Abstract
The transcription factor, SNAI2 is an inducer of the epithelial to mesenchymal transition (EMT) which mediates cell migration during development and tumor invasion. SNAI2 can also promote the generation of mammary epithelial stem cells from differentiated luminal cells when overexpressed. How SNAI2 regulates these critical and diverse functions is unclear. Here we show that the levels of SNAI2 expression are important for epidermal cell fate decisions. The expression of SNAI2 was found to be enriched in the basal layer of the interfollicular epidermis where progenitor cells reside and extinguished upon differentiation. Loss of SNAI2 resulted in premature differentiation whereas gain of SNAI2expression inhibited differentiation. SNAI2 controls the differentiation status of epidermal progenitor cells by binding to and repressing the expression of differentiation genes with increased binding leading to further transcriptional silencing. Thus, the levels of SNAI2 binding to genomic targets determines the differentiation status of epithelial cells with increased levels triggering EMT and dedifferentiation, moderate (physiological) levels promoting epidermal progenitor function, and low levels leading to epidermal differentiation.
Keywords: Stem Cell, Progenitor Cell, Differentiation, Epidermis, Skin Differentiation, SNAI2, SLUG, SNAIL, EMT, transcription factor, GRHL3, KLF4, metastasis, epithelium, ChIP-Seq, epidermal differentiation, EMT
Introduction
Adult stem and progenitor cells replenish the tissue they reside in for the life of the animal[1]. These cells must balance proliferation and differentiation to maintain homeostasis as well as respond to injury. The molecular mechanisms underlying how adult stem cells maintain themselves in an undifferentiated state is still unclear and is an active area of research. The epidermis is an ideal tissue to decipher the mechanisms of self-renewal and differentiation since it is a tissue that has a rapid turnover as well as being able to regenerate the entire human epidermis in-vitro[2,3]. The basal layer which is the deepest layer of the epidermis contains stem and progenitor cells that upon differentiation migrate upwards to form the spinous layer. As differentiation proceeds, the cells continue to migrate upwards to form the granular layer, which is characterized by the assembly of the cornified envelope proteins underneath the plasma membrane. During the final stages of differentiation, transglutaminase is activated which crosslinks the cornified envelope proteins while lipids are extruded into the extracellular space. This insoluble sac of proteins and lipids forms the water impermeable barrier known as the stratum corneum[4]. We and others have recently shown that both transcriptional and post-transcriptional mechanisms are necessary for maintaining self-renewal of epidermal stem and progenitor cells. Transcriptional mechanisms include epigenetic factors such as DNMT1, EZH2, UHRF1, CBX4, and ACTL6A which act to suppress differentiation gene expression through epigenetic mechanisms[3,5-8]. Post-transcriptional mechanisms include RNA degradation enzymes such as the exosome complex which promotes self-renewal by targeting and degrading pro-differentiation mRNAs in progenitor cells[9]. The switch to a differentiated state requires the upregulation and actions of transcription factors such as grainy head-like 1 and 3 (GRHL1 and GRHL3), zinc finger protein (ZNF750), CCAAT/enhancer-binding protein (CEBPA and CEBPB), and kruppel-like factor 4 (KLF4) [10-15].
Recently, it was reported that overexpression of epithelial to mesenchymal transition transcription factors (EMT-TFs) could convert differentiated luminal cells into a mammary epithelial stem cell state[16,17]. The epithelial to mesenchymal transition is the process by which epithelial cells lose cell-cell adhesion and gain cell motility during the conversion to a mesenchymal state[18]. This process is mediated through a family of transcription factors including SNAIL, TWIST, and ZEB[19]. The SNAIL family of zinc-finger transcription factors contain a highly conserved carboxyl terminus containing between four to six C2H2 zinc fingers which mediate specific binding to a consensus six base motif of CAGGTG (A subset of the E-box, CANNTG)[20,21]. Upon binding to the motif, SNAIL family members mediate transcriptional repression through the SNAG domain found in the amino terminus. The SNAIL family is composed of three members including SNAIL (SNAI1), SLUG (SNAI2), and SMUG (SNAI3). EMT-TFs have been shown to regulate development such as the ingression of the cells from the surface of the embryo to form organs[22]. In adults, SNAI2 has been shown to promote wound healing[23]. During cancer progression, EMT-TFs mediate the invasion and metastatic properties of epithelial tissues[24]. Because of EMT-TFs role in promoting cell motility and invasion, it was surprising that overexpression of EMT-TFs could promote both stem cell and cancer stem cell maintenance in the mammary epithelium[16,17]. The underlying mechanisms remain unclear. It is also not known whether promoting the stem cell state by EMT-TFs is unique to mammary stem cells or is applicable to other epithelial cells such as those found in the epidermis.
Here we investigate the function of the EMT-TF, SNAI2 in human epidermal cells. We find that the levels of SNAI2 expression dictate the differentiation status of the epidermis. High levels of SNAI2 leads to dedifferentiation and an EMT phenotype while moderate (physiological) levels of SNAI2 binding leads to sustained progenitor cell function. In contrast, low levels of SNAI2 seen during epidermal differentiation result in expression of differentiation genes due to loss of SNAI2 repressive activity at their promoters. These results provide an explanation for how EMT-TFs can regulate both EMT and the differentiation status of epithelial cells.
Results
SNAI2 expression is enriched in epidermal progenitor cells and downregulated during differentiation
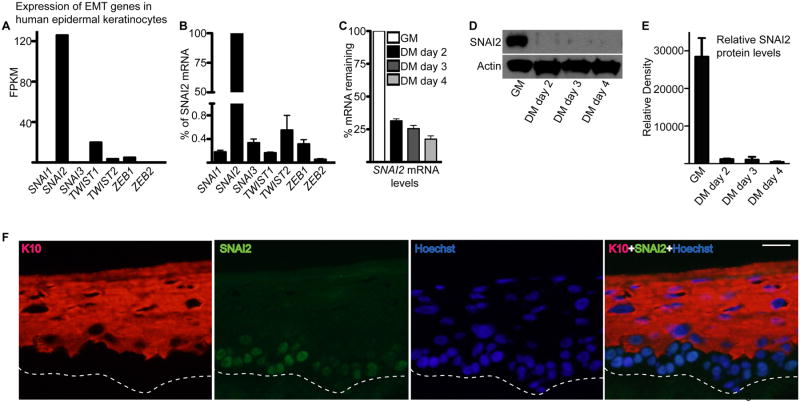
To identify EMT transcription factors important for maintaining progenitor cells in an undifferentiated state, we searched for genes whose transcripts levels were highly expressed in undifferentiated human epidermal keratinocytes. Of the SNAIL, TWIST, and ZEB family of EMT-TF, only SNAI2 was highly expressed while the others were undetectable or minimally expressed (Fig. 1A-B). Interestingly, SNAI2 was also downregulated on the transcript as well as protein level during epidermal differentiation (Fig. 1C-E). In adult human epidermis, SNAI2 protein expression was confined to the nucleus of the basal layer cells and diminished in the outer differentiated layers of the epidermis that is marked by the differentiation protein keratin 10: K10 (Fig. 1F). This suggests SNAI2 may play a role in preventing the differentiation of the progenitor cells residing in the basal layer.
Figure 1. EMT-TF, SNAI2 is highly expressed in epidermal progenitor cells and downregulated during differentiation.
(A) RNA-seq analysis on the expression of EMT-TF family members in epidermal keratinocytes. Expression levels are represented as reads per kilobase per million (RPKM). Data was derived from the ENCODE RNA seq analysis of human epidermal keratinocytes. (B) RT-qPCR for expression of EMT genes. Due to the high levels of SNAI2 expression, the expression of all other EMT genes were calculated as a percentage of SNAI2 expression. (C) RT-qPCR for expression of SNAI2 in progenitor cells (cultured in growth medium: GM) and differentiated cells (cultured in differentiation medium: DM). Expression levels were normalized to GADPH. Error bars=mean with SEM. (D) Western blot for protein levels of SNAI2 in progenitor (GM) and differentiated cells (DM). Actin was used as a loading control. (E) Quantitation of Western blots shown in (D). Signals were normalized to actin loading control and quantitated using ImageJ. N=3 independent experiments. (F) SNAI2 staining in adult human epidermis. SNAI2 staining is shown in green and differentiation protein keratin 10 (K10) is shown in red. Hoechst staining in blue marks the nuclei. Scale bar=25μm; dashed lines denote basement membrane zone.
The expression levels of SNAI2 determine the differentiation status of epidermal tissue and cultured cells
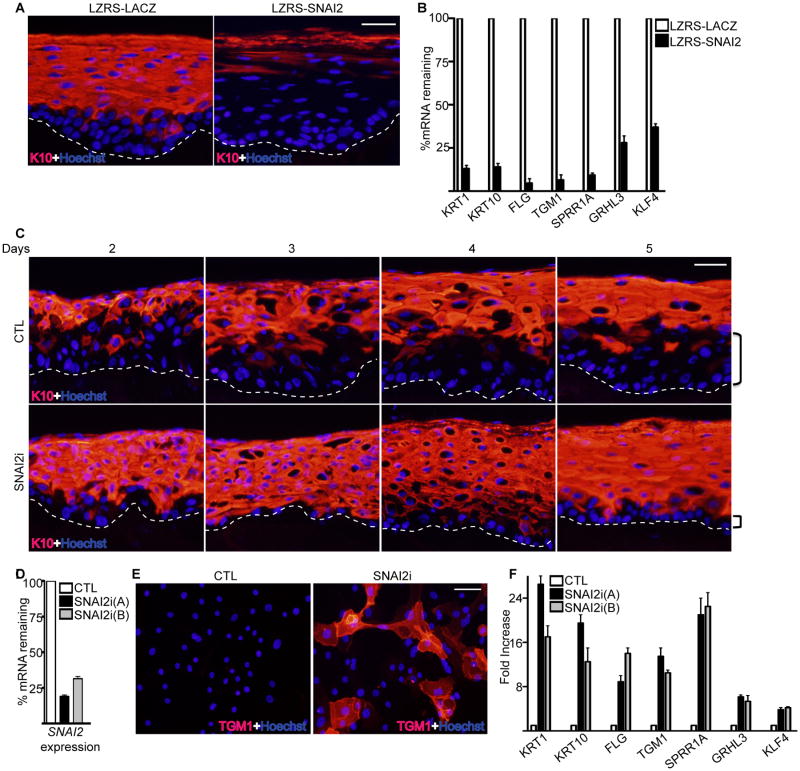
To test for a potential functional role for SNAI2 in the epidermis, SNAI2 was constitutively expressed through a retrovirus in organotypic human epidermal tissue. This regenerated human tissue recapitulates epidermal stratification and differentiation in a three dimensional context through the use of primary human epidermal keratinocytes and intact extracellular matrix in the form of human dermis[2,3]. Ectopic expression of SNAI2 prevented the expression of differentiation protein K10 as well as differentiation genes KRT1, FLG, TGM1, SPRR1A, GRHL3, and KLF4 (Fig. 2A-B). Overexpressed SNAI2 could be seen throughout the epidermis whereas endogenous SNAI2 was mainly localized to the basal layer (Supporting Information Fig. S1). Increased expression of SNAI2 in cultured primary epidermal progenitor cells resulted in an EMT phenotype with the cells acquiring a spindle shaped appearance and downregulation of epithelial adhesion genes such as CDH1, CLDN1, CLDN7, GJB6 and upregulation of mesenchymal genes such as VIM (Supporting Information Fig. S2A-B) [19]. The progenitor cells also became dedifferentiated due to decreased expression of basal levels of KLF4 and GRHL3 (Supporting Information Fig. S2B). Conversely, depletion of SNAI2 using shRNAs resulted in faster induction and more robust expression of differentiation protein K10 during the time course of epidermal tissue regeneration (Fig. 2C). Importantly, the basal layer was much smaller in the SNAI2i tissue with at most 1 cell layer whereas in control tissue there were several layers of undifferentiated basal layer cells (Fig. 2C). The knockdown of SNAI2 was validated with the absence of SNAI2 staining in the basal layer of SNAI2i epidermis (Supporting Information Fig. S1). SNAI2 depletion in cultured cells resulted in premature expression of differentiation protein TGM1, increased cell adhesion and differentiation gene expression similar to cells undergoing calcium induced differentiation (Fig. 2D-F and Supporting Information Fig. S2C-D). These results suggest that the levels of SNAI2 are critical for the differentiation status of epidermal cells with higher levels inhibiting and lower levels promoting differentiation.
Figure 2. The levels of SNAI2 controls epidermal differentiation.
(A) Epidermal progenitor cells transduced with the LZRS retrovirus encoding either LACZ controls (LZRS-LACZ) or SNAI2 (LZRS-SNAI2) were used to regenerate human epidermis by placing the cells on devitalized human dermis. Keratin 10 (K10) staining shown in red marks the differentiated epidermal layers. Hoechst staining in blue marks the nuclei. The dashed lines denote basement membrane zone (Scale bar=40μm; n=3 regenerated human epidermis per group). (B) RT-qPCR for expression of differentiation genes from samples isolated from (A). Expression levels were normalized to GADPH. Error bars=mean with SEM. (C) Epidermal cells were transduced with retroviruses expressing shRNAs targeting either control (CTL) or SNAI2 (SNAI2i) and used to regenerate human epidermis. Tissue was harvested days 2-5 after placement of cells onto dermis to determine the kinetics of epidermal differentiation with SNAI2 loss. K10 staining is shown in red. Brackets denote size of undifferentiated basal layer. Scale bar=40μm; n=3 regenerated human epidermis per group. (D) RT-qPCR for expression of SNAI2. Cells were knocked down for SNAI2 using two distinct shRNAs. (E) Control and SNAI2i cultured primary epidermal progenitor cells were stained for differentiation protein transglutaminase I (TGM1: shown in red). Scale bar=20μm. (F) RT-qPCR for expression of differentiation genes from CTL and SNAI2i cultured cells. Knockdown of SNAI2 using two distinct shRNAs [SNAI2i (A) and SNAI2i (B)] is shown. Expression levels were normalized to GADPH. Error bars=mean with SEM.
SNAI2 controls a gene expression program that represses differentiation and cell adhesion
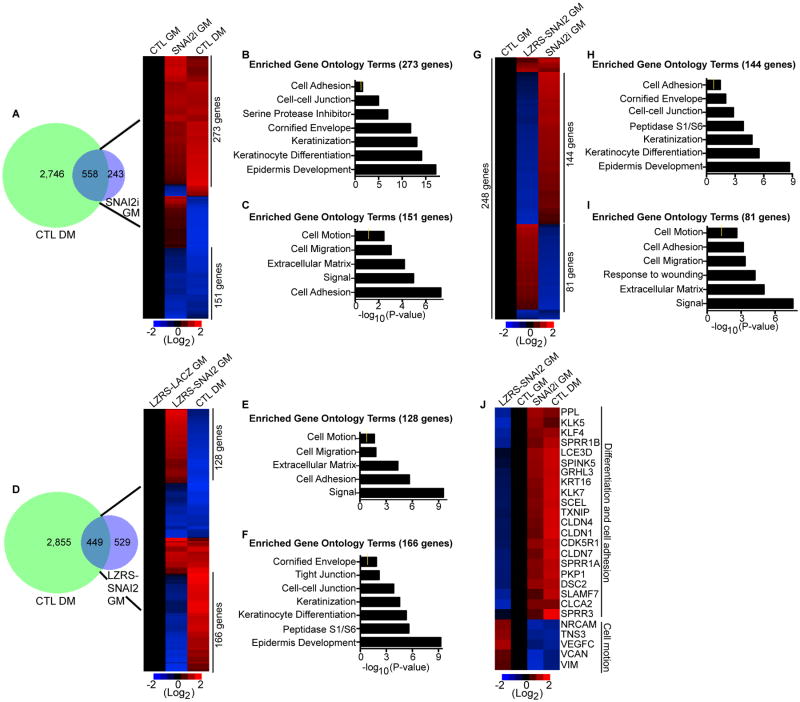
To determine how the levels of SNAI2 impacts the differentiation status of epidermal cells, global gene expression profiling was performed on control and SNAI2 knockdown cells. In keratinocytes cultured in growth medium (GM), SNAI2 knockdown altered the expression of 801 genes (≥ 2 fold change; ≤ 5% FDR) with 490 genes upregulated and 311 genes downregulated (Fig. 3A and Supporting Information Table S1A). Comparison with our previously generated data set of genes that changed during calcium-induced differentiation (DM: differentiation medium) revealed a significant overlap of 558 genes (∼70%, Fig. 3A and Supporting Information Table S1B-C)[5]. The vast majority (76%: 424/558) of the genes in the overlapped SNAI2i gene signature (SNAI2i GM) were regulated in the same direction as cells undergoing calcium induced differentiation (CTL DM) (Fig. 3A). 273 genes were upregulated in differentiated and SNAI2i cells with enriched gene ontology (GO) terms such as cell adhesion, cornified envelope, keratinization, and keratinocyte differentiation (Fig. 3A-B). 151 co-downregulated genes were enriched in terms such as cell motion, cell migration, and extracellular matrix (Fig. 3A and 3C). These results suggest that loss of SNAI2 mimics calcium induced epidermal differentiation through the increased expression of cell adhesion and differentiation genes and the downregulation of cell motility genes. To determine if increased levels of SNAI2 had the opposite effect as SNAI2 depletion, global gene expression profiling was performed on control LACZ and SNAI2 overexpressing epidermal progenitor cells. Increased SNAI2 expression resulted in the differential expression of 978 genes with 517 genes upregulated and 461 genes downregulated (Fig. 3D and Supporting Information Table S2A). 449 genes overlapped with the differentiation gene expression signature (Fig. 3D and Supporting Information Table S2B). Interestingly a majority of the overlapped genes (65%: 294/449) were oppositely regulated as the differentiation signature (Fig. 3D). 128 genes were upregulated in SNAI2 overexpressing keratinocytes while being downregulated during calcium induced differentiation (Fig. 3D). These genes were enriched in GO terms such as cell motion, cell migration, and extracellular matrix (Fig. 3E). 166 genes were downregulated in SNAI2 overexpressing cells and upregulated during differentiation with cornified envelope, cell-cell junction, and keratinocyte differentiation GO terms (Fig. 3D and 3F). This suggests that high levels of SNAI2 promote dedifferentiation and increased cell motility. To identify the genes that are the most susceptible to the levels of SNAI2 as well as potentially being direct targets, the SNAI2 depleted and overexpressed signatures were overlapped with each other. 248 genes overlapped with over 90% (225/248) of the genes oppositely regulated between the two data sets (Fig. 3G and Supporting Information Table S2C). 144 genes were downregulated in SNAI2 overexpressing cells and upregulated in SNAI2i cells with epidermal differentiation GO terms (Fig. 3G-H). These genes included differentiation induced structural (LCE3D, SCEL, SPRR1B), cell adhesion (DSC2, CLDN1, CLDN4) and transcription factor (GRHL3, KLF4) genes (Fig. 3J). 81 genes enriched for cell motion and cell migration were downregulated in SNAI2 depleted cells and upregulated in SNAI2 overexpressing cells (Fig. 3G and 3I). These include genes downregulated during differentiation such as VIM and VCAN (Fig. 3J). These results suggest that the levels of SNAI2 are critical to the differentiation status of epidermal cells. Decreased levels of SNAI2 lead to increased differentiation due to higher cell adhesion, keratinization, and cornified envelope gene expression while increased levels of SNAI2 promote cell motility and dedifferentiation.
Figure 3. SNAI2 represses the differentiation gene expression program.
(A) Overlap (left panel) of the differentiation gene signature (CTL DM: 3,304 genes change) with the genes that change upon knockdown of SNAI2 in cells cultured in growth medium (SNAI2i GM: 801 genes change). The differentiation gene signature (DM) is the differentially expressed genes between cells grown in low calcium (growth medium:GM) to cells grown in high calcium (differentiation medium:DM). Heat map (right panel) of the 558 genes that overlap. Differentiated control samples (CTL DM) were compared to control (CTL GM) and SNAI2i (SNAI2i GM) samples. Heat map is shown in red (induced genes) and blue (repressed genes) on a log2-based scale. (B) Gene ontology analysis of genes with increased expression that are co-regulated by SNAI2i GM and CTL DM samples. Yellow mark in bar graphs demark p value=0.5. (C) Gene ontology analysis of co-regulated genes with decreased expression. (D) Overlap (left panel) of CTL DM with the genes that change upon overexpression of SNAI2. LZRS-SNAI2 cells were cultured in growth medium (LZRS-SNAI2 GM). Heat map (right panel) of the 449 genes that overlap. Differentiated samples (CTL DM) were compared to control LZRS-LACZ GM and LZRS-SNAI2 GM samples. (E-F) Gene ontology analysis of genes oppositely regulated between LZRS-SNAI2 GM and CTL DM samples. (G) Heat map of the 248 genes that overlap between LZRS-SNAI2 GM and SNAI2i GM samples. (H-I) Gene ontology analysis of genes oppositely regulated between LZRS-SNAI2 GM and SNAI2i GM samples. (J) Heat map of differentiation, cell adhesion, and cell motility genes.
SNAI2 expression dictates the levels of SNAI2 binding across the genome
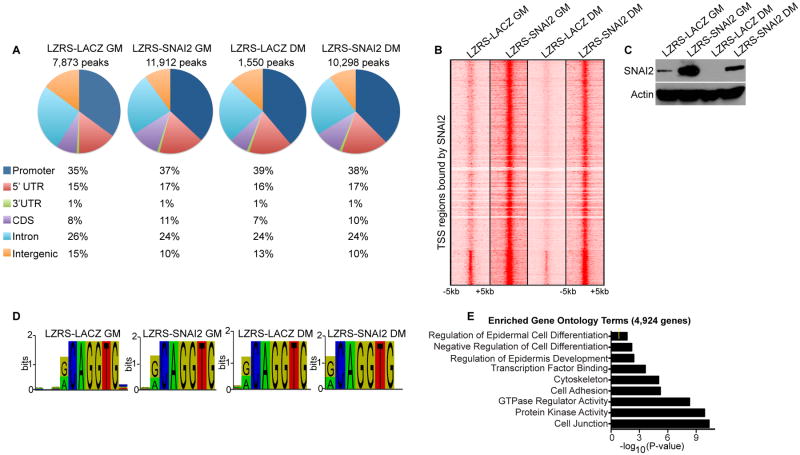
To determine the direct targets of SNAI2, we performed chromatin immunoprecipitations (ChIP) combined with deep sequencing (ChIP-Seq) using a SNAI2 antibody on progenitor cells overexpressing LACZ or SNAI2 and differentiated cells overexpressing LACZ or SNAI2. ChIP-Seq was also performed on control shRNA and SNAI2i progenitor cells to determine the specificity of the antibody. Depletion of SNAI2 by shRNAs resulted in a dramatic loss of SNAI2 binding on the genomic level as well as on single genes such as GRHL1, CDH1, and ITGA3 (Supporting Information Fig. S3A-D). Validation by qPCR also showed that knockdown of SNAI2 resulted in a dramatic loss of SNAI2 binding to genes such as GRHL1 and ITGA3 as well as known target CDH1 [25] (Supporting Information Fig. S3E). We identified 7,873 SNAI2 bound peaks in control LACZ expressing progenitor cells which were reduced to 1,550 peaks in differentiated LACZ epidermal cells (Fig. 4A and Supporting Information Table S3A-B). This suggests that during differentiation, 80% of the SNAI2 binding sites disappear due to the downregulation of SNAI2 (Fig. 4A and Fig. 1D-E). Overexpression of SNAI2 during differentiation (which blocks epidermal differentiation) restored the SNAI2 binding sites to 10,298 peaks, which is similar to the 11,912 peaks found in progenitor cells overexpressing SNAI2 (Fig. 4A and Supporting Information Table S3C-D). 50-55% of the SNAI2 bound peaks centered in regions around the transcriptional start site (TSS)(Fig. 4A-B). This included the promoter (35-39%) and 5′ UTR (15-17%) sequence of genes for all 4 conditions tested (Fig. 4A). A heat map of SNAI2 bound regions covering -5KB to +5KB from the TSS of genes revealed that higher expression levels of SNAI2 correlated with higher levels of binding to each site (Fig. 4B-C). SNAI2 overexpression in progenitor cells (LZRS-SNAI2 GM) had the highest levels of binding across all bound genes (Fig. 4B-C). Physiological levels of SNAI2 expression in progenitor cells (LZRS-LACZ GM) had moderate levels of SNAI2 binding which decreased further upon differentiation (LZRS-LACZ DM) (Fig. 4B-C). Remarkably, overexpression of SNAI2 in differentiation conditions (LZRS-SNAI2 DM) restored the binding of SNAI2 to similar levels as LZRS-SNAI2 GM cells (Fig. 4B-C). These results suggest that the expression levels of SNAI2 correlate with the amount of SNAI2 binding across the TSS regions of genes (Fig. 4B-C). To investigate whether the levels of SNAI2 expression resulted in changes to its binding motif, de novo motif search was used for each of the four conditions. The consensus core motif of CAGGTG was identified in all conditions which is identical to the motif previously reported for SNAI2 (discovered by in-vitro selection methods and transfection experiments) [26] (Fig. 4D). To define the genes that SNAI2 directly regulates, the 7,873 SNAI2 bound peaks in control LACZ progenitor cells (LZRS-LACZ GM) were mapped back to 4,924 genes (Supporting Information Table S4A). These genes were enriched for GO terms such as regulation of epidermal cell differentiation, negative regulation of cell differentiation, cell adhesion, and cell junction which correlates with SNAI2's role in preventing epidermal differentiation and promoting cell motility (Fig. 4E).
Figure 4. SNAI2 expression correlates with the levels of SNAI2 binding across the genome.
(A) Distribution of SNAI2 binding sites in epidermal progenitor and differentiated cells. Chromatin immunoprecipitation followed by high throughput sequencing (ChIP-Seq) was performed using a SNAI2 antibody on the following 4 samples: LZRS-SNAI2 GM and LZRS-LACZ GM (SNAI2 or control LACZ overexpressing cells cultured in growth medium); LZRS-LACZ DM and LZRS-SNAI2 DM (LACZ or SNAI2 overexpressing cells in differentiation medium). The total number of SNAI2 bound peaks were identified and their distributions across the genome calculated as a percentage for each sample. (B) Heat map of transcriptional start site (TSS) regions bound by SNAI2 in the four conditions tested. Each row shows the +/- 5kb centered regions around the TSS. (C) Western blot analysis for the expression of SNAI2 in the four conditions tested. Actin was used as a loading control. (D) SNAI2 binding motif found enriched under each condition. (E) Gene ontology analysis of the 4,924 genes that SNAI2 binds to in LACZ expressing progenitor cells (LZRS-LACZ GM). The 7,873 SNAI2 binding peaks identified in LZRS-LACZ GM samples were first mapped back to 4,924 genes and then gene ontology analysis performed. Yellow mark in bar graphs demark p value=0.5.
The levels of SNAI2 binding to differentiation genes determine the differentiation status of epidermal cells
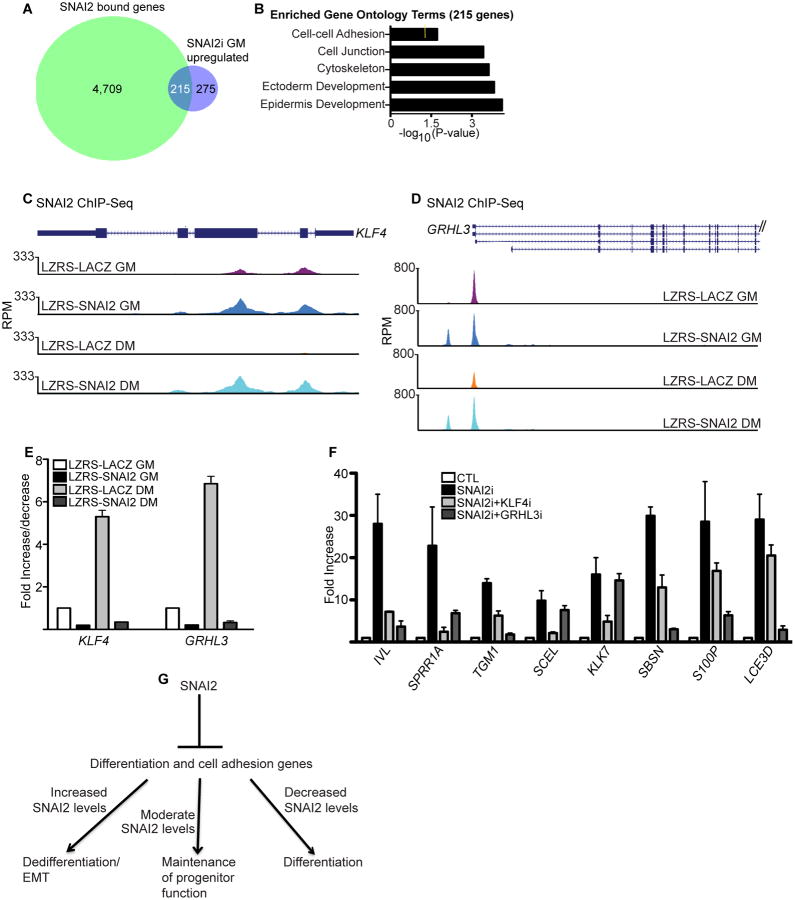
Since SNAI2 is a known transcriptional repressor, we compared SNAI2 bound genes with the 490 genes upregulated in SNAI2i cells because these are likely to be the genes directly repressed by SNAI2 binding[21]. 215 genes were found in the overlap which are likely to be directly regulated by SNAI2 binding levels (Fig. 5A and Supporting Information Table S4B). These genes were enriched for GO terms such as epidermis development and cell junction (Fig. 5B). Examination of the gene list showed that SNAI2 directly bound to the genomic regions of differentiation associated cell adhesion genes such as CLDN1 and GJB6 as well as differentiation promoting transcription factors such as KLF4 and GRHL3 (Fig. 5C-D and Supporting Information Fig. S4A-B). Knockdown of SNAI2 decreased binding to those regions demonstrating the specificity of SNAI2 binding to KLF4, GRHL3, CLDN1, and GJB6 genomic regions (Supporting Information Fig. S3F). The levels of SNAI2 binding to the transcriptional start site (+/- 2Kb) of these genes dictate the expression of these mRNAs (Fig. 5E and Supporting Information Fig. S4C). The higher the levels of SNAI2 binding to these genes the lower the expression level. SNAI2 overexpressing progenitor cells (LZRS-SNAI2 GM) had the most binding which correlated with the lowest expression of these genes (Fig. 5C-E and Supporting Information Fig. S4A-C). Physiological levels of SNAI2 binding in progenitor cells (LZRS-LACZ GM) resulted in moderate levels of expression. During differentiation (LZRS-LACZ DM) where SNAI2 binding to the TSS regions of these genes was decreased due to the downregulation of SNAI2 protein, the expression of these genes were increased to their highest levels (Fig. 4C, 5C-E and Supporting Information Fig. S4A-C). However, overexpression of SNAI2 in differentiated cells (LZRS-SNAI2 DM) restored the binding and blocked the expression of those genes (Fig. 5C-E and Supporting Information S4A-C).
Figure 5. The levels of SNAI2 binding to KLF4 and GRHL3 genomic regions determine the differentiation status of epidermal cells.
(A) Overlap of the 4,924 SNAI2 bound genes with the 490 genes that increase in expression upon SNAI2 knockdown. (B) Gene ontology analysis of the 215 genes found in the overlap. Yellow mark in bar graphs demark p value=0.5. (C-D) Gene tracks showing SNAI2 occupancy at regions around the TSS of KLF4 and GRHL3. The x axis denotes genomic position and y axis shows signal strength (reads per million, RPM). Binding of SNAI2 is shown for four conditions: LZRS-SNAI2 GM and LZRS-LACZ GM (SNAI2 or control LACZ overexpressing cells cultured in growth medium); LZRS-LACZ DM and LZRS-SNAI2 DM (LACZ or SNAI2 overexpressing cells in differentiation medium). (E) RT-qPCR for expression of KLF4 and GRHL3 in the four conditions described in (C-D). (F) RT-qPCR for differentiation gene expression in control (CTL), SNAI2i, and cells knocked down for both SNAI2 and KLF4 or SNAI2 and GRHL3. Expression levels were normalized to GADPH. Error bars=mean with SEM. (G) Model of SNAI2 mediated control of the differentiation status of epidermal cells.
Our results suggests that SNAI2 may directly regulate up to 43.9% (215/490) of the induced genes in SNAI2i cells. The other 56.1% may potentially be regulated indirectly by SNAI2 through direct regulation of other transcription factors. Candidate factors include KLF4 and GRHL3 which have been shown to be necessary for the transcriptional activation of the epidermal differentiation program[11,12]. To determine if the premature differentiation phenotype seen in SNAI2i cells is mediated in part by increased KLF4 or GRHL3 levels, SNAI2 and KLF4 or SNAI2 and GRHL3 were simultaneously knocked down. The mRNA levels of differentiation induced genes SPRR1A, IVL, and TGM1 were increased in SNAI2i cells but were restored similar to control levels in KLF4 and SNAI2 or GRHL3 and SNAI2 double knockdown cells (Fig. 5F). Furthermore both GRHL3 and KLF4 had specific target differentiation genes as double knockdown of GRHL3 and SNAI2 prevented the increase in SBSN, S100P and LCE3D gene expression levels seen in SNAI2i cells alone; whereas, KLF4 and SNAI2 knockdown had minimal impacts (Fig. 5F). Similarly double knockdown of KLF4 and SNAI2 prevented the increases in KLK7 and SCEL differentiation gene levels while GRHL3 and SNAI2 knockdown had no impact (Fig. 5F). These results suggest that while SNAI2 directly blocks the expression of a portion of the differentiation induced genes, it also prevents premature differentiation of progenitor cells by repressing the expression of KLF4 and GRHL3.
SNAI2's regulation of KLF4 and GRHL3 suggest that it may regulate other differentiation promoting transcription factors. Supporting this, SNAI2 binding was found in the TSS of both CEBPA and GRHL1 (Supporting Information S5A-B). CEBPA is necessary for the commitment to epidermal differentiation and GRHL1 regulates the expression of desmosomes during the differentiation process[10,14,15]. The amount of SNAI2 binding correlated with the expression levels of both genes with decreased binding leading to higher gene expression levels (Supporting Information S5A-C). These results suggest that SNAI2 regulates a subset of the differentiation program through differentiation associated transcription factors.
Discussion
Prior studies on the function of SNAI2 in the skin have revealed a role for SNAI2 in mediating epidermal homeostasis[21]. Microarray analysis on SNAI2 knockout mouse epidermis revealed alterations in epidermal differentiation, adhesion, motility, and angiogenesis although which genes are direct targets of SNAI2 remain unknown[27]. Studies in human epidermal keratinocytes have shown that conditional activation of SNAI2 can repress the expression of cell adhesion genes such as α3 and β1 integrins[28]. Furthermore SNAI2 is necessary for epidermal growth factor mediated re-epithelialization[29]. It is clear that SNAI2 has a functional role in epidermal differentiation, adhesion, motility, and wound repair but the underlying mechanisms of how SNAI2 controls these functions is unclear. Here, we report the mechanism by which the EMT-TF, SNAI2, controls the differentiation and adhesion status of epidermal cells. In the epidermis, SNAI2 expression is restricted to progenitor cells and its expression is extinguished upon induction of differentiation. Constitutive expression of SNAI2 in regenerated human epidermis inhibits differentiation. Overexpression of SNAI2 in cultured epidermal progenitor cells resulted in dedifferentiation and decreased cell adhesion. Inhibition of SNAI2 had the opposite effect and resulted in faster kinetics of differentiation and premature differentiation of the basal layer of the epidermis. Global gene expression profiling showed that cells with decreased SNAI2 expression resembled cells undergoing calcium induced differentiation (∼70% overlap with the differentiation gene expression signature) with increases in differentiation associated cell adhesion, keratinization, and cornified envelope formation. In contrast, increased expression of SNAI2 served to dedifferentiate the cells as SNAI2 overexpression still overlapped significantly with the differentiation signature but with the genes regulated in the opposite direction.
Genome-wide mapping of SNAI2 binding sites showed that approximately half of SNAI2 bound peaks were found in the promoter or 5′ UTR of genes. Analysis of the SNAI2 binding sites within +/- 5KB of TSS showed that the levels of SNAI2 expression correlated with the amount of binding to those regions. Physiological levels of SNAI2 in progenitor cells led to moderate amounts of binding allowing for maintenance of progenitor function. Upon differentiation where SNAI2 expression is downregulated, the binding decreased (7,873 peaks were identified in progenitor cells which decreased to 1,550 peaks upon differentiation) which resulted in expression of differentiation associated genes. Overexpression of SNAI2 in differentiation conditions restored the binding sites and blocked differentiation. Increased SNAI2 binding to the same sites due to SNAI2 overexpression in progenitor cells lead to dedifferentiation due to further downregulation of SNAI2 target genes. This suggests that the levels of SNAI2 binding determine the levels of differentiation gene expression. This phenomenon is similar to what has been recently described for c-Myc. In tumor cells elevated expression of c-Myc led to increased binding at promoter regions of active genes and resulted in further increases in gene expression which the authors coined as transcriptional amplification[30,31]. Our results show the opposite effect in which increasing levels of SNAI2 binding to canonical CAGGTG sites leads to further transcriptional silencing.
Many of the SNAI2 bound genes were cell adhesion and differentiation genes. 215 epidermal differentiation genes are likely to be directly regulated by SNAI2 since loss of SNAI2 expression lead to increased expression of these SNAI2 bound genes. These include differentiation induced cell adhesion genes such as GJB6 and CLDN1 as well as transcription factors such as GRHL3, KLF4, CEBPA, and GRHL1. Interestingly, double knockdown of either GRHL3 or KLF4 with SNAI2 can prevent the increases in differentiation gene expression seen in SNAI2 knockdown cells. These results suggest that at least part of the differentiation program (differentiation genes not directly regulated by SNAI2) is mediated through SNAI2 control of KLF4 and GRHL3 expression. Normal physiological levels of SNAI2 allow moderate levels of binding to its target genes (ie: KLF4, GRHL3, GRHL1, CEBPA, GJB6, and CLDN1) which maintains progenitor function by suppressing the differentiation program. This allows just the right amount of expression of cell adhesion and differentiation genes to maintain progenitor status (Fig. 5G). Upon induction of differentiation, SNAI2 is downregulated which causes the loss of SNAI2 binding. This alleviates SNAI2 mediated repression of target genes allowing increased cell adhesion, keratinization, and cornified envelope formation all features associated with differentiation (Fig. 5G). In contrast, elevated expression of SNAI2 which has been reported in numerous tumors as well as during RAS mediated epidermal tumorigenesis may result in additional binding to the same genes (ie KLF4, GRHL3, GRHL1, CEBPA, and cell adhesion genes)[32,33]. This increased binding results in transcriptional silencing amplification which causes an EMT phenotype and dedifferentiation due to further suppression of cell adhesion and differentiation genes. Similarly, SNAI2 expression is increased in epidermal cells bordering wounds which promotes a transient EMT phenotype to allow migration to the wound to promote reepithelialization[23]. Supporting this, wound healing is reduced in SNAI2 knockout mice[23]. In summary, the levels of SNAI2 binding to target genes are crucial in determining epidermal cell fate.
Conclusion
In conclusion, our study provides an explanation for how EMT-TFs mediate EMT, progenitor cell maintenance, and differentiation. The levels of SNAI2 expression determine the amount of SNAI2 binding to differentiation and adhesion genes across the genome. This in turn dictates the expression levels of those genes. High levels of expression as seen in cancer cells leads to increased SNAI2 binding to differentiation and adhesion genes leading to an EMT phenotype. Moderate (physiological) levels of binding promote the right amount of differentiation and cell adhesion gene expression to maintain progenitor function. Finally, low levels of SNAI2 binding as seen during epidermal differentiation results in the alleviation of SNAI2 mediated gene repression leading to full expression of the differentiation program.
Materials and Methods
The Supplemental Materials and Methods section which includes a list of retroviral constructs, siRNAs, antibodies, and primer sequences is available online.
Tissue culture
Primary human epidermal cells (keratinocytes) were derived from newborn foreskin. Cells were cultured in growth medium (GM) composed of KSF-M (Life Technologies) containing epidermal growth factor (EGF) and bovine pituitary extract (BPE). Cells cultured in growth growth medium are also referred to as progenitor cells. Cells were induced to differentiate (DM) by the addition of 1.2 mM calcium for 3 days in full confluence. Amphotrophic phoenix cells were maintained in DMEM and 10% fetal bovine serum.
Gene transfer
Amphotrophic phoenix cells were transfected with 3 ug of each retroviral construct to either knockdown or overexpress genes. Transfections were done in 6 well plates using Fugene 6 (Roche). Viral supernatants were collected 48 hours post transfection. These supernatants were placed on primary human keratinocytes and centrifuged for 1 hour at 1000rpm with polybrene (5ug/ml). Cells were transduced a total of 2 times and selected using puromycin (2ug/ml) after the last transduction with shRNA retroviral constructs. Cells were transduced once for LZRS retroviral overexpression constructs.
Gene knockdown and overexpression
ShRNA retroviral constructs were generated by cloning oligos into the pSuper Retro vector[34]. The full-length open reading frame of SNAI2 was cloned into the LZRS retroviral vector.
Western blotting and immunofluorescence
40 ug of the cell lysates were used for immunoblotting. For immunofluorescence experiments, 7 μm thick epidermal sections from adult human skin or organotypic cultures were used.
Quantitative reverse transcriptase-PCR analysis
Total RNA from cells was extracted using the GeneJET RNA purification kit (Thermo Scientific). One ug of total RNA was reverse transcribed and quantitative PCR was performed. Samples were normalized to GAPDH.
Gene expression profiling
Cells overexpressing SNAI2 or control LACZ were harvested 7 days post transduction. Cells knocked down for SNAI2 or control were harvested one week after the last infection. Microarray analysis using Affymetrix HG-U133 2.0 plus arrays was performed on duplicate samples. Significantly changed genes were identified as previously described[5]. Data was deposited in GEO with accession number: GSE55269.
Regenerated human epidermis
1 million control or SNAI2 knockdown cells were seeded on devitalized human dermis and raised to the air/liquid interface in order to induce differentiation and stratification. This method of regenerating human epidermis was previously described [3]. Tissue was harvested at the indicated time points in the figures.
Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
ChIP was performed as previously described[3]. 10 million cells were used for ChIP for each antibody used. 4ug of antibody was used for each pull down experiment. Experiments were performed in triplicates. QPCR Results were represented as a percent of input DNA. For ChIP-Seq, the ChIP DNA library was prepared using the TruSeq sample prep (Illumina). Experiments were performed in duplicates. Sequencing was done on a Hi-Seq System (Illumina) using single 1×100 reads. Reads were mapped back to the human genome assembly (GRCh37/hg19). Peaks were called using the Partek Genomics Suite (Partek Incorporated) with FDR ≤ 0.001. Data was deposited in GEO with accession number: GSE55421.
RNA Seq Data Analysis
RNA seq data from the ENCODE project (ENCBS563ENC: CSHL RNA seq) for human epidermal keratinocytes was used to determine expression levels of EMT-TFs[35]. Reads were mapped back to the human genome assembly (GRCh37/hg19) and expression generated using TopHat and Cufflinks[36,37].
Supplementary Material
Acknowledgments
The authors would like to thank M. Noutsou and N. Ling for pre-submission review and helpful discussions. We thank the UCSD Skin Genomics Core for bioinformatics analysis. This work is supported by the American Cancer Society Research Scholars Grant (RSG-12-148-01-DDC) to G.L. Sen and the UCSD Dermatologist Investigator Training Program (1T32 -AR062497–01) to D.S. Mistry.
Footnotes
Author Contributions: Conceived and designed the experiments: GLS and DSM. Performed the experiments: DSM, YC, YW. Analyzed the data: GLS, DSM. Wrote the paper: GLS.
References
- 1.Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, et al. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12:193–203. doi: 10.1016/j.stem.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luis NM, Morey L, Mejetta S, Pascual G, Janich P, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9:233–246. doi: 10.1016/j.stem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Mistry DS, Chen Y, Sen GL. Progenitor Function in Self-Renewing Human Epidermis is Maintained by the Exosome. Cell Stem Cell. 2012;11:127–135. doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mlacki M, Darido C, Jane SM, Wilanowski T. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One. 2014;9:e89247. doi: 10.1371/journal.pone.0089247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 12.Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- 13.Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, et al. ZNF750 Is a p63 Target Gene that Induces KLF4 to Drive Terminal Epidermal Differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilanowski T, Caddy J, Ting SB, Hislop NR, Cerruti L, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008;27:886–897. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez RG, Garcia-Silva S, Moore SJ, Bereshchenko O, Martinez-Cruz AB, et al. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 16.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 19.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 20.Sou PW, Delic NC, Halliday GM, Lyons JG. Snail transcription factors in keratinocytes: Enough to make your skin crawl. Int J Biochem Cell Biol. 2010;42:1940–1944. doi: 10.1016/j.biocel.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Shirley SH, Hudson LG, He J, Kusewitt DF. The skinny on Slug. Mol Carcinog. 2010;49:851–861. doi: 10.1002/mc.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 23.Hudson LG, Newkirk KM, Chandler HL, Choi C, Fossey SL, et al. Cutaneous wound reepithelialization is compromised in mice lacking functional Slug (Snai2) J Dermatol Sci. 2009;56:19–26. doi: 10.1016/j.jdermsci.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, et al. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 26.Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 27.Newkirk KM, MacKenzie DA, Bakaletz AP, Hudson LG, Kusewitt DF. Microarray analysis demonstrates a role for Slug in epidermal homeostasis. J Invest Dermatol. 2008;128:361–369. doi: 10.1038/sj.jid.5700990. [DOI] [PubMed] [Google Scholar]
- 28.Turner FE, Broad S, Khanim FL, Jeanes A, Talma S, et al. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem. 2006;281:21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- 29.Kusewitt DF, Choi C, Newkirk KM, Leroy P, Li Y, et al. Slug/Snai2 is a downstream mediator of epidermal growth factor receptor-stimulated reepithelialization. J Invest Dermatol. 2009;129:491–495. doi: 10.1038/jid.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie Z, Hu G, Wei G, Cui K, Yamane A, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuter JA, Ortiz-Urda S, Kretz M, Garcia J, Scholl FA, et al. Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell. 2009;15:477–488. doi: 10.1016/j.ccr.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alves CC, Carneiro F, Hoefler H, Becker KF. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front Biosci (Landmark Ed) 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]
- 34.Sen G, Wehrman TS, Myers JW, Blau HM. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat Genet. 2004;36:183–189. doi: 10.1038/ng1288. [DOI] [PubMed] [Google Scholar]
- 35.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.