mtb) cell wall.
Components of the Mtb cell wall are ligands for PRRs, including peptidoglycan, LAM and its variants, mycolic acids, and lipoproteins. Secreted proteins, c-diAMP, and extracellular DNA are also recognized by host PRRs.
The biosynthesis of the major cell wall components of Mtb has been recently reviewed (1, 2). Phosphatidyl inositol (PI) forms the backbone for the majority of the cell wall components including LM, LAM and ManLam, which are synthesized by sequential additions of mannoses and arabinoses to PI. The length and degree of branching of the sugar residues on PI dictates the nature of the product. Since they are formed on a PI backbone, these molecules are embedded in the plasma membrane or outer membrane by their lipid moiety (3). PDIM is a surface exposed bioactive lipid that requires the sequential action of several polyketide synthases, and ultimately is exported by a specific transporter, MmpL7 (4, 5). Finally, mycolic acids are long chain fatty acids that are a major component of the mycobacterial cell wall, and when conjugated to a trehalose sugar residue, become cord factor, the major cell surface lipid of Mtb. Ultimately, this complex lipid coat can shield Mtb from host defenses, but also presents an array of possible ligands for the host to recognize. Amongst all these lipids and lipoproteins are also cell wall associated proteins and embedded secretion machines that facilitate secretion of putative effector molecules including some molecules like ESAT-6 that may lead to phagosomal rupture (6, 7). In that way, Mtb can communicate with the host cytoplasm, but this also leads to exposure of mycobacterial PAMPs to cytoplasmic receptors.
Surface receptors
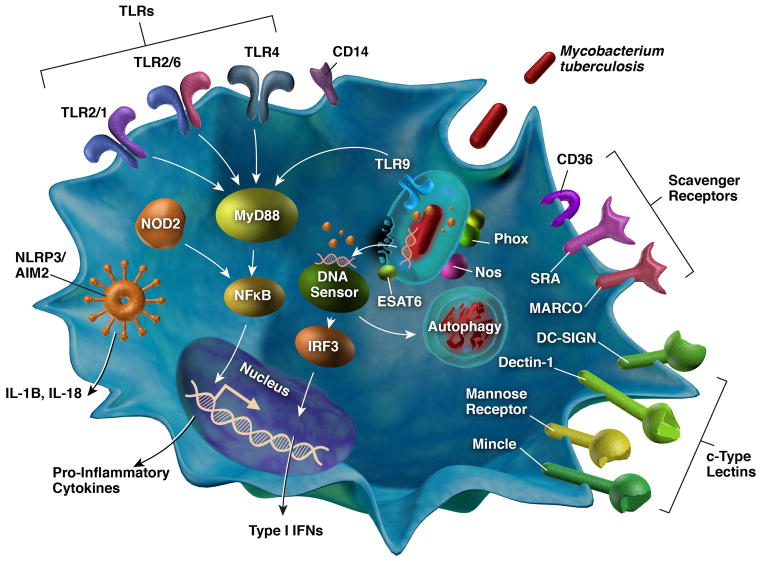
Surface expressed PRRs of host cells are the first to encounter bacteria as they contact the plasma membrane. Toll-like receptors (TLRs), c-type lectin receptors, and scavenger receptors are the main families of surface receptors that interact with Mtb (Fig. 2). Within each family are receptors capable of recognizing PAMPs on the surface of and secreted from Mtb. In most cases, the cell types responsible for detecting Mtb are macrophages and DCs, which are the main niche for the bacilli. Other cells such as neutrophils and lung epithelial cells are also able to detect Mtb.
Fig. 2. Pattern recognition receptors important for the sensing of mycobacterial PAMPs.
This schematic shows the major receptors involved in recognition of Mtb and their localization to the surface or the cytoplasm of host cells. PRRs on the surface of cells include TLRs, scavenger receptors, and c-type lectin receptors. Cytoplasmic receptors recognize mycobacterial components, such as secreted proteins and DNA that access the cytoplasm through phagosomal membrane damage. Major signaling pathways are shown including NFκB- and IRF3-dependent cytokine secretion, inflammasome-mediated IL-1β secretion, and the activation of autophagy.
Toll-like receptors
TLRs are a conserved family of leucine-rich repeat proteins that recognize a wide variety of PAMPs from all types of invading microbes. Of the known human receptors, TLR1, TLR2, TLR4, TLR5, and TLR6 are surface expressed (8). Each TLR has a specific subset of PAMPs that it can bind; however, ligands for each TLR continue to be discovered. In the TLR family, Mtb is primarily detected by TLR2, TLR4, and TLR9, with a dominant role for TLR2 (9, 10). Following recognition of Mtb PAMPs by TLRs, an intracellular signaling cascade is activated that recruits the adaptor protein myeloid differentiation primary response protein 88 (MyD88) to the intracellular domains of TLRs (11). MyD88 then serves as the scaffolding for recruitment of IL-1 receptor-associated kinases (IRAK), TNF receptor-associated factor 6 (TRAF), TGFβ-activated protein kinase 1 (TAK1), and mitogen-activated protein (MAP) kinase. This signaling cascade activates the transcription factor NFκB to translocate to the nucleus (8, 12), driving production of multiple pro-inflammatory cytokines including tumor necrosis factor (TNF), interleukin-1β (IL-1β) and interleukin-12 (IL-12). Secreted TNF and IL-12 then stimulate IFN-γ production from neighboring natural killer (NK) and T cells. A cycle is thus established whereby IFN-γ activates macrophages to enhance antigen presentation and promote anti-mycobacterial effector mechanisms such as the production of reactive nitrogen intermediates (RNI), reactive oxygen intermediates (ROI), phagolysosome fusion and acidification (13), and autophagy (14).
TLR2
The primary ligands for TLR2 are lipid-containing moieties such as lipoteichoic acid (LTA) from Gram-positive bacteria and acylated lipopeptides from a variety of bacterial, fungal, and parasitic organisms. Consistent with TLR2’s affinity for lipids, the main mycobacterial cell wall ligands for TLR2 are lipoglycans and lipids, namely uncapped lipoarabinomannan (LAM) and LAM precursors lipomannan (LM) and mannoyslated phosphatidylinositol (PIM). Treatment of mouse RAW264.7 macrophages with PIM or LAM leads to ERK1/2 phosphorylation and TNFα production (15). This activation is likely through TLR2, as Chinese hamster ovary (CHO) cells overexpressing TLR2 but not TLR4 produce TNFα in response to PIM or LAM (15). Similarly, LM isolated from the virulent M. tuberculosis strain H37Rv and M. bovis var Bacillus Calmette–Guérin (BCG) activates murine bone marrow derived macrophages (BMDMs) to produce TNF, IL-12, and RNI in a TLR2-dependent manner (16). The total lipid fractions of Mtb can also activate TLR2 signaling (11), the major component of which is trehalose-6,6′ dimycolate (TDM), also known as cord factor (17). In vitro, TDM treatment of HEK293 cells leads to stimulation of an NFκB luciferase reporter only in the presence of TLR2 and its co-receptors CD14 and MARCO (17). Furthermore, bone marrow macrophages from TLR2/TLR4−/− mice are unable to produce TNF-α in response to TDM treatment (17).
TLR2 recognition of Mtb extends beyond lipoglycan binding. TLR2 can recognize lipoproteins that are either attached to or secreted from the mycobacterial cell wall. Some lipoproteins are released as part of Mtb membrane vesicles, and these vesicles are enriched in lipoproteins LpqH (also known as 19-kDa lipoprotein) and LprG (18). TLR2 binds LpqH, thus leading to TLR2 translocation to lipid rafts where a signaling cascade is triggered (19). Activation of TLR2 by LpqH also induces macrophage apoptosis, which may aid in containment of intracellular Mtb (20). Finally, membrane vesicles isolated from BCG potently activated pro-inflammatory cytokine production in WT BMDMs, but not those isolated from TLR2−/− mice (18).
In addition to mycobacterial cell wall lipids and lipoproteins, many other Mtb-secreted proteins may be TLR2 ligands. The virulence factor ESAT-6 induces TGF-β and IL-6 production in DCs, and cytokine production is lost in TLR2−/− cells (21). Likewise, recombinant TB10.4 from Mycobacterium bovis, a low molecular weight antigen similar to ESAT-6, induces NFκB translocation and pro-inflammatory cytokine production in a TLR2-dependent manner in RAW264.7 cells (22). Other secreted proteins such as Rv0577 and hsp70 induce pro-inflammatory cytokine production in DCs and BMDMS, respectively. In both cases, pro-inflammatory cytokine secretion is dependent on TLR2 (23, 24). Lastly, PE_PGRS33 can induce apoptosis in RAW264.7 macrophage through interaction with TLR2 and the production of TNF-α (25).
The broad ligand specificity of TLR2 suggests a vital role for TLR2 activation in controlling Mtb infection. Indeed, some murine models of Mtb infection have underscored the importance of TLR2. While TLR2-deficient mice are not defective in pro-inflammatory cytokine production, they succumb faster to low-dose aerosol infections (26, 27). In contrast, a later report showed no defect in pro-inflammatory cytokine production or survival rates in TLR2/TLR4/TLR9-deficient mice (28). Different Mtb strains used for infection, differences in the route and dosage used for infection, and differences in genetic backgrounds or microbiomes of knockout mice may all contribute to the lack of a consistent phenotype in these knockout mouse studies.
Despite the lack of agreement of mouse studies, TLR2 is clearly a key signaling molecule whose function extends beyond triggering pro-inflammatory cytokine production and inducing apoptosis. One such alternate pathway is the activation of autophagy, a fundamental mechanism whereby cytoplasmic contents such as damaged organelles, protein aggregates, and invading microbes are engulfed in a double membrane structure and directed to the lysosome for degradation (29, 30). The role of autophagy in controlling intracellular infection has been extensively reviewed (30–32), and autophagy is an important host defense mechanism against Mtb (33–35). Treatment of human primary monocytes with LpqH isolated from H37Ra increases the number of cells with LC3 puncta, an indication of autophagy activation (36). LpqH-mediated LC3 puncta formation is inhibited by an α-TLR2 monoclonal antibody or knockdown of TLR2, suggesting that autophagy induction in response to LpqH requires TLR2 activation (36).
In humans, the TLR2 G2258A polymorphism causes impaired TLR2 tyrosine phosphorylation (37) and is associated with increased susceptibility to Mtb infection in European cohorts (38, 39). However, this polymorphism has not been associated with increased susceptibility in other ethnic groups (40–43).
TLR2: the bacterial view
While TLR2 activation in macrophages is important to control Mtb infection, it may not always be beneficial for host cells as Mtb has developed mechanisms to exploit TLR2 activation for its own benefit. Secreted Mtb proteins such as hsp60 and PPE18 induce the anti-inflammatory cytokine IL-10 in human THP-1 cells (44, 45). Production of IL-10 is blocked by an anti-TLR2 antibody (44, 45) or by an siRNA against TLR2 (44). Likewise, PPE18 is unable to induce IL-10 when cells express a dominant negative TLR2 (45). Interestingly, hsp60-dependent IL-10 production is blocked by monodansylcadaverine (MDC), a clathrin-mediated endocytosis inhibitor, suggesting that hsp60 may leverage TLR2 internalization to induce IL-10 (44). Another mycobacterial secreted effector ESAT-6 also binds TLR2 and rather than induce IL-10 inhibits IL-12 production in RAW macrophages (46). This inhibition is lost upon pre-treatment of macrophages with an anti-TLR2 but not anti-TLR4 antibody (46).
In addition to skewing the balance between pro- and anti-inflammatory cytokines, Mtb antigens also modulate other immune activities. For example, lipid fractions from the Beijing strain and to a lesser extent H37Rv decrease surface expression of HLA-DR in human macrophages (47). Furthermore, purified lipoproteins LpqH, LprA, and LprG interact with TLR2 to inhibit IFN-γ-mediated MHC-II antigen presentation (48–50). As a consequence, IFN-γ activated THP-1 cells treated with LpqH or LprG cannot process and present purified antigens (48, 49) or antigens from whole Mtb to T cells (48). Similarly, acylated LprA inhibits IFN-γ MHC-II expression in murine macrophages, and consequently antigen presentation, in a TLR2-dependent manner (50). Inhibition of antigen processing is lost when THP-1 cells are pre-treated with an anti-TLR2 antibody (48, 49). Taken together, these observations illustrate that TLR2 is a high value target for Mtb to modulate the host environment.
TLR2 promiscuity
The ability of TLR2 to bind multiple, structurally distinct ligands and elicit different signals begs the question of how one protein can have such promiscuous ligand binding that ultimately leads to different outcomes. TLR2 has the unique ability to heterodimerize with TLR1 or TLR6, as well as bind other co-receptors such as CD14 (51). One possibility is that the interaction of TLR2 with other binding partners diversifies ligand recognition. For example, both genetic and structural studies have shown that the TLR2/1 heterodimer recognizes triacylated lipoproteins, whereas TLR2/6 recognizes diacylated lipoproteins (51–53). Furthermore, the coreceptors CD14 and CD36 can facilitate TLR2 activation in response to triacylated lipopeptides and diacylated lipopeptides, respectively (54, 55). Other proposed TLR2 co-receptors include Dectin-1, mannose binding lectin, scavenger receptors, integrin B3, and CXCR4 (56, 57).
While co-receptors can broaden TLR2 ligand recognition, it is possible that TLR2 alone can bind multiple ligands. Biochemical studies with recombinant human TLR2 ectodomain (hTLR2ED) showed that TLR2 can bind ligands in the absence of co-receptors (58). hTLR2ED binds both Pam2CSK4 and Pam3CSK4, as well as LTA from S. aureus and synthetic Pim2 and Pim4 (58). The authors note that TLR2 lacks conserved asparagines and phenylalanines in the central LRR repeats that may provide TLR2 more flexiblity, thus allowing it to accommodate multiple lipoprotein ligands (58). Indeed, exposure to several ligands simultaneously or sequentially, as would be expected during a genuine infection, is likely to generate multiple signaling inputs and outputs.
TLR4
TLR4 is best known for its recognition of lipopolysaccharide (LPS) from Gram-negative bacteria. In Mtb, TLR4 recognizes cell wall lipids, glycoproteins, and secreted proteins. As discussed above, the LAM precursor LM induces pro-inflammatory cytokines in macrophages. LM is a mixture of molecules, and its tetra-acylated form (Ac4LM) functions as a specific TLR4 activator since BMDMs produce TNF and RNI in the presence of Ac4LM in a TRL4-dependent manner (59).
A number of mycobacterial proteins signal through TLR4, including multiple heat shock proteins, an Mtb 50S ribosomal protein Rv0652, and the 38-kDA glycoprotein of Mtb H37Rv (24, 60–62). Importantly, besides inducing MyD88-mediated signaling like other TLRs, stimulation of TLR4 can also activate the MyD88-independent TIR-domain containing adapter-inducing interferon-β (TRIF) pathway. In this pathway, TRIF upregulates IRF3 to induce IFN-β secretion (8), and both IRF-3 and IFN-β play crucial roles in Mtb pathogenesis (63, 64). Consistently, whereas some Mtb strains activate mainly TLR2, others also activate TLR4, resulting in different cytokine profiles characterized by varying production of IFN-β and consequently varying bacterial virulence (65).
In vivo studies in mice have yielded conflicting results on the importance of TLR4 in controlling Mtb infection. Although TLR4-deficient mice are not more susceptible to infection than wildtype mice in two studies (66, 67), another found that TLR4-deficient mice have decreased survival and higher bacterial numbers compared to wildtype controls (68). As noted above, differences in mouse husbandry, genetic backgrounds, Mtb strains, and route of infection likely account for these discrepant results. While no current studies have identified polymorphisms in TLR4 associated with TB susceptibility in HIV-negative individuals, a TLR4 SNP has been identified that is associated with increased susceptibility to Mtb in HIV-infected individuals (69).
TLR9
TLR9 is located on the phagosomal membrane and recognizes undermethylated CG motifs (CpG) in bacterial DNA, including DNA from Mtb (Fig. 2). Treatment of primary macrophages with DNA isolated from BCG induces production of TNF-α, which can then be blocked by a TLR9 inhibitor or methylation of the DNA (70). Mice deficient in TLR9 are more susceptible to a low dose aerosol infection compared to wildtype controls (26). Although there is no significant difference in bacterial load in vivo, when BMDMs from TLR9−/− mice are infected with Mtb in vitro, they produce less IL-12p40 than wildtype BMDM (26). Furthermore, IL-12p40 production is completely lost in TLR2/TLR9−/− cells infected with Mtb, suggesting a cooperative effect of TLR2 and TLR9 activation on cytokine production (26). In humans, multiple studies have found an association between TLR9 polymorphisms and susceptibility to Mtb (71–73), suggesting that TLR9 plays an important early role in sensing Mtb in primary human infection.
TLR4 and TLR9: the bacterial view
Mtb evades and manipulates both TLR4 and TLR9 activation. PIM purified from BCG inhibits the ability of LPS, the canonical TLR4 ligand, to induce TNF by murine BMDMs (74). Synthetic PIM1 inhibits MyD88-dependent cytokine release, NFκB nuclear translocation, and TRIF-dependent CD40 and CD86 surface expression in BMDMs (74). Together these data suggest that PIM is able to inhibit MyD88 and TRIF-dependent signaling, likely downstream of TLR4, in response to LPS. Similarly, a di-acylated LM (Ac2LM) inhibits LPS-induced TNF, IL12p40 and RNI production in wildtype and TLR2−/− BMDMs, suggesting the inhibition is independent of TLR2 (59).
As noted earlier, cytokine production by macrophages is Mtb strain specific and demonstrates TLR4 dependence. The strain 02–171 of the Beijing lineage induces wildtype and TLR2−/− but not TLR4−/− BMDMs to produce more TNF and IL-6 than H37Rv. Importantly, BMDMs infected with Mtb Beijing 02–171 also transcribe and secrete significantly more IFN-β than those infected with H37Rv in a TLR4-dependent manner (65), which is deleterious to the host. Similarly, TLR9 recognition of Mtb may be strain dependent. Murine and human macrophages treated with DNA isolated from virulent mycobacteria H37Rv produce less TNF-α than macrophages treated with DNA from attenuated mycobacteria H37Ra, suggesting that virulent mycobacteria can evade TLR9 activation in macrophages (70).
Mtb may broadly evade TLR activation through the membrane lipid PDIM. In a zebrafish model of M. marinum infection, macrophages are recruited to the bacteria even in the absence of MyD88, suggesting that TLR signaling is not required for this process (75). However, infection of zebrafish with a mutant for PDIM transport (ΔmmpL7) leads to decreased macrophage recruitment in the absence of MyD88 (75) indicating that in the absence of PDIM, TLR signaling does play a role in macrophage recruitment. Macrophages that are recruited to ΔmmpL7 bacteria showed higher NOS2 expression compared to macrophages recruited to wild type bacteria (75). These data suggest that the absence of PDIM leads to increased TLR activation. In support of this, co-infection of zebrafish with wildtype and Δmmpl7 or crushed, heat-killed M. marinum attenuates the growth of wildtype bacteria (75). Together, these observations suggest that PDIM on the mycobacterial surfaces masks PAMPs from TLR recognition.
C-type lectin receptors
C-type lectin receptors are a family of plasma membrane molecules that recognize conserved carbohydrate moieties on the surface of pathogens (76). In the immune response, they function as both cell adhesion molecules and pathogen recognition receptors (77). Mtb PAMPs are recognized by C-type lectin receptors including mannose receptor (MR), DC-specific intercellular adhesion molecule-3 grabbing nonintegrin (DC-SIGN), Mincle, and Dectin-1 (78).
Mannose receptors
Phagocytosis of Mtb by human macrophages is mediated primarily through MRs (79), suggesting an important role for these receptors in initiating Mtb host cell entry. Notably, two of the most abundant mycobacterial cell wall products, LAM and ManLAM, are ligands for mannose receptor C type 1 and 2 (MRC1 and MRC2) (80). Engagement of MRs triggers the production of the anti-inflammatory cytokines IL-10, IL-1R antagonist, and IL-1R type II, as well as inhibits the production of IL-12 (81). The nuclear receptor peroxisome proliferator-activated receptor γ (PPAR-γ) is also activated, which further inhibits the inflammatory response (82). Additionally, MR binding by Mtb inhibits phagosome-lysosome fusion in the macrophage, promoting Mtb infection (83). Taken together, LAM/ManLAM-mediated entry via MRs would appear to be detrimental to the host. However, MR-deficient mice are not more resistant to Mtb infection, as bacterial growth is similar to wildtype mice and MR-deficient mice show similar lung pathology following aerosol infection (84). Nonetheless, human polymorphisms in MRs have been associated with increased susceptibility to Mtb infection (85, 86), suggesting that the mechanisms of Mtb sensing and entry (at least as it relates to MRs) may differ between mice and humans.
DC-SIGN
DC-SIGN is a type II transmembrane receptor found on the surface of DCs and some macrophage populations in humans (87, 88). DC-SIGN recognizes LAM on the mycobacterial surface, and binding of Mtb to DC-SIGN expressing cells can be blocked by yeast mannan and LAM derived from Mtb, suggesting that DC-SIGN specifically recognizes the mannosyl residues of Mtb LAM (89). While there are 8 genetic homologs of human DC-SIGN found in mice, there is not a clear direct ortholog, complicating the ability to study its role in mouse models (88). However, the murine DC-SIGN homolog SIGNR3 can recognize LM and ManLAM as well as the lipoprotein LpqH (90). Mice deficient for SIGNR3 have increased bacterial growth in their lungs at 21 days post infection compared to wildtype mice, but this increased growth does not affect mouse survival, suggesting that SIGNR3 is involved in early but not late control of Mtb infection (90).
In humans, DC-SIGN promoter polymorphisms (336Aand 871G) have been associated with an increased risk of tuberculosis in South African cohorts (91, 92); however, no association between these promoter polymorphisms and Mtb susceptibility have been found in Tunisian (93) or Chinese populations (94). Consistent with the later studies, two recent independently conducted meta-analyses did not find an association between either polymorphism and Mtb susceptibility (95, 96).
Dectin-1
Dectin-1 is a type II transmembrane receptor found on the surface of macrophages, DCs, and neutrophils. While the role of dectin-1 in the production of inflammatory cytokines in response to fungal β-glucans is well-characterized (97), recent studies suggest an additional role for dectin-1 in the recognition of Mtb in response to an as yet uncharacterized ligand. Mtb infection induces the expression of dectin-1 on airway epithelial cells in a TLR2-dependent manner (98), and TLR2-mediated TNF production in murine BMDMs is dependent on dectin-1 following infection with avirulent mycobacteria (99). In the later case, blocking dectin-1 with a monoclonal antibody prior to infection with Mycobacterium smegmatis, Mycobacterium avium, and the avirulent Mtb H37Ra strain, but not with virulent Mtb H37Rv prevents TNF production. Likewise, BMDMs deficient for dectin-1 are unable to produce TNF-α or ROS or control bacterial load during M. bovis infection (100). In splenic dendritic cells, Mtb infection initiates production of IL-12p40 and IL-12p70 in a dectin-1 dependent but TLR2-independent mechanism (101). Likewise, dectin-1 activation in monocyte-derived DCs promotes a Th1/Th17 response and the production of IL-1, IL-23, TNF, and IL-6 (102). Although dectin1−/− mice infected with Mtb have a lower number of pulmonary CFU, this apparent defect in bacterial survival in the lungs does not translate to an effect on mouse survival. There are also no significant differences in inflammatory cytokine levels following Mtb infection when compared to WT mice, suggesting a possible redundant role in cytokine production (103). Since dectin-1, in conjunction with TLR2, upregulates cytokine expression in airway epithelial cells when challenged with Mtb (98), it is tempting to suggest that dectin-1’s role might be to bind Mtb on epithelial cells early in the context of mycobacterial infection.
Mincle
Mincle is an inducible C-type lectin receptor expressed on the surface of macrophages that recognizes TDM (104), the most abundant glycolipid in the Mtb cell wall. TDM activates macrophages to produce inflammatory cytokines and RNI in a Mincle-dependent manner in vitro (104, 105). In neutrophils, Mincle expression is induced by TDM, and neutrophil recruitment and adhesion to TDM-challenged sites is dependent on Mincle (106). However, the role of Mincle in vivo remains controversial. In one study, BMDMs from Mincle-deficient mice produced less GCSF and TNF in response to BCG and H37Rv infection, yet Mincle-deficient mice developed a normal granulomatous response and demonstrated no defect in controlling Mtb infection, suggesting other receptors may compensate for Mincle deficiency in vivo (107). In another study, pro-inflammatory cytokine levels were decreased in BAL fluid from Mincle−/− mice challenged with BCG as compared to wildtype mice. Concordantly, bacterial burden in lungs, draining lymph nodes, and spleens of Mincle−/− mice were higher than wildtype mice (108). The discovery of a constitutively expressed Mincle homolog macrophage C-type lectin (MCL) may explain the discrepancies in Mincle responses to Mtb. TDM can bind to MCL and induces it to signal through the FcRγ. Mice deficient for MCL are impaired for cytokine expression in response to TDM. Finally MCL induces Mincle expression, suggesting the two constitute a feedback loop to detect and control Mtb infection (109).
C-type lectin receptors: the bacterial view
C-type lectin receptors can be exploited by Mtb to provide a more amenable environment. Treatment of human DCs with ManLAM decreases IL-12 expression, but increases IL-10 expression and this outcome can be reversed by siRNA knockdown of DC-SIGN (110). ManLAM binding to DCs prevents their maturation in response to either Mtb or LPS, and DC maturation is restored in the presence of blocking antibody to DC-SIGN (111). Thus, Mtb may be able to engage DC-SIGN to promote an anti-inflammatory state. On macrophages, ManLAM binds to MR, and this interaction contributes to phagosome maturation arrest (83). Furthermore, ManLAM binding to MR activates PPARγ and BMDMs knocked down for PPARγ are able to control Mtb infection better than control cells, suggesting that induction of PPARγ through ManLAM-MR interaction is beneficial to Mtb survival (82).
Scavenger receptors
Scavenger receptors (SRs) are expressed on the surface of and secreted from monocytes and macrophages. SRs are capable of recognizing various ligands including self and non-self lipoproteins and perform many versatile functions within the immune system (112). Both class A and class B SRs are involved in Mtb recognition (78) including CD36, macrophage receptor with collagenous structure (MARCO), scavenger receptor A (SRA), and the secreted SR known as apoptosis inhibitor of macrophages (AIM).
CD36
CD36 is a class B SR expressed broadly across cell types including monocytes/macrophages and DCs (113). CD36 recognizes and internalizes oxidized LDL and plays a role in the clearance of apoptotic cells (113). With regards to innate immunity, CD36 is a sensor of bacterial diglycerides including lipoteichoic acid (LTA) from Gram-positive bacteria (114). This function of CD36 likely aids in the delivery of lipoproteins to TLR2 heterodimers (57).
CD36 recognizes mycobacterial lipoglycans including ManLAM and LM. In vitro, ManLAM treatment increases the expression of TNFα in LPS-induced macrophages and this increase is partially blocked by an antibody to CD36 (115). Likewise, macrophages isolated from CD36 mutant mice produce less TNFα than wildtype macrophages in response to LM isolated from M. smegmatis (116). Despite this, CD36−/− macrophages control the growth of both Mtb and M. marinum in vitro (117). Similarly, mice deficient in CD36 are better able to control bacterial burden early after intraperitoneal (IP) infection with BCG (117).
MARCO
MARCO is a class A SR expressed by macrophages upregulated in response to bacterial infection (118). MARCO ligand specificity overlaps with other class A SRs including bacterial cell wall components and LDL (118). Expression of TLR2, CD14, and MARCO leads to NFκB activation in response to TDM-coated beads in HEK293 cells, suggesting that MARCO works as a co-receptor for TLR2 (17). Consequently, peritoneal macrophages isolated from MARCO−/− mice are deficient for pro-inflammatory cytokine production in response to TDM or Mtb (17). Polymorphisms in MARCO are associated with susceptibility to pulmonary tuberculosis in Gambian (119) and Chinese Han populations (120). Alternatively, a polymorphism in a possible regulatory element of MARCO is associated with resistance to infection (119). This could suggest that expression levels of MARCO are responsible for conferring resistance or susceptibility to Mtb infection.
Scavenger receptor A
SRA is another class A receptor with the ability to bind and internalize endogenous proteins as well as microbial ligands, like LPS from Gram-negative bacteria and LTA from Gram-positive bacteria (112). SRA can associate with TLRs and enhance their endocytosis of microbial PAMPs (57, 112).
In the context of Mtb, CHO-K1 cells expressing SRA-I or SRA-II are able to bind TDM-coated beads (17). TDM induces TNF in peritoneal macrophages, but macrophages from SRA−/− mice are impaired for TNF production. Double knockout of MARCO with SRA impairs TNF production in response to TDM even further (17). Interestingly, mice deficient for SRA survive significantly longer than wildtype controls after aerosol infection with Mtb (121). However, SRA-deficient mice inoculated via intranasal route control bacterial burden similarly to wildtype mice (84). In contrast, mice doubly deficient for SRA and CD36 have an early impairment in controlling bacterial burden, but have no significant differences in survival compared to wildtype mice (84). These differences may be due to an aerosol (121) compared to intranasal infection route (84).
AIM
AIM is a SR that is secreted by tissue macrophages and was first described for its ability to promote macrophage survival. More recently it has been implicated in pathogen recognition (122). AIM levels are increased in the serum of Mtb infected mice (123). Stable expression of AIM in THP-1 macrophages leads to increased macrophage viability and decreased bacterial load upon infection with Mtb (123). AIM expression enhances Mtb-induced cathelicidin production, with a concomitant increase in Beclin-1 mRNA and LC3 conversion, both hallmarks of autophagy activation (123). These data suggest that AIM may provide a protective role in Mtb infection by the induction of autophagy.
Scavenger receptors: the bacterial view
SR recognition of Mtb can be detrimental to host cells. While SRA can bind TDM-coated beads, BMDMs that express high levels of SRA produce less TNF in response to TDM-beads than cells expressing high levels of MARCO (17). Furthermore, while mouse alveolar macrophages produce TNF in response to TDM, alveolar macrophages pretreated with an SRA monoclonal antibody, or those isolated from SRA−/− mice produce significantly more TNF in response to TDM (124). Together these data suggest SRA may play an anti-inflammatory role in the recognition of Mtb mycolic acids.
Mycobacteria may preferentially utilize CD36 for uptake and entry into cells. RNAi knockdown of a CD36 family member in Drosophila S2 cells inhibits Mycobacterium fortuitum infection and prevents uptake of M. smegmatis (125). Furthermore, expression of CD36 in HEK293 cells makes them permissive to M. fortuitum infection (125). While differences in bacterial uptake are not observed in CD36−/− murine macrophages, CD36−/− mice are able to control bacterial burden better than wildtype littermates (117). The absence of CD36 may lead to an anti-inflammatory environment. CD36−/− macrophages produce more IL-10 than CD36+/+ macrophages upon infection with M. marinum or BCG (117). Overall, these data suggest that CD36 contributes to mycobacterial survival.
Other cell surface receptors
Complement receptor 3
Opsonization of Mtb by complement cleavage fragment C3b and its proteolytically inactive form iC3b leads to complement receptor 3 (CR3)-mediated phagocytosis of Mtb (126). CR3 contains multiple binding sites that allows it to bind iC3b, intracellular adhesion molecule-1 (ICAM-1), and a variety of bacterial products including mycobacterial antigen 85C and mycobacterial oligosaccharides like LAM (127). While CR3-deficient macrophages display decreased uptake of Mtb, there is no effect on the induction of anti-microbial effector mechanisms or bacterial survival in vitro (128). Additionally, CR3−/− mice exhibit no defects in the control of Mtb infection (129), suggesting CR3 does not play a critical role in the immune response to mycobacteria.
CD14
The ability of CD14 to bind LPS from Gram-negative bacteria and lipoteichoic acids and peptidoglycan from Gram-positive bacteria is well described (130, 131). However, CD14 can also bind LAM (131) and chaperonin 60.1 (132) to mediate uptake of Mtb (133). CD14−/− mice do not exhibit differences in the protective response to Mtb up to 32 weeks post infection (66, 134). However after 32 weeks, while no differences in bacterial numbers are observed, CD14−/− mice display decreased pulmonary inflammation and overall increased survival compared to wildtype control mice (134), suggesting a role for CD14 in the chronic inflammatory response seen during Mtb infection.
Intracellular receptors
Microbes that communicate with or exist within the cytoplasm are not immune to recognition from the host. Mtb has long been considered a pathogen with a purely intraphagosomal existence, but recent data have highlighted that Mtb not only directly communicates with the cytoplasm (64) but also may completely escape the phagosome to reside in the cytoplasm (135). In that respect, it is no surprise that mycobacterial PAMPs engage intracellular PRRs either in the cytoplasm or attached to the cytoplasmic face of organelles. These include leucine-rich repeat (LRR)-containing nucleotide binding and oligomerization domain (NOD) proteins, NOD-like receptors (NLRs), and DNA/RNA sensing retinoid acid-inducible gene I (RIG-I)-like receptors (RLRs). Activation of cytoplasmic PRRs leads to NFκB-dependent and independent pro-inflammatory cytokine production, autophagy activation, and in some cases cell death (136–138). To date, the intracellular PRRs that recognize Mtb PAMPs include NOD2, NLRP3, AIM2, and an as yet unidentified DNA sensor (78, 139).
NODs
NODs are a cytoplasmic family of over 20 different pattern recognition receptors. Generally, NOD1 and NOD2 recognize bacterial cell wall peptidoglycan, leading to the NFκB-dependent production of pro-inflammatory cytokines (140). Of the Mtb ligands, NOD2 recognizes the Mtb cell wall component mycolyarabinogalactan-peptidoglycan (141). In vitro, macrophages from NOD2 deficient mice produce decreased levels of TNF, IL-12p40, RANTES, and NO following Mtb infection, although no defects in control of Mtb replication are observed (141). Studies in NOD2 deficient mice show no difference in bacterial numbers in the first few months after infection (141, 142). However, NOD2−/− mice have higher bacterial numbers six months post infection, reduced activation of antigen specific T cells, and succumb to infection sooner than wildtype mice (142). While no differences in IL-12p40 or iNOS expression are seen in NOD2-deficient animals, IFN-γ and TNF concentrations in BAL are significantly reduced (142). Similarly, knockdown of NOD2 in human monocyte derived macrophages leads to decreased secretion of TNF and IL-1β following infection. Consequently, and in contrast to mouse macrophages, NOD2 silencing in human macrophages leads to increased intracellular bacterial growth (143). Treatment of Mtb infected human alveolar macrophages with MDP (a NOD2 ligand) improves their ability to control Mtb infection (144) by increasing TNF and IL-6 production and increasing autophagocytosis of Mtb as measured by LC3 colocalization with intracellular bacteria (144). These latter results suggest a link between NOD2 activation and induction of autophagy, leading to enhanced control of Mtb intracellular growth.
A NOD2 polymorphism that predicts defective NOD2 activity has been identified in an African American cohort that is associated with increased susceptibility to Mtb disease (145). However, two other polymorphisms in NOD2 identified in the same cohort were correlated with decreased susceptibility (145). Studies in a Chinese cohort have identified an additional NOD2 polymorphism associated with an increased risk of Mtb, although this polymorphism was not associated with an increased risk of disease in Uyghur or Kazak populations (146, 147). Thus, although NOD2 activity is important in macrophages and in mice, whether NOD2 contributes directly to human immunity to Mtb remains unknown.
NLRP3
NLR family, pyrin domain-containing 3 (NLRP3) is a vital protein for the formation of a multiprotein complex termed the NLRP3 inflammasome. The pyrin domain of the NLR interacts with the pyrin domain of apoptosis-associated speck-like protein containing a CARD (ASC), thus bridging the NLR domain of NLRP3 to caspase-1, a cysteine protease (140). Activation of caspase-1, and to a lesser extent, caspase-11 leads to the cleavage of pro-IL-1β and pro-IL-18, allowing for their secretion (148). NLRP3 mediated inflammasome activation follows Mtb infection (149) and caspase-1 activation and production of IL-1β in both human and mouse macrophages is dependent on NLRP3 in vitro (150). Human macrophages with a gain of function mutation in NLRP3 are better able to control Mtb growth (151). However, NLRP3−/− mice show no differences in IL-1β production, bacterial burden, or survival following Mtb infection compared to wild type mice (150).
A specific NLRP3 ligand remains elusive, and in fact, a ligand may not exist that binds directly to NLRP3. However, caspase-1 activation and IL-1β production by Mtb requires a functional Mtb ESX-1 secretion system (139, 152). Furthermore, treatment of THP-1 macrophages with ESAT-6, a pore forming molecule (7), is sufficient to activate caspase-1 for IL-1β production (152). Although ESAT-6 may interact directly with the inflammasome (152) membrane damage alone also triggers inflammasome activation (153, 154). Pore formation, membrane damage and phagocytosis lead to potassium depletion in the cytosol (155), and Mtb activation of inflammasomes requires potassium depletion (139). Finally, NLRP3 can be activated by the DExD/H-box RNA helicase family member DHX33 that senses viral and bacterial cytosolic RNA (156). Whether DHX33 is involved in the sensing of an Mtb ligand has yet to be determined.
AIM2 and DNA sensing
Binding of the cytosolic protein absent in melanoma 2 (AIM2) to double stranded DNA (dsDNA) can activate the inflammasome, leading to production of IL-1β and IL-18 (157, 158). Although Mtb DNA reaches the cytoplasm in an ESX-1-dependent manner (64) and Mtb DNA co-localizes with AIM2 in infected cells (159), activation of the AIM2-inflammasome pathway only occurs following infection with avirulent Mtb. Infection of BMDCs with M. smegmatis or avirulent H37Ra leads to AIM2 dependent- IL-1β secretion, but H37Rv induced IL-1β production is not dependent on AIM2 (160). Thus, it has been suggested that virulent mycobacteria inhibit AIM2 activation in vitro (160). In contrast, in vivo work suggests a role for AIM2-inflammasome activation in the response to virulent Mtb. AIM2−/− mice have impaired IL-1β and IL-18 production, increased bacterial burden, abnormal granuloma formation, and decreased survival compared to wildtype mice following infection (159). In light of the recent discovery that extracellular DNA can induce type I interferon signaling in macrophages and mice infected with Mtb (64), it is likely that the enhanced susceptibility of AIM2−/− mice reflects excess accumulation of intracellular DNA leading to a pathologic exuberant inflammatory response.
While Mtb DNA may not act directly through AIM2, it may be sensed through other cytoplasmic receptors. The cytosolic surveillance pathway (CSP) is activated in response to cytoplasmic bacteria and bacterial or viral products that gain access to the cytoplasm through damage to the phagosomal membrane (64, 161). While not all CSP sensors are known, they converge on activation of the endoplasmic reticulum protein stimulator of IFN genes (STING). Activated STING dimerizes, translocates to the Golgi, and facilitates tank-binding kinase-1 (TBK) activation, resulting in the phosphorylation of the transcription factor IRF3. Phosphorylated IRF3 then dimerizes and translocates to the nucleus where it induces IFN-β production (162, 163). Mtb infection of murine BMDMs (63) as well as human THP-1 macrophages and primary human monocytes (164) leads to production of IFN-β. This early IFN-β production is absent during macrophage infection by a Mtb mutant lacking the region of difference 1 (RD1), where the ESAT-6 secretion system (ESX)-1 is located (164) or by a mutant lacking ESAT-6 (esxA) (63). Furthermore, esxA is required for Mtb to induce IFN-β production in the lungs and spleen of mice challenged by aerosol infection in the early stages of infection (63). These data suggest that Mtb induced IFN-β production is mediated in part by the ESX-1 secretion system.
In mouse macrophages, ESX-1-dependent IFN-β production requires the CSP as IRF3−/−, TBK1−/−, or STING−/− BMDM produce no IFN-β in response to Mtb infection (64). Activation of the CSP by Mtb is likely through exposure to mycobacterial DNA as IFN-β production is decreased in macrophages over-expressing the cytoplasmic nuclease TREX-1 and, similarly, TREX-1 knockdown led to higher levels of INF-β (64). Knockdown of a putative DNA sensor IFI204 in murine BMDMs leads to a decrease but not complete loss of IFN- β production when challenged with Mtb (64). This finding suggests that IFI204 can activate the CSP in response to Mtb but may not be the only DNA sensor Mtb engages during infection.
A dsDNA sensor was discovered recently that can activate the CSP through production of cyclic GMP-AMP (cGAMP) with subsequent cGAMP-mediated activation of STING (165, 166). The cGAMP synthase (cGAS) is important for activating the CSP in response to DNA viruses and retroviruses (167). In vitro, cGAS also plays a role in IFN-β production in response to the intracellular pathogens Chlamydia trachomatis (168) and Listeria monocytogenes (169). Some bacteria such as L. monocytogenes, and Legionella pneumophila can produce cyclic dinucleotides such as cyclic-di-AMP and cyclic di-GMP, which are capable of bypassing cGAS to activate STING directly (170, 171). Whether Mtb, which requires STING to induce IFN-β in macrophages (64), also secretes cyclic dinucleotides is controversial. Although one study found no evidence of cyclic nucleotide secretion or a role for cyclic nucleotide synthesis in Mtb mediated IFN-β production (64), another recent study demonstrated that infection of macrophages with Mtb deleted for a cyclic di-AMP phosphodiesterase (cnpB) that secreted more c-di-AMP resulted in higher levels of IFN-β production by macrophages (172). Although this could be due to c-di-AMP mediated activation of STING, an alternative explanation is that the excess c-di-AMP produced by mutant Mtb affects mycobacterial cell wall homeostasis, as has been observed for L. monocytogenes (173), which then affects dsDNA release and activation of cGAS. Thus, how Mtb activates the CSP awaits analysis of the role of cGAS in Mtb infection. Based on its role in activating STING, we predict that cGAS will be essential for Mtb to activate the CSP.
Intracellular receptors: the bacterial view
Although cytoplasmic Mtb components can be sensed by intracellular PRRs to activate host defenses, activation of the CSP can also benefit the bacterium. As noted above, CSP induces production of Type-I IFNs (34, 63), and this can promote bacterial survival (64). Mice infected with the hypervirulent Mtb Beijing isolates HN878 or W4 express more IFN-α in the lungs than when infected with a less virulent strain NHN5 (174, 175). Mtb-infected mice given IFN-α/β intranasally fail to control bacterial burden in lungs and succumb to infection significantly quicker than untreated mice or those treated with IFN-γ (174). Consequently, deletion of IFNAR, which abrogates signaling in response to Type-I IFN, protects mice from aerosol infection with both hypervirulent and virulent strains of Mtb (175).
The protection afforded to Mtb by IFN-β may be due to its antagonistic effects on IFN-γ. Differential expression of IFNs leads to distinct manifestations of Mycobacterium leprae. Self-healing tuberculoid lesions (T-lep) are associated with high levels of IFN-γ and antimicrobial peptide production, whereas disseminated lepromatous (L-lep) lesions are associated with low levels of INF-γ but high levels of IFN-β and IL-10 (176). IFN-γ-treated monocytes upregulate antimicrobial peptides and are able to inhibit M. leprae growth by about 35% (176). However, INF-γ-mediated antimicrobial peptide production and M. leprae restriction are completely abrogated by the addition of IFN-β or IL-10, suggesting that IFN-β antagonizes the antimicrobial activity of IFN-γ (176). Finally, Type I IFN-inducible gene transcripts are over-represented in the blood of patients with active, but not latent Mtb, suggesting a role for type I IFN production in human tuberculosis (177).
Conclusions
A large number of mycobacterial PAMPs bind to and activate host immune receptors. These PAMPs span the full repertoire of chemical structures, from lipids and lipoproteins to carbohydrates, nucleotides and proteins. Such variety necessitates an equally diverse set of host receptors located at both the cell surface and the cytoplasm. When reduced to the level of individual molecules and receptors, clear patterns and signaling mechanisms emerge. However, the real world interactions of mycobacteria with human cells are much more complex, as multiple PAMPs and their receptors are engaged simultaneously. Thus, future work in determining the nature of Mtb-host interactions may require a systems biology approach. To that end, leveraging new high-throughput genetic approaches may facilitate a broader understanding of how Mtb is sensed by host cells. Furthermore, as this review has highlighted, mouse and human studies of identical receptors can be discrepant. Thus, increasing the use of primary human tissues and cells will likely yield a more precise understanding of how Mtb is sensed, with greater application to human disease.
Acknowledgments
We thank David Greenberg for reviewing the manuscript and Richard Howdy (Visually Medical) for artwork. This work was supported by NIH grants R01 AI099439, R21 AI111023, U19 AI109725, and T32 AI007520. M.U.S. is a Disease Oriented Clinical Scholar at UT Southwestern.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Kaur D, Guerin ME, Skovierová H, Brennan PJ, Jackson M. Chapter 2: Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol. 2009;69:23–78. doi: 10.1016/S0065-2164(09)69002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol Rev. 2011;35:1126–1157. doi: 10.1111/j.1574-6976.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitarque S, Larrouy-Maumus G, Payré B, Jackson M, Puzo G, Nigou J. The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis (Edinb) 2008;88:560–565. doi: 10.1016/j.tube.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox JS, Chen B, McNeil M, Jacobs WR. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 5.Jain M, Cox JS. Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 2005;1:e2. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jonge MI, et al. ESAT-6 from Mycobacterium tuberculosis Dissociates from Its Putative Chaperone CFP-10 under Acidic Conditions and Exhibits Membrane-Lysing Activity. Journal of Bacteriology. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J, et al. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda K. Toll-like receptors in innate immunity. Int Immunol. 2004;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 9.Basu J, Shin D-M, Jo E-K. Mycobacterial signaling through toll-like receptors. Front Cell Infect Microbiol. 2012;2:145. doi: 10.3389/fcimb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Zeng J, Xie J. Navigating through the maze of TLR2 mediated signaling network for better mycobacterium infection control. Biochimie. 2014;102:1–8. doi: 10.1016/j.biochi.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc Natl Acad Sci USA. 1999;96:14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 13.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 14.Fabri M, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones BW, et al. Different Toll-like receptor agonists induce distinct macrophage responses. J Leukoc Biol. 2001;69:1036–1044. [PubMed] [Google Scholar]
- 16.Quesniaux VJ, et al. Toll-like receptor 2 (TLR2)-dependent-positive and TLR2-independent-negative regulation of proinflammatory cytokines by mycobacterial lipomannans. J Immunol. 2004;172:4425–4434. doi: 10.4049/jimmunol.172.7.4425. [DOI] [PubMed] [Google Scholar]
- 17.Bowdish DME, et al. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000474. doi: 10.1371/journal.ppat.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prados-Rosales R, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin D-M, et al. Mycobacterium tuberculosis lipoprotein-induced association of TLR2 with protein kinase C zeta in lipid rafts contributes to reactive oxygen species-dependent inflammatory signalling in macrophages. Cell Microbiol. 2008;10:1893–1905. doi: 10.1111/j.1462-5822.2008.01179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López M, Sly LM, Luu Y, Young D, Cooper H, Reiner NE. The 19-kDa Mycobacterium tuberculosis protein induces macrophage apoptosis through Toll-like receptor-2. J Immunol. 2003;170:2409–2416. doi: 10.4049/jimmunol.170.5.2409. [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee S, et al. Early Secreted Antigen ESAT-6 of Mycobacterium tuberculosis Promotes Protective T Helper 17 Cell Responses in a Toll-Like Receptor-2-dependent Manner. PLoS Pathog. 2011;7:e1002378. doi: 10.1371/journal.ppat.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, et al. Recombinant TB10.4 of Mycobacterium bovis induces cytokine production in RAW264. 7 macrophages through activation of the MAPK and NF-κB pathways via TLR2. Mol Immunol. 2014;62:227–234. doi: 10.1016/j.molimm.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Byun E-H, et al. Mycobacterium tuberculosis Rv0577, a novel TLR2 agonist, induces maturation of dendritic cells and drives Th1 immune response. The FASEB Journal. 2012;26:2695–2711. doi: 10.1096/fj.11-199588. [DOI] [PubMed] [Google Scholar]
- 24.Bulut Y, et al. Mycobacterium Tuberculosis Heat Shock Proteins Use Diverse Toll-like Receptor Pathways to Activate Pro-inflammatory Signals. Journal of Biological Chemistry. 2005;280:20961–20967. doi: 10.1074/jbc.M411379200. [DOI] [PubMed] [Google Scholar]
- 25.Basu S, et al. Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J Biol Chem. 2007;282:1039–1050. doi: 10.1074/jbc.M604379200. [DOI] [PubMed] [Google Scholar]
- 26.Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drennan MB, et al. Toll-like receptor 2-deficient mice succumb to Mycobacterium tuberculosis infection. Am J Pathol. 2004;164:49–57. doi: 10.1016/S0002-9440(10)63095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hölscher C, et al. Containment of aerogenicMycobacterium tuberculosis infection in mice does not require MyD88 adaptor function for TLR2, -4 and -9. Eur J Immunol. 2008;38:680–694. doi: 10.1002/eji.200736458. [DOI] [PubMed] [Google Scholar]
- 29.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009 doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moy RH, Cherry S. Antimicrobial autophagy: a conserved innate immune response in Drosophila. J Innate Immun. 2013;5:444–455. doi: 10.1159/000350326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes LC, Dikic I. Autophagy in Antimicrobial Immunity. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Castillo EF, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA. 2012;109:E3168–76. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 36.Shin D-M, et al. Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell Microbiol. 2010;12:1648–1665. doi: 10.1111/j.1462-5822.2010.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Song C, Snyder GA, Sundberg EJ. R753Q polymorphism inhibits Toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response. Journal of Biological. 2012 doi: 10.1074/jbc.M112.375493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalgic N, et al. Arg753Gln polymorphism of the human Toll-like receptor 2 gene from infection to disease in pediatric tuberculosis. Hum Immunol. 2011;72:440–445. doi: 10.1016/j.humimm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Ogus AC, et al. The Arg753Gln polymorphism of the human Toll-like receptor 2 gene in tuberculosis disease. European Respiratory Journal. 2004;23:219–223. doi: 10.1183/09031936.03.00061703. [DOI] [PubMed] [Google Scholar]
- 40.Wang J-J, et al. Meta-analysis on the associations of TLR2 gene polymorphisms with pulmonary tuberculosis susceptibility among Asian populations. PLoS ONE. 2013;8:e75090. doi: 10.1371/journal.pone.0075090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS ONE. 2007;2:e1318. doi: 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez D, Lefebvre C, Rioux J. Evaluation of Toll-like receptor and adaptor molecule polymorphisms for susceptibility to tuberculosis in a Colombian population. Int J Immunogenet. 2012;39:216–223. doi: 10.1111/j.1744-313X.2011.01077.x. [DOI] [PubMed] [Google Scholar]
- 43.Selvaraj P, Harishankar M, Singh B, Jawahar MS, Banurekha VV. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis (Edinb) 2010;90:306–310. doi: 10.1016/j.tube.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Parveen N, Varman R, Nair S, Das G, Ghosh S, Mukhopadhyay S. Endocytosis of Mycobacterium tuberculosis heat shock protein 60 is required to induce interleukin-10 production in macrophages. Journal of Biological Chemistry. 2013;288:24956–24971. doi: 10.1074/jbc.M113.461004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair S, et al. The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. The Journal of Immunology. 2009;183:6269–6281. doi: 10.4049/jimmunol.0901367. [DOI] [PubMed] [Google Scholar]
- 46.Pathak SK, et al. Direct extracellular interaction between the early secreted antigen ESAT-6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat Immunol. 2007;8:610–618. doi: 10.1038/ni1468. [DOI] [PubMed] [Google Scholar]
- 47.Rocha-Ramírez LM, et al. Mycobacterium tuberculosis lipids regulate cytokines, TLR-2/4 and MHC class II expression in human macrophages. Tuberculosis. 2008;88:212–220. doi: 10.1016/j.tube.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Gehring AJ, Rojas RE, Canaday DH, Lakey DL, Harding CV, Boom WH. The Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits gamma interferon-regulated HLA-DR and Fc gamma R1 on human macrophages through Toll-like receptor 2. Infect Immun. 2003;71:4487–4497. doi: 10.1128/IAI.71.8.4487-4497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gehring AJ, Dobos KM, Belisle JT, Harding CV, Boom WH. Mycobacterium tuberculosis LprG (Rv1411c): A Novel TLR-2 Ligand That Inhibits Human Macrophage Class II MHC Antigen Processing. The Journal of Immunology. 2004;173:2660–2668. doi: 10.4049/jimmunol.173.4.2660. [DOI] [PubMed] [Google Scholar]
- 50.Pecora ND, Gehring AJ, Canaday DH, Boom WH, Harding CV. Mycobacterium tuberculosis LprA is a lipoprotein agonist of TLR2 that regulates innate immunity and APC function. J Immunol. 2006;177:422–429. doi: 10.4049/jimmunol.177.1.422. [DOI] [PubMed] [Google Scholar]
- 51.Drage MG, et al. TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell Immunol. 2009;258:29–37. doi: 10.1016/j.cellimm.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeuchi O, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 53.Jin MS, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Nakata T, et al. CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cell Microbiol. 2006;8:1899–1909. doi: 10.1111/j.1462-5822.2006.00756.x. [DOI] [PubMed] [Google Scholar]
- 55.Hoebe K, et al. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 56.Oliveira-Nascimento L, Massari P, Wetzler LM. The Role of TLR2 in Infection and Immunity. Frontiers in Immunology. 2012;3:1–17. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Bergenhenegouwen J, et al. TLR2 & Co: a critical analysis of the complex interactions between TLR2 and coreceptors. J Leukoc Biol. 2013;94:885–902. doi: 10.1189/jlb.0113003. [DOI] [PubMed] [Google Scholar]
- 58.Jimenez-Dalmaroni MJ, et al. Soluble human TLR2 ectodomain binds diacylglycerol from microbial lipopeptides and glycolipids. Innate Immun. 2014 doi: 10.1177/1753425914524077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doz E, et al. Acylation determines the toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor-, and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J Biol Chem. 2007;282:26014–26025. doi: 10.1074/jbc.M702690200. [DOI] [PubMed] [Google Scholar]
- 60.Cehovin A, et al. Comparison of the moonlighting actions of the two highly homologous chaperonin 60 proteins of Mycobacterium tuberculosis. Infect Immun. 2010;78:3196–3206. doi: 10.1128/IAI.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim K, et al. Mycobacterium tuberculosis Rv0652 stimulates production of tumour necrosis factor and monocytes chemoattractant protein-1 in macrophages through the Toll-like receptor 4 pathway. Immunology. 2012;136:231–240. doi: 10.1111/j.1365-2567.2012.03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung SB, et al. The Mycobacterial 38-Kilodalton Glycolipoprotein Antigen Activates the Mitogen-Activated Protein Kinase Pathway and Release of Proinflammatory Cytokines through Toll-Like Receptors 2 and 4 in Human Monocytes. Infect Immun. 2006;74:2686–2696. doi: 10.1128/IAI.74.5.2686-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 64.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carmona J, et al. Mycobacterium tuberculosis Strains Are Differentially Recognized by TLRs with an Impact on the Immune Response. PLoS ONE. 2013;8:e67277. doi: 10.1371/journal.pone.0067277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reiling N, et al. Cutting Edge: Toll-Like Receptor (TLR)2- and TLR4-Mediated Pathogen Recognition in Resistance to Airborne Infection with Mycobacterium tuberculosis. The Journal of Immunology. 2002;169:3480–3484. doi: 10.4049/jimmunol.169.7.3480. [DOI] [PubMed] [Google Scholar]
- 67.Shim TS, Turner OC, Orme IM. Toll-like receptor 4 plays no role in susceptibility of mice to Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2003;83:367–371. doi: 10.1016/s1472-9792(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 68.Abel B, et al. Toll-like receptor 4 expression is required to control chronic Mycobacterium tuberculosis infection in mice. J Immunol. 2002;169:3155–3162. doi: 10.4049/jimmunol.169.6.3155. [DOI] [PubMed] [Google Scholar]
- 69.Pulido I, Leal M, Genebat M, Pacheco YM, Sáez ME, Soriano-Sarabia N. The TLR4 ASP299GLY polymorphism is a risk factor for active tuberculosis in Caucasian HIV-infected patients. Curr HIV Res. 2010;8:253–258. doi: 10.2174/157016210791111052. [DOI] [PubMed] [Google Scholar]
- 70.Kiemer AK, et al. Attenuated activation of macrophage TLR9 by DNA from virulent mycobacteria. J Innate Immun. 2009;1:29–45. doi: 10.1159/000142731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres-García D, et al. Variants in toll-like receptor 9 gene influence susceptibility to tuberculosis in a Mexican population. J Transl Med. 2013;11:220. doi: 10.1186/1479-5876-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kobayashi K, et al. Association of TLR polymorphisms with development of tuberculosis in Indonesian females. Tissue Antigens. 2012;79:190–197. doi: 10.1111/j.1399-0039.2011.01821.x. [DOI] [PubMed] [Google Scholar]
- 73.Velez DR, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet. 2010;127:65–73. doi: 10.1007/s00439-009-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doz E, et al. Mycobacterial phosphatidylinositol mannosides negatively regulate host Toll-like receptor 4, MyD88-dependent proinflammatory cytokines, and TRIF-dependent co-stimulatory molecule expression. J Biol Chem. 2009;284:23187–23196. doi: 10.1074/jbc.M109.037846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cambier CJ, et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cambi A, Koopman M, Figdor CG. How C-type lectins detect pathogens. Cell Microbiol. 2005;7:481–488. doi: 10.1111/j.1462-5822.2005.00506.x. [DOI] [PubMed] [Google Scholar]
- 77.Cambi A, Figdor CG. Dual function of C-type lectin-like receptors in the immune system. Curr Opin Cell Biol. 2003;15:539–546. doi: 10.1016/j.ceb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 78.Killick KE, Ní Cheallaigh C, O’Farrelly C, Hokamp K, MacHugh DE, Harris J. Receptor-mediated recognition of mycobacterial pathogens. Cell Microbiol. 2013;15:1484–1495. doi: 10.1111/cmi.12161. [DOI] [PubMed] [Google Scholar]
- 79.Schlesinger LS. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. The Journal of Immunology. 1993;150:2920–2930. [PubMed] [Google Scholar]
- 80.Martinez-Pomares L. The mannose receptor. J Leukoc Biol. 2012;92:1177–1186. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 81.Chieppa M, et al. Cross-linking of the mannose receptor on monocyte-derived dendritic cells activates an anti-inflammatory immunosuppressive program. J Immunol. 2003;171:4552–4560. doi: 10.4049/jimmunol.171.9.4552. [DOI] [PubMed] [Google Scholar]
- 82.Rajaram MVS, Brooks MN, Morris JD, Torrelles JB, Azad AK, Schlesinger LS. Mycobacterium tuberculosis activates human macrophage peroxisome proliferator-activated receptor gamma linking mannose receptor recognition to regulation of immune responses. J Immunol. 2010;185:929–942. doi: 10.4049/jimmunol.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kang PB. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. Journal of Experimental Medicine. 2005;202:987–999. doi: 10.1084/jem.20051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Court N, et al. Partial redundancy of the pattern recognition receptors, scavenger receptors, and C-type lectins for the long-term control of Mycobacterium tuberculosis infection. The Journal of Immunology. 2010;184:7057–7070. doi: 10.4049/jimmunol.1000164. [DOI] [PubMed] [Google Scholar]
- 85.Zhang X, et al. The novel human MRC1 gene polymorphisms are associated with susceptibility to pulmonary tuberculosis in Chinese Uygur and Kazak populations. Mol Biol Rep. 2013;40:5073–5083. doi: 10.1007/s11033-013-2610-7. [DOI] [PubMed] [Google Scholar]
- 86.Zhang X, et al. Polymorphic allele of human MRC1 confer protection against tuberculosis in a Chinese population. Int J Biol Sci. 2012;8:375–382. doi: 10.7150/ijbs.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soilleux EJ, et al. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J Leukoc Biol. 2002;71:445–457. [PubMed] [Google Scholar]
- 88.Garcia-Vallejo JJ, Van Kooyk Y. The physiological role of DC-SIGN: a tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 89.Tailleux L, et al. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanne A, et al. A murine DC-SIGN homologue contributes to early host defense against Mycobacterium tuberculosis. J Exp Med. 2009;206:2205–2220. doi: 10.1084/jem.20090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barreiro LB, Neyrolles O, Babb CL, Tailleux L, Quach H. Promoter variation in the DC-SIGN–encoding gene CD209 is associated with tuberculosis. PLoS medicine. 2006 doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vannberg FO, et al. CD209 genetic polymorphism and tuberculosis disease. PLoS ONE. 2008;3:e1388. doi: 10.1371/journal.pone.0001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ben-Ali M, et al. Promoter and neck region length variation of DC-SIGN is not associated with susceptibility to tuberculosis in Tunisian patients. Hum Immunol. 2007;68:908–912. doi: 10.1016/j.humimm.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Zheng R, et al. Relationship between polymorphism of DC-SIGN (CD209) gene and the susceptibility to pulmonary tuberculosis in an eastern Chinese population. Hum Immunol. 2011;72:183–186. doi: 10.1016/j.humimm.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang K, et al. Association between CD209-336A/G and-871A/G polymorphisms and susceptibility of tuberculosis: a meta-analysis. PLoS ONE. 2012 doi: 10.1371/journal.pone.0041519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miao R, Li J, Sun Z, Li C, Xu F. Association between the CD209 promoter -336A/G polymorphism and susceptibility to tuberculosis: a meta-analysis. Respirology. 2012;17:847–853. doi: 10.1111/j.1440-1843.2012.02185.x. [DOI] [PubMed] [Google Scholar]
- 97.Öhman T, et al. Dectin-1 pathway activates robust autophagy-dependent unconventional protein secretion in human macrophages. The Journal of Immunology. 2014;192:5952–5962. doi: 10.4049/jimmunol.1303213. [DOI] [PubMed] [Google Scholar]
- 98.Lee H-M, Yuk J-M, Shin D-M, Jo E-K. Dectin-1 is Inducible and Plays an Essential Role for Mycobacteria-Induced Innate Immune Responses in Airway Epithelial Cells. J Clin Immunol. 2009;29:795–805. doi: 10.1007/s10875-009-9319-3. [DOI] [PubMed] [Google Scholar]
- 99.Yadav M, Schorey JS. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108:3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Das R, et al. Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110:E2997–3006. doi: 10.1073/pnas.1301128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rothfuchs AG, et al. Dectin-1 interaction with Mycobacterium tuberculosis leads to enhanced IL-12p40 production by splenic dendritic cells. J Immunol. 2007;179:3463–3471. doi: 10.4049/jimmunol.179.6.3463. [DOI] [PubMed] [Google Scholar]
- 102.Zenaro E, Donini M, Dusi S. Induction of Th1/Th17 immune response by Mycobacterium tuberculosis: role of dectin-1, Mannose Receptor, and DC-SIGN. J Leukoc Biol. 2009;86:1393–1401. doi: 10.1189/jlb.0409242. [DOI] [PubMed] [Google Scholar]
- 103.Marakalala MJ, et al. The Syk/CARD9-coupled receptor Dectin-1 is not required for host resistance to Mycobacterium tuberculosis in mice. Microbes Infect. 2011;13:198–201. doi: 10.1016/j.micinf.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schoenen H, et al. Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. The Journal of Immunology. 2010;184:2756–2760. doi: 10.4049/jimmunol.0904013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee W-B, et al. Neutrophils Promote Mycobacterial Trehalose Dimycolate-Induced Lung Inflammation via the Mincle Pathway. PLoS Pathog. 2012;8:e1002614. doi: 10.1371/journal.ppat.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heitmann L, Schoenen H, Ehlers S, Lang R. Mincle is not essential for controlling Mycobacterium tuberculosis infection. Immunobiology. 2013;218:506–516. doi: 10.1016/j.imbio.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 108.Behler F, et al. Role of Mincle in alveolar macrophage-dependent innate immunity against mycobacterial infections in mice. The Journal of Immunology. 2012;189:3121–3129. doi: 10.4049/jimmunol.1201399. [DOI] [PubMed] [Google Scholar]
- 109.Miyake Y, et al. C-type lectin MCL is an FcRγ-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity. 2013;38:1050–1062. doi: 10.1016/j.immuni.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 110.Wu T, et al. Interaction between mannosylated lipoarabinomannan and dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin influences dendritic cells maturation and T cell immunity. Cell Immunol. 2011;272:94–101. doi: 10.1016/j.cellimm.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 111.Geijtenbeek TBH, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003;197:7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol. 2013;13:621–634. doi: 10.1038/nri3515. [DOI] [PubMed] [Google Scholar]
- 113.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martin C, Chevrot M, Poirier H, Passilly-Degrace P, Niot I, Besnard P. CD36 as a lipid sensor. Physiol Behav. 2011;105:36–42. doi: 10.1016/j.physbeh.2011.02.029. [DOI] [PubMed] [Google Scholar]
- 115.Józefowski S, Sobota A, Paw owski A, Kwiatkowska K. Mycobacterium tuberculosis lipoarabinomannan enhances LPS-induced TNF-α production and inhibits NO secretion by engaging scavenger receptors. Microb Pathog. 2011;50:350–359. doi: 10.1016/j.micpath.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 116.Jimenez-Dalmaroni MJ, et al. Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS ONE. 2009;4:e7411. doi: 10.1371/journal.pone.0007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hawkes M, et al. CD36 deficiency attenuates experimental mycobacterial infection. BMC Infect Dis. 2010;10:299. doi: 10.1186/1471-2334-10-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kraal G, van der Laan L, Elomaa O. The macrophage receptor MARCO. Microbes Infect. 2000;2:313–316. doi: 10.1016/s1286-4579(00)00296-3. [DOI] [PubMed] [Google Scholar]
- 119.Bowdish DM, et al. Genetic variants of MARCO are associated with susceptibility to pulmonary tuberculosis in a Gambian population. BMC Med Genet. 2013;14:47. doi: 10.1186/1471-2350-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ma M-J, et al. Genetic variants in MARCO are associated with the susceptibility to pulmonary tuberculosis in Chinese Han population. PLoS ONE. 2011;6:e24069. doi: 10.1371/journal.pone.0024069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sever-Chroneos Z, Tvinnereim A, Hunter RL, Chroneos ZC. Prolonged survival of scavenger receptor class A-deficient mice from pulmonary Mycobacterium tuberculosis infection. Tuberculosis. 2011;91:S69–S74. doi: 10.1016/j.tube.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martinez VG, et al. The macrophage soluble receptor AIM/Api6/CD5L displays a broad pathogen recognition spectrum and is involved in early response to microbial aggression. Cellular & Molecular Immunology. 2014;11:343–354. doi: 10.1038/cmi.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanjurjo L, et al. The scavenger protein apoptosis inhibitor of macrophages (AIM) potentiates the antimicrobial response against Mycobacterium tuberculosis by enhancing autophagy. PLoS ONE. 2013;8:e79670. doi: 10.1371/journal.pone.0079670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ozeki Y, et al. Macrophage scavenger receptor down-regulates mycobacterial cord factor-induced proinflammatory cytokine production by alveolar and hepatic macrophages. Microb Pathog. 2006;40:171–176. doi: 10.1016/j.micpath.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 125.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 126.Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- 127.Velasco-Velázquez MA, Barrera D. Macrophage— Mycobacterium tuberculosis interactions: role of complement receptor 3. Microb Pathog. 2003;35:125–131. doi: 10.1016/s0882-4010(03)00099-8. [DOI] [PubMed] [Google Scholar]
- 128.Rooyakkers AWJ, Stokes RW. Absence of complement receptor 3 results in reduced binding and ingestion of Mycobacterium tuberculosis but has no significant effect on the induction of reactive oxygen and nitrogen intermediates or on the survival of the bacteria in resident and interferon-gamma activated macrophages. Microb Pathog. 2005;39:57–67. doi: 10.1016/j.micpath.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 129.Hu C, Mayadas-Norton T, Tanaka K, Chan J, Salgame P. Mycobacterium tuberculosis infection in complement receptor 3-deficient mice. J Immunol. 2000;165:2596–2602. doi: 10.4049/jimmunol.165.5.2596. [DOI] [PubMed] [Google Scholar]
- 130.Dziarski R, Ulmer AJ, Gupta D. Interactions of CD14 with components of gram-positive bacteria. Chem Immunol. 2000;74:83–107. doi: 10.1159/000058761. [DOI] [PubMed] [Google Scholar]
- 131.Pugin J, et al. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 132.Lewthwaite JC, et al. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60. 2 (Hsp 65) and contains a CD14-binding domain. Infect Immun. 2001;69:7349–7355. doi: 10.1128/IAI.69.12.7349-7355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peterson PK, et al. CD14 receptor-mediated uptake of nonopsonized Mycobacterium tuberculosis by human microglia. Infect Immun. 1995;63:1598–1602. doi: 10.1128/iai.63.4.1598-1602.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wieland CW, van der Windt GJW, Wiersinga WJ, Florquin S, van der Poll T. CD14 contributes to pulmonary inflammation and mortality during murine tuberculosis. Immunology. 2008;125:272–279. doi: 10.1111/j.1365-2567.2008.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van der Wel N, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 136.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 137.Moretti J, Blander JM. Insights into phagocytosis-coupled activation of pattern recognition receptors and inflammasomes. Current Opinion in Immunology. 2014;26:100–110. doi: 10.1016/j.coi.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 139.Briken V, Ahlbrand SE, Shah S. Mycobacterium tuberculosis and the host cell inflammasome: a complex relationship. Front Cell Infect Microbiol. 2013;3:62. doi: 10.3389/fcimb.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kanneganti T-D, Lamkanfi M, Núñez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 141.Gandotra S, Jang S, Murray PJ, Salgame P, Ehrt S. Nucleotide-binding oligomerization domain protein 2-deficient mice control infection with Mycobacterium tuberculosis. Infect Immun. 2007;75:5127–5134. doi: 10.1128/IAI.00458-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Divangahi M, et al. NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. The Journal of Immunology. 2008;181:7157–7165. doi: 10.4049/jimmunol.181.10.7157. [DOI] [PubMed] [Google Scholar]
- 143.Brooks MN, et al. NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell Microbiol. 2011;13:402–418. doi: 10.1111/j.1462-5822.2010.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Juárez E, et al. NOD2 enhances the innate response of alveolar macrophages to Mycobacterium tuberculosis in humans. Eur J Immunol. 2012;42:880–889. doi: 10.1002/eji.201142105. [DOI] [PubMed] [Google Scholar]
- 145.Austin CM, Ma X, Graviss EA. Common nonsynonymous polymorphisms in the NOD2 gene are associated with resistance or susceptibility to tuberculosis disease in African Americans. J Infect Dis. 2008;197:1713–1716. doi: 10.1086/588384. [DOI] [PubMed] [Google Scholar]
- 146.Pan H, Dai Y, Tang S, Wang J. Polymorphisms of NOD2 and the risk of tuberculosis: a validation study in the Chinese population. Int J Immunogenet. 2012;39:233–240. doi: 10.1111/j.1744-313X.2011.01079.x. [DOI] [PubMed] [Google Scholar]
- 147.Zhao M, et al. A novel single nucleotide polymorphism within the NOD2 gene is associated with pulmonary tuberculosis in the Chinese Han, Uygur and Kazak populations. BMC Infect Dis. 2012;12:91. doi: 10.1186/1471-2334-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 149.Abdalla H, et al. Mycobacterium tuberculosis infection of dendritic cells leads to partially caspase-1/11-independent IL-1β and IL-18 secretion but not to pyroptosis. PLoS ONE. 2012;7:e40722. doi: 10.1371/journal.pone.0040722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.TeKippe E, Allen IC, Hulseberg PD, Sullivan JT. Granuloma formation and host defense in chronic Mycobacterium tuberculosis infection requires PYCARD/ASC but not NLRP3 or caspase-1. PLoS ONE. 2010;5:e12320. doi: 10.1371/journal.pone.0012320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Eklund D, et al. Human gene variants linked to enhanced NLRP3 activity limit intramacrophage growth of Mycobacterium tuberculosis. J Infect Dis. 2014;209:749–753. doi: 10.1093/infdis/jit572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mishra BB, et al. Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell Microbiol. 2010;12:1046–1063. doi: 10.1111/j.1462-5822.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- 153.Ogura Y, Sutterwala FS, Flavell RA. The Inflammasome: First Line of the Immune Response to Cell Stress. Cell. 2006;126:659–662. doi: 10.1016/j.cell.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 154.Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 155.Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL. K+ Efflux Is the Common Trigger of NLRP3 Inflammasome Activation by Bacterial Toxins and Particulate Matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mitoma H, et al. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fernandes-Alnemri T, Yu JW, Datta P, Wu J. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bürckstümmer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 159.Saiga H, et al. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol. 2012;24:637–644. doi: 10.1093/intimm/dxs062. [DOI] [PubMed] [Google Scholar]
- 160.Shah S, et al. Cutting edge: Mycobacterium tuberculosis but not nonvirulent mycobacteria inhibits IFN-β and AIM2 inflammasome-dependent IL-1β production via its ESX-1 secretion system. The Journal of Immunology. 2013;191:3514–3518. doi: 10.4049/jimmunol.1301331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci. 2011;68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Novikov A, et al. Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1β production in human macrophages. The Journal of Immunology. 2011;187:2540–2547. doi: 10.4049/jimmunol.1100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang Y, et al. The DNA Sensor, Cyclic GMP-AMP Synthase, Is Essential for Induction of IFN-β during Chlamydia trachomatis Infection. The Journal of Immunology. 2014;193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hansen K, et al. Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Abdul-Sater AA, et al. The overlapping host responses to bacterial cyclic dinucleotides. Microbes Infect. 2012;14:188–197. doi: 10.1016/j.micinf.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang J, Bai Y, Zhang Y, Gabrielle VD, Jin L, Bai G. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol. 2014;93:65–79. doi: 10.1111/mmi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Witte CE, Whiteley AT, Burke TP, Sauer J-D, Portnoy DA, Woodward JJ. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio. 2013;4:e00282–13. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Manca C, et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc Natl Acad Sci USA. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Manca C, et al. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J Interferon Cytokine Res. 2005;25:694–701. doi: 10.1089/jir.2005.25.694. [DOI] [PubMed] [Google Scholar]
- 176.Teles RMB, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339:1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Berry MPR, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]