Abstract
Rationale
A better understanding of the mechanism underlying skeletal muscle repair is required to develop therapies that promote tissue regeneration in adults. Hedgehog signaling has been shown previously to be involved in myogenesis and angiogenesis: 2 crucial processes for muscle development and regeneration.
Objective
The objective of this study was to identify the role of the hedgehog transcription factor Gli3 in the crosstalk between angiogenesis and myogenesis in adults.
Methods and Results
Using conditional knockout mice, we found that Gli3 deficiency in endothelial cells did not affect ischemic muscle repair, whereas in myocytes, Gli3 deficiency resulted in severely delayed ischemia-induced myogenesis. Moreover, angiogenesis was also significantly impaired in HSA-CreERT2; Gli3Flox/Flox mice, demonstrating that impaired myogenesis indirectly affects ischemia-induced angiogenesis. The role of Gli3 in myocytes was then further investigated. We found that Gli3 promotes myoblast differentiation through myogenic factor 5 regulation. In addition, we found that Gli3 regulates several proangiogenic factors, including thymidine phosphorylase and angiopoietin-1 both in vitro and in vivo, which indirectly promote endothelial cell proliferation and arteriole formation. In addition, we found that Gli3 is upregulated in proliferating myoblasts by the cell cycle–associated transcription factor E2F1.
Conclusions
This study shows for the first time that Gli3-regulated postnatal myogenesis is necessary for muscle repair–associated angiogenesis. Most importantly, it implies that myogenesis drives angiogenesis in the setting of skeletal muscle repair and identifies Gli3 as a potential target for regenerative medicine.
Keywords: angiogenesis, pathological, hedgehogs, ischemia, muscle, skeletal, regeneration
Skeletal muscle regeneration typically occurs as a result of trauma or diseases, such as congenital myopathies or ischemic diseases. It is a complex biological process that involves overlapping phases of inflammation, new tissue formation, and remodeling. New tissue formation involves the generation of new muscle fibers, formed by proliferation and fusion of resident muscle precursor cells called satellite cells.1 Concomitantly, the newly generated muscle is vascularized, primarily through the process of angiogenesis. Some studies have suggested that angiogenesis and myogenesis are tightly regulated, as muscle satellite cells and endothelial cells (ECs) are close neighbors.2 It has been shown previously that ECs release myogenic growth factors, including insulin-like growth factor-1 (IGF-1), hepatocyte growth factor, and fibroblast growth factor 2, and that differentiating muscle cells produce angiogenic factors, including vascular endothelial growth factor A (VEGFA).3
Researchers have administered angiogenic factors either as recombinant proteins or via gene therapy to improve skeletal muscle repair and limb revascularization in the setting of hindlimb ischemia (HLI) with promising results.4,5 However, large clinical trials conducted to date have shown that administration of a single growth factor has limited effects.5 A recent preclinical study that combined administration of a proangiogenic factor with a myogenic factor demonstrated enhanced ischemic muscle recovery,6 suggesting that promoting angiogenesis along with muscle regeneration may be a more suitable therapeutic strategy. As such, a better understanding of the mechanisms underlying skeletal muscle repair may help to optimize future clinical interventions.
Embryonic signaling pathways, including the hedgehog pathway, provide promising new targets for regenerative medicine.7–9 Hedgehog signaling has been shown to be involved in postnatal muscle regeneration after mechanical crush or cardiotoxin injection in mice, by regulating VEGFA, stromal cell–derived factor-1, and IGF-1 expression, as well as satellite cell proliferation.10 When sonic hedgehog (Shh), one of the hedgehog ligands, was administered either as a recombinant protein or via gene therapy, it promoted neovascularization of ischemic tissues by promoting both angiogenesis11 and endothelial progenitor cell recruitment.12 Studies conducted to elucidate the cellular mechanisms responsible for these findings have shown that Shh induces overexpression of several proangiogenic growth factors, including VEGFA and angiopoietin-1 (Ang1), by fibroblasts and cardiomyocytes.11,12 More recently, our group reported that desert hedgehog promotes angiogenesis by maintaining peripheral nerve–derived angiogenic factors in ischemic muscle.13
Together, these data provide evidence for a role of the hedgehog pathway in postnatal muscle regeneration and neovascularization. However, despite evidence for the in vivo effects of hedgehog signaling in the adult, little is known about the molecular mechanisms by which these effects occur. As such, we sought to investigate the role of Gli3, a transcription factor mediating hedgehog signaling, in adult muscle regeneration.
Gli3 was shown recently to be upregulated in regenerating adult skeletal muscle, and Gli3 knockdown was shown to impair angiogenesis in mouse models of HLI and myocardial infarction (MI).14 Moreover, Gli3 is known to regulate embryonic myogenesis and to mediate Shh-induced somite specification by regulating Myogenic factor 5 (Myf5) expression.15,16 Collectively, these observations suggest that Gli3 has an important role in both angiogenesis and myogenesis and could be a key factor regulating muscle regeneration after an ischemic injury.
The present study further characterizes the action of Gli3 in the setting of adult skeletal muscle regeneration using Gli3 conditional knockout mice (bred with Tie-Cre and HSA-CreERT2 mice). We demonstrate that Gli3 is overexpressed during myoblasts proliferation and that it is necessary for Myf5 expression and myogenic differentiation. Moreover, our data demonstrate for the first time that Gli3 expression is regulated by the myogenic factor IGF-1 through E2F1 using E2F1−/− mice. Finally, this study demonstrates that the process of myogenesis is required for angiogenesis.
Methods
Mice
C57BL/6 mice were obtained from Charles River Laboratories and bred in our animal facility. Gli3 Floxed (Gli3Flox) mice,17 under the Swiss Webster genetic background, were kindly provided by Dr A.L. Joyner. Tie2-Cre mice,18 HSA-CreERT2 mice,19 and Rosa26R mice were obtained from Jackson laboratories. Gli3Flox mice were genotyped as previously described.17 E2F1 knockout mice were generated and genotyped as described previously.20
Mice were handled in accordance with the guidelines established by the National Institute of Medical Research (INSERM) and approved by the local Animal Care and Use Committee of Bordeaux University. Cre recombinase of HSA-CreERT2 mice was activated by intraperitoneal injection of 1 mg tamoxifen for ≥5 consecutive days.
HLI Model and Assessments
HLI was performed as previously described21 in 8- to 12-week-old mice.
For histological assessment and gene expression analysis, mice were euthanized and tibialis anterior muscles were harvested and cut in half. The lower half was fixed in methanol, paraffin-embedded, and cut into 6-μm sections and the upper half was snap-frozen in liquid nitrogen. Each group included ≥6 animals. Capillary and arteriole densities were evaluated using sections stained for the expression of CD31 and α-smooth muscle actin, respectively. Muscle repair and myogenesis were assessed after hematoxylin and eosin staining of muscle sections as previously described.13
Mice were perfused with LacZ fix solution for x-gal staining, before muscles were harvested.
Immunostaining
ECs were identified with rat anti-CD31 antibodies, Gli3 was stained with goat anti-Gli3 antibodies, and skeletal myocytes were identified with rabbit anti-desmin antibodies. Primary antibodies were resolved with Alexa-Fluor–conjugated secondary antibodies and counter-stained with 4′,6-diamidino-2-phenylindole, for immunofluorescent analysis or with biotin-conjugated secondary antibodies, streptavidin-HRP complex, DAB substrate, and counterstained with hematoxylin.
Quantitative Reverse Transcription Polymerase Chain Reaction
RNA was isolated using Tri Reagent per manufacturer’s instructions from 3×105 cells or skeletal muscle previously snap-frozen in liquid nitrogen and homogenized. Total RNA was reverse transcribed with M-MLV reverse transcriptase (Promega), and amplification was performed on a DNA Engine Opticon 2 (MJ Research Inc) using B-R SYBER Green SuperMix (Quanta Biosciences). Primer sequences are reported in Online Table I.
The relative expression of each mRNA was calculated by the comparative threshold cycle method and normalized to hypoxanthine guanine phosphoribosyl transferase mRNA expression.
Western Blot Analysis
Expression of Gli3 was evaluated by SDS-PAGE using goat anti-Gli3 antibodies (R&D systems). Expression of Ang1 was evaluated using rabbit anti-Ang1 antibodies (Santa-Cruz), and expression of thymidine phosphorylase (TYMP) was evaluated using anti-mouse monoclonal anti–PD-ECGF antibodies (Santa-Cruz). Equal protein loading was confirmed using monoclonal anti–α-tubulin antibodies (Sigma).
Cell Culture
Cell culture and in vitro assays are described in the Detailed Methods in the Online Data Supplement.
Statistics
Results are reported as mean±SEM. Comparisons between groups were analyzed for significance with the non-parametric Mann–Whitney test. Differences between groups were considered to be significant when P≤0.05; *P≤0.05; **P ≤0.01; and ***P≤0.001.
Results
Gli3 Is Overexpressed in Regenerating Skeletal Muscle
To study skeletal muscle regeneration, we used the HLI model, in which the ischemic tibialis anterior muscle undergoes complete necrosis (Figure 1A) as early as 24 hours after surgery. Muscle regeneration begins at approximately day 4 from the edge of the muscle and by day 10, the muscle is totally recolonized by new muscle fibers.
Figure 1. Gli3 is overexpressed in regenerating skeletal muscle.

Unilateral hindlimb ischemia (HLI) was induced in 12-week-old C57BL/6 mice. A, Cross-sections of tibialis anterior skeletal muscle harvested at the indicated time points were stained with hematoxylin and eosin. B, The expression of Gli3 mRNA in both hindlimbs was measured from day 0 (D0) to day 21 (D21) via real-time reverse transcription polymerase chain reaction, n=6 mice in each group. C, Cross-sections of tibialis anterior skeletal muscle coimmunostained for the expression of Gli3 (green) and markers for endothelial cells (CD31) or myocytes (desmin; red) as labeled. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue). ***P≤0.001. NS indicates not significant.
Ischemia-induced angiogenesis was shown to be impaired recently in Gli3 heterozygote mice.14 To characterize the mechanism of action of Gli3, C57BL/6 wild-type mice underwent HLI surgery, and both ischemic and control skeletal muscles were harvested at 0, 2, 5, 7, 10, 14, and 21 days after surgery. We confirmed that Gli3 mRNA was strongly overexpressed in the ischemic muscle from days 2 to 21, with maximal expression between days 5 and 7 (+1460%; Figure 1B).
Gli3 has been shown to be expressed, at least in part, by ECs in vivo and to regulate EC function in vitro,14 and we first verified whether Gli3 was expressed by ECs in the regenerating ischemic skeletal muscle. As shown in Figure 1C (top), Gli3 was expressed by ECs identified by CD31 staining 5 days after HLI was induced, supporting the notion that EC Gli3 expression may play a role in ischemia-induced angiogenesis.
EC-Specific Knockout of Gli3 Does Not Impair Ischemia-Induced Angiogenesis
To investigate the role of endothelial Gli3 in vivo, we bred Gli3 conditional knockout mice (Gli3Flox) with Tie2-Cre mice to disrupt Gli3 expression in EC specifically. We first verified that Cre recombinase was active in skeletal muscle ECs of adult mice by breeding Tie2-Cre mice with Rosa26R mice. Recombination of the Rosa26R allele was verified after LacZ staining of skeletal muscle sections (Figure 2A). We further verified that Gli3 deficiency was not compensated either by Gli1 or by Gli2 upregulation (Online Figure IA and IB).
Figure 2. Endothelial Cell Gli3 is not required for ischemia-induced angiogenesis.
A, X-gal staining was performed on cross-sections of skeletal muscle from Tie2-Cre; Rosa26R mice. B to E, Unilateral HLI was induced in 12-week-old Tie2-Cre; Gli3Flox/Flox mice (n=6) and in their control littermates (Ctrl, Tie2-Cre and Gli3Flox/Flox) (n=16), and mice were euthanized 10 days later. B, Sections of tibialis anterior muscle were stained with hematoxylin and eosin. C, Repaired surface areas were measured using Sigma Scan Pro software. D, Capillary density was evaluated by staining muscle sections for CD31 expression (brown) and reported as the number of CD31-positive vessels per HPF (E). NS indicates not significant.
Tie2-Cre; Gli3Flox/Flox mice and their control littermates (wild-type, Tie2-Cre, and Gli3Flox/Flox) underwent surgery to induce HLI and were euthanized 10 days later. Skeletal muscle repair was evaluated after hematoxylin and eosin staining (Figure 2B). The repaired surface area, measured as previously described,22 was not different between Tie2-Cre; Gli3Flox/Flox and control mice (Figure 2C). Angiogenesis was quantified after CD31 staining (Figure 2D), as shown in Figure 2E, and there was no difference in capillary density between the Tie2-Cre; Gli3Flox/Flox and control mice (Figure 2E). We further confirmed that the total number of ECs was the same in Tie2-Cre; Gli3Flox/Flox and control regenerating skeletal muscles, by measuring the expression of the ECs marker Cadherin-5 (Online Figure IIA). Moreover, ischemic foot perfusion of Tie2-Cre; Gli3Flox/Flox mice was equivalent to that of control mice (Online Figure IIB and IIC), 10 days after HLI surgery.
Together these results demonstrate that Gli3 expressed by ECs is not essential for ischemia-induced angiogenesis and muscle repair.
Myocyte-Specific Knockout of Gli3 Severely Impairs Postnatal Myogenesis
Gli3 has been shown previously to regulate muscle specification in embryos through regulation of Myf5,16 and we hypothesized that it may regulate myogenesis in the setting of ischemic skeletal muscle repair. We first verified that Gli3 was expressed in skeletal muscle cells, by staining with both anti-Gli3 and anti-desmin antibodies to identified muscle cells. As shown in Figure 1C (bottom), we found that Gli3 was expressed by desmin-positive myocytes 10 days after HLI surgery was performed.
To investigate the role of muscle cell–expressed Gli3, Gli3Flox mice were bred with HSA-CreERT2 mice. Mice were administered tamoxifen daily from days 3 to 9 and euthanized at day 10. Cre recombinase activity in skeletal muscle cells was verified by breeding HSA-CreERT2 mice with Rosa26R mice, and recombination of the Rosa26R allele was assessed after LacZ staining of skeletal muscle sections (Figure 3A). We also verified that knockout of Gli3 was not compensated by Gli1 or Gli2 overexpression in HSA-CreERT2; Gli3Flox/Flox (Online Figure IC and ID).
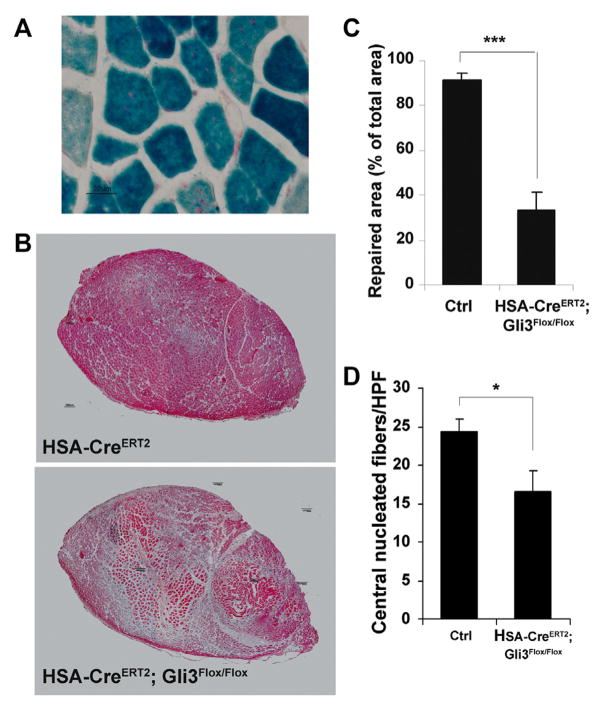
Figure 3. Skeletal muscle regeneration is impaired in HSA-CreERT2; Gli3Flox/Flox mice.
A, X-gal staining was performed on cross-sections of skeletal muscle of HSA-CreERT2; Rosa26R mice. B to D, Unilateral HLI was induced in 12-week-old HSA-CreERT2; Gli3Flox/Flox mice (n=9) and in their control littermates (Ctrl, HSA-CreERT2 and Gli3Flox/Flox; n=10), and mice were euthanized 10 days later. B, Sections of tibialis anterior muscle were stained with hematoxylin and eosin. C, Repaired surface areas were measured using Sigma Scan Pro software. D, Myogenesis was quantified as the number of central nucleated muscle fibers per HPF. ***P≤0.001; *P≤0.05.
We then performed HLI surgery in HSA-CreERT2; Gli3Flox/Flox mice and their control littermates (wild-type, HSA-CreERT2, and Gli3Flox/Flox mice). Skeletal muscle repair was evaluated after hematoxylin and eosin staining (Figure 3B). The repaired surface area was significantly smaller in HSA-CreERT2; Gli3Flox/Flox (33.40±7.96%) compared with control mice (91.66±3.09%; P=0.001; Figure 3C). Impaired muscle repair was confirmed by quantifying the density of newly formed muscle fibers (ie, central nucleated muscle fibers) in the ischemic skeletal muscle of HSA-CreERT2; Gli3Flox/Flox and control mice. The number of central nucleated muscle fibers per HPF was significantly lower in HSA-CreERT2; Gli3Flox/Flox compared with control mice (Figure 3D).
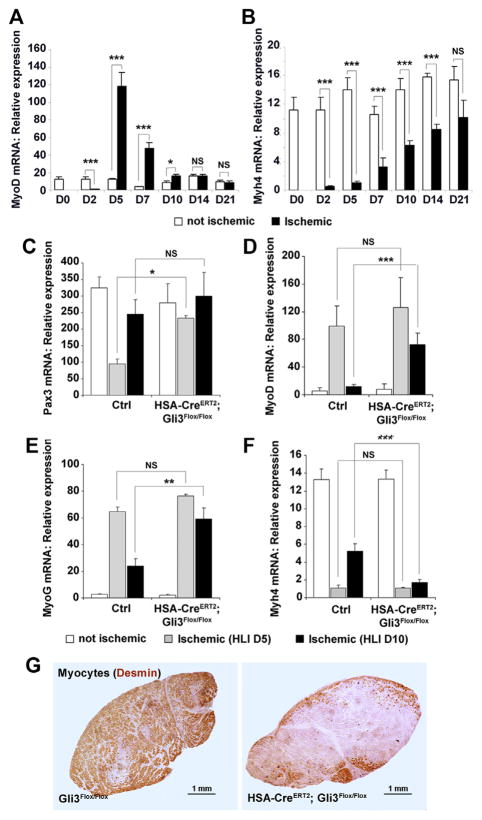
Myogenesis was further assessed by measuring the mRNA expression level of Pax3 (a satellite cell marker), myogenic differentiation 1 (MyoD; an early marker of muscle differentiation), myogenin (an intermediate marker of muscle differentiation), and myosin, heavy polypeptide 4, skeletal muscle (Myh4; a terminal marker of muscle differentiation). In wild-type mice, MyoD expression is strongly elevated 5 and 7 days after HLI surgery, which corresponds to the phase of myoblast activation and amplification (Figure 4A), whereas Myh4 mRNA expression consistently increase from days 2 to 21 (Figure 4B) along with the progression of muscle regeneration. As shown in Figure 4C, although Pax3 was significantly downregulated in the regenerating muscle of control mice 5 days after HLI surgery, its expression remained high in HSA-CreERT2; Gli3Flox/Flox mice. MyoD and myogenin expression strongly increased both in the regenerating ischemic muscle of HSA-CreERT2; Gli3Flox/Flox and control mice, 5 days after HLI was induced; nevertheless, 10 days after HLI, they failed to decrease in HSA-CreERT2; Gli3Flox/Flox mice in contrast to control mice (Figure 4D and 4E). These results strongly suggest that differentiation of HSA-CreERT2; Gli3Flox/Flox myoblasts is impaired. As a consequence, 10 days after HLI surgery, myosin, heavy polypeptide 4, skeletal muscle (Myh4) mRNA was significantly lower in HSA-CreERT2; and Gli3Flox/Flox (Figure 4F) mice. Consistently, although the ischemic tibialis anterior skeletal muscle of control mice was almost completely recolonized by desmin-positive new muscle fibers, HSA-CreERT2; Gli3Flox/Flox ischemic muscle still mainly included dead muscle fibers (Figure 4G) demonstrating that myogenesis is delayed in HSA-CreERT2; Gli3Flox/Flox compared with control mice.
Figure 4. Myogenesis is delayed in HSA-CreERT2; Gli3Flox/Flox mice.
A to B, Unilateral hindlimb ischemia (HLI) was induced in 12-week-old C57BL/6 mice. The expression of (A) myogenic differentiation 1 (MyoD) and (B) myosin, heavy polypeptide 4, skeletal muscle (Myh4) mRNA in both hind limbs was measured from day 0 (D0) to day 21 (D21) via real-time reverse transcription polymerase chain reaction (RT-PCR), n=6 mice in each group. C to G, Unilateral HLI was induced in 12-week-old HSA-CreERT2; Gli3Flox/Flox mice (n=5 and 9) and in their control littermates (Ctrl, HSA-CreERT2; Gli3Flox/Flox; n=5 and 10). Mice were euthanized 5 and 10 days later. The expression of (C) Pax3, (D) MyoD, (E) myogenin (MyoG), and (F) myosin heavy chain 4 (MYH4) mRNA in both hindlimbs was measured via real-time RT-PCR. G, Myogenesis was evaluated by staining muscle sections for desmin expression (brown). *P≤0.05; ***P≤0.001. NS indicates not significant.
Taken together these data demonstrate that Gli3 expressed by muscle cells is essential for muscle regeneration in adults. More specifically, Gli3 is required for myoblasts differentiation into myocytes but does not seem to be required for myoblast activation.
Gli3 Promotes Myoblast Differentiation
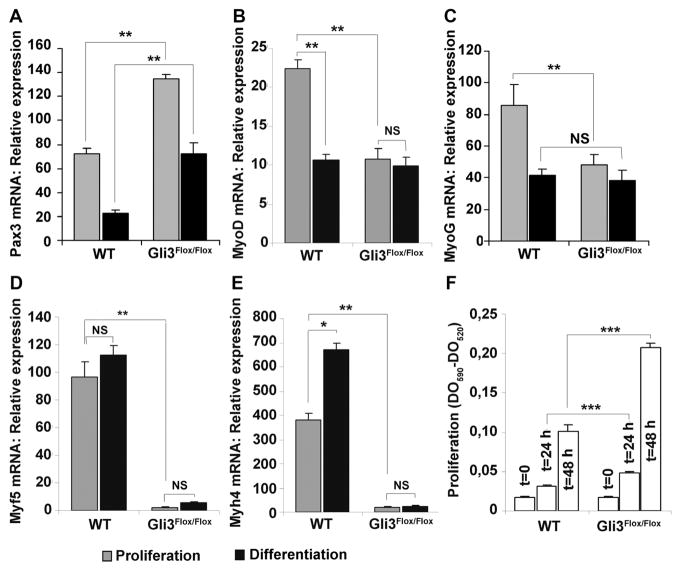
To further confirm the role of Gli3 in myogenesis, myoblats were isolated from Gli3Flox/Flox and wild-type mice. Knockout of Gli3 in Gli3Flox/Flox myoblasts was induced by Cre recombinase encoding lentivirus transduction. We measured the expression of several myoblast differentiation markers in Gli3 knockout and wild-type myoblasts; and as shown in Figure 5A–5E, although the expression of Pax3, the premyoblast marker, was higher in Gli3 knockout myoblasts, the mRNA expression of MyoD, myogenin, Myf5, and Myh4 was significantly downregulated in Gli3 knockout compared with wild-type myoblasts (grey bars). MyoD and myogenin were downregulated by ≈50%, and both Myf5 and Myh4 by >95%. We then induced myoblast differentiation by culturing cells in 5% horse serum containing Dulbecco’s modified Eagle’s medium (differentiation medium) for 48 hours. Expression of Pax3, MyoD, and myogenin was significantly reduced in wild-type myoblasts, after cells were cultured in differentiation medium (Figure 5A–5C), whereas Myh4 significantly increased (Figure 5E). In contrast, in Gli3 knockout myoblasts, MyoD, myogenin, and Myh4 expression did not change (Figure 5B, 5C, and 5E). Together these results demonstrate that Gli3 is necessary for adult myoblast differentiation, and they support previously published results showing Gli3 is necessary for Myf5 expression in embryos.15
Figure 5. Gli3 regulates myoblast differentiation.
A to E, LV-Cre-transduced, Gli3Flox/Flox or wild-type (WT) primary cultured myoblasts were cultured in proliferation medium (20% FBS containing Dulbecco’s modified Eagle’s medium [DMEM]/F10) or differentiation medium (5% horse serum containing DMEM) for 48 hours. The expression of (A) Pax3, (B) myogenic differentiation 1 (MyoD), (C) myogenin (MyoG), (D) myogenic factor 5 (Myf5), and (E) myosin, heavy polypeptide 4, skeletal muscle (Myh4) mRNA was measured via real-time reverse transcription polymerase chain reaction. F, LV-Cre-transduced, Gli3Flox/Flox or WT primary cultured myoblasts were cultured in proliferation medium for the indicated time points. Proliferation was evaluated with the MTT assay. *P≤0.05; **P≤0.01; and ***P≤0.001. NS indicates not significant.
We further characterized the role of Gli3 in myoblasts proliferation. As shown in Figure 5D, Gli3 knockout myoblasts proliferated more than wild-type myoblasts, which may because of the fact that they cannot differentiate. The role of Gli3 in myoblasts differentiation was confirmed in Gli3+/− myoblasts (Online Figure IIIA–IIIE).
Gli3 Knockout in Myocytes Indirectly Impairs Ischemia-Induced Angiogenesis
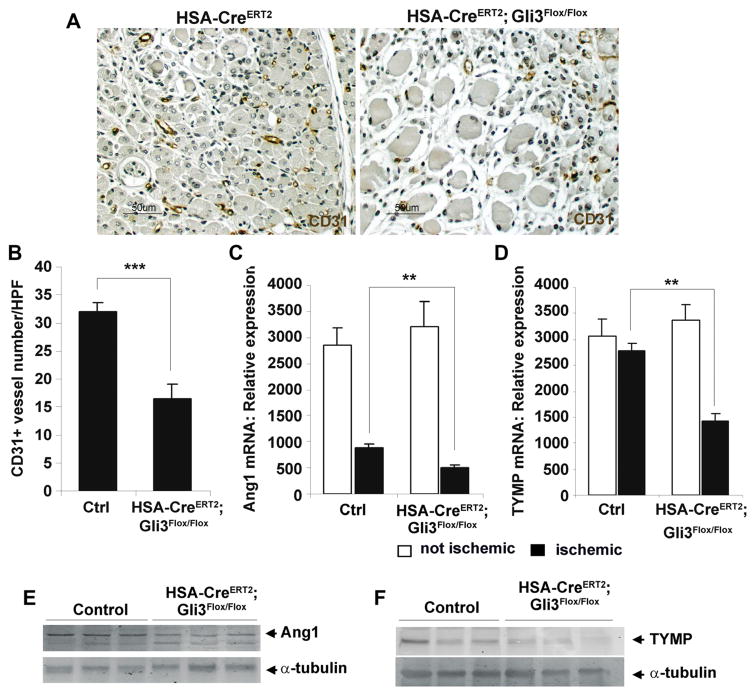
Differentiating myoblasts have been reported to regulate angiogenesis, at least in part, through VEGFA regulation by MyoD.3 We hypothesized that impaired myogenesis observed in HSA-CreERT2; Gli3Flox/Flox may indirectly affect angiogenesis. Thus, we measured capillary density in HSA-CreERT2; Gli3Flox/Flox and control mice, 10 days after HLI surgery (Figure 6A). Figure 6B shows that capillary density was significantly reduced in HSA-CreERT2; Gli3Flox/Flox compared with control mice (16±3 CD31+ vessels/HPF in HSA-CreERT2; Gli3Flox/Flox versus 32±2 vessels in control mice; P=0.001). Moreover, decreased capillary density in HSA-CreERT2; Gli3Flox/Flox was associated with a decreased α-smooth muscle actin–positive arteriole density (Online Figure IVA) and resulted in a significant reduction in the ischemic foot perfusion compared with control mice (Online Figure IVB and IVC). The present data thus demonstrate for the first time that myogenesis regulates angiogenesis in the setting of skeletal muscle repair.
Figure 6. Muscle Gli3 regulates ischemia-induced angiogenesis.
A and B, Unilateral hindlimb ischemia was induced in 12-week-old HSA-CreERT2; Gli3Flox/Flox mice (n=9) and in their control littermates (Ctrl, HSA-CreERT2 and Gli3Flox/Flox; n=10), and mice were euthanized 10 days later. A, Capillary density was evaluated by staining muscle sections for CD31 expression (brown) and reported as the number of CD31-positive vessels per HPF (B); (C) Ang1; and (D) thymidine phosphorylase (TYMP) mRNA in both hindlimbs was measured via real-time reverse transcription polymerase chain reaction. E, Ang1 and (F) TYMP protein expression was measured by Western blot analysis in the ischemic limb of HSA-CreERT2; Gli3Flox/Flox mice and control mice. **P≤0.01; ***P≤0.001.
To further confirm the role of Gli3 in regulating ischemia-induced angiogenesis, Gli3 was overexpressed or not, via adenoviral transduction, in the skeletal muscle of 12-week-old wild-type mice. As shown in Online Figure VA and VB, 7 days after HLI surgery, central nucleated muscle fiber density was significantly increased in the regenerating ischemic skeletal muscle of Ad-Gli3–treated mice compared with control Ad-treated mice. And consistently with the data obtained in HSA-CreERT2; Gli3Flox/Flox, both capillary density (Online Figure VA and VC) and ischemic limb perfusion (Online Figure VD and VE) were also significantly increased in Ad-Gli3–treated mice compared with control Ad-treated mice.
Gli3 Regulates Ang1 and Thymidine Phosphorylase Expression in Myoblasts
To test the hypothesis that Gli3 might regulate expression of proangiogenic factors in myoblasts, we compared their expression in Gli3 knockout and wild-type myoblasts. As shown in Online Figure VIA, there was no difference in VEGFA expression in Gli3 knockout versus wild-type myoblasts, but Ang1 and TYMP were both significantly downregulated in Gli3 knockout myoblasts (Online Figure VIB and VIC). We further confirmed their downregulation by Western blot (Online Figure VID and VIE) and verified that Ang1 and TYMP were also downregulated in regenerating skeletal muscle of HSA-CreERT2; Gli3Flox/Flox mice in vivo. mRNA expression of Ang1 and TYMP was quantified by real-time reverse transcription polymerase chain reaction, and both were significantly lower in the regenerating skeletal muscle of HSA-CreERT2; Gli3Flox/Flox compared with control mice (Figure 6C and 6D). Moreover, downregulation of Ang1 and TYMP at the protein level was confirmed by Western blot analysis (Figure 6E and 6F).
TYMP is a potent mitogen for ECs,23 and we next evaluated the effect of Gli3 knockout and wild-type myoblast-conditioned medium on human umbilical vein endothelial cell proliferation in vitro. As shown in Online Figure VIIA, the proliferation of human umbilical vein endothelial cells when cultured in Gli3 knockout myoblast-conditioned medium was significantly reduced when compared with human umbilical vein endothelial cells cultured in wild-type myoblast-conditioned medium. The decreased EC proliferation was confirmed in HSA-CreERT2; Gli3Flox/Flox in vivo. As shown Online Figure VIIB and VIIC, the number of bromodeoxyuridine (BrdU)-positive ECs was significantly reduced in the ischemic regenerating skeletal muscle of HSA-CreERT2; Gli3Flox/Flox mice compared with control mice.
Together these data demonstrate that Gli3 regulates Ang1 and TYMP expression in myoblasts and indirectly promotes EC proliferation and angiogenesis.
Gli3 Expression Is Associated With Cell Proliferation
We next sought to investigate mechanisms regulating Gli3 mRNA expression in muscle cells. In contrast to Gli1 and Gli2, Gli3 is not regulated at the mRNA level by Shh,24 suggesting that a mechanism independent of hedgehog signaling may regulate Gli3 mRNA expression in muscle cells. Interestingly, we found that Gli3 expression is higher in proliferating myoblasts compared with more differentiated myotubes at both the mRNA (Online Figure VIIIA) and protein level (Online Figure VIIIB), which is consistent with the Gli3 expression profile observed in the setting of ischemic muscle repair. Indeed, Gli3 expression is maximal between days 5 and 7 after HLI surgery, which corresponds to the myoblast activation and amplification step (MyoD upregulation). We next sought to determine whether Gli3 accumulates in proliferating cells in vivo. BrdU was administered for 24 hours before mice were euthanized to identify proliferating cell. As shown in Figure 7A, Gli3 (green) was primarily detected in BrdU-positive cells (red), demonstrating that Gli3 expression is associated with cell proliferation during the process of muscle regeneration.
Figure 7. E2F1 regulates Gli3 expression in myoblasts.
A, Unilateral hindlimb ischemia (HLI) was induced in 12-week-old C57BL/6 mice. Bromodeoxyuridine (BrdU) was injected at day 4 and mice were euthanized at day 5. Cross-sections of the tibialis anterior skeletal muscle were coimmunostained for the expression of Gli3 (green) and BrdU (red) as labeled. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue). B, Wild-type (WT) primary cultured myoblasts were cotransfected with c-Myc, Ets1, Myb, or E2F1 encoding vectors together with pGL3 basic vector in which 1940 bp of Gli3 promoter has been cloned and pHook-LacZ. Luciferase activity was measured 48 hours later and normalized to β-galactosidase activity. C, Unilateral HLI was induced in 12-week-old E2F1−/− mice (n=6) and their WT littermates (n=6). Mice were euthanized 5 days later. The expression of Gli3 mRNA in both hindlimbs was measured via real-time reverse transcription polymerase chain reaction (RT-PCR). D, Serum-starved WT primary cultured myoblasts were treated with 50 ng/mL fibroblast growth factor 2 (FGF2), 20 ng/mL insulin-like growth factor-1 (IGF-1), 20% FBS or BSA alone for 24 hours. Gli3 mRNA expression was measured via real-time RT-PCR. *P≤0.05; **P≤0.01; and ***P≤0.001. NS indicates not significant.
Gli3 Expression Is Regulated by E2F1
To identify transcription factors regulating Gli3 mRNA expression in proliferating myoblasts, we performed a systematic analysis of the Gli3 promoter. A 1940-bp long fragment of the promoter was cloned upstream of the luciferase reporter gene and 5′ deletions of the promoter were made. Gene reporter assays revealed that the −876 to −793 and −476 to −205 Gli3 promoter fragments include cis-regulating elements that positively regulate Gli3 transcription (Online Figure IXA). MatInspector (Genomatix) sequence analysis of these fragments revealed potential Myb, c-Myc, nuclear factor of activated T cells, E2F, Myt1, and Ets1 binding sequences.
To investigate whether one of these transcription factors may regulate Gli3 mRNA expression in proliferating myoblasts, we compared mRNA expression of each of these transcription factors in proliferating and differentiated myoblasts. We found that Myb, E2F1, Ets1, c-Myc, and to a lesser extent nuclear factor of activated T cell-c3 were significantly upregulated in proliferating myoblasts, whereas there was no difference in E2F4, nuclear factor of activated T cell-5, and Myt1 mRNA expression (Online Figure XA–XH). To test whether those transcription factors regulate Gli3 transcription, myoblasts were cotransfected with plasmids encoding c-Myc, Ets1, Myb, or E2F1 together with the pGL3 vector expressing luciferase under the control of 1940 bp of Gli3 promoter. As shown in Figure 7B, only E2F1 was able to activate the Gli3 promoter, which includes several potential E2F binding sites (Online Figure IXB). E2F1 seemed to activate the −476 to −205 region of Gli3 promoter (Online Figure IXC) that includes 2 potential E2F binding sites.
To further investigate whether Gli3 is regulated by E2F1 in the setting of skeletal muscle repair, we induced HLI in E2F1 knockout mice and in their wild-type littermates and euthanized them 5 days later. As shown in Figure 7C, Gli3 mRNA expression was significantly lower in the ischemic regenerating skeletal muscle of E2F1 knockout mice compared with their wild-type littermates, confirming that Gli3 expression is regulated by E2F1 in the setting of skeletal muscle regeneration.
As E2F1 is a cell cycle–associated transcription factor, we hypothesized that Gli3 expression could be regulated by myogenic growth factors for myoblasts. As shown in Figure 7D, Gli3 mRNA is indeed upregulated in FBS- or in IGF-1–treated myoblasts. This result identifies a novel mechanism of regulation of Gli3, which is more likely independent on hedgehog signaling in myoblasts.
Ang1 and TYMP Expression in Cardiomyocytes Is Not Dependent on Gli3
Finally, we wished to verify whether Gli3 regulation of proangiogenic factors could be expended to cardiomyocytes in the setting of MI. To this aim, HSA-CreERT2; Gli3Flox/Flox mice and their Gli3Flox/Flox littermates were submitted to MI surgery. As shown in Online Figure XIA and XIB, in contrast to the skeletal muscle, the expression of Ang1 and TYMP was not affected by the absence of Gli3 in cardiomyocytes. Nevertheless, according to previous investigation demonstrating the role of hedgehog signaling in cardiomyocytes survival25 and differentiation,26 we found that the expression of the cardiomyocyte markers NK2 homeobox 5, myocyte enhancer factor 2C, and cardiac myosin heavy polypeptide 6 was significantly decreased in the border zone of the infarcted area of HSA-CreERT2; Gli3Flox/Flox hearts compared with Gli3Flox/Flox hearts (Online Figure XIC–XIE).
Discussion
Hedgehog signaling has been shown previously to promote tissue regeneration in adults, specifically in the setting of ischemia- or cardiotoxin-induced muscle injury.10–12 It has been proposed to act on a wide variety of cells, including fibroblasts,11 endothelial progenitor cells,12 and ECs.27 More recently, Shh was shown to promote muscle regeneration, at least in part, by promoting myogenesis.10,28 In the current article, we further characterize the role of hedgehog signaling in the regulation of postnatal myogenesis and identify the downstream hedgehog transcription factor, Gli3, as a key regulator of this process. We used conditional knockout mice, for the first time, to investigate the role of Gli3 in skeletal muscle cell lineage specifically.
To date, the role and expression pattern of Gli3 have been studied almost exclusively during development, particularly during neural and limb skeletal development.22,29–31 In the establishment of skeletal muscle, Gli3, together with Gli2, has been shown to be a primary mediator of Shh-induced regulation of the Myf5 epaxial somite enhancer.15,16 Our data indicate that, in the setting of ischemic skeletal muscle regeneration, Gli3 is upregulated in proliferating myoblasts and is necessary for Myf5 expression and adult myoblast differentiation.
Muscle regeneration requires newly generated muscle to be vascularized, and some studies have suggested that angiogenesis and myogenesis are tightly regulated, as muscle satellite cells and ECs are close neighbors.2 Moreover, ECs release myogenic growth factors, including IGF-1, hepatocyte growth factor, and fibroblast growth factor 2, and differentiating muscle cells produce angiogenic factors including VEGFA. Our data demonstrate that Gli3 knockout in muscle cells impairs ischemia-induced angiogenesis, demonstrating for the first time that myogenesis is necessary for angiogenesis in the setting of muscle repair. More specifically, we found that Gli3 knockout in myocytes resulted in decreased EC proliferation and impaired arteriole formation; accordingly, we found that Gli3 expression in myoblasts is required for the expression of the proangiogenic factors TYMP and Ang1 known to promote EC proliferation23 and vessel maturation, respectively.32 VEGFA was not downregulated in Gli3-deficient myoblasts in which MyoD was only softly impaired. Those results, thus, confirm the role of Gli3 in angiogenesis previously described14 and identify the mechanism by which Gli3 regulates angiogenesis.
Our previous data demonstrated that constitutive Gli3 deficiency impaired reparative angiogenesis and consequently cardiac function after MI14; nevertheless, the present study shows that in contrast to skeletal muscle cells, Gli3 does not seem to regulate Ang1 or TYMP expression in cardiomyocytes in the setting of MI. Although hedgehog signaling has been shown widely to regulate Ang1 expression,11,33–35 a specific study designed to investigate molecular mechanism involved in the regulation of Ang1 and TYMP expression by hedgehog signaling and Gli transcription factors needs to be performed. Gene regulation by Gli transcription factors is indeed complex and depends on the context.36 It is noteworthy that, in contrast to the skeletal muscle, the heart muscle barely regenerates; comparing the interactions between vascular cells and muscle cells in the setting of HLI and MI would be indeed interesting. The present study demonstrates the crucial role of myogenesis in driving angiogenesis in the setting of HLI. To date, in the setting of MI, conversely, angiogenesis is proposed to drive cardiomyocytes behavior.37 Nevertheless, cardiac progenitor cells when delivered as a cell therapy were shown to secrete proangiogenic factors including VEGFA.38 Moreover, in embryos, cardiomyocyte-derived VEGFA was shown to be necessary for the development of the coronary vasculature,39 suggesting that cardiomyocytes may also participate in angiogenesis in ischemic condition in adults.
Interestingly, we found that in the setting of skeletal muscle differentiation both in vivo and in vitro, Gli3 is regulated at the mRNA level, where as it is typically known to be regulated post-translationally by proteolytic cleavage. In the absence of hedgehog proteins, Gli3 is cleaved (Nterm-Gli3) and acts as a transcriptional repressor. When cells are stimulated, full-length Gli3 is translocated in the nucleus and activates gene transcription via the classical Gli cis-regulating element 5′-ACCCACCAG-3′.40 Few studies have identified transcription factors regulating Gli3 mRNA expression. Gli3 mRNA was shown to be regulated by β-catenin in the neural tube of chick embryos,41 HoxD13 was shown to regulate Gli3 transcription in the setting of limb development42 and recently Gli3 was shown to be a transcriptional target of the Notch transcription factor N1ICD/RBPJ.43 Finally, 1 study has reported that in human colorectal carcinoma cell lines, Gli3 mRNA expression is dependent on Smo activity, molecular mechanisms involved in this regulation were suggested to be independent on canonical hedgehog signaling.44 We found that, in skeletal myoblasts, Gli3 transcription is regulated by the cell cycle–associated transcription factor E2F1, and that it is a downstream target of the myogenic factor IGF-1. Moreover, we confirmed Gli3 regulation by E2F1, in vivo, in the setting of skeletal muscle regeneration in adults. Interestingly, muscle regeneration was shown to be impaired in both IGFR-knockout mice45 and E2F1 knockout mice,46 suggesting that Gli3 may be a downstream effector of the IGF-R/E2F1 pathway.
IGF-1 was recently shown to be cooperating with Shh in the setting of myoblasts differentiation. Both factors were shown to promote MAPK ERK1/2 and p38 phosphorylation synergistically.47 Our data identify a new cross-talk between IGF-1 and hedgehog signaling through Gli3.
Morphogens involved in embryonic development, and more specifically in muscle differentiation, were shown to be reactivated in adults and regulate muscle repair in adults. Notch signaling has been shown to be involved in satellite cell activation and cell fate determination during postnatal myogenesis,48 the hedgehog pathway has been shown to be reactivated after muscle injury in adults and to promote myogenesis,10 and Wnt signaling has been shown to induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration.49 Taken together, these data identify morphogens as potential targets for regenerative medicine. By characterizing molecular mechanism upregulating Gli3, a mediator of hedgehog signaling, we have identified a potentially novel target to regulate hedgehog signaling activation and consequently muscle repair.
Most importantly, this study implies that regulating myogenesis in addition to angiogenesis is essential when designing strategies to promote skeletal muscle revascularization and regeneration.
Supplementary Material
Novelty and Significance.
What Is Known?
Skeletal muscle regeneration involves both angiogenesis and myogenesis. Large clinical trials to promote skeletal muscle repair have thus far been focused on stimulating angiogenesis and have shown that administration of a single proangiogenic factor have limited effects.
Reactivation of embryonic signaling pathways participates in ischemic tissue repair in adults.
Hedgehog transcription factor, Gli3, is known to promote somite specification in embryos.
What New Information Does This Article Contribute?
Gli3 is required for postnatal myogenesis by inducing expression of myogenic factor 5.
Myogenesis drives angiogenesis in the setting of skeletal muscle regeneration through Gli3-dependent angiogenic growth factor production.
Gli3 is regulated by the cell cycle–associated transcription factor E2F1.
In conclusion, the present study identifies Gli3 as a novel target to promote myogenesis. Moreover, it implies that regulating myogenesis in addition to angiogenesis is essential when designing strategies to promote skeletal muscle revascularization and regeneration.
Acknowledgments
We thank Jérôme Guignard (INSERM U1034, Pessac) for his excellent technical assistance in the animal facility and Christelle Boullé for administrative assistance. We thank A.L. Joyner for providing Gli3Flox mice. CreERT2 mice are licensed by GIE-CERBM (ICS-IGBMC).
Sources of Funding
This study was supported by grants from the Fondation de la Recherche Médicale, program on cardiovascular aging (DCV20070409258); the Conseil Régional d’Aquitaine (action inter-régionale Aquitaine-Midi Pyrénées); the Communauté de Travail des Pyrénées and the ANR program (ANR-07-PHYSIO-010-02 to A.-P. Gadeau); and The National League against Cancer (M.-A. Renault). C. Chapouly and S. Vandierdonck are supported by fellowships from the CHU de Bordeaux. This work was also supported by the National Institute of Health grants (HL093439 and HL113541 to G. Qin).
Nonstandard Abbreviations and Acronyms
- Ang1
angiopoietin-1
- EC
endothelial cell
- HLI
hindlimb ischemia
- IGF-1
insulin-like growth factor-1
- MI
myocardial Infarction
- Myf5
myogenic factor 5
- Myh4
myosin, heavy polypeptide 4, skeletal muscle
- MyoD
myogenic differentiation 1
- Shh
sonic hedgehog
- TYMP
thymidine phosphorylase
- VEGFA
vascular endothelial growth factor A
Footnotes
Disclosures
None.
References
- 1.Ten Broek RW, Grefte S, Von den Hoff JW. Regulatory factors and populations involved in skeletal muscle regeneration. J Cell Physiol. 2010;224:7–16. doi: 10.1002/jcp.22127. [DOI] [PubMed] [Google Scholar]
- 2.Christov C, Chrétien F, Abou-Khalil R, Bassez G, Vallet G, Authier FJ, Bassaglia Y, Shinin V, Tajbakhsh S, Chazaud B, Gherardi RK. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell. 2007;18:1397–1409. doi: 10.1091/mbc.E06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan BA, Walshe TE, Mitchell DC, Havumaki JS, Saint-Geniez M, Maharaj AS, Maldonado AE, D’Amore PA. Coordinated vascular endothelial growth factor expression and signaling during skeletal myogenic differentiation. Mol Biol Cell. 2008;19:994–1006. doi: 10.1091/mbc.E07-09-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grounds MD. Muscle regeneration: molecular aspects and therapeutic implications. Curr Opin Neurol. 1999;12:535–543. doi: 10.1097/00019052-199910000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Roncalli J, Tongers J, Renault MA, Losordo DW. Biological approaches to ischemic tissue repair: gene- and cell-based strategies. Expert Rev Cardiovasc Ther. 2008;6:653–668. doi: 10.1586/14779072.6.5.653. [DOI] [PubMed] [Google Scholar]
- 6.Borselli C, Storrie H, Benesch-Lee F, Shvartsman D, Cezar C, Lichtman JW, Vandenburgh HH, Mooney DJ. Functional muscle regeneration with combined delivery of angiogenesis and myogenesis factors. Proc Natl Acad Sci USA. 2010;107:3287–3292. doi: 10.1073/pnas.0903875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh M. WNT signaling in stem cell biology and regenerative medicine. Curr Drug Targets. 2008;9:565–570. doi: 10.2174/138945008784911750. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz i Altaba A, Mas C, Stecca B. The gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahlgren C, Lendahl U. Notch signaling and its integration with other signaling mechanisms. Regen Med. 2006;1:195–205. doi: 10.2217/17460751.1.2.195. [DOI] [PubMed] [Google Scholar]
- 10.Straface G, Aprahamian T, Flex A, Gaetani E, Biscetti F, Smith RC, Pecorini G, Pola E, Angelini F, Stigliano E, Castellot JJ, Jr, Losordo DW, Pola R. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J Cell Mol Med. 2009;13:2424–2435. doi: 10.1111/j.1582-4934.2008.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pola R, Ling LE, Silver M, Corbley MJ, Kearney M, Blake Pepinsky R, Shapiro R, Taylor FR, Baker DP, Asahara T, Isner JM. The morphogen Sonic hedgehog is an indirect angiogenic agent upregulating two families of angiogenic growth factors. Nat Med. 2001;7:706–711. doi: 10.1038/89083. [DOI] [PubMed] [Google Scholar]
- 12.Kusano KF, Pola R, Murayama T, et al. Sonic hedgehog myocardial gene therapy: tissue repair through transient reconstitution of embryonic signaling. Nat Med. 2005;11:1197–1204. doi: 10.1038/nm1313. [DOI] [PubMed] [Google Scholar]
- 13.Renault MA, Chapouly C, Yao Q, Larrieu-Lahargue F, Vandierdonck S, Reynaud A, Petit M, Jaspard-Vinassa B, Belloc I, Traiffort E, Ruat M, Duplàa C, Couffinhal T, Desgranges C, Gadeau AP. Desert hedgehog promotes ischemia-induced angiogenesis by ensuring peripheral nerve survival. Circ Res. 2013;112:762–770. doi: 10.1161/CIRCRESAHA.113.300871. [DOI] [PubMed] [Google Scholar]
- 14.Renault MA, Roncalli J, Tongers J, Misener S, Thorne T, Jujo K, Ito A, Clarke T, Fung C, Millay M, Kamide C, Scarpelli A, Klyachko E, Losordo DW. The Hedgehog transcription factor Gli3 modulates angiogenesis. Circ Res. 2009;105:818–826. doi: 10.1161/CIRCRESAHA.109.206706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson CP., Jr Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev. 2002;16:114–126. doi: 10.1101/gad.940702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott A, Gustafsson M, Elsam T, Hui CC, Emerson CP, Jr, Borycki AG. Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development. 2005;132:345–357. doi: 10.1242/dev.01537. [DOI] [PubMed] [Google Scholar]
- 17.Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135:2093–2103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 19.Schuler M, Ali F, Metzger E, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in skeletal muscles of the mouse. Genesis. 2005;41:165–170. doi: 10.1002/gene.20107. [DOI] [PubMed] [Google Scholar]
- 20.Qin G, Kishore R, Dolan CM, et al. Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF. Proc Natl Acad Sci USA. 2006;103:11015–11020. doi: 10.1073/pnas.0509533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couffinhal T, Silver M, Zheng LP, Kearney M, Witzenbichler B, Isner JM. Mouse model of angiogenesis. Am J Pathol. 1998;152:1667–1679. [PMC free article] [PubMed] [Google Scholar]
- 22.te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, Zeller R. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- 23.Finnis C, Dodsworth N, Pollitt CE, Carr G, Sleep D. Thymidine phosphorylase activity of platelet-derived endothelial cell growth factor is responsible for endothelial cell mitogenicity. Eur J Biochem. 1993;212:201–210. doi: 10.1111/j.1432-1033.1993.tb17651.x. [DOI] [PubMed] [Google Scholar]
- 24.Cohen MM., Jr The hedgehog signaling network. Am J Med Genet A. 2003;123A:5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- 25.Lavine KJ, Kovacs A, Ornitz DM. Hedgehog signaling is critical for maintenance of the adult coronary vasculature in mice. J Clin Invest. 2008;118:2404–2414. doi: 10.1172/JCI34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voronova A, Coyne E, Al Madhoun A, Fair JV, Bosiljcic N, St-Louis C, Li G, Thurig S, Wallace VA, Wiper-Bergeron N, Skerjanc IS. Hedgehog signaling regulates MyoD expression and activity. J Biol Chem. 2013;288:4389–4404. doi: 10.1074/jbc.M112.400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renault MA, Roncalli J, Tongers J, Thorne T, Klyachko E, Misener S, Volpert OV, Mehta S, Burg A, Luedemann C, Qin G, Kishore R, Losordo DW. Sonic hedgehog induces angiogenesis via Rho kinase-dependent signaling in endothelial cells. J Mol Cell Cardiol. 2010;49:490–498. doi: 10.1016/j.yjmcc.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palladino M, Gatto I, Neri V, Stigliano E, Smith RC, Pola E, Straino S, Gaetani E, Capogrossi M, Leone G, Hlatky L, Pola R. Combined therapy with sonic hedgehog gene transfer and bone marrow-derived endothelial progenitor cells enhances angiogenesis and myogenesis in the ischemic skeletal muscle. J Vasc Res. 2012;49:425–431. doi: 10.1159/000337921. [DOI] [PubMed] [Google Scholar]
- 29.Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- 30.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 31.Walterhouse D, Ahmed M, Slusarski D, Kalamaras J, Boucher D, Holmgren R, Iannaccone P. gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev Dyn. 1993;196:91–102. doi: 10.1002/aja.1001960203. [DOI] [PubMed] [Google Scholar]
- 32.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 33.Fujii T, Kuwano H. Regulation of the expression balance of angiopoietin-1 and angiopoietin-2 by Shh and FGF-2. In Vitro Cell Dev Biol Anim. 2010;46:487–491. doi: 10.1007/s11626-009-9270-x. [DOI] [PubMed] [Google Scholar]
- 34.Lee SW, Moskowitz MA, Sims JR. Sonic hedgehog inversely regulates the expression of angiopoietin-1 and angiopoietin-2 in fibroblasts. Int J Mol Med. 2007;19:445–451. [PubMed] [Google Scholar]
- 35.Nakamura K, Sasajima J, Mizukami Y, et al. Hedgehog promotes neovascularization in pancreatic cancers by regulating Ang-1 and IGF-1 expression in bone-marrow derived pro-angiogenic cells. PLoS One. 2010;5:e8824. doi: 10.1371/journal.pone.0008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: Implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 37.Cochain C, Channon KM, Silvestre JS. Angiogenesis in the infarcted myocardium. Antioxid Redox Signal. 2013;18:1100–1113. doi: 10.1089/ars.2012.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuura K, Honda A, Nagai T, Fukushima N, Iwanaga K, Tokunaga M, Shimizu T, Okano T, Kasanuki H, Hagiwara N, Komuro I. Transplantation of cardiac progenitor cells ameliorates cardiac dysfunction after myocardial infarction in mice. J Clin Invest. 2009;119:2204–2217. doi: 10.1172/JCI37456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giordano FJ, Gerber HP, Williams SP, VanBruggen N, Bunting S, Ruiz-Lozano P, Gu Y, Nath AK, Huang Y, Hickey R, Dalton N, Peterson KL, Ross J, Jr, Chien KR, Ferrara N. A cardiac myocyte vascular endothelial growth factor paracrine pathway is required to maintain cardiac function. Proc Natl Acad Sci U S A. 2001;98:5780–5785. doi: 10.1073/pnas.091415198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Martí E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–247. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- 42.Cao D, Jin C, Ren M, Lin C, Zhang X, Zhao N. The expression of Gli3, regulated by HOXD13, may play a role in idiopathic congenital talipes equinovarus. BMC Musculoskelet Disord. 2009;10:142. doi: 10.1186/1471-2474-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Hibbs MA, Gard AL, Shylo NA, Yun K. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells. 2012;30:741–752. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, James RM, Peter A, Lomas C, Cheung F, Harrison DJ, Bader SA. Functional Smoothened is required for expression of GLI3 in colorectal carcinoma cells. Cancer Lett. 2004;207:205–214. doi: 10.1016/j.canlet.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 45.Heron-Milhavet L, Mamaeva D, LeRoith D, Lamb NJ, Fernandez A. Impaired muscle regeneration and myoblast differentiation in mice with a muscle-specific KO of IGF-IR. J Cell Physiol. 2010;225:1–6. doi: 10.1002/jcp.22218. [DOI] [PubMed] [Google Scholar]
- 46.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem. 2003;278:8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 47.Madhala-Levy D, Williams VC, Hughes SM, Reshef R, Halevy O. Cooperation between Shh and IGF-I in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/Akt pathways requires Smo activity. J Cell Physiol. 2012;227:1455–1464. doi: 10.1002/jcp.22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 49.Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.