Abstract
Hairpin polyamides (PAs) are an important class of sequence-specific DNA minor groove binders, and frequently employ a flexible motif, β-alanine (β), to reduce the molecular rigidity to maintain the DNA recognition register. To better understand the diverse effects β can have on DNA-PA binding affinity, selectivity, and especially kinetics, which have rarely been reported, we have initiated a detailed study for an eight-heterocyclic hairpin PA and its β derivatives with their cognate and mutant sequences. With these derivatives, all internal pyrroles of the parent PA are systematically substituted with single or double βs. A set of complementary experiments have been conducted to evaluate the molecular interactions in detail: UV-melting, biosensor-surface plasmon resonance, circular dichroism and isothermal titration calorimetry. The β substitutions generally weaken the binding affinities of these PAs with cognate DNA, and have large and diverse influences on PA binding kinetics in a position- and number-dependent manner. The DNA base mutations have also shown positional effects on binding of a single PA. Besides the β substitutions, the monocationic Dp group [3-(dimethylamino) propylamine] in parent PA has been modified into a dicationic Ta group (3, 3'-Diamino-N-methyldipropylamine) to minimize the frequently observed PA aggregation with ITC experiments. The results clearly show that the Ta modification not only maintains the DNA binding mode and affinity of PA, but also significantly reduces PA aggregation and allows the complete thermodynamic signature of eight-ring hairpin PA to be determined for the first time. This combined set of results significantly extends our understanding of the energetic basis of specific DNA recognition by PAs.
Introduction
Polyamides (PA) are N-methylpyrrole (Py) and N-methylimidazole (Im) based cations that derive from the natural product AT base-pair specific minor groove binders distamycin and netropsin.1–4 The discovery that distamycin could form a stacked dimer in wider DNA minor grooves and recognize both strands of the double helix led to extensive developments and new applications of PAs.5, 6 Much of the advancement with PAs is due to the research on DNA recognition rules for stacked heterocycle-pairs for hairpin PAs developed by Dervan and coworkers.3, 7, 8 The enhancement of PA affinity and specificity through covalent linking of PAs into hairpin structures was a major improvement over the original noncovalent, stacked dimers.7, 9 However, as PAs increase in the number of heterocycles, they go out of register with the DNA base pairs, and both affinity and selectivity are lost.10, 11 To overcome this size and rigidity barrier, flexible inserts such as a β-alanine (β) in place of one or more heterocycles can help to reset the heterocycle-base pair register. A number of studies have indicated that β inserts can improve the affinity and specificity of hairpin PAs for their target DNA sequence.12–15
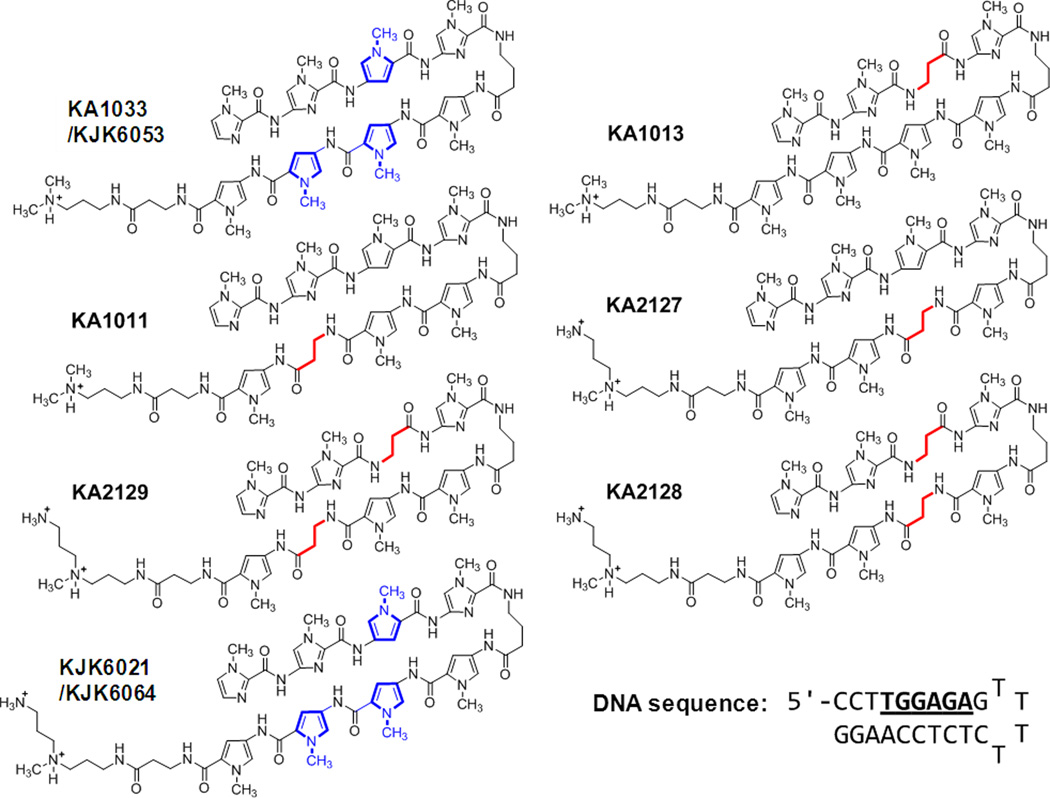
There are, however, some questions about optimum positioning for substitution of Py by β inserts and depending on the DNA and PA sequences, β substitution can decrease the binding affinity of PA molecules for DNA target sites.16–19 The role of β in DNA complex formation by PAs, is thus, more complicated than anticipated. Clearly, it is essential to understand the influence of the number and position of β substitutions on PA affinity and specificity for rational design of the most effective, biologically active agents. The binding affinity and selectivity of hairpin PAs with their target sequences have been extensively investigated by several groups using footprinting and other methods.16, 20–23 However, the kinetics and detailed thermodynamics of PA-DNA interaction have received much less attention,13, 18, 24, 25 To aid in establishing the diverse effects of β inserts we have initiated a detailed study of the binding thermodynamics, selectivity and kinetics for an eight-heterocyclic PA, KA1033, and five β-containing derivatives with a DNA recognition sequence of the Human COX2 promoter, 5’-TTGGAGA-3’ (Fig. 1). Among the five derivatives, three (KA1011, KA1013 and KA2127) have a single β substitution in different positions, and the other two (KA2128 and KA2129) have double βs in the positions covering the single β inserts. We note that the PA KA1033 and an alkylator attached analog of KA1013 were evaluated with other DNA sequences in early studies of PA-DNA interaction.26, 27 With these derivatives, all internal Py positions of the parent PA KA1033 are substituted with β groups (Fig. 1). Five mutant DNA sequences with one or two base pair mutations in different positions have also been studied to evaluate the positional role or sequence context of DNA base pairs in the PA-DNA interactions. Both PA modifications and DNA base mutations show that there is a large position-dependent effect of single and double β substitution placement on PA-DNA binding affinity and kinetics.
Fig. 1.
Parent polyamide, KA1033, and both β- and Ta- containing analogs along with their cognate DNA sequence are shown. Internal pyrroles are shown in blue and β inserts are in red.
There is, however, an additional dimension to the kinetics and thermodynamics of PA interactions, namely strong PA-PA interactions that are independent of DNA binding, including intermolecular hydrogen bonding that involves amide NH-groups and acceptors such as carbonyl oxygens, and π-π stacking of the pyrrole and imidazole rings. These interactions, perhaps enhanced by the flat, crescent shape of PA segments, can lead to aggregation.28, 29 Aggregation of hairpin PAs is dependent on a number of parameters not described in the literature, but which we have been characterizing, and compounds similar to or larger in size than KA1033 are particularly prone to aggregate (unpublished work by us and colleagues C. M. Dupureur and M. R. Nichols). As shown below, even in the crystal structures of dimeric PA building blocks, the hydrogen bonding responsible for much of the aggregation is readily seen as intermolecular interactions. In an effort to minimize PA aggregation and obtain thermodynamics of PA-DNA binding, we increased charge-charge repulsion between PA molecules by replacing the monocationic Dp group (3-(dimethylamino) propylamine) in KA1033 with the dicationic Ta group (3, 3'-Diamino-N-methyldipropylamine) in KJK6021. Two PAs having the same structure as KA1033 and KJK6021 but different anionic counterions, KJK6053 and KJK6064, have also been synthesized and studied. The results presented here show that the counterions used do not affect the PA binding affinity and thermodynamics with cognate and mutant DNAs under our conditions. Modification of Dp to Ta, however, not only maintains the DNA binding affinity and binding mode of PAs, but also significantly reduces PA aggregation. Presumably, due to aggregation of eight-ring PAs, no ITC experiments have been reported for PAs of this size.29 The Ta substitution allows the eight-ring PA-DNA binding enthalpy to be determined through ITC and allows the very first complete thermodynamic signature of eight-ring hairpin PA-DNA binding to be determined. The results significantly extend the understanding of the molecular basis of specific DNA recognition by PAs.
Results
Qualitative Comparison of PA Binding Affinity by Thermal Melting Study
Thermal melting provides a qualitative and relative comparison of compound binding affinities with DNA.30 KA1033 (ImImPyIm-γ-PyPyPyPy-β-Dp, Fig. 1) is an eight-ring hairpin PA, while KJK6021 has the same heterocycle sequence but with a Ta group replacing the Dp of KA1033 at the C-terminal tail. The binding affinities of these two PAs and five β-substituted analogs (Fig. 1) with their cognate (5’-TGGAGA-3’) and five mutant DNA sequences (Fig. S1A) were compared by thermal melting at a 1:1 molar ratio. The normalized DNA melting curves with and without PAs are shown in Fig. S1B. The ΔTm values, the Tm of the PA-DNA complex minus the Tm of free DNA, for each PA with each DNA are listed in Table 1.
Table 1.
ΔTm values of PAs with cognate site TGGAGA and five mutant sequences*.
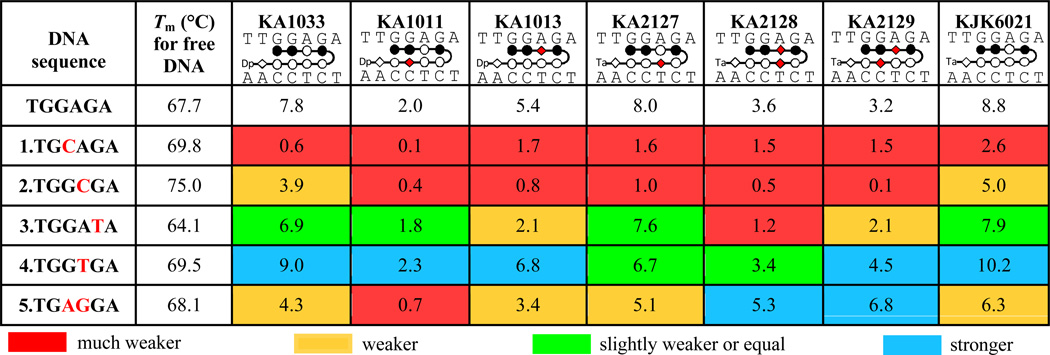 |
The error of these ΔTm values is ± 0.2 °C based on experimental reproducibility. The color for ΔTm is given based on the comparison with the value for PA-cognate DNA complex: < 50% (red), 50% ~ 80% (orange), 80% ~ 100% (green), and > 100% (blue) of ΔTm for PA-cognate DNA complex.
For the cognate sequence (TGGAGA), the parent PA, KA1033, increased the DNA Tm by 7.8 °C, and the ΔTm for KJK6021, the Ta group containing analog, is slightly higher. Three single β-substituted PAs increased the DNA Tm by very different amounts: KA1013 (ImImβIm-γ-PyPyPyPy-β-Dp), which has a β before the γ loop, has a lower ΔTm than KA1033 (5.4 °C), while KA2127 (ImImPyIm-γ-PyβPyPy-β-Ta), which has a Ta tail and a β in the same stacking position as KA1013, but after the γ loop (Table 1), has a slightly lower ΔTm (8.0 °C) than the parent with Ta. KA1011 (ImImPyIm-γ-PyPyβPy-β-Dp), which has a β substitution just next to that in KA2127, has a significantly lower ΔTm (2.0 °C). Two double β-substituted PAs, KA2128 (ImImβIm-γ-PyβPyPy-β-Ta) and KA2129 (ImImβIm-γ-PyPyβPy-β-Ta), both have much lower ΔTm values than the Ta parent. The ΔTm value for KA2129 is approximately the average of its two single β analogs KA1011 and KA1013. But the ΔTm for KA2128 is lower than either of its single β analogs KA1013 and KA2127.
For the five mutant sequences embracing either single or double base pair mutations in different positions, PAs display quite diverse binding affinities as indicated by the significantly changed ΔTm values (Table 1). In order to obtain a more clear comparison of binding, the ΔTm values have been classified with four colors (red, orange, green and blue), and the color for each ΔTm is given based on the value relative to the ΔTm with cognate DNA: <50% (red), 50% ~ 80% (orange), 80% ~ 100% (green), and >100% (blue) of the ΔTm for PA-cognate DNA complex. It is clear that all PAs bind more weakly to mutants 1–3, while PA complexes with mutants 4 and 5 have both weakening and enhancing effects. The unexpectedly large sequence-dependent variations of the ΔTm values are discussed in detail below.
Quantitative Determination of PA Binding Affinity and Kinetics by Biosensor-Surface Plasmon Resonance (SPR)
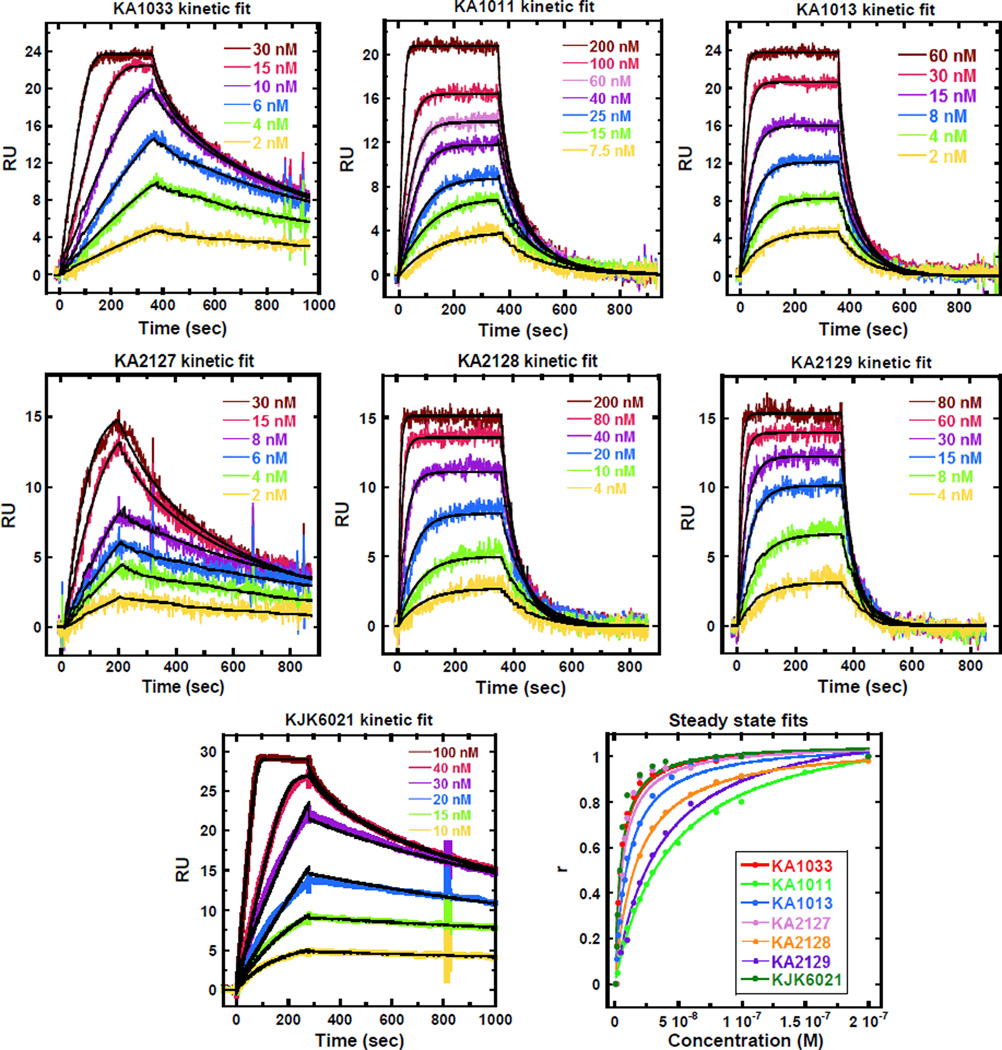
Biosensor SPR is a sensitive technique to monitor the progress of reactions in real time. It provides kinetics and equilibrium binding affinities as well as stoichiometries and cooperativity values of biomolecular interactions.31, 32 To quantitatively evaluate the interactions of hairpin PAs with DNA, and also to provide details about the positional effects of the internal β substitutions, SPR experiments were performed for KA1033 and derivatives with their target DNA, TGGAGA. The kinetic rate constants obtained from global kinetic fits (Fig. 2A) and the equilibrium binding constants (KD) measured from both global kinetic and steady state fits (Fig. 2B) of the sensorgrams for different PAs are listed in Table 2. The KD values measured by both fits are quite comparable and the values measured by kinetic fit are used for the following comparisons.
Fig. 2.
(A) SPR sensorgrams (color) and global kinetic fits (black overlays) for KA1033 and derivatives with their cognate DNA TGGAGA at 25 °C with 50 mM NaCl. (B) Steady state fits for SPR sensorgrams.
Table 2.
SPR analysis of kinetic rate constants and equilibrium affinities for PAs binding to their cognate site TGGAGA at 25 °C with 50 mM NaCl*. ΔTm values with cognate DNA are included for comparison.
| PA | Kinetics | KD (nM) | |||
|---|---|---|---|---|---|
|
ka (×106M−1s−1) |
kd (×10−3s−1) |
Kinetic fit | Steady state | ΔTm (°C) | |
| KA1033 | 4.7 ± 0.7 | 12 ± 1 | 2.5 ± 0.4 | 2.8 ± 0.3 | 7.8 |
| KA1011 | 2.6 ± 0.3 | 96 ± 6 | 36.4 ± 0.4 | 36.6 ± 0.1 | 2.0 |
| KA1013 | 3.7 ± 0.4 | 45 ± 7 | 12.1 ± 0.4 | 10.3 ± 0.5 | 5.4 |
| KA2127 | 1.8 ± 0.3 | 4.7 ± 0.3 | 2.6 ± 0.2 | 2.9 ± 0.2 | 8.0 |
| KA2128 | 0.7 ± 0.2 | 13 ± 2 | 18.6 ± 0.8 | 17.5 ± 0.1 | 3.6 |
| KA2129 | 1.1 ± 0.2 | 29 ± 5 | 26.3 ± 2.2 | 24.0 ± 0.5 | 3.2 |
| KJK6021 | 1.3 ± 0.2 | 2.6 ± 0.2 | 2.0 ± 0.2 | 2.2 ± 0.3 | 8.8 |
Errors listed are the standard errors for the fits.
KA1033, the parent PA, has a very strong binding affinity with the cognate sequence (KD = 2.5 ± 0.4 nM). This strong binding can be attributed to the fast association (ka) and slow dissociation rates (kd) (Table 2). In the following discussion, all comparisons are made with the properties of KA1033 unless explicitly stated otherwise. An analogue eight-ring PA with a Ta tail, KJK6021, has around four-times lower ka and kd, and a similar KD (2.0 ± 0.2 nM) to KA1033. These results indicate that the replacement of the Dp group by a Ta influences the kinetics, but does not significantly affect the overall binding affinity due to ka and kd compensation. The single β substitution of the Py on the N-terminal side of KA1033 results in a slightly lower ka and a higher kd, with a 5-fold decrease in the binding affinity of KA1013 with the cognate DNA (KD = 12.1 ± 0.4 nM). For KA1011, the single β substitution on the C-terminal side of the γ loop not only slows its association, but also increases its dissociation by 8-fold and therefore causes a 15-fold weaker binding affinity (KD = 96 ± 6 nM). KA2127, another single C-terminal β-substituted PA, has both faster ka and kd and a KD (2.6 ± 0.2 nM) comparable to KJK6021. The results of these three PAs show that a single β inserted in different positions can have diverse effects on PA binding kinetics, but in this PA sequence and for the cognate DNA target, β generally accelerates PA dissociation and weakens the PA binding affinity.
The two double-β-substituted PAs, KA2128 and KA2129, have Ta instead of Dp groups at the C-terminus. Relative to their eight-ring analog KJK6021, both KA2128 and KA2129 have slightly slower association to DNA but much faster dissociation and around 10-times weaker DNA-binding affinities (Table 2). KA2129, has an average affinity (KD = 26.3 ± 2.2 nM) of its corresponding single β analogs, KA1011 and KA1013, while the affinity of KA2128 is weaker than either of the two single β analog (KA1013 and KA2127). The order of binding affinity obtained by SPR is in good agreement with that from the DNA melting study. All of the experiments have been repeated twice, and the results are reproducible within experimental errors.
Evaluation of PA Binding Mode and DNA structural Changes by CD
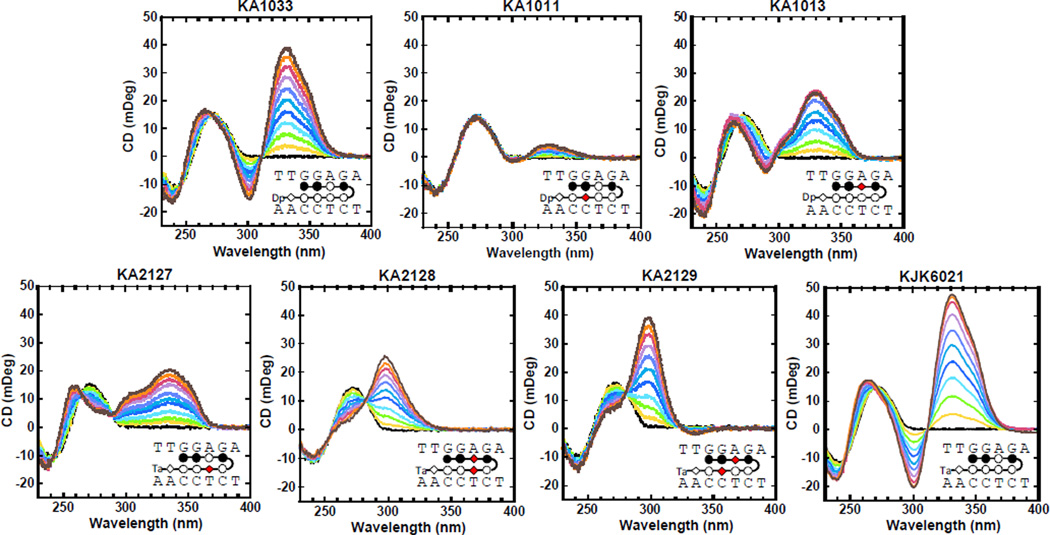
CD spectroscopy monitors the asymmetric environment of ligands when they bind to DNA and thus can provide information about the ligand binding mode.33 In Fig. 3, the CD results for the PA-DNA complexes are characterized by large, positive induced signals from around 300 to 370 nm, where the PAs absorb and DNA signals do not interfere. The large positive induced CD signals indicate a DNA minor groove binding mode as expected for hairpin PAs.34 The titrations were conducted from 0:1 to 2:1 molar ratios of PA to hairpin DNA. The signal and pattern of the induced CD by KJK6021 are very similar to KA1033 which suggests that these two PAs target the DNA minor groove in a very similar manner. This again indicates that the change of Dp to Ta does not significantly affect the PA binding affinity or the structural influence of PA on DNA.
Fig. 3.
CD spectra for TGGAGA with all PAs at 25 °C. Molar ratios of PA to DNA hairpin are from 0 to 2.0 as the induced CD signal for the PAs increases.
Thermodynamic Study of PA-DNA Binding
Effects of salt concentration and temperature
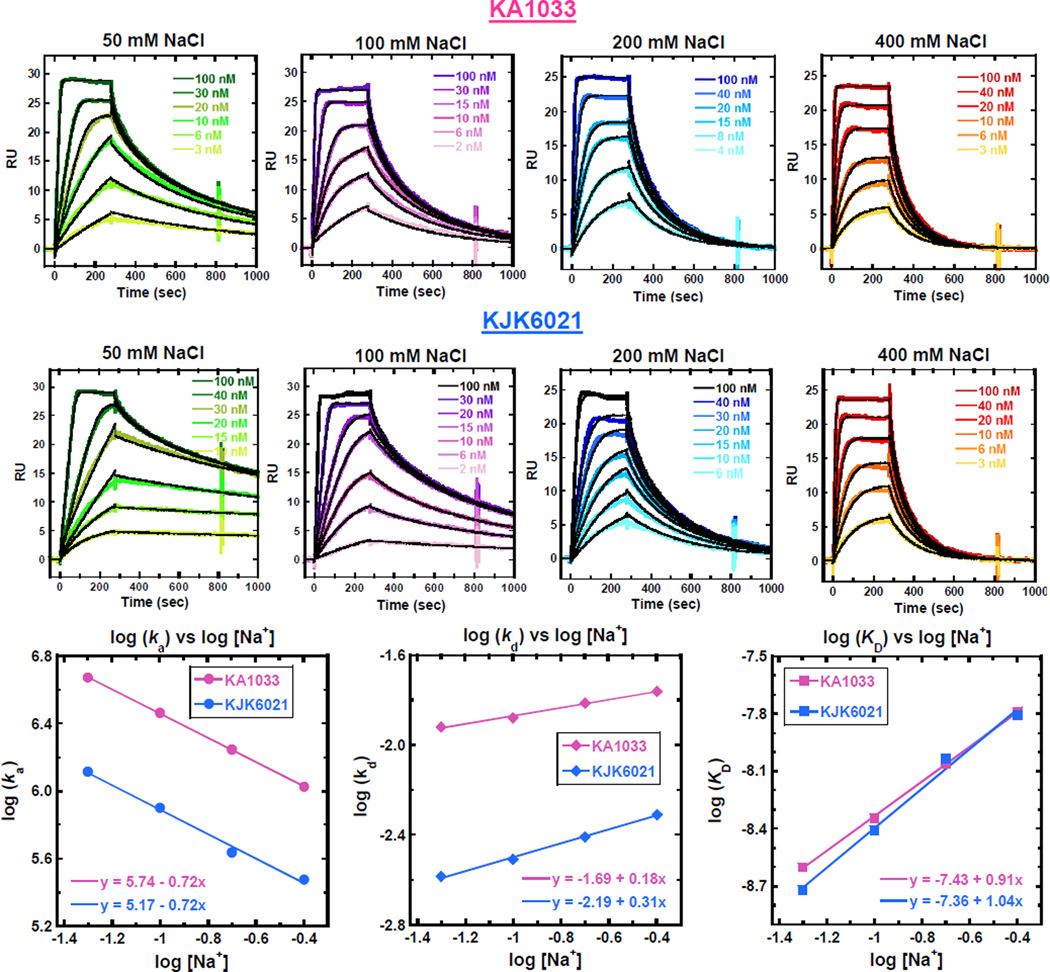
From the qualitative thermal melting studies and quantitative SPR experiments to CD spectra evaluation, the Ta group containing KJK6021 exhibits binding similar to KA1033. However, the replacement of the monocationic Dp by a dicationic Ta group increases the number of positive charges of PA. According to the counterion condensation theory, the charges of minor groove binders affect the release of counterions on binding to DNA.29, 35 In order to evaluate the thermodynamic signatures for PAs with different charges, SPR experiments were conducted for KA1033 (+1) and KJK6021 (+2) with their cognate sequence TGGAGA under several salt concentrations at 25 °C. The corresponding sensorgrams are shown in Fig. 4A, and the rate constants and equilibrium constants determined from 1:1 global kinetic fits of the sensorgrams are listed in Table 3. Overall, as the salt concentrations increase, the ka decreases while the kd increases and thus the KD decreases for both PAs, as expected.36 The KD values for KJK6021 under different salt concentrations are quite comparable with those for KA1033, but the ka and kd of KJK6021 binding are both much lower than for KA1033.
Fig. 4.
(A) SPR sensorgrams (color) and global kinetic fits (black overlays) for KA1033 and KJK6021 with cognate DNA TGGAGA at 25 °C as a function of salt concentration. (B) Salt concentration dependence of kinetic constants and binding affinities for KA1033 and KJK6021. All values are calculated based on data in Table 3.
Table 3.
SPR analysis of kinetic rate constants and equilibrium affinities for KA1033 and KJK6021 binding to TGGAGA at 25 °C with different salt concentrations*.
| KA1033 | KJK6021 | |||||
|---|---|---|---|---|---|---|
| [NaCl] (mM) |
ka (×106M−1s−1) |
kd (×10−3s−1) |
KD (nM) |
ka (×105 M−1s−1) |
kd (×10−3s−1) |
KD (nM) |
| 50 | 4.7 ± 0.7 | 12 ± 1 | 2.5 ± 0.4 | 13 ± 2 | 2.6 ± 0.2 | 2.0 ± 0.2 |
| 100 | 2.9 ± 0.4 | 13 ± 2 | 4.5 ± 0.4 | 8.0 ± 0.5 | 3.1 ± 0.2 | 3.9 ± 0.3 |
| 200 | 1.8 ± 0.3 | 15 ± 2 | 8.7 ± 0.5 | 4.3 ± 0.3 | 3.9 ± 0.6 | 9.0 ± 0.5 |
| 400 | 1.1 ± 0.3 | 17 ± 1 | 16 ± 0.8 | 3.0 ± 0.3 | 4.9 ± 0.5 | 16 ± 0.4 |
Errors listed are the standard errors for the 1:1 global kinetic fits.
The logarithm values for ka, kd and KD for both PAs have been plotted as a function of salt concentration in Fig. 4B. The linear change of the log values for KA1033 [slopes = −0.72 for log(ka), +0.18 for log(kd) and +0.91 for log(KD)] are as predicted by the counterion condensation theory for one charge interaction,36, 37 and as previously observed on binding of another monocationic hairpin PA (KA1039).29 The slopes of the linear change for KJK6021 are −0.72, +0.31 and 1.04 for log(ka), log(kd) and log(KD), respectively. The salt concentration dependencies of ka and KD for KJK6021 are similar to KA1033, but its kd is slightly more salt concentration dependent than KA1033 (slope 0.31 vs. 0.18). Based on counterion condensation theory, the slopes of log(KD) indicate that approximately one cation has been displaced by both PAs on DNA binding. The number of phosphate contacts (Z) between PAs and DNA can be determined by the slope/Ψ, where Ψ is the fraction of phosphate shielded by condensed counterions and is 0.88 for double stranded B-DNA35: Z = 0.91/0.88=1.03 for KA1033 and Z = 1.04/0.88=1.18 for KJK6021. Thus there is about one phosphate affected by binding both PAs.
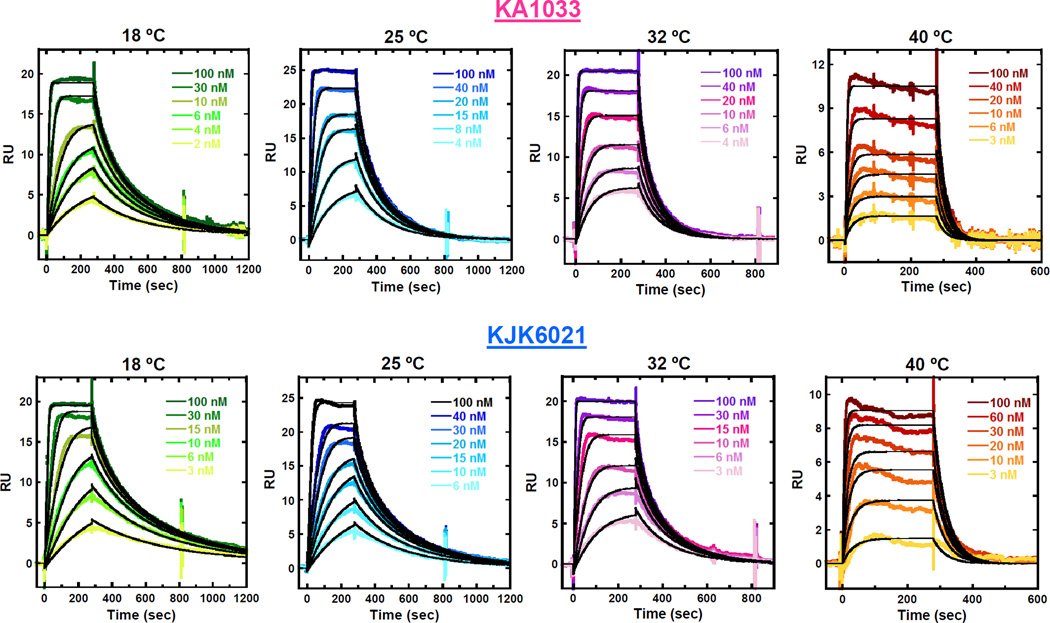
The effects of temperature on PA binding affinity and kinetics have also been investigated by SPR experiments. The sensorgrams of KA1033 and KJK6021 with 200 mM NaCl-HEPES buffer from 18 °C to 40 °C are shown in Fig. 5. This salt concentration was chosen due to both PAs having stronger binding affinity than under higher salt concentration, and less mass transfer limitations than under lower salt concentrations with SPR experiments. The kinetic rate constants and equilibrium constants determined from 1:1 global kinetic fits of the sensorgrams are listed in Table 4. Based on the equilibrium binding constants, the binding Gibbs free energy (ΔG) at different temperatures was also calculated (ΔG = -RT ln Keq, where R is 1.987 cal mol−1 K−1 and T is 298 K). The data clearly show that as the temperatures increase, both ka and kd increase, and the overall binding affinity KD decreases for both PAs. However, ΔG values for both PAs binding are essentially temperature independent, and quite large and negative (−11.0 ± 0.2 kcal mol−1).
Fig. 5.
SPR sensorgrams (color) and global kinetic fits (black overlays) for KA1033 and KJK6021 with cognate DNA TGGAGA at different temperatures. The SPR-determined binding affinities and kinetics are listed in Table 4.
Table 4.
SPR analysis of kinetic rate constants and equilibrium affinities as a function of temperature for KA1033 and KJK6021 binding to TGGAGA with 200 mM NaCl*.
| KA1033 | ||||
|---|---|---|---|---|
| Temp. (°C) |
ka(×106 M−1s−1) |
kd (×10−3s−1) |
KD (nM) |
ΔG(kcal mol−1) |
| 18 | 1.4 ± 0.2 | 6.9 ± 0.3 | 5.0 ± 0.4 | −11.1 |
| 25 | 1.8 ± 0.4 | 15 ± 2 | 8.7 ± 0.5 | −11.0 |
| 32 | 2.2 ± 0.3 | 30 ± 4 | 14 ± 0.5 | −11.0 |
| 40 | 3.0 ± 0.6 | 52 ± 6 | 18 ± 0.8 | −10.8 |
| KJK6021 | ||||
|
Temp. (°C) |
ka(×105 M−1s−1) |
kd (×10−3s−1) |
KD (nM) |
ΔG(kcal mol−1) |
| 18 | 2.9 ± 0.3 | 2.0 ± 0.2 | 6.8 ± 0.5 | −10.9 |
| 25 | 5.0 ± 0.4 | 4.5 ± 1.0 | 9.0 ± 0.4 | −11.0 |
| 32 | 8.7 ± 0.6 | 11 ± 2 | 13 ± 0.8 | −11.0 |
| 40 | 18 ± 1 | 25 ± 4 | 14 ± 0.06 | −11.0 |
Errors listed are the standard errors for the 1:1 global kinetic fits.
Enthalpy of binding
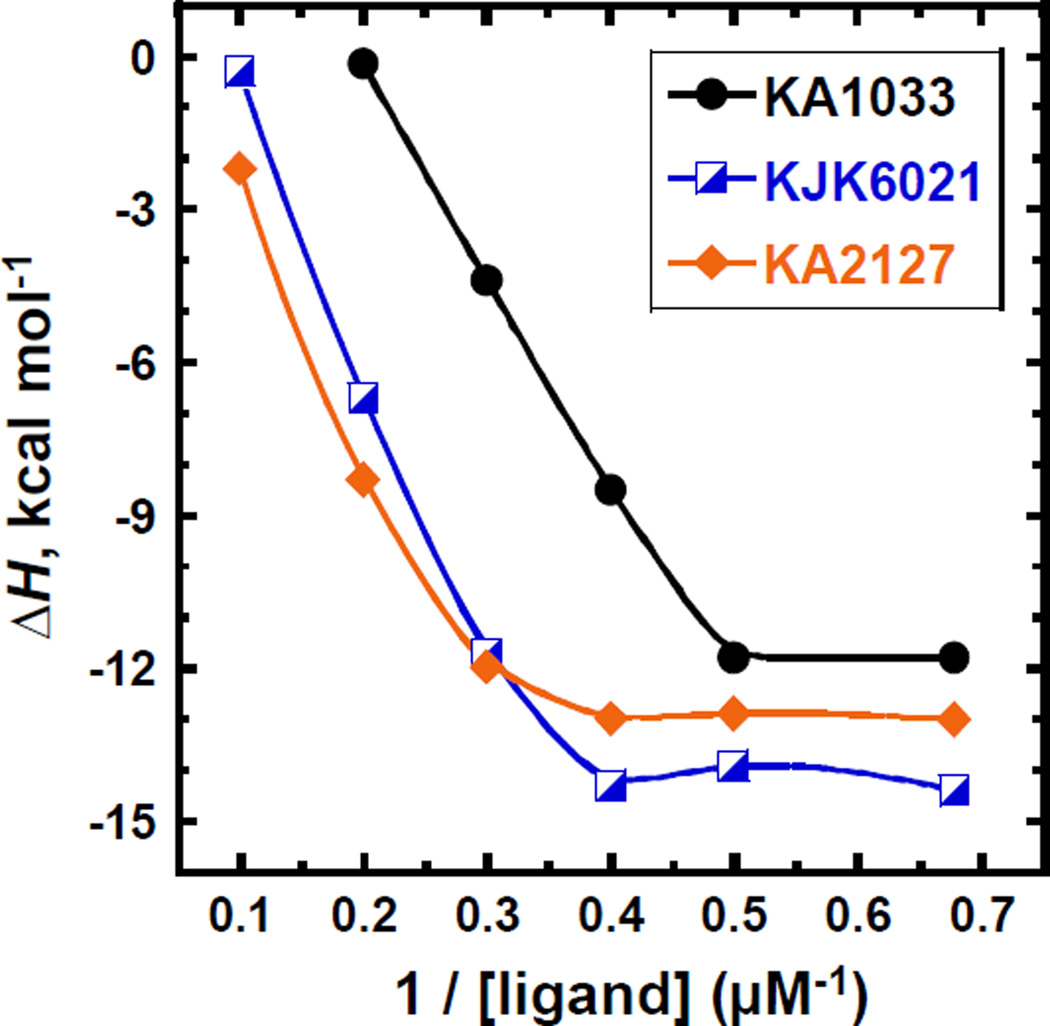
Besides the effects of salt concentration and temperature on the kinetics and affinity of PA binding, the determination of binding enthalpy is very critical for a more detailed energetic understanding of the PA-DNA interactions. However, as previously observed, the eight-ring hairpin PAs aggregate under typical ITC standard concentrations, such as 50 µM and generally prohibit the binding enthalpy assessment.17, 29 Therefore, several quite low ligand concentrations, down to 1.5 µM, have been used in the ITC experiments for KA1033, KJK6021 and KA2127. The ITC titrations for the three PAs are shown in Figs. S2-4, and the binding enthalpies (ΔH) obtained at each PA concentration are listed in Table S1 ESI†, and are plotted versus the reciprocal of PA concentrations in Fig. 6. For these three PAs, the ΔH values first become much more negative, and then reach a constant value as the PA concentrations decrease. The ΔH values for KA2127 and KJK6021 at each concentration are generally more negative than that for KA1033, and reach a plateau (−13.0 ± 0.4 kcal mol−1 and −14.1 ± 0.2 kcal mol−1, respectively) at 0.4 µM−1 (2.5 µM). The ΔHs obtained from 2.5 µM, 2.0 µM and 1.5 µM (0.4 µM−1, 0.5 µM−1 and 0.67 µM−1) of KA2127 and KJK6021 agree very well within experiment error and indicate that we have obtained the actual binding enthalpies. The ΔHs for KA1033 are consistent (−11.8 ± 0.3 kcal mol−1) at 2.0 µM and 1.5 µM (0.5 µM−1 and 0.67 µM−1) which suggest the actual binding enthalpy for KA1033 has also been reached with these two concentrations. These results indicate that with the decreasing sample concentration in ITC, concentration-dependent PA aggregation can be reduced to an insignificant amount. Once the PA aggregation has been totally overcome, the actual binding enthalpy can be determined.
Fig. 6.
Binding enthalpies of different concentrations of KA1033 and KJK6021 with 10 µM TGGAGA hairpin DNA measured by ITC at 25 °C.
Besides the modification of Dp to Ta, the counterion used during isolation of KA1033 (Dp) is also different with that for KJK6021 (Ta): trifluoroacetate (∙CF3COO−, TFA) with KA1033 while formate (∙HCOO−) is used for KJK6021. In order to evaluate the effects of counterions on PA binding affinities and thermodynamics, two PAs in the same structure as KA1033 or KJK6021 but with switched counterions, KJK6053 (ImImPyIm-γ-PyPyPyPy-β-Dp, formate) and KJK6064 (ImImPyIm-γ-PyPyPyPy-β-Ta, TFA), respectively, have been studied with UV-melting experiments. The thermal melting curves for these four PAs with cognate and mutant DNA sequences are shown in Fig. S5, and the ΔTm values are listed in Table S2, ESI†. The melting curves for KJK6053 and KJK6064 complexes agree very well with those for their counterion analogs KA1033 and KJK6021, and the corresponding ΔTm values are also quite comparable for PAs of the same structure (KJK6053 vs. KA1033, KJK6064 vs. KJK6021). ITC experiments have also been conducted with 2.5 µM of KJK6053 and KJK6064 into 10 µM of cognate DNA hairpin, and compared with the titrations of KA1033 and KJK6021 at the same PA concentration (Fig. S6, ESI†). The ΔH is −8.3 ± 0.3 kcal mol−1 for KJK6053 binding and −14.0 ± 0.3 kcal mol−1 for KJK6064 binding. Both of these values are consistent with the ΔH values measured for KA1033 and KJK6021. These results suggest that under our conditions (50 mM NaCl in the buffer) the formate and TFA counterions do not significantly affect the affinity and thermodynamics of PAs binding.
Based on SPR binding free energy values and the ITC enthalpy values, the T ΔS values were calculated from ΔG = ΔH- T ΔS for KA1033, KA2127 and KJK6021 (Table 5). Comparison of the contributions of the enthalpy and entropy to the free energy shows that PA complex formation is dominated by the favorable binding enthalpies. This is the first complete thermodynamic study for DNA minor groove binding with eight-ring hairpin PAs.
Table 5.
Thermodynamic profiles for KA1033, KJK6021 and KA2127 with their cognate DNA TGGAGA with 50 mM NaCl at 25 °C*.
| ΔG (kcal/mol) |
ΔH (kcal/mol) |
TΔS (kcal/mol) |
|
|---|---|---|---|
| KA1033 | −11.7 | −11.8 | −0.1 |
| KJK6021 | −11.9 | −14.1 | −2.2 |
| KA2127 | −11.7 | −13.0 | −1.3 |
The TΔS are calculated by ΔG= ΔH - TΔS.
Discussion
Effects of DNA Sequence and PA Structure on Relative Binding Affinity
The relative binding affinities of KA1033 and derivatives with five mutant DNA sequences with either single or double base pair mutations in the PA binding site have been determined (Table 1). Through the comparison of ΔTm values of KA1033 (ΔTm = 7.8 °C) with mutants 1–3, it is clear that mutation to reverse the third base pair of the 5’-TGGAGA-3’ site (mutant 1, 5’- TGCAGA-3’, ΔTm = 0.6 °C) decreases the induced thermal stability much more significantly than complete changing of the fourth base pair (mutant 2, 5’- TGGCGA-3’, ΔTm=3.9 °C), while the mutant at the fifth base pair (mutant 3, 5’- TGGATA-3’, 6.9 °C) influences the binding the least. These mutation results are in agreement with the position-dependent effects of β substitution in PA, since KA1011, with β targeting the most sensitive third base pair has a much lower ΔTm than KA1013, with β targeting the less sensitive fourth base pair. Moreover, the ΔTm values for all PAs with mutants 1–3 suggest that all of these three single DNA base pair mutations can weaken the PA binding in the order of: 3rd base pair > 4th base pair > 5th base pair.
In mutant 4 (5’-TGGTGA-3’), the fourth base pair (A/T) of the 5’-TGGAGA-3’ site has been switched into T/A, and KA1033, KA1011, KA1013 and KA2129 display higher ΔTm values than with cognate DNA. This result means their Py/Py and β/Py pairs which target the fourth base pair in the binding site favor T/A base pair recognition slightly more than A/T base pair. On the contrary, KA2127 has a slightly lower ΔTm with mutant 4 than with cognate DNA (6.7 °C vs 8.0 °C) which suggests the Py/β pair in KA2127 prefers to target the A/T base pair. The ΔTm values for KA2128 with cognate DNA and mutant 4 are very similar (3.6 °C vs 3.4 °C) which indicates that the β/β pair does not have a strong binding preference to A/T or T/A base pair.14 These data clearly show that the incorporation of β inserts can have very diverse influences on the specificity of DNA recognition by PAs, and these effects strongly depend on the PA structure and DNA sequence.
Mutant 5 has base pair changes at both the third and fourth base pairs of the 5’-TGGAGA-3’ site (mutant 5, 5’- TGAGGA-3’) through switching the middle GA to AG. These mutations could either reverse the binding orientation of PAs, or significantly weaken the PAs binding if they keep the original binding orientation, as discussed above with mutants 1 and 2. Based on the recognition sequence, these results suggest that the original orientation is the usual N → C (ImImPyIm) for KA1033, for example, bound 5’ → 3’ to 5’- TGGAGA -3’, while with mutant 5, binding becomes reversed, with N → C (ImImPyIm) targeting 3’ → 5’ to 3’-AGGAGT -5’. It should be noted that although the results are consistent with this interpretation, more detailed experimental studies, such as NMR or X-ray analysis, would be necessary for a more definitive answer. The ΔTm values with mutant 5 for KA1033, KJK6021 and their single β analogs, KA1011, KA1013 and KA2127, are all lower than the ΔTm for the cognate sequence, but greater than ΔTm with mutants 1 and 2. This suggests that the two base pair mutations altered the PA binding into the reverse orientation (3’ → 5’). Interestingly, two double β-substituted PAs, KA2128 and KA2129, display an opposite behavior in that they prefer mutant 5 over cognate binding and probably have the reverse binding orientation, as indicated by the higher ΔTm values than with cognate DNA. The flexibility from the double β inserts might play a role in the preferred binding for KA2128 and KA2129. The β-modulated binding orientation of PA has also often been observed by Dervan and coworkers using different powerful methods.38–40 In summary, as previously found with other PA-DNA complexes,17 the different contributions of the base pairs in the targeting site cause distinct affinity reductions for the β-substituted PAs and different effects on binding affinities for the PA-bound DNA mutants. Moreover, the possibility to modulate PA binding orientation through incorporating double β inserts provides possible additional ideas for rational drug design. More studies of additional β substituted PAs will be required before we can define the complex rules for their effects on KD in different DNA sequences.
Effects of Dp and Ta Groups and Internal β on PA-DNA Thermodynamics
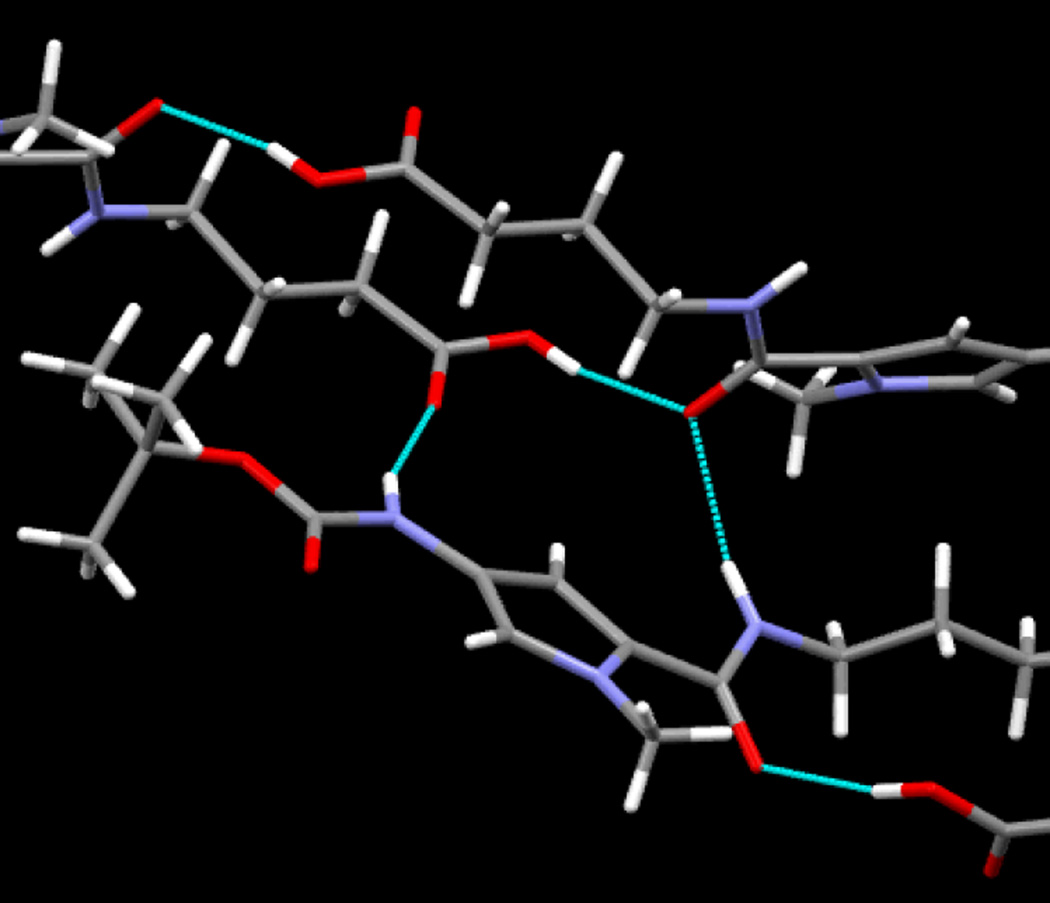
ITC titrations have been previously performed for another eight-ring hairpin PA KA1002 and its single β-inserted analogs, but their binding enthalpies could not be determined, presumably due to the significant compound aggregation at current ITC concentration requirements.17 The binding enthalpy of a six-ring hairpin PA was first published in 1996,41 and in the following eighteen years, there was no reports on larger PAs. In the experiments reported here we have been able to determine direct ITC binding enthalpies for eight-ring PA, for the first time. KA1039, a six-ring hairpin derivative of KA1002, shows much less aggregation than KA1002 in ITC and thus allows its ΔH to be determined through using diluted samples.17, 29 Here, in order to reduce the aggregation and obtain the thermodynamic signatures of eight-ring hairpin PAs, the monocationic Dp group in KA1033 has been modified into a dicationic Ta (KJK6021), and studies on both full heterocycle and β-substituted (KA2127) hairpins have been conducted (Figs. S2-4, ESI†). The results (Fig. 6) clearly confirm that PA aggregation depends on compound charge and concentration, and can be minimized as the PA concentrations decrease and the charge increases. KA1033 shows a constant ΔH when its concentration is around 2 µM or less, while KA2127 and KJK6021 reach a constant ΔH with concentrations equal or less than 2.5 µM. Through the comparison of the curves in Fig. 6, in the range where all PAs aggregate (10 µM – 2.5 µM), the ΔH of single β substituted PA KA2127 is always closer to the constant binding enthalpy than its full heterocyclic analog KJK6021. The observed ΔH of the Ta group containing KJK6021 is always closer to the actual value than the Dp group containing KA1033. These data show PAs aggregate in order of KA2127< KJK6021< KA1033, and suggest that the additional positive charge from the Ta group and the extra flexibility from β help to reduce PA aggregation. To see a molecular scale picture of the amide hydrogen bonds that we believe, along with π-stacking, constitute strong tendencies toward aggregation, the crystal structures of two dimer building blocks are instructive (Fig. 7 and Figs. X-ray 2, X-ray 4 and X-ray 6, ESI†). We clearly see from Fig. 7 that, for Boc-Py-β-COOH·0.5dioxane, an amide NH to carbonyl and related intermolecular H-bonds dominate intermolecular interactions in the solid state. Structural details and more extensive H-bonding views are provided in ESI†. This structure makes it easy to understand why the increasing size of PAs with associated increased intermolecular interactions can lead to PA aggregation. At about an eight-ring PA size, the concentration for PAs to aggregation is lower than that used in ITC experiments which has prevented previous determination eight-ring PA-DNA binding enthalpies.
Fig. 7.
Close-up view of intermolecular NH and OH hydrogen bonding in the crystal structure of Boc-Py-β-COOH•0.5 dioxane. The Boc-amine-NH donates an H-bond to the carboxylic acid carbonyl of an adjacent molecule, while the amide carbonyl accepts H-bonds from the carboxylic acid OH group and amide NH of an adjacent molecule.
To obtain a better understanding of the effects of counterions on PA binding affinity and thermodynamics, KJK6053 and KJK6064 have been investigated and compared with their analogs KA1033 and KJK6021 which have the same structure but different counterions. The quite comparable binding affinities with cognate and mutant DNA sequences through UV-melting (Fig. S5, ESI†), and the constant binding enthalpies obtained through ITC (Fig. S6, ESI†) for these PAs suggest that the counterions, TFA and formate, do not have significant influence on PA-DNA interaction under the experimental conditions. However, it is still worthwhile to note that the counterions always play critical roles for the biological activities for synthetic small molecules.42, 43
The effects of Dp and Ta groups on PA-DNA complex formation have also been systematically investigated as a function of salt concentration by SPR. Under a large range of salt concentrations (from 50 mM to 400 mM NaCl, Table 3), relative to Dp, the more flexible Ta group slows down both association and dissociation rates of PA. But the effects on kinetics partially cancel so that Ta gives only a small affinity increase. Moreover, the salt concentration dependencies of kinetics and affinity for the dication KJK6021 are quite similar with the monocation KA1033 (Fig. 4B), and this is different than a classical two charge interaction as predicted by the counterion condensation theory.35, 36 The two positively charged amines in Ta are connected on the same linker (Fig. 1) and, as a result of this closer spacing, they may have strong electronic interactions with only one phosphate on DNA backbone, and thus, release only one counterion on PA binding.
The SPR results for KA1033 and KJK6021 as a function of temperature show that the temperature dependences of kinetics for KJK6021 binding are greater than that for KA1033 (Fig. 5B). This suggests that the relatively longer Ta group becomes more dynamic than the Dp tail as the temperature increases, and contributes to the larger kinetic changes of KJK6021. Interestingly, the changes in Ta kinetics are again partially cancelled as a result of the temperature dependences of binding affinities for KJK6021 and KA1033. The binding free energies for both Dp and Ta complexes are also quite similar and are essentially temperature independent in the range that could be studied (Table 4). This signature of temperature on DNA minor groove binding free energy has been observed with other minor groove complexes.29
In summary, based on the UV melting data, CD titrations and SPR studies, Dp and Ta groups have very similar effects on DNA recognition by PAs, but the dicationic Ta group is capable of reducing PA aggregation in ITC experiments. In the presence of a Ta group, an additional β can further decrease the PA aggregation (Fig. 6). The thermodynamic profiles (Table 5) suggest that complex formation of KA1033, KJK6021 and KA2127 on the TGGAGA site are strongly driven by enthalpy which is opposite to most of the classic minor groove binders that target AT-rich sites.44 The presence of GC base pairs in the target binding site changes the geometry and hydration status of the minor groove relative to that of AT-rich sites, and the stacking of Im and Py rings and numerous H-bonds between PA-DNA convert minor groove binding thermodynamics from entropy driven with most of the AT-rich cases into enthalpy driven. This is the first calorimetrically determined binding enthalpy for eight-ring hairpin PAs and provides an increased understanding of PA-DNA interaction thermodynamics, including a shift in enthalpic vs. entropic control of binding.
Effects of β Inserts and PA Modification on Binding Affinity and Kinetics
In order to extend our understanding of the effects of different numbers internal of β building blocks on DNA recognition, comprehensive binding affinity studies of PAs were conducted with SPR. The results show that β substitutions for heterocycles in this sequence (Fig. 1) generally weaken the binding affinities of PAs with their cognate DNA. Together with previous studies of other eight-ring PA-DNA interactions16, 17 the weakening effect that single and double β substitutions can have on PA binding affinity is further confirmed. Moreover, the detailed evaluations in this work substantially demonstrate the large and diverse influences of β substitutions on the association and dissociation rates of PA-DNA interaction in a position-dependent manner (Table 2). The parent PA, KA1033, is a strong binder with the highest association rate among all PAs reported here, from which we infer an easily-optimized shape for the minor groove binding site. PAs, especially those containing internal βs, have numerous additional conformations attainable in solution because of the possibilities for rotation about single bonds. The γ-turn and C-terminal groups also have several rotatable bonds, and Ta has more than Dp. However, on optimized binding DNA, PAs must adopt a less dynamic conformation with a crescent shape to match the minor groove structure. In addition, stacked PA heterocycles and numerous H-bonds between PAs and DNA must be appropriately aligned in the minor groove complex. The necessary molecular rearrangements and alignments both require time, and consequently affect the association rates of PAs. The ka values for single internal β-containing PAs, KA1011, KA1013 and KJK6021 are slower than that for the all heterocycle-based KA1033. In addition, the ka values for double internal β-containing PAs (KA2128 and KA2129) are even slower than that for KJK6021. This can be explained by the increased flexibility of the β group and the increased number of conformations in solution that do not have a shape to match the DNA minor groove. Reorientation to bind DNA requires additional time relative to PAs with only heterocycles and thus slows the kinetics of association. It should also be noted that another single β-inserted PA, KA2127, has a slightly faster ka than KJK6021, which means the presence of some β inserts can also increase the PA association rates. In such cases, the added β inserts must adjust in more rapid alignment with functional groups in the DNA minor groove. These very diverse effects that β inserts can have on PA association rates might also correlate with the torsion angles of PA, as suggested by a study of other eight-ring PAs from the Sugiyama group.45 Additional kinetics studies on β containing PAs will be required to establish the molecular basis for these differences.
For the dissociation rates, the order for Dp containing PAs is KA1011 > KA1013 > KA1033, and the order for Ta containing PAs is KA2129 > KA2128 > KA2127 > KJK6021 (Table 2). Based on the decreasing order of dissociation rates, it is clear to see that the double β inserts speed up the PA dissociation more than single β inserts. This is the first instance whereby the number and position of β are shown to have such a large and diverse effect on association and dissociation rates and affinities of PAs in the same binding site. These results emphasize that the β group does not have the same favorable contacts with the minor groove as a Py heterocycle. For a β group to increase the affinity of a PA for DNA, it must increase other interactions in the PA, through resetting the PA-DNA interaction register, that are lost due to a switch of the heterocycle to a β. Additional studies of the correlation between β inserts and PA binding are under investigation with other PA sequences and DNAs. Overall, all of the current results are sufficient to validate the various modulation capabilities that the β group can have on PA binding affinity and kinetics, which confirms the β group as an important module in PA design.
Conclusions
In conclusion, the energetic effects of positional substitution of Py by single and double βs on hairpin PA-DNA interaction have been investigated in detail. As previously reported with other PAs, reduced binding affinity has been observed for all of the β-modified PAs. The SPR results clearly illustrate that the β substitution can have large, position- and number-dependent effects on association and dissociation rates, and also the binding affinities of PAs in the same DNA binding site. Comparison of mutant DNA sequences with all PAs also shows the position-dependent contributions of DNA base pairs in the PA binding and the preference of binding orientation for PAs. A modification of the positively charged functional Dp group into a Ta group, rather than a change in the heterocycles, preserves the PA binding mode and affinity but reduces PA aggregation in ITC. This made a thermodynamic study possible for eight-ring hairpin PAs for the first time, and it took eighteen years to advance our understanding of the energetic basis of PA-DNA interactions from six-ring to eight-ring PA. All of the above information significantly extends our knowledge of PA-DNA complexes and takes our understanding of the kinetics and thermodynamics of DNA-PA molecular recognition an important step forward.
Materials and Methods
DNA and Compounds
DNA sequences were purchased from Integrated DNA Technologies, Inc. (Coralville, IA USA). The synthesis and verification of KA1011 and KA1013 were previously reported,18 and that of the other PAs in Fig. 1 are presented in ESI†. Synthesis was generally based on the reported solid-phase Boc approach.46 The main changes to standard procedures were the use of PyBOP (benzotriazol-1-yl-oxy-tripyrrolidinophosphonium hexafluorophosphate) as the coupling agent and the extensive use of dimer building blocks. Dimer building blocks have long been used in solid and solution phase chemistry,10, 46–48 but their use in solid phase PA chemistry has often been limited to cases where the poorly nucleophilic amine of the imidazole amino acid does not couple well to an incoming active ester.46 We, however, used dimers at essentially every step in order to improve the solid phase yield and cut the number of solid-phase coupling steps in half. Many of these dimers are commercially available, including Boc-Py-PyCOOH and Boc-β-PyCOOH, though they are not always available, so we provide several procedures for dimer syntheses and scale up, and characterization data where appropriate (see ESI†). Furthermore, both Boc-Py-βCOOH and Boc-β-PyCOOH have appeared in the literature but were not characterized.49, 50 One standard synthesis of dimers46 uses the water-soluble carbodiimide EDC, but we found that the much less expensive combination of carbonyldiimidazole (CDI) and imidazolium hydrochloride described by Woodman et al.51 worked well to form the Boc-Py-βCOOH and Boc-β-PyCOOH dimers (both methods gave 60–70% isolated yields, much lower than the >90% yields reported48 and reproduced in our lab for other dimers with a range of coupling agents, but reagents for the CDI method are 1/10th the cost of EDC). The use of pre-formed HOBt ester Boc-Py-OBt has also been reported for dimer synthesis,48 and this is a very efficient method; it gave a 97% yield of Boc-Py-βCOOH on a 20g scale (see ESI† for scale-up of Boc-Py-PyCOOH and Boc-Py). CDI and Boc-β-Ala-OH were purchased from Chem-Impex Int’l (Wood Dale, IL USA). Imidazolium hydrochloride, β-alanine ethyl ester hydrochloride, dimethyl sulfoxide, trifluoroacetic acid, acetic anhydride, N, N-diisopropylethylamine, piperidine, 3-(dimethylamino)-1-propylamine, and 3,3’-diamino-N-methyl-dipropylamine were purchased from Aldrich (St. Louis, MO USA). Boc-Py-ImCO2H, O2NPy-COCCl3 and Boc-Py-OBt ester were from Oakwood Chemical (West Columbia, SC USA). N, N-dimethylformamide and dichloromethane were from Applied Biosystems (Foster City, CA USA). Boc-Py-PyCO2H was from A Chemtek (Worcester, MA USA), Boc-γ-Abu-OH and PyBOP from Novabiochem (Billerica, MA USA), and Boc-β-PAM resin (0.23 meq/g) from Peptides Int’l (Louisville, KY USA). Boc-β-ImCO2H was prepared via PyBOP coupling/saponification according to the literature48 as was desIm-ImCO2H.10
X-ray crystal structures of the Boc-Py-βCOOH and Boc-β-PyCOOH dimers (as dioxane and methanol solvates, and as the dioxane solvate) were obtained as a possible aid to molecular mechanics parameters, and ended up being quite instructive regarding intermolecular interactions (see the text below on aggregation, and ESI†). New compounds were characterized by elemental analysis, HPLC/MS, HRMS, and NMR, including 1H, 13C and 2-D methods. We changed from trifluoroacetate (TFA) salts to formate salts by changing running buffer of the standard chromatographic methods46 used in preparative HPLC from 0.1% trifluoroacetic acid to 0.2% formic acid.
Biosensor-Surface Plasmon Resonance (SPR)
SPR measurements were performed with four-channel Biacore 2000 and BiaT200 optical biosensor systems (GE Healthcare, Inc. Piscataway, NJ). The process of DNA immobilization is previously described.31, 52 Briefly, a 5’-biotin labeled hairpin duplex containing PA binding site (5’-biotin-CCTTGGAGAGTTTTCTCTCCAAGG, Fig. 1) was immobilized onto an SA chip. Filtered and degassed HEPES buffer containing 10 mM HEPES, 1 mM EDTA and varying NaCl (from 50 mM to 400 mM) at pH 7.4 with 0.05% v/v surfactant P20 was used in SPR experiments.
For the determination of PA binding affinity and kinetics, a series of PA samples in different concentrations (1 nM to 1 µM) was injected over the DNA sensor-chip until a steady state was obtained. Then, PA-free experimental buffer was flowed to measure PA dissociation and this was followed by chip regeneration with a pH 2.5 glycine solution, and baseline stabilization for the next cycle with multiple buffer injections. The flow rate of the entire cycle for all PAs in Fig. 2A is 25 µL / min, but 100 µL / min for KA2127 and 75 µL / min for KJK6021. A flow rate of 75 µL / min is used in a KA1033 and KJK6021 comparison (Figs. 4–5). Kinetic analyses were performed by global fitting of the binding results for the entire concentration series using a standard 1:1 kinetic model with integrated mass transport-limited binding parameters as described previously.53, 54 Steady-state analyses were also conducted and data were fit with a one site model: r = (Keq×Cf) / (1+ Keq× Cf), where r represents the moles of bound PA per mole of DNA hairpin duplex, Keq is equilibrium binding constants, and Cf is the free PA concentration in equilibrium with the complex. The observed steady-state response unit, RUobs, at saturation of binding site divided by the calculated response per bound PA, RUcal, gives the binding stoichiometry (n) of PA, n= RUobs/RUcal.
UV-Thermal Melting
DNA melting (Tm) curves were recorded with a Cary 300 UV visible spectrophotometer (Varian Inc., Palo Alto, CA) equipped with a thermoelectrically controlled cell holder. The absorbance of the free DNAs (Fig. S1, ESI†) and PA-DNA complexes were measured at 260 nm from 25 °C to 95 °C with a heating rate of 0.5 °C/min. The DNA concentration was 3 µM in hairpin, and PA in a 1:1 molar ratio of DNA was added. The Tm curves were normalized to give equimolar DNA concentrations. All the thermal melting experiments were conducted three times with HEPES buffer containing 10 mM HEPES, 50 mM NaCl and 1 mM EDTA at pH 7.4.
Circular Dichroism (CD)
CD spectra were collected using a Jasco J-810 spectrometer (Jasco Inc., Easton, MD) from 400 nm to 230 nm at 25 °C. The spectra were averaged over four scans with a scan speed of 50 nm / min and a buffer blank correction. A 5 µM DNA solution was first scanned and the PAs, at increasing concentration ratios, were then titrated into the same cuvette and the complexes were scanned under the same conditions. The same hairpin DNA and buffer as in the melting study were used.
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a MicroCal VP-ITC (GE Healthcare, Inc., Piscataway, NJ) with VP-2000 software for instrument control and Origin 7.0 for data analysis. The sample cell was filled with 10 µM of target DNA TGGAGA in HEPES buffer, and 20 injections of 10 µL of PA solution at variable concentrations were performed incrementally. A delay of 300 sec was used between each injection to ensure the equilibration of baseline. The heat for each injection was obtained by integration of the peak area as a function of time. The heats of dilution, determined by injecting compound into the sample cell containing only buffer, were subtracted from those in PA/DNA titrations to obtain the corrected binding-induced enthalpy changes. Because all the PAs bind quite strongly to the cognate sequence in this work, the heat/mole of added compound is essentially constant in the initial titration region where all added compound is bound to DNA. The ΔH can be determined by a linear fit as shown in Figs. S2-4. HEPES buffer containing 10 mM HEPES, 50 mM NaCl and 1 mM EDTA at pH 7.4 was used for ITC experiments.
Supplementary Material
Acknowledgements
The authors thank NIH NIAID AI064200 to W.D.W and NIH NIAID AI083803 to J.K.B for support, NSF MRI 0959360 for 600 MHz NMR, NSF MRI 0420497 for the purchase of an ApexII diffractometer, and the Center for Diagnostics and Therapeutics for a fellowship to S.W. The authors thank Carol Wilson for manuscript proofreading.
Footnotes
Electronic Supplementary Information (ESI) available: experimental details and characterization data (Tables S1-S3 and Figs S1-S9) and Xray crystallographic information (Tables X-ray 1-18 and Figs X-ray 1-6). See DOI: 10.1039/b000000x/
Contributor Information
James K. Bashkin, Email: bashkinj@umsl.edu.
W. David Wilson, Email: wdw@gsu.edu.
Notes and references
- 1.Neidle S. Nat. Prod. Rep. 2001;18:291. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 2.Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. Proc. Natl. Acad. Sci. U.S.A. 1985;82:1376. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dervan PB. Bioorg. Med. Chem. 2001;9:2215. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 4.Wemmer DE. Annu. Rev. Bioph. Biom. 2000;29:439. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- 5.Pelton JG, Wemmer DE. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5723. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wemmer DE, Geierstanger BH, Fagan PA, Dwyer TJ, Jacobsen JP, Pelton JG, Ball GE, Leheny AR, Chang W-H, Bathini Y, Lown JW, Rentzeperis D, Marky LA, Singh S, Kollman P. Structural Biology: The state of the art. Vol. 2. New York: Adenine Press; 1994. pp. 301–323. [Google Scholar]
- 7.Mrksich M, Parks ME, Dervan PB. J. Am. Chem. Soc. 1994;116:7983. [Google Scholar]
- 8.Parks ME, Baird EE, Dervan PB. J. Am. Chem. Soc. 1996;118:6147. [Google Scholar]
- 9.Woods CR, Ishii T, Wu B, Bair KW, Boger DL. J. Am. Chem. Soc. 2002;124:2148. doi: 10.1021/ja0122039. [DOI] [PubMed] [Google Scholar]
- 10.Wetzler M, Wemmer DE. Org. Lett. 2010;12:3488–3490. doi: 10.1021/ol1013262. [DOI] [PubMed] [Google Scholar]
- 11.Kelly JJ, Baird EE, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6981. doi: 10.1073/pnas.93.14.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Bando T, Sugiyama H. J. Am. Chem. Soc. 2006;128:8766. doi: 10.1021/ja0580587. [DOI] [PubMed] [Google Scholar]
- 13.Trauger JW, Baird EE, Mrksich M, Dervan PB. J. Am. Chemi. Soc. 1996;118:6160. [Google Scholar]
- 14.Turner JM, Swalley SE, Baird EE, Dervan PB. J. Am. Chem. Soc. 1998;120:6219. [Google Scholar]
- 15.Wang CCC, Ellervik U, Dervan PB. Bioorg. Med. Chem. 2001;9:653. doi: 10.1016/s0968-0896(00)00282-0. [DOI] [PubMed] [Google Scholar]
- 16.Bashkin JK, Aston K, Ramos JP, Koeller KJ, Nanjunda R, He G, Dupureur CM, Wilson WD. Biochimie. 2013;95:271. doi: 10.1016/j.biochi.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Nanjunda R, Aston K, Bashkin JK, Wilson WD. Biochemistry. 2012;51:9796. doi: 10.1021/bi301327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupureur CM, Bashkin JK, Aston K, Koeller KJ, Gaston KR, He G. Anal. Biochem. 2012;423:178. doi: 10.1016/j.ab.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeller KJ, Harris GD, Aston K, He G, Castaneda CH, Thornton MA, Edwards TG, Wang S, Nanjunda R, Wilson WD, Fisher C, Bashkin JK. Med. Chem. 2014;4:338. doi: 10.4172/2161-0444.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trauger JW, Dervan PB. Methods Enzymol. 2001;340:450. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 21.Murty M, Sugiyama H. Biol. Pharm. Bull. 2004;27:468. doi: 10.1248/bpb.27.468. [DOI] [PubMed] [Google Scholar]
- 22.Henry JA, Le NM, Nguyen B, Howard CM, Bailey SL, Horick SM, Buchmueller KL, Kotecha M, Hochhauser D, Hartley JA, Wilson WD, Lee M. Biochemistry. 2004;43:12249. doi: 10.1021/bi048785z. [DOI] [PubMed] [Google Scholar]
- 23.Bando T, Minoshima M, Kashiwazaki G, Shinohara KI, Sasaki S, Ohtsuki A, Murakami M, Nakazono S, Suglyarna H, Fujimoto J. Bioorg. Med. Chem. 2008;16:2286. doi: 10.1016/j.bmc.2007.11.064. [DOI] [PubMed] [Google Scholar]
- 24.Baliga R, Baird EE, Herman DM, Melander C, Dervan PB, Crothers DM. Biochemistry. 2001;40:3. doi: 10.1021/bi0022339. [DOI] [PubMed] [Google Scholar]
- 25.Han YW, Kashiwazaki G, Morinaga H, Matsumoto T, Hashiya K, Bando T, Harada Y, Sugiyama H. Bioorg. Med. Chem. 2013;21:5436. doi: 10.1016/j.bmc.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Walker WL, Landaw EM, Dickerson RE, Goodsell DS. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5634. doi: 10.1073/pnas.94.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CCC, Dervan PB. Tetrahedron. 2007;63:6146. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hargrove AE, Raskatov JA, Meier JL, Montgomery DC, Dervan PB. J. Med. Chem. 2012;55:5425. doi: 10.1021/jm300380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Kumar A, Aston K, Binh N, Bashkin JK, Boykin DW, Wilson WD. Chem. Commun. 2013;49:8543. doi: 10.1039/c3cc44569k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson WD, Tanious TA, Fernandez-Saiz M, Rigl CT. Methods Mol. Biol. 1997;90:219. doi: 10.1385/0-89603-447-X:219. [DOI] [PubMed] [Google Scholar]
- 31.Nanjunda R, Munde M, Liu Y, Wilson WD. Methods for Studying Nucleic Acid/Drug Interactions. Boca Raton: CRC Press; 2011. pp. 91–122. [Google Scholar]
- 32.Wang S, Munde M, Wang SM, Wilson WD. Biochemistry. 2011;50:7674. doi: 10.1021/bi201010g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodger A, Norden B. New York: Oxford University Press; 1997. [Google Scholar]
- 34.Lyng R, Rodger A, Norden B. Biopolymers. 1992;32:1201. doi: 10.1002/bip.360320910. [DOI] [PubMed] [Google Scholar]
- 35.Anderson CF, Record MT, Jr, Lohman TM. Q. Rev. Biophys. 1978;11:103. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 36.Wilson WD, Krishnamoorthy CR, Wang YH, Smith JC. Biopolymers. 1985;24:1941. doi: 10.1002/bip.360241008. [DOI] [PubMed] [Google Scholar]
- 37.Lohman TM, DeHaseth PL, Record MT., Jr Biophys. Chem. 1978;8:281. doi: 10.1016/0301-4622(78)80011-8. [DOI] [PubMed] [Google Scholar]
- 38.Meier JL, Yu AS, Korf I, Segal DJ, Dervan PB. J. Am. Chem. Soc. 2012;134:17814. doi: 10.1021/ja308888c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rucker VC, Melander C, Dervan PB. Helv. Chim. Acta. 2003;86:1839. [Google Scholar]
- 40.Kang JS, Meier JL, Dervan PB. J. Am. Chem. Soc. 2014;136:3687. doi: 10.1021/ja500211z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilch DS, Poklar N, Gelfand CA, Law SM, Breslauer KJ, Baird EE, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8306. doi: 10.1073/pnas.93.16.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prakash AS. J. Excipients Food Chem. 2011;2:28. [Google Scholar]
- 43.Loeser E, Sutton P, Skorodinsky A, Lin M, Yowell G. Drug Dev. Ind. Pharm. 2012;38:357. doi: 10.3109/03639045.2011.604330. [DOI] [PubMed] [Google Scholar]
- 44.Chaires JB. Arch. Biochem. Biophys. 2006;453:26. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 45.Han Y-W, Matsumoto T, Yokota H, Kashiwazaki G, Morinaga H, Hashiya K, Bando T, Harada Y, Sugiyama H. Nucleic Acids Res. 2012;40:11510. doi: 10.1093/nar/gks897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baird EE, Dervan PB. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- 47.Xiao JH, Yuan G, Huang WQ, Chan ASC, Lee KLD. J. Org. Chem. 2000;65:5506. doi: 10.1021/jo000135n. [DOI] [PubMed] [Google Scholar]
- 48.Chenoweth DM, Harki DA, Dervan PB. J. Am. Chem. Soc. 2009;131:7175. doi: 10.1021/ja901307m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howard P, Masterson L, Roffery J. U.S. Patent WO2009060208. 2009 May 14;
- 50.Krutzik PO, Chamberlin AR. Bioorg. Med. Chem. Lett. 2002;12:2129. doi: 10.1016/s0960-894x(02)00359-1. [DOI] [PubMed] [Google Scholar]
- 51.Woodman EK, Chaffey JGK, Hopes PA, Hose DRJ, Gilday JP. Org. Process Res. Dev. 2009;13:106. [Google Scholar]
- 52.Liu Y, Wilson WD. Methods Mol. Biol. 2010;613:1. doi: 10.1007/978-1-60327-418-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karlsson R. J. Mol. Recognit. 1999;12:285. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<285::AID-JMR469>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 54.Morton TA, Myszka DG. Methods Enzymol. 1998;295:268. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.