Summary
For decades, proteins were thought to be the sole or at least the dominant source of antigens for T cells. Studies in the 1990s demonstrated that CD1 proteins and mycobacterial lipids form specific targets of human αβ T cells. The molecular basis by which T-cell receptors (TCRs) recognize CD1-lipid complexes is now well understood. Many types of mycobacterial lipids function as antigens in the CD1 system, and new studies done with CD1 tetramers identify T-cell populations in the blood of tuberculosis patients. In human populations, a fundamental difference between the CD1 and major histocompatibility complex systems is that all humans express nearly identical CD1 proteins. Correspondingly, human CD1 responsive T cells show evidence of conserved TCRs. In addition to natural killer T cells and mucosal-associated invariant T (MAIT cells), conserved TCRs define other subsets of human T cells, including germline-encoded mycolyl-reactive (GEM) T cells. The simple immunogenetics of the CD1 system and new investigative tools to measure T-cell responses in humans now creates a situation in which known lipid antigens can be developed as immunodiagnostic and immunotherapeutic reagents for tuberculosis disease.
Keywords: CD1, mycolyl lipids, T cells, T-cell receptor, Mycobacterium tuberculosis
Lipids as T-cell antigens
Immunologists have understood for decades that homogenized Mycobacterium tuberculosis cell walls, known as complete Freund’s adjuvant, stimulate unusually strong immune responses. Efforts to elucidate the mechanisms of Freund’s adjuvant have emphasized the roles immunostimulatory lipids, including phosphatidylinositolmannoside (PIM), lipoarabinomannan (LAM) and mycolyl glycolipids (1). These and other mycobacterial lipids have long been known to activate macrophages through innate receptors, such as Toll-like receptor 2 (TLR) and Mincle (2–4). Although some innate receptors are also present on T and B cells, the most distinctive receptors of the adaptive immune system are the recombining receptors for antigen: the T-cell receptors (TCRs) and B-cell receptors. Therefore, the discovery of TCR-mediated recognition of mycobacterial lipids that are displayed by human CD1 proteins changed several general views about the role of lipids in control of immune response (5, 6).
Whereas lipids were thought solely to activate innate receptors, these studies proved that rearranged TCRs specifically respond to lipids. Second, whereas T cells were thought to solely or mainly recognize peptide antigens bound to T cells, studies of CD1 and mycobacteria expanded the range of natural T-cell antigens to include lipids (6), glycolipids (7), phospholipids (8), sulfolipids (9), and lipopeptides (10). Third, unlike the invariant, germline-encoded receptors of the innate system, TCRs are formed by somatic rearrangements and appear as millions of combinations in a single individual. Such extreme receptor diversity is usually considered the hallmark of T cells, as key effectors in acquired immunity. However, studies of T-cell response to CD1d and CD1b show marked conservation of TCRs responding to CD1-lipid complexes (11, 12). These findings raise questions about whether TCRs are always diverse and represent effectors of acquired immunity or instead can also exist as ‘innate T cells’. This review focuses on human T-cell activation by mycobacterial lipids via the TCR as it contacts CD1-lipid complexes. We highlight the newest studies of measurement of populations of human T cells in tuberculosis patients using newly developed CD1 tetramers. CD1 proteins do not vary in structure from person to person. The simple population genetics of CD1 genes appears to enable a response that is shared among individual patients, enhancing the prospects for using lipid antigens as a new approach to immunodiagnosis and immunomodulation.
Mammalian CD1 genes
CD1 proteins are related in structure to major histocompatibility complex (MHC) class I molecules in that both consist of a membrane-anchored heavy chain associated with a β2 microglobulin (13). The heterodimer folds to form a hollow groove or cleft that binds antigen (14). Another shared feature is that the MHC class I and CD1 loci are polygenic. The number of CD1 genes per genome varies between two in mice and thirteen in horses (15). The human locus contains five distinct CD1 genes, which in this field are known as isoforms: CD1a, CD1b, CD1c, CD1d, and CD1e. CD1 genes in all mammals are named according to their human orthologs. For example, bovine genomes encode five genes that most closely resemble CD1b, and these genes are named CD1b1, CD1b2, CD1b3, CD1b4, and CD1b5. Muroid rodents, including common strains of experimental mice, encode only two copies of the CD1d gene. In contrast, nearly all other mammalian genomes encode larger numbers of CD1 genes, including orthologs of CD1a, CD1b, or CD1c. Rabbits, guinea pig, cattle, pig, dog, horse encode from six to 13 CD1 genes (15–20). Like for MHC class I and class II loci, CD1 pseudogenes are present in most mammalian genomes (21), so the number of genes actually expressed is not always known, although the natural expression and function of non-human CD1 genes has been examined in several species, including those used to study tuberculosis, such as guinea pigs and cattle (17, 18, 22). However, that most mammals have generated and then retained relatively large, polygenic CD1 loci suggests that the different isoforms have distinct functions.
Animal models of CD1
Mycobacterium tuberculosis is primarily a pathogen of humans. Yet zebrafish, mice, guinea pigs, rabbits, cynomolgus monkeys, rhesus macaques, common marmoset, and cattle have all been used as models and mimic certain aspects of human tuberculosis. Consideration of the naturally occurring CD1 proteins in these various species provides insights into which of these experimental hosts measure the contribution of CD1 to mycobacterial infection outcomes (Fig. 1). Like all jawed fish, the zebrafish has MHC genes, but it has no CD1 genes (23–25). Mice and rats have CD1d-encoding genes, but lack CD1a, CD1b, CD1c, and CD1e-encoding genes (26). As discussed later, most known mycobacterial antigens are presented by those genes lacking in the mouse. The absent or incomplete set of CD1 genes in these common and tractable animal models has provided a rationale for creation of new trans-species models. Mice that are transgenic for human CD1a, CD1b, CD1c, and CD1e have been created and used for experiments addressing immune recognition of mycobacterial lipids (27). Also, humanized severe combined immunodeficiency (SCID) mice reproduce certain CD1 expression patterns seen in humans, and spleen cells of these mice can present mycobacterial lipids to human T-cell lines in vitro (28).
Fig. 1. Characteristics of the five CD1 isoforms and distribution among animal models.
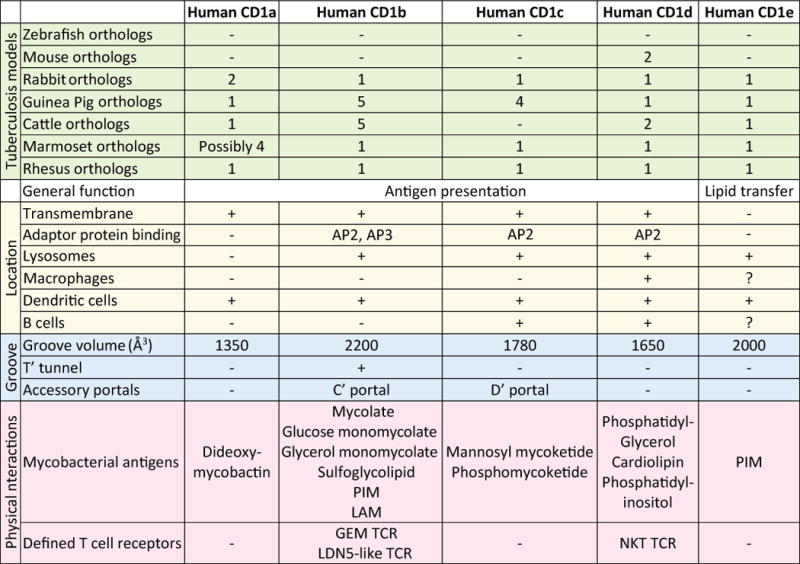
The numbers of orthologous CD1 genes are given for animals that are commonly used as model for tuberculosis (green). The expression and functions of these genes have only been studied in humans, cows, guinea pigs, and non-human primates. Gene numbers are based on published papers and, when no information is available, on annotated genes in the genomes (available at www.ensembl.org). Subcellular location and cellular expression profile (yellow), groove characteristics (blue), and interacting antigens and T-cell receptors (pink) are shown for each human CD1 isoform.
Considering animals with naturally evolved CD1a, CD1b, or CD1c proteins, guinea pigs, and rabbits express at least one ortholog of each human CD1 isoform. These animals are relatively small, commercially available, and the conditions for experimental mycobacterial infections have been well established. In guinea pigs, experimental immunizations with crude mycobacterial lipid extracts have been performed and some protection against disease was demonstrated (29). In addition, in vitro assays showed CD1-specific T-cell responses against mycobacterial lipids and patterns of CD1 expression in lymphoid organs that mimic certain patterns seen in humans (29–31). Cows are both a model for tuberculosis as well as a natural host for M. bovis, which causes bovine tuberculosis. Even though CD1c genes are not present in the bovine genome, all other isoforms are present, which makes this animal a useful model (18, 32). Bovine CD1b-dependent responses against mycobacterial lipids during natural disease and upon experimental immunization have been demonstrated (33, 34). Disadvantages of the bovine model are the high costs for housing the animals in clean environments, and the difficulties of buying animals that have not been exposed to mycobacteria. Monkey models have considerable costs, but the marmoset and rhesus monkey CD1 proteins resemble their human counterparts more closely than the CD1 proteins of other widely used tuberculosis models (35). Overall, animals that best replicate the human system of five CD1 genes are less tractable experimentally, creating a situation in which much CD1 research is carried out in humans, which is the central topic of this review.
CD1 proteins have distinct functions
A central question dominating the cell biological study of CD1 presentation is whether the five types of CD1 proteins have essentially the same or instead possess distinct functions. Porcelli pointed out that the sequence homology of the human CD1 heavy chains is low in absolute terms (36). Studies conducted over the past decade now clearly establish distinct structures and functions of each CD1 isoform in cells. A clear example is human CD1e, which unlike other CD1 proteins, undergoes cleavage of the heavy chain in the luminal side of the stalk so that it is not membrane bound. Membrane anchoring at the cell surface is the essential feature of antigen-presenting molecules. Instead, De La Salle and colleagues (37–39) have shown that CD1e is a soluble protein that is detectable within the luminal and extracellular compartments, where it serves to bind and transfer lipids from membranes to other CD1 proteins. For example, the first data to establish CD1e’s core function found that it assists in the extraction of PIM from membranes for action by mannosidases and presentation by CD1b proteins (39). In fact, intact CD1e proteins influence lipid antigen display by several human CD1 proteins (40). In contrast to CD1e, CD1a, CD1b, CD1c, and CD1d are membrane anchored antigen-presenting molecules for T cells. Yet each type of human CD1 antigen-presenting molecule differs with regard to pocket architecture, subcellular trafficking patterns and expression patterns across different tissues. These differences are summarized in Fig. 1, and they strongly point to distinct immunological functions for each type of human CD1 protein.
Patterns of tissue expression
Consistent with its known function in allowing T-cell responses to viruses present in nearly any cell, MHC class I is expressed on nearly all nucleated cells outside the central nervous system. MHC II is expressed in a more restricted pattern, mainly on professional antigen-presenting cells (APCs), such as macrophages, dendritic cells, B cells, and thymic epithelial cells. This pattern of expression matches its known function relating to capture of exogenous antigens derived from other cell types or extracellular pathogens that enter into the endosomal pathway. CD1 genes were discovered in the thymus, where they are expressed on thymocytes (rather than thymic epithelium) (41). Like MHC class II, CD1 proteins are expressed on professional APCs in the periphery but are even more restricted in their expression (42). Individual CD1 isoforms are present on different subtypes of APCs, as summarized in Fig. 1. In the periphery, the only cell type that is well established to express CD1b is the myeloid dendritic cell (DC). Some studies of human DCs now use CD1b as a defining lineage marker (43, 44). In addition to being expressed by DCs, CD1a is the only isoform that is expressed at high density on epidermal Langerhans cells (42). Both CD1c and CD1d are co-expressed on a subpopulation of blood-derived B cells and marginal zone B cells, as well as on DCs (42). CD1d is much more broadly expressed than other CD1 isoforms as it can be found on the cell types listed above as well as macrophages and gastrointestinal epithelial cells (42, 45). The prominent expression of CD1 proteins on professional APCs that routinely acquire exogenous antigens from other cell types is consistent with the proposed role of CD1 proteins in antibacterial response. Yet, the differing patterns seen for each human CD1 protein, which are also seen in non-human species such as guinea pigs, again points to differing functions for each CD1 isoform. The differing patterns of CD1 expression in macrophages and DCs are considered in detail because these two cell types play central roles in the host response to tuberculosis.
Two groups of CD1 proteins
Calabi and Milstein assigned CD1 proteins to two groups in the 1980s based on sequence homology, with CD1d comprising group 1, and CD1a, CD1b, and CD1c in group 2 (46). This classification scheme is still widely used, in part because newer studies show that the group 1 and group 2 CD1 proteins have fundamentally different mechanisms of gene regulation in macrophages and DCs. First, macrophages express group 1 but not group 2 CD1 proteins, while myeloid DCs express both group 1 and 2 CD1 proteins. Further, group 1 and 2 CD1 transcription show opposite patterns of response to the same bacterial stimulus (47, 48). Both findings suggest distinct immunological roles for group 1 and 2 proteins, with CD1d functioning on a constitutive basis, and group 1 CD1 proteins being controlled by antecedent activation of innate receptors (49). Given the importance of macrophages and DCs in host response to mycobacteria and the direct evidence for mycobacterial control of CD1 expression (44, 48, 50), these differing patterns of CD1 gene and protein regulation are considered in detail.
CD1 gene regulation in DCs
In humans monocytes and macrophages constitutively express CD1d, but not CD1a, CD1b, or CD1c (51, 52). Blood monocytes constitutively express CD1d, but do not detectably transcribe the other CD1 isoforms or express protein (48). In vitro activation of monocytes with stimuli that promote their differentiation into DCs, such as granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4), show the concordant appearance of CD1a, CD1b, and CD1c proteins in 2 to 3 days. In vivo differentiation of monocytes into tissue DCs cannot be observed directly, but these in vitro systems do likely mimic aspects of the natural differentiation process in tissue because DCs in vivo express CD1a, CD1b, and CD1c proteins at high levels (43, 44, 50). Importantly, this process is driven by mycobacteria themselves. Cellular upregulation of CD1 in vitro is dependent on Toll-like receptor (TLR) activation and is stronger when concomitant inflammasome (NLRP3) activation is present, leading to cleaved bio-active IL1-β, which is sufficient to strongly induce CD1a, CD1b, and CD1c expression (2, 44, 47, 48). In vivo, CD1a, CD1b, and CD1c proteins are strongly upregulated at the site of tuberculoid leprosy lesions (50) and in the bronchoalveolar lavage specimens of tuberculosis patients (53). Also, it is notable that the mycobacterial signals that drive up CD1a, CD1b, and CD1c expression reduce the expression of human CD1d (48).
These patterns suggest a general model in which CD1a, CD1b, and CD1c proteins are rarely expressed within peripheral tissues in the absence of inflammation. Infection or other signals that lead to IL-1β cleavage cause local CD1a, CD1b, and CD1c expression near the site of infection and promote CD1-mediated host responses. Thus, as contrasted with both MHC class I and II, which are constitutively expressed on most professional APCs, the inducible nature and more limited cell type expression of CD1a, CD1b, and CD1c suggests a strong role of pathogen-induced gene regulation and the resulting T-cell response. In this model, M. tuberculosis enters tissues and triggers CD1 expression locally via IL1-β and sheds lipid antigens, while in proximity to cells that are in the process of inducing cell surface CD1 proteins. Thus, the restricted expression of group 1 CD1 is locally reversed by the pathogen during infection in ways that can promote the capture and display of foreign lipids (49). Group 1 CD1-reactive T cells are primed by DCs, so they might carry out antimicrobial effects via granulysin or cytokine-mediated activation of macrophages. In particular, CD1b and mycobacterial mycolyl-lipid reactive T cells secrete interferon-γ and tumor necrosis factor at high levels (12).
Distinct subcellular lipid capture pathways
After translation in the endoplasmic reticulum (ER), CD1 proteins exit to the cell surface via the secretory pathway and then are internalized into the endosomal network and recycle back to the surface. The spatially separate secretory and endosomal compartments provide differing antigens, chaperone proteins and pH environments and so can be considered to form the basis of two distinct modes of lipid antigen capture (Fig. 2A). During the first passage toward the cell surface, CD1 proteins bind self cellular lipids. Subsequently, CD1 proteins are internalized and recycle through the endocytic network, where contact foreign and exogenous self antigens that are internalized into endosomes via receptor mediated endocytosis and phagocytosis. Thus, many studies have now coalesced into an integrated model by which the secretory and endosomal pathways have separate functions in antigen loading (54–56), and this review focuses on the intersection of mycobacterial lipids with CD1 proteins the endosomal pathway.
Fig. 2. Antigen uptake and loading in the endosomal pathway.
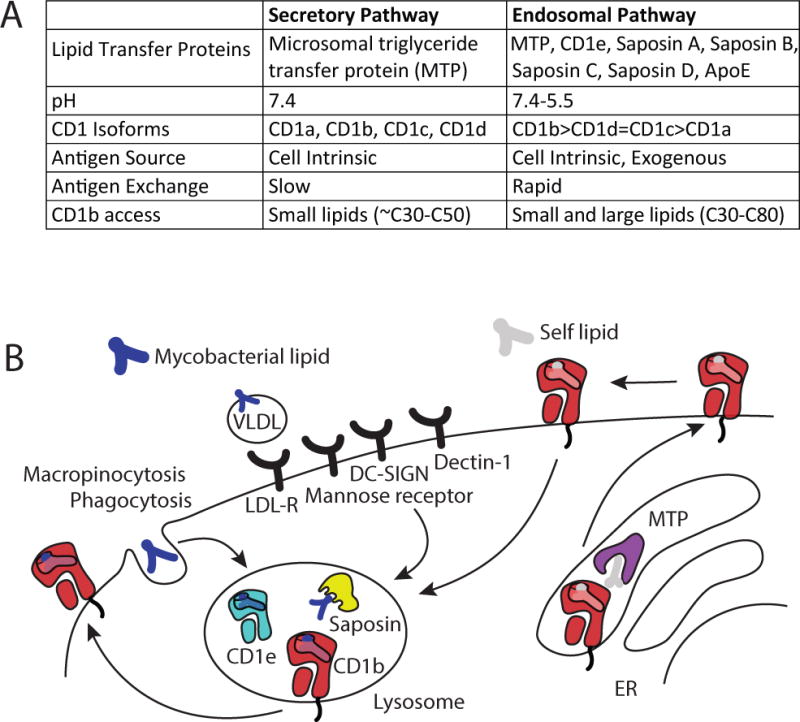
The secretory and endosomal pathways have differing characteristics that influence antigen presentation by CD1 proteins (A). After an initial appearance at the cell surface with an endogenous self-lipid, presentation of exogenous lipids by CD1b proteins is accomplished by the trafficking of CD1b proteins and lipid antigens into the lysosome, which has a low pH, contains lipid binding proteins and is enriched in lipids that bind cell surface receptors.
During the time that newly synthesized CD1 molecules are co-translationally folded they are exposed to self lipids in the ER and then transit the golgi and arrive at the cell surface (57). Loading of lipids onto CD1 heavy chains likely occurs co-translationally (58), and bacterial lipids are not substantially imported into these compartments. Therefore, the secretory pathway predominantly handles capture of self lipids derived from within the CD1-expressing APC (cell intrinsic antigens), including hundreds of short chain (C30–C40) distinct phospholipids and sphingolipids (59–61).
CD1 recycling through endosomes
The short cytoplasmic tails of CD1 proteins contain functionally important signaling motifs that contain tyrosine (Y), followed by spacer (X), and hydrophobic (Z) residues. At the cell surface, these cytoplasmic YXXZ motifs bind to adaptor protein complexes (AP), which mediate sorting into endocytic compartments that are bound for endosomes (AP-2) or lysosomes (AP-3) (56, 62–64). Then CD1 proteins recycle back to the cell surface for a second round of antigen display. Each human CD1 protein uses the secretory and endosomal pathways to a varying extent, depending on whether its cytoplasmic tail encodes a YXXZ motif that binds to two (CD1b), one (CD1c, CD1d), or zero (CD1a) types of AP complexes. Human CD1b binds both AP-2 and AP-3 and so is most efficiently delivered to the late endosomes. Accordingly, CD1b antigen-binding and cellular function is blocked by deletion of tail motifs or drugs that neutralize endosomal pH (62–64). Human CD1c and CD1d bind AP-2 but not AP-3 and so enter endosomes but show somewhat less penetration into LAMP1+ compartments and dependence of antigen display on low pH (65, 66). Human CD1a spends most of its time at the cell surface with a minor pool that enters sorting endosomes and early endosomes (64, 67), and CD1a readily loads antigen at the surface without a requirement for low pH (68). The differing subcellular localizations again highlights the different function of each human CD1 protein type and creates a situation in which antigens can be sampled in the ER, on the cell surface, and from phagocytosed particles or whole organisms.
Antigen uptake into DCs
Whole bacteria, fragments of bacteria, lipids aggregated with lipid binding proteins or in apolipoproteins can be taken up by CD1-expressing APCs via phagocytosis or endocytosis and interact with endosomal CD1 molecules. Alternatively, lipid antigens can interact with CD1 proteins at the cell surface, a process that occurs efficiently for CD1a (68) and inefficiently for CD1b (64, 69). Because M. tuberculosis inhibits acidification (70) and presentation of certain mycobacterial lipids, such as long chain mycolyl lipids, requires low pH for CD1b binding (9, 69), the natural acidification blockade from live intracellular M. tuberculosis might impair lipid antigen loading onto CD1b. However, mycobacterial lipids can be presented efficiently in a pH range of 6. 2 to 7. 4 by CD1a (64) and to a lesser extent CD1c (66). Lipids and glycolipids from mycobacteria that reside in phagolysosomes can escape and distribute to other subcellular compartments (71–73). Also, mycobacteria often grow in extracellular spaces in ways that allow shedding and uptake of antigens into CD1b-containing compartments that lack live bacteria. Therefore, it is likely that the effects of mycobacterial inhibition of endosomal acidification on lipid presentation do not completely block lipid display. In vitro and in vivo experimental data support this conclusion, as cells with live M. tuberculosis can present lipid antigens recognized by group 1 CD1 proteins (10, 74), and tuberculosis patients show responses to CD1b-mediated mycolyl glycolipids (12, 75). Therefore, while acidification blockade is likely relevant to other aspects of the host immune response, it is not a global inhibitor of CD1-mediated lipid presentation.
Several studies now identify cellular pathways that contribute to mycobacterial lipid uptake into dendritic cells (Fig. 2B). In addition to antigen sampling by macropinocytosis, several receptors mediate internalization of mycobacterial lipid antigens, or lipid-containing complexes, which leads to highly efficient antigen presentation. The LDL receptor binds complexes of ApoE and mycobacterial lipid antigens, and leads to increased T-cell responses (76). The mannose receptor (CD206) (77, 78) and DC-SIGN (79) bind LAM and other phospholipids and also shuttle their cargo efficiently to the antigen presentation route. In addition, dectin-1 (80), the non-opsonic ligand binding part of complement receptor 3 (CD11b/CD18) (81), and DC-SIGN (79) bind mycobacterial polysaccharide structures other than LAM, and will likely internalize lipid antigens along with the polysaccharide ligands, when they are in complexes or larger particles. Langerin (CD207) has been shown to efficiently mediate antigen presentation to CD1a-restricted T cells when mycobacterial antigen complexes were fed to Langerhans cells (82). Once lipid antigens have entered the endocytic pathway, some lipids control their own fate based on receptor-independent interactions, which likely relate to lipid-lipid interactions in membranes. For example, dialkylindocarbocyanine (DiI) analogs, mycolyl glycolipids, and fluorescently labeled phosphatidylcholine analogs with longer and more rigid tails preferentially traffic to late endosomes, while analogs with shorter lipid tails and unsaturations in the lipid tail stay in early endosomes (69, 83).
Cellular processing of lipid antigens
Antigen processing can be understood as a global cellular mechanism by which antigen precursors, which are not recognizable by T cells, are rendered recognizable by TCRs. For MHC systems, the key event is the covalent cleavage of peptide bonds to generate 8-mer to 20-mer peptides from full-length proteins. Thus, the term antigen processing is sometimes used as a jargon term that is shorthand for covalent trimming of antigens. In this narrow sense of antigen processing, cells do process large glycolipids into a recognizable form as first shown by analysis of the synthetic, artificial lipid α-digalactosyl ceramide (84). Also, molecular trimming occurs for at least two natural mycobacterial antigens. Hexamannosylated phosphatidylinositol (PIM6) needs to be partially deglycosylated before it can be presented by CD1b to T cells (39, 85), and mannosyl phosphomycoketide is deglycosylated before it can be recognized by DN6 T cells as phosphomycoketide (86). However, unlike the situation with peptide antigen processing for MHC proteins, covalent trimming is not a universal aspect of lipid antigen presentation. The vast majority of known glycolipid antigens can bind to CD1 proteins and be recognized by T cells in assays that lack enzymes or intact APCs. For glucose monomycolate (GMM), intact unmodified lipids have been eluted from antigenic CD1b complexes formally proving recognition of unmodified antigens (87).
In addition to antigen trimming, lipid antigen processing substantially involves transfer of lipids from aggregates or membrane structures into a monomeric form for binding to CD1 proteins. CD1 grooves are directly exposed to aqueous solvent, yet normally bind lipids with low aqueous solubility. Therefore, most or all lipids require some cellular help in disaggregating and transferring from membranes, bacterial cell walls or lipid-protein complexes prior to insertion into the groove. Thus, a key function of the endosomal pathway is to bring CD1 molecules, exogenous antigenic lipids, and lipid binding proteins together, and if necessary, activate the lipid binding proteins through covalent cleavage into biologically active subunits. For example, prosaposin is cleaved into its active components (saposin A, saposin B, saposin C, saposin D), which contribute to lipid loading into CD1 and the final range of lipids that are displayed by cells (88–90). Other functionally active lipid transfer proteins known in the CD1 system are CD1e (39, 85), apolipoprotein E (76) and microsomal triglyceride transfer protein (91).
Mycolyl lipid antigens
The first known antigen for the CD1 system was CD1b presentation of free mycolic acid (6). Subsequently, glucose monomycolate (7), and glycerol monomycolate (92) were identified as CD1b presented antigens (Fig. 3). Compared to all conventional membrane lipids, the mycobacterial mycolates show unusually long chain length (~ C70–80) and such lipids are not cleaved within APCs (87). This long chain length approximately matches the unusually large volume of CD1b (2300Å3), which is more than 50% larger than grooves in other CD1 proteins (figure 1). The crystal structure of glucose monomycolate bound to CD1b shows how the long carbon chains of mycobacterial mycolates fill up the four pockets of CD1b (93). The α-branch lies in the C′ pocket, and the C50 meromycolate chain spans the ATF superchannel, which is formed from the combined length and end to end organization of the A′, T′, and F′ pockets and is only present in CD1b. The size match of the large CD1b groove with the unusually long length of mycolyl lipids predicts that only CD1b will present these characteristic mycobacterial lipids. However, shorter mycolates such as those found in Rhodococcus and Nocardia (C32-C54) are also presented by CD1b, most likely by binding the smaller antigen together with a scaffold lipid to fill the unused space in the CD1b groove (60). The GMM antigens were discovered using T cells from a leprosy patient, which gave rise to the clone LDN5. Initially, LDN5 was considered a unique reagent found only in this patient, but recent studies discuss below suggest that many humans express a TCR similar to that found in LDN5, so that GMM-reactive LDN5-like T cells may be common in humans (94).
Fig. 3. Classes of CD1-presented mycobacterial lipid antigens.
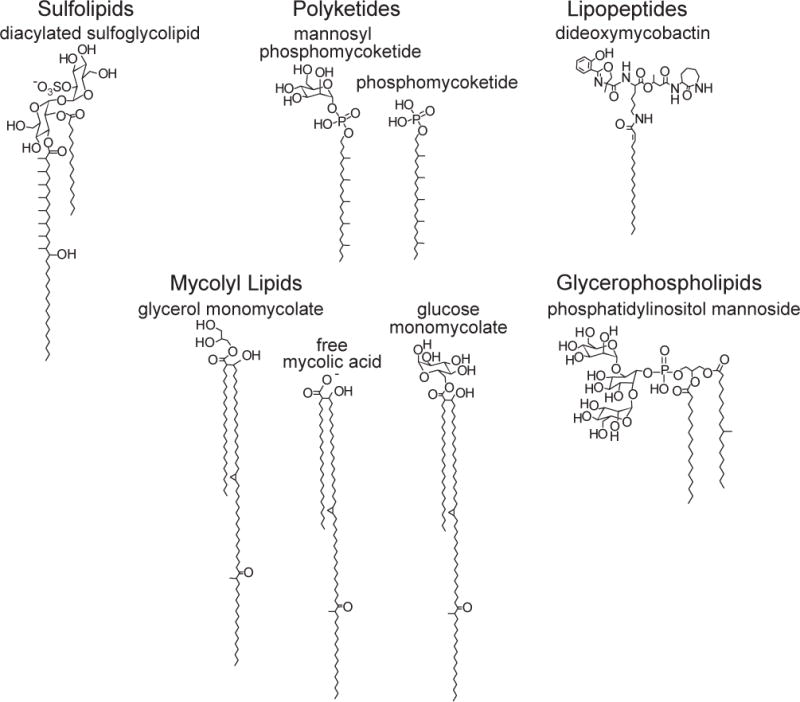
Diacylated sulfoglycolipid is presented by CD1b (9); polyketide antigens are presented by CD1c (86, 102); the lipopeptide dideoxymycobactin is presented by CD1a (10); mycolic acid derivatives (6, 7, 92) and phosphatidylinositol mannoside (39) are presented by CD1b.
Sulfolipid antigens
Discovery of antigens produced only by M. tuberculosis and not by other mycobacterial species might be of special value for immunodiagnostic purposes because any detected T responses in patients would not be confounded by BCG vaccination or exposure to environmental mycobacteria. Among antigens discussed here, only the sulfoglycolipids are specifically expressed by M. tuberculosis. T-cell stimulation by sulfoglycolipid antigens is mediated by CD1b and like other CD1b-presented antigens, is dependent on internalization and endosomal acidification of the APC (9). Further based on crystal structures of CD1b-sulfolipid complexes (95) and functional studies of antigen analogs (96), the sulfolipid system has been useful for discerning the role of glycolipid structure on CD1b and TCR binding. Using semisynthetic sulfolipids, it was shown that chain length and chain composition influence potency of antigenic sulfolipids (96, 97). As with short chain mycolates, sulfoglycolipid antigens that do not completely fill up the antigen binding groove can bind together with scaffold lipids that bind in addition to the antigenic lipid to stabilized the groove (95, 98–101).
Mycoketide antigens
CD1c presents two structurally related polyketide lipids, phosphomycoketide and mannosyl phosphomycoketide (86, 102) (Fig. 3). Mycoketides are a recently discovered class of mycobacterial lipid that is produced by polyketide synthase 12 (Pks12) (103). Synthetic agonists (104–106) showed that the branching alkyl chains introduced by the action of Pks12 contribute to recognition (107). The crystal structure of CD1c bound to mannosyl phosphomycoketide shows how branched polyketide descends like a spiral staircase into the A′ pocket of CD1a, so that the methyl branches fill up the larger outer surface of the spiral groove (101). Mycoketides, like most CD1 presented antigens, promote mycobacterial growth in cells (108), and they are selectively produced by mycobacteria of medical importance like M. tuberculosis, M. bovis, and M. avium, but not by fast growing environmental mycobacteria and other non-mycobacterial pathogens (103). Also, a recent study shows that altered pks12 genes are selected in drug-resistant mycobacteria (109) and that mycoketides might play a role in drug tolerance (110, 111). These findings suggest that there is selective evolutionary pressure on M. tuberculosis to produce mycoketide lipids, which leads to the speculation that mycoketides could not be readily jettisoned as a means of immune evasion.
Lipopeptide antigens
The chemical structure of what is currently the only known mycobacterial antigen that is presented by CD1a, was reported to be a lipopeptide (10, 112). The molecule dideoxymycobactin (DDM) (Fig. 3) is a trace metabolite that is produced by non-ribosomal peptide synthases as an intermediate in mycobactin synthesis. The larger pathway is essential for non-heme iron capture, and it promotes growth of M. tuberculosis in vivo. The discovery of an acyl-peptide antigen presented by CD1a invited the speculation that CD1a-lipopeptide presentation resembles MHC-peptide antigen presentation, with the peptide displayed on top of CD1a. However, the co-crystal of CD1a with bound DDM, it became clear that the peptidic part is bent into a U shape in the antigen-binding groove and so has a compact structure, as contrasted with the nearly linear and extended conformation of peptides presented by MHC class I (113). Small modifications of the polyketide-peptide head group and lipid tail ablate its recognition by the TCR, demonstrating that the T-cell response is peptide-specific (10, 114).
Phosphatidylinositol mannoside (PIM) antigens
PIM and its more highly glycosylated derivatives lipomannan (LM) and lipoarabinomannan (LAM) are exclusively synthesized by the genera Mycobacterium, Corynebacterium, and Rhodococcus (115–117). Both CD1b and CD1d mediate recognition of glycosylated phosphatidylinositols by T cells (8, 39, 118–120). Synthetic and semisynthetic PIM variants were instrumental in determining the structural requirements for CD1 binding and antigenicity (118, 121). One study suggests that tetramannosylated PIM (PIM4) is presented by CD1d to human and murine T cells (120), whereas dimannosylated PIM (PIM2) is recognized by CD1b-restricted T cells (39). In the crystal structure of murine CD1d bound to PIM2, the mannosyl groups are well ordered and have many interactions with CD1d, suggesting that they may form a stable surface for recognition by the TCR (121). Even though it is clear that presentation of LAM and LM is dependent on internalization and endosomal acidification, the need for trimming of its glycans for the TCR to recognize the such complex has been postulated (8). LAM is a 15–20 kD lipoglycan polymer, and it is easy to imagine that such big molecules might need to be trimmed before they can form a stable docking surface for the TCR on the surface of CD1b. One study shows that LAM and LM-specific clones do not recognize the core structures that LAM and LM share with PIM (122), and LAM loaded tetramers have not been produced.
Lipids produced by mycobacteria and the host
The lipids discussed above are produced by mycobacteria and are lacking in mammalian host, so can be considered foreign antigens. For certain immunodiagnostic applications, antigens that are more broadly expressed might a source of false positive response. If lipids are developed as vaccines, it may not be necessary that an antigenic lipid is highly specific for M. tuberculosis. Therefore it is notable that T-cell responses occur that recognized lipids that are shared by host and pathogen, including the glycerophospholipids cardiolipin (CL), phosphatidylglycerol (PG), and phosphatidylinositol (PI), which are presented by CD1d to NKT cells (123). PG and PI have also been identified as autoantigens for human and murine NKT cells (124, 125) and are also found in mycobacteria. Liganded CD1d crystal structures show how glycosylated and non-glycosylated PI bind CD1d (121, 126). CD1b molecules also bind phospholipids, but the responding cell populations are unknown (98, 127). Even though all other known CD1-presented antigens have one or two lipid tails, CL has four, two of which bind in the antigen binding groove of CD1d, and the other two are most likely solvent exposed (128).
Non-polymorphic CD1 genes
An important difference between MHC class I genes and CD1 proteins is that the former are polymorphic and the latter are not (129, 130). CD1 proteins are not literally non-polymorphic, as polymorphisms in CD1a and CD1d have been described, but the differences are minimal, and the affected amino acids are not located in or near the antigen binding groove or the surface that is recognized by the T-cell receptor (131). No polymorphisms that clearly affect the ability to present antigen have been detected to date, which justifies the common description of CD1 genes as non-polymorphic. Thus, all humans should be considered to share the same set of CD1 proteins with nearly identical structures. Of note, genetic polymorphisms outside the coding region exist that affect CD1 expression levels on DCs, and these interindividual differences in CD1 expression might be important in disease. For example, a human CD1a allele has been described that is located outside the CD1a coding sequence, but is correlated with altered CD1a expression (132). Also, low group 1 CD1 expression has long been recognized as a correlate of lepromatous leprosy (50). Therefore, altered CD1 expression remains a candidate mechanism for immunological disease causation in humans.
This overall situation for human CD1 genes contrasts with MHC class I and II in which the odds of any two humans having substantially the same MHC haplotype are extremely low (Fig. 4). There is not a single MHC molecule that is shared by the majority of humans. Among the MHC class I molecules, HLA-C is the least polymorphic, yet the frequencies of the three most frequent alleles in human populations are not high: 18% (C*04:01), 12% (C*07:01), and 10% (C*16:01) among African Americans, and rates are 16% (C*07:01), 13% (C*07:02), and 11% (C*04:01) among Caucasian Americans (from the Bone Marrow Registry datasets available at the Allele Frequency Net Database at http://www.allelefrequencies.net/). These numbers also illustrate that the allelic frequencies differ among races.
Fig. 4. Polymorphism in MHC and CD1 genes and its implications for technology development.
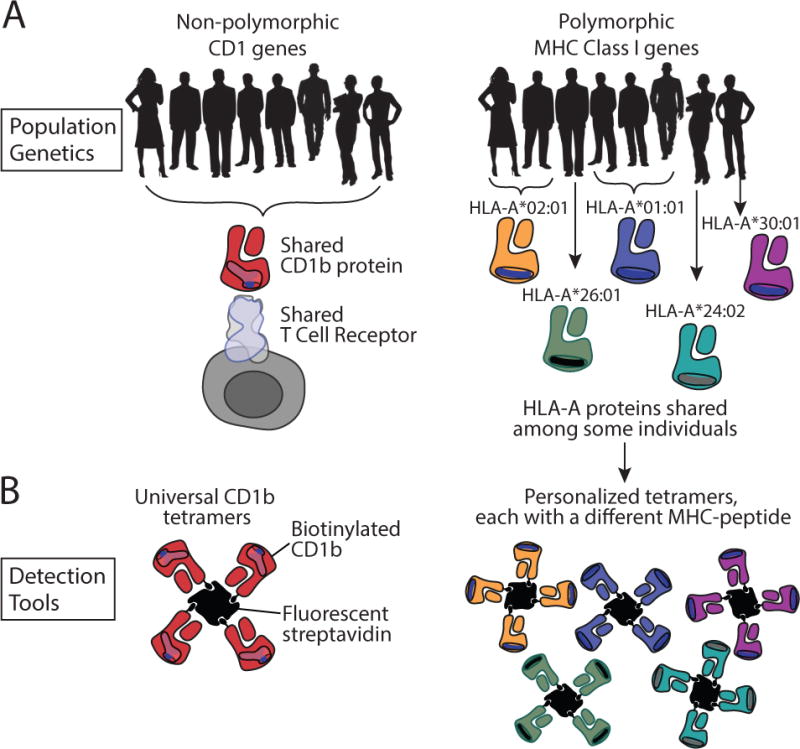
(A). Among the human population, CD1 antigen-presenting molecules are non-polymorphic (left), while MHC genes class I (right) and MHC class II molecules (not shown) are polymorphic. T-cell receptors that are shared among the whole human population exist, and they are specific for non-polymorphic antigen presenting molecules (left). (B). Tetramers made of non-polymorphic antigen presenting molecules can be used for any individual human being, whereas tetramers made of MHC class I or class II proteins need to match the specific allele that is present in a specific blood donor.
This situation (Fig. 4) has potentially far-reaching functional and experimental implications for human clinical research and technology development related to CD1 and lipid antigens. Inbred mouse strains overcome complications related to the polymorphic nature of MHC class I and II proteins, so that meaningful research can be done in mice with unlimited T cells and APCs that are genetically matched. However, basic immunological research in humans is severely complicated by MHC polymorphism, because research units cannot generate an unlimited supply of MHC-matched APCs and T cells. However, the non-polymorphic nature of CD1 eliminates the need for genetic matching to get similar responses from unrelated donors to the same lipid antigen. For clinical research, this means that the genetic background of the donor of the T cells is irrelevant for functional assays that involve ex vivo interaction of T cells with recombinant or cell surface-expressed CD1 proteins. Any reagent that stimulates T cells via antigen presentation by CD1 molecules can be used for in vitro assays using T cells of any human, regardless of HLA type. APCs from anonymously donated cells can be routinely used to stimulate T-cell clones from unrelated donors, which has allowed successful efforts to test and discover lipid antigens in allogeneic systems, which would not be possible for MHC-restricted T-cell clones.
The relatively simple population genetics of the CD1 system has practical implications for technology development. HLA-A2 tetramers can only be used to study HLA-A2+ donors, and there is no way to carry out tetramer-based studies of immunodominance in all humans. If all humans encode the same CD1b protein, then only 1 CD1b tetramer should be adequate to study the response to any defined lipid antigen in any human being. Efforts to map immunodominant peptide antigens in any population to develop subunit vaccines and immunodiagnostic reagent that could be used worldwide must consider the prevalent HLA types in any given population in which the test or vaccine might be applied. For example, a major effort is underway to move intradermal PPD testing into an ex vivo format that would allow antigen recall testing in a single visit. The increasingly used QuantiFERON®–TB Gold In-Tube test and the T-SPOT®. TB test rely on peptides presented by MHC proteins. As MHC proteins are polymorphic, the discovery of and validation of antigens that bind as many human MHC proteins possible was necessary, and the lack of truly universal peptides is a hindrance to ideal test characteristics. Developing lipids as targets for immunodiagnosis and immunization would not face this important issue. A corollary of this basic immunogenetic principle of the lack of need for matching humans at their CD1 loci creates a situation in which ‘universal APCs’ (APCs that can be used with any donor’s T cells) could be made by transfection of only one type of CD1 gene into a cell line that does not stimulate MHC-dependent T-cell responses (133, 134). Like CD1, MR1 proteins are nonpolymorphic and present mycobacterial small molecules to mucosa-associated invariant T (MAIT) cells (135–137), so these ideas apply to MAIT-based immunomodulation as well, as highlighted by Gold and Lewinsohn in this issue.
CD1 tetramers
Until recently, most published work in the human group 1 CD1 system was performed using individual T-cell clones. Clones represent the best immunological reagents for isolation of individual antigenic lipids, establishing TCR structures and evaluating antigen processing in dendritic cells. However, in considering the biological functions of T cells in host response, T-cell clones studied in isolation may not represent the larger population of CD1-restricted T cells. Also, months-long culture of T-cell clones allows a drift of phenotype and function, and other changes in natural effector mechanisms or homing receptors. Also, the outgrowth of clones in vitro is somewhat unpredictable so it is difficult to enumerate CD1 and lipid-reactive T cells based solely on limiting dilution studies. Human CD1 transgenic mice and humanized SCID mice have been developed recently and represent model systems for answering biological questions (27, 28). For study of tuberculosis disease in humans, these limitations provided a strong rationale for developing new tools that track T cells as populations, rather than clones, and emphasize phenotypic information gathered in the ex vivo state rather than after weeks of culture.
Altman and Davis (138) invented tetramer technology, which uses tetramerized antigen-presenting molecules loaded with immunodominant antigens to stain individual T cells expressing antigen-specific TCRs. Tetramers are assembled by binding biotinylated antigen presenting molecules with fluorescently labeled streptavidin molecules in a 4:1 molecular ratio (Fig. 4B). Higher order multimers, some of which are known as dextramers, can be created on carbohydrate backbones (139). As contrasted with monomers, the multimerized state of MHC and CD1 tetramers confers high avidity binding based on multiple low affinity interactions with several TCRs on one cell. Tetramers of MHC, MR1, and CD1 are extremely useful, because they can be fluorescently tagged and then used to label single antigen-specific T cells among millions of other, freshly isolated T cells. Subsequently these cells can be counted, rapidly sorted by flow cytometry for enrichment of the particular T cell needed in functional assays, or subjected to multi-parameter flowcytometric analysis for detailed phenotyping. Thus, tetramers allow study T cells directly ex vivo without the risk of changing their phenotype through long-term culture.
Mouse (140–142) and human (143) CD1d tetramers loaded with α-galactosyl ceramide have been used to quantitate and isolated NKT cells for more than a decade. The antigen that was used in the early CD1d tetramer studies, α-galactosylceramide, is not synthesized by M. tuberculosis. T cells that are specific for α-galactosylceramide have nevertheless been shown to play a role in limiting tuberculosis in mice (144). However, most of the M. tuberculosis-derived lipid antigens are presented by CD1a, CD1b, and CD1c. Development of the first CD1a, CD1b, and CD1c tetramers awaited isolation of chemically defined lipid antigens and their production or synthesis in high yield. In the past three years, human group CD1 tetramers bound to mycobacterial lipids have been reported, including CD1a-DDM (75), CD1b-GMM (145), and CD1c-phosphomycoketide (86). In addition, CD1a dextramers stain CD1a-DDM reactive T cells (75).
In general terms, tetramers add two facets to CD1 research. Prior to their validation, direct interactions of TCRs with CD1a, CD1b, and CD1c were inferred rather than measured. Tetramer binding proves the existence ternary interaction of TCR with lipid complexes involving CD1a, CD1b, and CD1c. As contrasted with functional studies of T-cell activation, the direct biophysical association of TCR-lipid-CD1 allows insight into the final structure of the loaded antigen in ways that address questions of antigen processing and discovering the precise structures of lipid epitopes. A second research direction is that tetramers enhance the ability to study the numbers and types of lipid reactive T cells in the ex vivo state using cohorts of randomly selected human patients. Because of MHC polymorphism, the use of MHC class I tetramers is limited to alleles that are commonly expressed in the population, and only patients that express the individual MHC protein of interest can be recruited into studies (Fig. 4B). In contrast, tetramers made from non-polymorphic CD1 proteins can be used with T cells from any human. To date CD1b tetramers have been used in small cohorts of patients to show that T cells, which were previously known only at the clonal level, also exist as polyclonal polyclonal populations, defining two new T-cell types in the human repertoire: germline-encoded mycolyl reactive (GEM) T cells and LDN5-like T cells.
Tetramers dissect a phospholipid epitope
T-cell activation assays can show that an added molecule stimulates a T-cell response, but cannot distinguish whether the response occurs indirectly through costimulation, upregulation of antigen-presenting molecules, or through direct TCR binding to the added molecule when displayed by CD1. The direct physical contact of CD1-lipid with the TCR provides a means to unequivocally sort out the structure of the final processed lipid epitope that mediates response. For example, CD1c is known to mediate T-cell response to mycobacterial mannosyl-β1-mycoketides (102, 103, 107) and phosphomycoketide antigens (86). These two naturally occurring antigens differ only in the presence or absence of a mannosyl unit. Unexpected patterns of cross-reactivity were observed in two clones in which one clone recognized both antigens, and another recognized only the phosphomycoketide epitope when presented by APCs. As discussed above, some but not all antigens are processed through glycan cleavage that occurs after uptake into cells and before binding to the TCR. When used in antigen presentation assays by CD1c-expressing cells, mycobacterial mannosyl phosphomycoketide can be processed into phosphomycoketide, which makes it hard to unequivocally determine the true antigenic epitope for T-cell clones. However, when a clone that recognizes the mannosylated version in cells can only bind to CD1c tetramers loaded with the demannosylated version, the data argue strongly for processing and identify the epitope as the demannosylated molecule (86). Understanding the precise structure of the finally presented epitope is important for development of in vitro antigen response tests.
LDN5-like and GEM T cells
For years it was unclear if individual T-cell clones that recognized mycolic acid, sulfoglycolipid, DDM, GMM, or other mycobacterial lipids represented anecdotal evidence of an extremely rare response with limited value for in vivo biology or instead corresponded to new T-cell types that exist as a polyclonal populations among unrelated humans. For example, LDN5 was a clone derived from the skin of a leprosy patient that recognized CD1b and GMM (7), but it was unknown whether GMM-reactive T-cell clones generally circulate in the blood of humans. However, studies of using GMM-loaded CD1b tetramers identified two types of cells with moderate or very bright tetramer staining could be seen in random donors and among tuberculosis patients (12, 94, 145). The LDN5 clone expresses a TCR with the TRAV17 and TRBV4-1 variable regions, and the polyclonal T cells detected that stained with moderate intensity with the tetramer expressed the TRBV4-1 gene segment. Subsequently, multiple clones that expressed TRAV17 and TRBV4-1 were grown from cells that were sorted based on tetramer staining. Therefore, GMM-reactive T exist in humans as polyclonal populations and they even express TCRs that are similar to the first known GMM-reactive clone, LDN5, leading to their designation as LDN5-like T cells. Even more strikingly, the brightest CD1b tetramer+ T cells expressed nearly identical TCR α chains, comprised of a rearrangement of the TRAV1-2 variable region gene and the TRAJ9 joining region to give nearly identical CDR3 sequences. The extreme conservation of a CD1b and a GMM-reactive TCR α chain can be compared to those that define invariant NKT cells and mucosal associated invariant T (MAIT) cells (Fig. 5), which are separately reviewed in this issue.
Fig. 5.
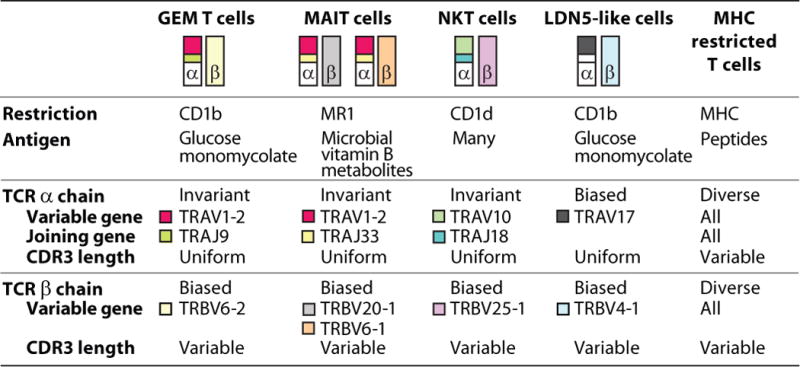
Human T cells with conserved T-cell receptors
Group 1 CD1 tetramers have led to several general insights into the existence and nature of CD1-restricted T cells in the human repertoire. T-cell clones recognizing mycobacterial lipids, which were considered to be rare or unique in terms of antigen specificity, have now been shown to represent populations that are encountered among many tuberculosis patients (12, 86, 94). Second, the concept of highly conserved, or invariant T cells was developed based on the discovery of CD1d-restricted NKT cells (11), and no patterns of TCR conservation among CD1a, CD1b, and CD1c-restricted T-cell clones were known. Therefore, it was presumed that CD1a, CD1b, and CD1c-reactive TCRs were highly diverse based on a limited number of TCRs derived from clones. However, the existence of conserved TCRs among individuals is now proven (12, 94). This outcome is consistent with the nonpolymorphic nature of CD1 proteins and implies a simple immunogenetic pattern among humans, creating new prospects for diagnosis based on exposure to mycobacterial lipids and detection of conserved TCRs. These studies raise the question of whether antibodies, polymerase chain reaction, or high throughput sequencing could be used to identify expanded GEM T cells and TB exposure in humans. Third, it is now possible to determine the actual frequencies of CD1a, CD1b, and CD1c-restricted T cells specific for mycobacterial antigens (75, 86, 145). The frequencies in blood are low when compared to virus-specific MHC class I-restricted cytotoxic T cells, but are comparable to the rates human NKT cells in blood. A formal statistical analysis on well documented tuberculosis-exposed and control cohorts has not yet been published, but the published data suggest that frequencies of CD1a, CD1b, and CD1c-restricted T cells specific for mycobacterial antigens are higher in blood from tuberculosis-exposed subjects compared to blood from healthy random bank donors. Last, certain conserved aspects of the phenotype of CD1 and mycobacteria-reactive T cells are becoming apparent. CD1b-reactive T cells have higher than expected rates of CD4 expression, raising the question of whether such T cells are infected by the human immunodeficiency virus (145). Also, CD1b and lipid reactive T cells express high levels of interferon-γ and TNF in the ex vivo state, consistent with a role in host defense against tuberculosis (12).
Technology development
Clearly, the potential of tetramers to contribute to our insights in the in vivo biology of human T cells that recognize mycobacterial lipids have not yet been exhausted. Whereas nearly all current studies focus on blood-derived T cells, the effector functions of T cells in the lungs, lymphoid and other tissues in which CD1 proteins are expressed and the tuberculosis disease process plays out have not yet been described in detail. Studies of T-cell response in large patient cohorts will be needed to answer the question of whether or not CD1 and mycobacterial lipid-reactive T cells show immunological memory, a key question that underlies the feasibility of lipid-based vaccination. The phenotypic hallmarks of T-cell memory as well as quantitative expansion of T-cell populations during and after tuberculosis disease can be addressed with tetramers. Transcriptional analysis of antigen-specific CD1a, CD1b, and CD1c-restricted T cells will shed light on transcription factors, stimulation pathways, and effector functions of these cells. The recently enhanced understanding of the presentation of mycobacterial lipid antigens creates a new opportunity for therapy and diagnosis. The known mycobacterial lipid antigens, which are now available in synthetic or pure forms, can be administered to CD1a, CD1b, and CD1c-expressing animals, such as primates and guinea pigs, to determine whether immune responses can be generated, how long they last, and whether they have any protective effect against tuberculosis. In addition, in humans infected with M. tuberculosis, lipid antigen-specific T cells can now be monitored with CD1 tetramers so that the immunophenotype, expansion, and trafficking can be determined. Lastly, the identification of interdonor-conserved TCRs will enable the detection and quantification of mycobacterial lipid-specific T cells by high throughput TCR sequencing.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (AI049313,AR048632 to D. B. M.), the Burroughs Wellcome Fund for Translational Research, the Bill and Melinda Gates Foundation, and Nederlands Wetenschappelijk Onderzoek (Meervoud 836. 08. 001 to I. V. R.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Russell DG, Mwandumba HC, Rhoades EE. Mycobacterium and the coat of many lipids. The Journal of cell biology. 2002;158:421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brightbill HD, et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guerin is due principally to trehalose mycolates. J Immunol. 2005;174:5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 5.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 6.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted αβ+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 7.Moody DB, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 8.Sieling PA, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 9.Gilleron M, et al. Diacylated Sulfoglycolipids Are Novel Mycobacterial Antigens Stimulating CD1-restricted T Cells during Infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moody DB, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 11.Fowlkes BJ, et al. A novel population of T-cell receptor αβ-bearing thymocytes which predominantly expresses a single V beta gene family. Nature. 1987;329:251–254. doi: 10.1038/329251a0. [DOI] [PubMed] [Google Scholar]
- 12.Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14:706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMichael AJ, Pilch JR, Galfre G, Mason DY, Fabre JW, Milstein C. A human thymocyte antigen defined by a hybrid myeloma monoclonal antibody. Eur J Immunol. 1979;9:205–210. doi: 10.1002/eji.1830090307. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 15.Dossa RG, Alperin DC, Hines MT, Hines SA. The equine CD1 gene family is the largest and most diverse yet identified. Immunogenetics. 2014;66:33–42. doi: 10.1007/s00251-013-0741-6. [DOI] [PubMed] [Google Scholar]
- 16.Hayes SM, Knight KL. Group 1 CD1 genes in rabbit. J Immunol. 2001;166:403–410. doi: 10.4049/jimmunol.166.1.403. [DOI] [PubMed] [Google Scholar]
- 17.Dascher CC, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- 18.Van Rhijn I, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 19.Eguchi-Ogawa T, et al. Analysis of the genomic structure of the porcine CD1 gene cluster. Genomics. 2007;89:248–261. doi: 10.1016/j.ygeno.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Looringh van Beeck FA, et al. Two canine CD1a proteins are differentially expressed in skin. Immunogenetics. 2008;60:315–324. doi: 10.1007/s00251-008-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiromatsu K, et al. Characterization of guinea-pig group 1 CD1 proteins. Immunology. 2002;106:159–172. doi: 10.1046/j.1365-2567.2002.01422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Perera TV, Ford HL, Dascher CC. Characterization of a divergent non-classical MHC class I gene in sharks. Immunogenetics. 2003;55:57–61. doi: 10.1007/s00251-003-0542-4. [DOI] [PubMed] [Google Scholar]
- 24.Miller MM, et al. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci U S A. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 26.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–418. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 27.Felio K, et al. CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. J Exp Med. 2009;206:2497–2509. doi: 10.1084/jem.20090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockridge JL, et al. Analysis of the CD1 antigen presenting system in humanized SCID mice. PloS one. 2011;6:e21701. doi: 10.1371/journal.pone.0021701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dascher CC, et al. Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. Int Immunol. 2003;15:915–925. doi: 10.1093/intimm/dxg091. [DOI] [PubMed] [Google Scholar]
- 30.Hiromatsu K, et al. Induction of CD1-restricted immune responses in guinea pigs by immunization with mycobacterial lipid antigens. J Immunol. 2002;169:330–339. doi: 10.4049/jimmunol.169.1.330. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe Y, et al. BCG vaccine elicits both T-cell mediated and humoral immune responses directed against mycobacterial lipid components. Vaccine. 2006;24:5700–5707. doi: 10.1016/j.vaccine.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen TK, et al. The bovine CD1D gene has an unusual gene structure and is expressed but cannot present α galactosylceramide with a C26 fatty acid. Int Immunol. 2013;25:91–98. doi: 10.1093/intimm/dxs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Rhijn I, et al. Low cross-reactivity of T-cell responses against lipids from Mycobacterium bovis and M. avium paratuberculosis during natural infection. Eur J Immunol. 2009;39:3031–3041. doi: 10.1002/eji.200939619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen TK, Koets AP, Santema WJ, van Eden W, Rutten VP, Van Rhijn I. The mycobacterial glycolipid glucose monomycolate induces a memory T cell response comparable to a model protein antigen and no B cell response upon experimental vaccination of cattle. Vaccine. 2009;27:4818–4825. doi: 10.1016/j.vaccine.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita D, et al. Trans-species activation of human T cells by rhesus macaque CD1b molecules. Biochem Biophys Res Commun. 2008;377:889–893. doi: 10.1016/j.bbrc.2008.10.075. [DOI] [PubMed] [Google Scholar]
- 36.Porcelli SA. The CD1 family: a third lineage of antigen-presenting molecules. Adv Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- 37.Maitre B, et al. The assembly of CD1e is controlled by an N-terminal propeptide which is processed in endosomal compartments. Biochem J. 2009;419:661–668. doi: 10.1042/BJ20082204. [DOI] [PubMed] [Google Scholar]
- 38.Angenieux C, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 39.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 40.Facciotti F, et al. Fine tuning by human CD1e of lipid-specific immune responses. Proc Natl Acad Sci U S A. 2011;108:14228–14233. doi: 10.1073/pnas.1108809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 42.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- 43.Schenk M, et al. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat Med. 2012;18:555–563. doi: 10.1038/nm.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krutzik SR, et al. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med. 2005;11:653–660. doi: 10.1038/nm1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somnay-Wadgaonkar K, et al. Immunolocalization of CD1d in human intestinal epithelial cells and identification of a beta2-microglobulin-associated form. Int Immunol. 1999;11:383–392. doi: 10.1093/intimm/11.3.383. [DOI] [PubMed] [Google Scholar]
- 46.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 47.Yakimchuk K, et al. Borrelia burgdorferi infection regulates CD1 expression in human cells and tissues via IL1-beta. Eur J Immunol. 2011;41:694–705. doi: 10.1002/eji.201040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roura-Mir C, et al. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J Immunol. 2005;175:1758–1766. doi: 10.4049/jimmunol.175.3.1758. [DOI] [PubMed] [Google Scholar]
- 49.Moody DB. TLR gateways to CD1 function. Nat Immunol. 2006;7:811–817. doi: 10.1038/ni1368. [DOI] [PubMed] [Google Scholar]
- 50.Sieling PA, et al. CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol. 1999;162:1851–1858. [PubMed] [Google Scholar]
- 51.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 53.Buettner M, et al. Inverse correlation of maturity and antibacterial activity in human dendritic cells. J Immunol. 2005;174:4203–4209. doi: 10.4049/jimmunol.174.7.4203. [DOI] [PubMed] [Google Scholar]
- 54.Moody DB, Porcelli SA. CD1 trafficking: invariant chain gives a new twist to the tale. Immunity. 2001;15:861–865. doi: 10.1016/s1074-7613(01)00250-3. [DOI] [PubMed] [Google Scholar]
- 55.Dascher CC, Brenner MB. CD1 antigen presentation and infectious disease. Contrib Microbiol. 2003;10:164–182. doi: 10.1159/000068136. [DOI] [PubMed] [Google Scholar]
- 56.Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 57.Briken V, Jackman RM, Dasgupta S, Hoening S, Porcelli SA. Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J. 2002;21:825–834. doi: 10.1093/emboj/21.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox D, et al. Determination of cellular lipids bound to human CD1d molecules. PloS one. 2009;4:e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang S, et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci U S A. 2011;108:19335–19340. doi: 10.1073/pnas.1112969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haig NA, Guan Z, Li D, McMichael A, Raetz CR, Xu XN. Identification of self-lipids presented by CD1c and CD1d proteins. The Journal of biological chemistry. 2011;286:37692–37701. doi: 10.1074/jbc.M111.267948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugita M, et al. Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs. Science. 1996;273:349–352. doi: 10.1126/science.273.5273.349. [DOI] [PubMed] [Google Scholar]
- 63.Jackman RM, et al. The tyrosine-containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity. 1998;8:341–351. doi: 10.1016/s1074-7613(00)80539-7. [DOI] [PubMed] [Google Scholar]
- 64.Sugita M, et al. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 65.Briken V, Jackman RM, Watts GF, Rogers RA, Porcelli SA. Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. J Exp Med. 2000;192:281–288. doi: 10.1084/jem.192.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugita M, van Der WN, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci U S A. 2000;97:8445–8450. doi: 10.1073/pnas.150236797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barral DC, et al. CD1a and MHC Class I Follow a Similar Endocytic Recycling Pathway. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manolova V, et al. Functional CD1a is stabilized by exogenous lipids. Eur J Immunol. 2006;36:1083–1092. doi: 10.1002/eji.200535544. [DOI] [PubMed] [Google Scholar]
- 69.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 70.Sturgill-Koszycki S, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 71.Xu S, et al. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J Immunol. 1994;153:2568–2578. [PubMed] [Google Scholar]
- 72.Schaible UE, Hagens K, Fischer K, Collins HL, Kaufmann SH. Intersection of group I CD1 molecules and mycobacteria in different intracellular compartments of dendritic cells. J Immunol. 2000;164:4843–4852. doi: 10.4049/jimmunol.164.9.4843. [DOI] [PubMed] [Google Scholar]
- 73.Beatty WL, Rhoades ER, Ullrich HJ, Chatterjee D, Heuser JE, Russell DG. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1:235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 74.Hava DL, et al. Evasion of peptide, but not lipid antigen presentation, through pathogen-induced dendritic cell maturation. Proc Natl Acad Sci U S A. 2008;105:11281–11286. doi: 10.1073/pnas.0804681105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kasmar A, et al. CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol. 2013;191:4499–4503. doi: 10.4049/jimmunol.1301660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van den Elzen P, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- 77.Prigozy TI, et al. The mannose receptor delivers lipoglycan antigens to endosomes for presentation to T cells by CD1b molecules. Immunity. 1997;6:187–197. doi: 10.1016/s1074-7613(00)80425-2. [DOI] [PubMed] [Google Scholar]
- 78.Kang PB, et al. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med. 2005;202:987–999. doi: 10.1084/jem.20051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tailleux L, et al. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197:121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yadav M, Schorey JS. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108:3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cywes C, Hoppe HC, Daffe M, Ehlers MR. Nonopsonic binding of Mycobacterium tuberculosis to complement receptor type 3 is mediated by capsular polysaccharides and is strain dependent. Infect Immun. 1997;65:4258–4266. doi: 10.1128/iai.65.10.4258-4266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hunger RE, et al. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Invest. 2004;113:701–708. doi: 10.1172/JCI19655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee S, Soe TT, Maxfield FR. Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J Cell Biol. 1999;144:1271–1284. doi: 10.1083/jcb.144.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prigozy TI, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291:664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 85.Cala-De Paepe D, et al. Deciphering the role of CD1e protein in mycobacterial phosphatidyl-myo-inositol mannosides (PIM) processing for presentation by CD1b to T lymphocytes. J Biol Chem. 2012;287:31494–31502. doi: 10.1074/jbc.M112.386300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ly D, et al. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med. 2013;210:729–741. doi: 10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng TY, et al. Role of lipid trimming and CD1 groove size in cellular antigen presentation. EMBO J. 2006;25:2989–2999. doi: 10.1038/sj.emboj.7601185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 89.Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 90.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaser A, et al. Microsomal triglyceride transfer protein regulates endogenous and exogenous antigen presentation by group 1 CD1 molecules. Eur J Immunol. 2008;38:2351–2359. doi: 10.1002/eji.200738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Layre E, et al. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol. 2009;16:82–92. doi: 10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 93.Batuwangala T, et al. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2004;172:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 94.Van Rhijn I, et al. TCR Bias and Affinity Define Two Compartments of the CD1b-Glycolipid-Specific T Cell Repertoire. J Immunol. 2014 doi: 10.4049/jimmunol.1400158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garcia-Alles LF, et al. Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci U S A. 2011;108:17755–17760. doi: 10.1073/pnas.1110118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guiard J, et al. Fatty acyl structures of mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol. 2009;182:7030–7037. doi: 10.4049/jimmunol.0804044. [DOI] [PubMed] [Google Scholar]
- 97.Gau B, et al. Simplified deoxypropionate acyl chains for Mycobacterium tuberculosis sulfoglycolipid analogues: chain length is essential for high antigenicity. Chembiochem. 2013;14:2413–2417. doi: 10.1002/cbic.201300482. [DOI] [PubMed] [Google Scholar]
- 98.Gadola SD, et al. Structure of human CD1b with bound ligands at 2. 3 A, a maze for alkyl chains. NatImmunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 99.Wu D, et al. Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc Natl Acad Sci U S A. 2006;103:3972–3977. doi: 10.1073/pnas.0600285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia-Alles LF, et al. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scharf L, et al. The 2. 5 a structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moody DB, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 103.Matsunaga I, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Crich D, Dudkin V. Confirmation of the connectivity of 4,8,12,16,20-pentamethylpentacosylphoshoryl beta-D-mannopyranoside, an unusual beta-mannosyl phosphoisoprenoid from Mycobacterium avium, through synthesis. J Am Chem Soc. 2002;124:2263–2266. doi: 10.1021/ja0123958. [DOI] [PubMed] [Google Scholar]
- 105.van Summeren RP, Moody DB, Feringa BL, Minnaard AJ. Total synthesis of enantiopure beta-D-mannosyl phosphomycoketides from Mycobacterium tuberculosis. J Am Chem Soc. 2006;128:4546–4547. doi: 10.1021/ja060499i. [DOI] [PubMed] [Google Scholar]
- 106.Li NS, Scharf L, Adams EJ, Piccirilli JA. Highly stereocontrolled total synthesis of beta-D-mannosyl phosphomycoketide: a natural product from Mycobacterium tuberculosis. The Journal of organic chemistry. 2013;78:5970–5986. doi: 10.1021/jo4006602. [DOI] [PubMed] [Google Scholar]
- 107.de Jong A, et al. CD1c presentation of synthetic glycolipid antigens with foreign alkyl branching motifs. Chem Biol. 2007;14:1232–1242. doi: 10.1016/j.chembiol.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sirakova TD, Dubey VS, Kim HJ, Cynamon MH, Kolattukudy PE. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect Immun. 2003;71:3794–3801. doi: 10.1128/IAI.71.7.3794-3801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Farhat MR, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsunaga I, Maeda S, Nakata N, Fujiwara N. The polyketide synthase-associated multidrug tolerance in Mycobacterium intracellulare clinical isolates. Chemotherapy. 2012;58:341–348. doi: 10.1159/000343311. [DOI] [PubMed] [Google Scholar]
- 111.Philalay JS, Palermo CO, Hauge KA, Rustad TR, Cangelosi GA. Genes required for intrinsic multidrug resistance in Mycobacterium avium. Antimicrob Agents Chemother. 2004;48:3412–3418. doi: 10.1128/AAC.48.9.3412-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rosat JP, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ αβ T cell pool. J Immunol. 1999;162:366–371. [PubMed] [Google Scholar]
- 113.Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 114.Young DC, et al. Synthesis of dideoxymycobactin antigens presented by CD1a reveals T cell fine specificity for natural lipopeptide structures. The Journal of biological chemistry. 2009;284:25087–25096. doi: 10.1074/jbc.M109.000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crellin P, Luo C, Morita YS. Metabolism of Plasma Membrane Lipids in Mycobacteria and Corynebacteria. Lipid Metabolism. 2013 [Google Scholar]
- 116.Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol Rev. 2011;35:1126–1157. doi: 10.1111/j.1574-6976.2011.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gibson KJ, Gilleron M, Constant P, Puzo G, Nigou J, Besra GS. Structural and functional features of Rhodococcus ruber lipoarabinomannan. Microbiology. 2003;149:1437–1445. doi: 10.1099/mic.0.26161-0. [DOI] [PubMed] [Google Scholar]
- 118.Ernst WA, et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8:331–340. doi: 10.1016/s1074-7613(00)80538-5. [DOI] [PubMed] [Google Scholar]
- 119.Sieling PA, et al. Evidence for human CD4+ T cells in the CD1-restricted repertoire: derivation of mycobacteria-reactive T cells from leprosy lesions. J Immunol. 2000;164:4790–4796. doi: 10.4049/jimmunol.164.9.4790. [DOI] [PubMed] [Google Scholar]
- 120.Fischer K, et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc Natl Acad Sci U S A. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. [DOI] [PubMed] [Google Scholar]
- 122.Torrelles JB, et al. Structural differences in lipomannans from pathogenic and nonpathogenic mycobacteria that impact CD1b-restricted T cell responses. J Biol Chem. 2011;286:35438–35446. doi: 10.1074/jbc.M111.232587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tatituri RV, et al. Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proc Natl Acad Sci U S A. 2013;110:1827–1832. doi: 10.1073/pnas.1220601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 125.Brigl M, et al. Conserved and heterogeneous lipid antigen specificities of CD1d-restricted NKT cell receptors. J Immunol. 2006;176:3625–3634. doi: 10.4049/jimmunol.176.6.3625. [DOI] [PubMed] [Google Scholar]
- 126.Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34:315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Girardi E, et al. Crystal structure of bovine CD1b3 with endogenously bound ligands. J Immunol. 2010;185:376–386. doi: 10.4049/jimmunol.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dieude M, et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted gammadelta T cells in the normal murine repertoire. J Immunol. 2011;186:4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martin LH, Calabi F, Milstein C. Isolation of CD1 genes: a family of major histocompatibility complex-related differentiation antigens. Proc Natl Acad Sci U S A. 1986;83:9154–9158. doi: 10.1073/pnas.83.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988;7:3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. 1999;54:122–127. doi: 10.1034/j.1399-0039.1999.540202.x. [DOI] [PubMed] [Google Scholar]
- 132.Seshadri C, Turner MT, Lewinsohn DM, Moody DB, Van Rhijn I. Lipoproteins are major targets of the polyclonal human T cell response to Mycobacterium tuberculosis. J Immunol. 2013;190:278–284. doi: 10.4049/jimmunol.1201667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.de Jong A, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15:177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wong EB, et al. Low levels of peripheral CD161++CD8+ mucosal associated invariant T (MAIT) cells are found in HIV and HIV/TB co-infection. PloS one. 2013;8:e83474. doi: 10.1371/journal.pone.0083474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gold MC, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 138.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 139.Casalegno-Garduno R, et al. Multimer technologies for detection and adoptive transfer of antigen-specific T cells. Cancer immunology, immunotherapy : CII. 2010;59:195–202. doi: 10.1007/s00262-009-0778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gadola SD, Dulphy N, Salio M, Cerundolo V. Vα24-JαQ-independent, CD1d-restricted recognition of α-galactosylceramide by human CD4(+) and CD8αβ (+) T lymphocytes. J Immunol. 2002;168:5514–5520. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
- 143.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chackerian A, Alt J, Perera V, Behar SM. Activation of NKT cells protects mice from tuberculosis. Infect Immun. 2002;70:6302–6309. doi: 10.1128/IAI.70.11.6302-6309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kasmar AG, et al. CD1b tetramers bind αβ T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208:1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]


