Abstract
Minor groove binding compounds have been shown to induce changes in global DNA conformation, allosterically inhibiting DNA-protein interactions necessary for transcriptional processes. Many minor groove binders are specific for AT-base pairs but have little preference over alternating AT or A-tract sequences. Few compounds, other than polyamides, show selectivity for mixed sequences with AT and GC base pairs. Electrospray ionization mass spectrometry (ESI-MS) can provide insight on the stoichiometry and relative affinities in minor groove recognition of different DNA sequences with a library of minor groove binders. A goal in our current research is to develop new compounds that recognize mixed sequences of DNA. In an effort to optimize screening for compounds that target mixed AT and GC base pair sequences of DNA, ESI-MS was used to study the competitive binding of compounds with a mixed set of DNA sequences. The method identified preferred binding sites, relative affinities, and concentration-dependent binding stoichiometry for the minor groove binding compounds netropsin and DB75 with AT-rich sequences, and DB293 with ATGA and AT-sites.
Keywords: Electrospray ionization mass spectrometry, mixed DNA sequences, selectivity, minor groove recognition, minor groove binders
Introduction
Non-polyamide minor groove (MG) binders target AT-rich sites with variable distinction among AT sequences [1]. Netropsin (Net) and DB75 (Figure 1) are MG binding compounds which bind with 1:1 stoichiometry to AT-rich sites [2, 3]. Polyamides can selectively target GC base pairs (bp) in a DNA sequence, but few non-polyamide MG binders target mixed sequence sites. DB293 (Figure 2) is the first dicationic diamidine to strongly recognize a sequence with mixed bp [4]. It binds in the MG of ATGA sequences as an antiparallel stacked dimer with positive cooperativity. Using the known interactions of Net, DB75, and DB293 as reference points, we are developing a mixed sequence method to screen DNA-MG binders using electrospray ionization spectrometry (ESI-MS). Because of its improved versatility, ESI-MS has widely increased its utility for studying biomacromolecules and is ideal for characterizing systems with non-covalent interactions.
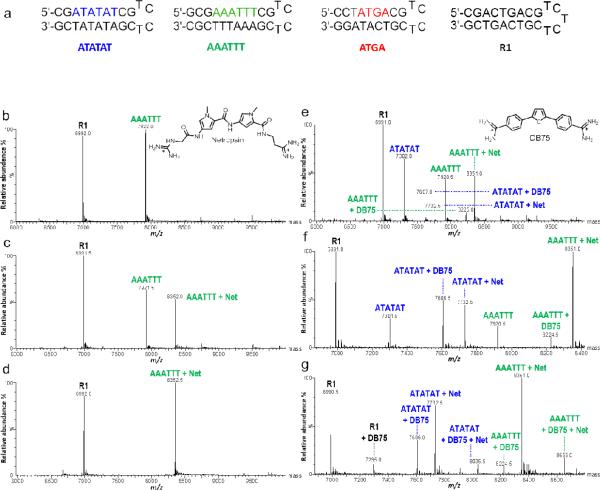
Fig. 1.
(a) DNA sequences used to simultaneously test binding of multiple sequences. ATATAT, AAATTT, ATGA, and R1. Concentration of DNA is 5μM. (b-d) Spectra of DNA and Net binding with increasing ligand concentration. Ligand to DNA ratios are [0:1], [1:1], and [2:1], respectively. (e-g) Spectra show the competitive binding of Net and DB75 with multiple DNA sequences. Respective ratios shown at [0.5:1], [1:1], and [2:1]. Free DNA is indicated by sequence name above corresponding peak (AAATTT, m/z 7,921.5) and ligand-DNA complex as “name + n” (AAATTT + Net, m/z 8,352.5)
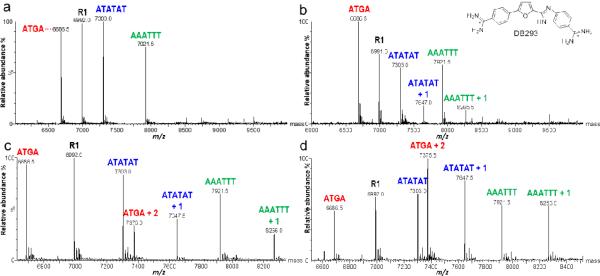
Fig. 2.
Spectra of mixed DNA sequences titrated with DB293. Unbound DNA indicated by sequence “name” above corresponding peak (ATGA, m/z 6,686.5) and ligand-DNA complex as “name + n” (ATGA + 2, m/z 7,375.0). DNA concentration is 5μM for each sequence. Molar ratios of [0:1], [1:1], [2:1], and [4:1] correspond to (a-d), respectively
We report a method that provides rapid screening of MG compound libraries for interactions, which can be easily characterized based on differences in structure and DNA sequence. To the best of our knowledge, this is the first example using ESI-MS to screen the competitive interaction of ligands with multiple DNA sequences in a single sample. Using this approach, we can begin to define a basis for specificity of drug binding at a target sequence. With enough DNAs, this method can screen target sites and complexes of interest. Information including relative binding affinity, stoichiometry, and binding cooperativity can be determined.
Experimental Protocol
Electrospray ionization mass spectrometry
Ligand and DNA stock solution preparation are described in the Electronic Supplementary Material. Titration experiments were performed with a mixed set of DNA in a solution with a total volume of 100 μL. DNAs were diluted to 5 μM each in 150 mM NH4OAc (pH 6.7) and stored at 4 °C. Titration ratios are expre ssed as compound-to-single-DNA, instead of compound-to-total-DNA concentration. For example, 20 μM DB293 to 5 μM ATGA is expressed as a [4:1] ratio. R1 was used as a reference because it contained no known target sequence.
ESI-MS experiments were performed using a Waters (Milford, MA) Micromass Q-TOF in negative ion mode using the MassLynx 4.1 software. Conditions were chosen based on published methods [5] and optimized (see Electronic Supplementary Material, Fig. S1). Capillary voltage, 2200 V; sample cone voltage, 30 V; extraction cone voltage, 3 V; source block temperature, 70 °C; desolvation temperature, 100 °C, and sample injection flow rate, 5 μL/min. A volatile solvent, such as MeOH, is often added to facilitate solution-to-gas phase transition, however, the addition of a solvent was not necessary using our conditions (see Electronic Supplementary Material). The instrument was flushed with 150 mM NH4OAc prior to sample injection. Scans were collected every 1.0 sec for 10 min with the final 2 min averaged. Raw spectra of the free DNA and DNA-complexes showed multiply charged species ranging 300 – 3000 m/z and the most abundant peaks belonged to -4, -5, and -6 charge states (see Electronic Supplementary Material, Fig. S1). Spectra were deconvoluted using the Maximum Entropy function which calculates the molecular ion (M) based on the equations below, where H is proton molecular weight, m’ and m” are specific m/z values, and z’ is the charge state for m’.
| (1) |
| (2) |
Results
Testing the ESI-MS method with known MG binders
Net is a well-understood MG binder which forms high affinity 1:1 complexes with the AT-rich sequences selected for our experiments [2]. Figure 1b is a deconvoluted spectrum of AAATTT and R1 sequences at a molar ratio of [0:1] Net:DNA and both DNA sequences have single peaks with similar intensity and minimal background noise. Adding Net to form a molar ratio of [1:1] Net to AAATTT led to a large decrease in peak intensity for the oligonucleotide compared to R1 and the appearance of a peak corresponding to a 1:1 Net:DNA complex, which indicates strong binding between Net and AAATTT. A [2:1] sample showed an additional decrease in parent peak intensity indicating that Net had completely saturated the binding site of the AAATTT. Even at a [2:1] ratio, no 2:1 species was observed. This indicated that binding is 1:1, as expected.
Similar to Net, DB75 is also a well-characterized MG binder that binds with high affinity as a monomer to AT-rich regions. Experiments were performed using both MG binders with ATATAT, AAATTT, and R1 to test competitive binding. Equal concentrations of DB75 and Net were added at a mole:mole ratio of [0.5:1] for a single ligand-to-DNA. At this ratio, the tallest peak corresponded to 1:1 AAATTT with Net. Smaller peaks show ATATAT+Net and ATATAT+DB75 complexes, but no binding is detected for AAATTT+DB75 (Figure 1e). An increase in ligand concentration to [1:1] showed a different pattern in complex formation where AAATTT+Net was the most abundant. Peak intensities increased to roughly 50% for ATATAT+DB75 and ATATAT+Net, while a new peak for AAATTT+DB75 was detected (Figure 1f). Upon further increasing ligand concentrations to [2:1], a new series of peaks appeared and free ATATAT or AAATTT decreased (Figure 1g). The tallest peak became AAATTT+Net and the intensity of ATATAT+Net surpassed ATATAT+DB75. The peaks corresponding to the parent nucleotides disappeared, indicating complete complexation. This increase in intensity for the ATATAT+Net complex likely occurred once all free AAATTT was consumed due to the high affinity of Net for AAATTT. Residual Net could then saturate the ATATAT binding site.
The high binding affinity of Net for AT sequences is apparent, as is the competitive binding of Net over DB75 for AT sequences. Surprisingly, as the concentration was increased, new peaks were detected for DB75+R1. DB75, known to bind strongly in the MG of AT sequences, has also been reported to intercalate weakly with GC-sites, which is likely the case for R1+DB75 at increased concentrations. The intercalated binding of DB75 occurs because the stronger binding Net saturates the AT-rich MG site. As the free concentration of DB75 reaches a high level, intercalation at the available GC-sites then ensues. All of the results with Net and DB75 are in good agreement with published results but also provide new insight into the interactions of these compounds with different DNA sequences.
Using DB293 to test monomer versus dimer binding
Previous work with DB293 indicates it forms a cooperatively stacked dimer with ATGA and a 1:1 complex with AATT. Thus the binding of provides a more complex ESI-MS test system [4]. Solutions of DB293 were mixed with R1, AAATTT and ATATAT, as well as ATGA (Figure 2). Initially, only monomer complexes with ATATAT and AAATTT are formed at a [1:1] ratio with little preference for either sequence. At a [2:1] ratio, a 1:1 species between both AT sequences and DB293 is again observed. For ATGA and DB293, no 1:1 complex is detected but a 2:1 species is observed. The lack of a 1:1 complex between DB293 and ATGA is indicative of strong cooperative dimer binding and is in agreement with biosensor-SPR results [4]. When the concentration of DB293 was doubled to a [4:1] ratio, the pattern remained the same. R1 showed no interaction with DB293 at any of the ligand concentrations. The peak at 7,375.5 m/z not only indicates a 2:1 complex for DB293 with ATGA, but having the highest intensity suggests a greater affinity for the cooperative dimer with ATGA over monomer formation with ATATAT and AAATTT sequences.
Discussion
Using a mixed set of DNA sequences, we can quickly and accurately evaluate relative affinities, stoichiometry, and cooperativity. These tests show ESI-MS not only detects binding using control DNAs and MG binders, but also competition among ligands and DNAs can be observed. ESI-MS can screen multiple sequences simultaneously, and alternative binding modes for MG binding compounds can be detected. Our intention for developing this method is to complement information obtained by other techniques such as footprinting or next-generation assays. The goal is to determine differences in binding for compounds with a closely related set of sequences having systematic variations. The DNA has a known binding site, in addition to sequences variants (e.g. ATAT, AATT, and AAAA), so we can focus on sequence specificity and compound selectivity. The strongest binding ligands will interact with available binding sites first, which makes this a true competitive assay.
Analyzing interactions between ligand and a single DNA is not efficient. One can study any number of interactions in the same time it takes to study one. The key is in ligands and sequences which form complexes with distinguishable molecular weights. By using sequences with like compositions, the sensitivity of DNA and DNA-complexes are also similar, thus, limitations such as response factors are overlooked (see Electronic Supplementary Material). This feature allows direct comparison of unbound DNA and complexes to determine the relative binding affinity. It further demonstrates that our method to investigate the competitive and selective binding between multiple, mixed DNA-ligand interactions in a single sample is not only more efficient but also provides important new information with DNA interactions.
Conclusion
We have developed a novel approach to observe competitive binding and to screen for DNA-ligand interactions. To the best of our knowledge, this is the first example using ESI-MS to simultaneously examine ligand binding with multiple DNA sequences. Our findings show excellent consistency between ESI-MS and other biophysical methods. This technique has many favorable features: it is rapid, convenient, and requires small amounts of sample. It allows direct comparison of relative binding affinities in addition to stoichiometry and cooperativity. Using this innovative method, one could theoretically screen dozens of sequences and obtain a large amount of information from a single sample, reducing reagents used and time spent cleaning between sample runs. More importantly, this method is not limited to DNA and small molecules, and can be applied to other biomacromoleculear interactions including proteins, RNA, and carbohydrates.
Supplementary Material
Acknowledgements
National Institutes of Health (NIH) AI064200 grant awarded to WDW and DWB, the Molecular Basis of Diseases Area of Focus (MBDAF) Fellowship to SRL, and Carol Wilson for help in manuscript preparation.
Abbreviations
- bp
base pairs
- DNA
deoxyribonucleic acid
- ESI-MS
electrospray ionization mass spectrometry
- Kd
dissociation constant
- MG
minor groove
- Net
netropsin
- m/z
mass-over-charge
- SPR
surface plasmon resonance
References
- 1.Abudaya A, Brown PM, Fox KR. Nucleic Acids Res. 1995;23:3385–3392. doi: 10.1093/nar/23.17.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wartell RM, Larson JE, Wells RD. J Biol Chem. 1974;249:6719–6731. [PubMed] [Google Scholar]
- 3.Laughton CA, Tanious F, Nunn CM, Boykin DW, Wilson WD, Neidle S. Biochemistry. 1996;35:5655–5661. doi: 10.1021/bi952162r. [DOI] [PubMed] [Google Scholar]
- 4.Bailly C, Tardy C, Wang L, Armitage B, Hopkins K, Kumar A, Schuster GB, Boykin DW, Wilson WD. Biochemistry. 1996;40:9770–9779. doi: 10.1021/bi0108453. [DOI] [PubMed] [Google Scholar]
- 5.Rosu F, De Pauw E, Gabelica V. Biochimie. 2008;90:1074–1087. doi: 10.1016/j.biochi.2008.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.