Abstract
Although individual pseudogenes have been implicated in tumor biology, the biomedical significance and clinical relevance of pseudogene expression have not been assessed in a systematic way. Here we generate pseudogene expression profiles in 2,808 patient samples of seven cancer types from The Cancer Genome Atlas RNA-seq data using a newly developed computational pipeline. Supervised analysis reveals a significant number of pseudogenes differentially expressed among established tumor subtypes; and pseudogene expression alone can accurately classify the major histological subtypes of endometrial cancer. Across cancer types, the tumor subtypes revealed by pseudogene expression show extensive and strong concordance with the subtypes defined by other molecular data. Strikingly, in kidney cancer, the pseudogene-expression subtypes not only significantly correlate with patient survival, but also help stratify patients in combination with clinical variables. Our study highlights the potential of pseudogene expression analysis as a new paradigm for investigating cancer mechanisms and discovering prognostic biomarkers.
Introduction
Pseudogenes are dysfunctional copies of protein-coding genes that have lost their ability to encode amino acids through the accumulation of deleterious mutations such as in-frame stop codons and frame-shift insertion/deletions1. In the human genome, there are pseudogene copies for many protein-coding genes: for example, the ENCODE project recently annotated ~15,000 human pseudogenes2. Importantly, a large fraction of pseudogenes are transcriptionally active2. Despite their huge number and prevalent occurrence in the genome, pseudogenes have long been considered as nonfunctional and assumed to evolve neutrally3. In recent years, a growing body of evidence has strongly suggested that individual pseudogenes play critical roles in human diseases such as cancer4,5. For example, NANOG and OCT4 are essential transcription factors for the maintenance of pluripotency in embryonic stem cells6,7, while their pseudogenes, NANOGP1 and POU5F1P1, are aberrantly expressed in human cancers8. Poliseno et al. (2010) showed that the pseudogenes of key cancer genes (e.g., PTENP1 and KRASP1) can regulate the expression of their wild-type (WT) cognate genes by sequestering miRNAs9. More recently, Kalyana-Sundaram and colleagues (2012) performed the first genome-wide characterization of pseudogene expression in human cancers using the RNA-seq approach and revealed a considerable number of pseudogenes with a lineage- or cancer-specific expression pattern10. These studies provide key insights into the potential role of transcribed pseudogenes in tumor biology. However, due to the limited number of patient samples surveyed in previous studies, the biomedical significance of pseudogene expression in cancer cannot be fully assessed. In particular, it remains unclear whether pseudogene expression can effectively characterize the tumor heterogeneity within a specific cancer type and represent a meaningful dimension for patient stratification. Therefore, it is essential to perform a systematic analysis across large patient sample cohorts to evaluate the potential clinical utility of pseudogene expression.
Taking advantage of large-scale RNA-seq transcriptomic data recently made available from The Cancer Genome Atlas (TCGA) project, we developed a computational pipeline and characterized the pseudogene expression profiles of a large number of patient samples in a wide range of cancer types. With this unprecedented dataset, we first identified differentially expressed pseudogenes among established tumor subtypes and demonstrated the predictive power in classifying clinical tumor subtypes of endometrial cancer. Then we examined the biomedical relevance of the tumor subtypes revealed by pseudogene expression and assessed the potential clinical utility of pseudogene-expression subtypes in terms of predicting patient survival. Taken together, our results indicate that expressed pseudogenes represent an exciting paradigm for investigating cancer-related molecular mechanisms and discovering effective prognostic biomarkers.
Results
Overview of pseudogene expression in multiple cancer types
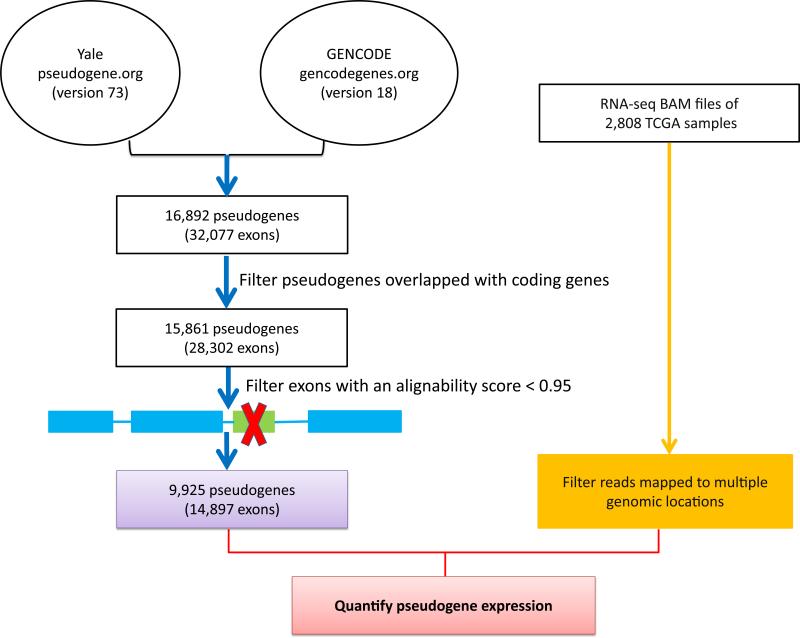
To comprehensively detect expressed pseudogenes and quantify their expression levels in human cancer, we developed a computational pipeline, as shown in Fig. 1. First, we combined the latest pseudogene annotations from the Yale Pseudogene database11 and the GENCODE Pseudogene Resource2 and filtered those pseudogenes overlapped with any known protein-coding genes. Second, to address the issue of potential cross-mapping between pseudogenes and their WT coding genes, we evaluated the sequence uniqueness of each exon of a pseudogene12, and only retained those pseudogenes containing exon(s) with sufficient alignability for further characterization (Methods). Third, we filtered those reads mapped to multiple genomic locations from TCGA BAM files. Through analyzing more than 378 billion RNA-seq reads, we measured the expression levels of 9,925 pseudogenes (based on the regions of high sequence uniqueness) in 2,808 samples of seven cancer types (Table 1). These cancer types included breast invasive carcinoma (BRCA)13, glioblastoma multiforme (GBM)14, kidney renal clear cell carcinoma (KIRC)15, lung squamous cell carcinoma (LUSC)16, ovarian serous cystadenocarcinoma (OV)17, colorectal carcinoma (CRC)18, and uterine corpus endometrioid carcinoma (UCEC)19.
Figure 1. A computational pipeline to quantify the expression of pseudogenes from TCGA RNA-seq data.
First, we combined the latest pseudogene annotations from the Yale Pseudogene database and the GENCODE Pseudogene Resource and filtered those pseudogenes that overlapped with any known protein-coding genes. Second, we evaluated the sequence uniqueness of each exon of a pseudogene, and only retained those pseudogenes containing exon(s) with sufficient alignability for further characterization. Third, we filtered those reads mapped to multiple genomic locations from TCGA BAM files.
Table 1.
Summary of TCGA RNA-seq datasets used in this study
| Cancer type | # of Nontumor samples | # of Tumor samples | Sequencing strategy | # of mappable reads | # of detectable pseudogenes |
|---|---|---|---|---|---|
| BRCA | 105 | 837 | Paired-end | 161 M | 747 |
| KIRC | 67 | 448 | Paired-end | 166 M | 712 |
| LUSC | 17 | 220 | Paired-end | 171 M | 813 |
| OV | 0 | 412 | Paired-end | 170 M | 670 |
| GBM | 0 | 154 | Paired-end | 106 M | 875 |
| CRC | 0 | 228 | Single-end | 22 M | 168 |
| UCEC | 4 | 316 | Single-end | 26 M | 181 |
Among the seven cancer types we surveyed, five datasets (BRCA, GBM, LUSC, KIRC, and OV) had been obtained through a paired-end sequencing strategy, while the other two (CRC and UCEC) had resulted from a single-end sequencing strategy. Moreover, samples in the paired-end group had many more mappable reads than those in the single-end group (Table 1, Supplementary Fig. 1). We detected more expressed pseudogenes (with an average Reads Per Kilobase per Million [RPKM]20 cutoff≥0.3, as in the literature21,22) in the paired-end group (OV: 670, KIRC 712, LUSC 813, BRCA: 747, and GBM, 875) than in the single-end group (UCEC, 649 and CRC, 741) (Table 1). Both the larger numbers of sequenced reads and the higher read mapping accuracy in the paired-end group contributed to this difference. Indeed, the two groups showed distinct global patterns of pseudogene expression (Supplementary Fig. 2). For each cancer type, we observed generally weak correlations between the expression level of pseudogenes and their WT genes, which is consistent with the previous study10 (Supplementary Fig. 3). In general, the expression correlation between a pseudogene and its WT coding gene could be affected by three factors: (i) the sequence similarity between the pseudogene/gene pair; (ii) the molecular mechanisms through which the pseudogene functions; and (iii) the detection sensitivity given the setting of RNA-seq experiments. Considering the potential confounding factors (e.g., sequencing strategy and read coverage) for quantifying the pseudogene expression, we performed the cross-tumor analyses for these two groups separately. As observed in Kalyana-Sundaram et al. (2012)10, we detected some tumor-lineage-specific pseudogenes (296 from the paired-end group and 41 from the single-end group, Supplementary Fig. 4). In addition, for three cancer types with available RNA-seq data from nontumor tissue samples, we identified differentially expressed pseudogenes between tumor and nontumor samples (54 in BRCA, 110 in KIRC and 138 in LUSC, Supplementary Fig. 5).
Supervised analysis of pseudogene expression on tumor subtypes
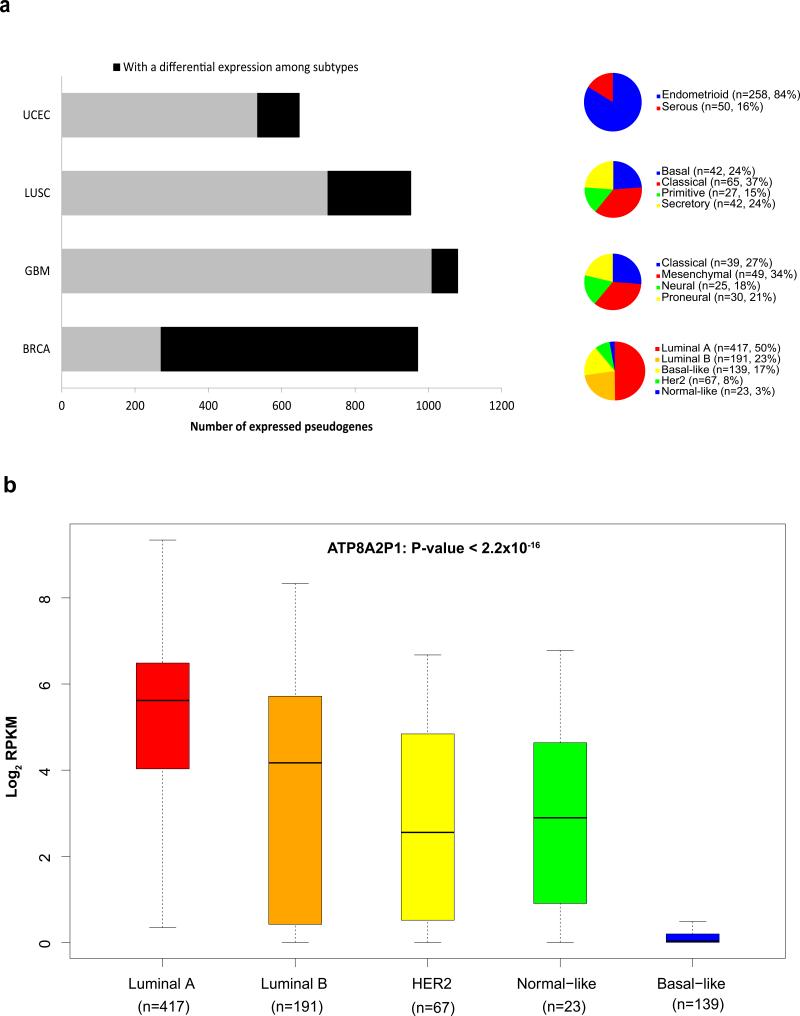
However, the tumor-lineage-specific or cancer-specific pseudogenes identified above may only reflect biological characteristics unique to distinct tissue types rather than key biological factors involved in tumorigenesis. Therefore, it is more critical and informative to examine the expression patterns of pseudogenes among tumor subtypes within a disease. For several cancer types with established tumor subtypes, we performed the supervised analysis and revealed substantial numbers of pseudogenes with significant differential expression: 48 in UCEC (endometrioid vs. serous)23, 138 in LUSC (basal, classical, primitive and secretory)16, 71 in GBM (classical, mesenchymal, neural and proneural)24 and 547 in BRCA (PAM50 subtypes: luminal A, luminal B, basal-like, Her2-enriched and normal-like)25 (Methods, Fig. 2a, Supplementary Data 1). This analysis not only reveals a large number of pseudogenes with potential biomedical significance, but also provides new insights into known oncogenic pseudogenes. For example, ATP8A2P1 has been reported to play a growth regulatory role and to be expressed in a BRCA-specific manner10. Through the analysis of the large BRCA sample cohort, we further demonstrated that this pseudogene shows significant expression variation across subtypes, with the highest level in luminal A and the lowest level in the basal-like subtype (ANOVA P < 2.2×10−16, Fig. 2b).
Figure 2. Identification of differentially expressed pseudogenes among established tumor subtypes.
(a) Numbers of significantly differentially expressed pseudogenes in multiple cancer types. For each cancer type, the whole bar represents the number of expressed pseudogenes (mean RPKM≥0.3) in the analysis; the black part represents the number of expressed pseudogenes with a detected significance for differential expression among tumor subtypes (t-test or single-factor ANOVA, corrected P < 0.05); and the pie chart shows the sample numbers and percentages in each cancer type. (b) The box plot for the expression pattern of ATP8A2P1 in 837 BRCA samples based on PAM50 subtypes: luminal A (n = 417), luminal B (n = 191), basal-like (n = 139), Her2-enriched (n = 67), and normal-like (n = 23). The boxes show the median ± 1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile range from the box boundaries.
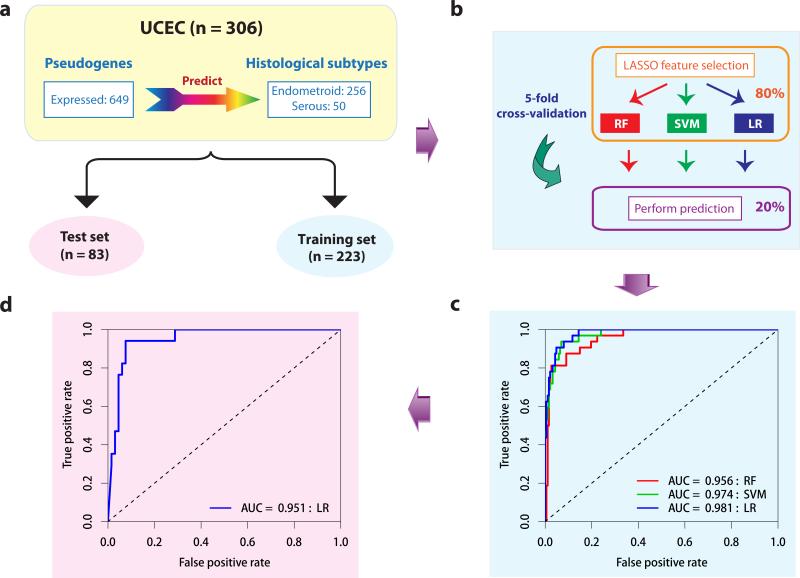
Among the tumor subtypes we surveyed, endometrioid and serous endometrial tumors are two major histological subtypes for UCEC, which are defined independently from the molecular data. Importantly, these two subtypes have distinct pathological characteristics and clinical behaviors. Early-stage endometrioid cancers are often treated with adjuvant radiotherapy, whereas serous tumors are usually treated with chemotherapy26. Therefore, subtype classification is crucial for selecting appropriate therapy. To assess the clinical utility of pseudogene expression in UCEC, we applied a rigorous machine-learning approach to assess the power of expressed pseudogenes in classifying these two subtypes. First, we divided the TCGA UCEC samples into training and test sets according to their tissue source sites (Fig. 3a). Second, within the training set, we applied three well-established machine learning algorithms (random forest [RF]27; support vector machine [SVM]28; and logistic regression [LR]) and evaluated their performance based on the area under the receiver operating characteristics curve (AUC score) through 5-fold cross validation (Methods, Fig. 3b). Strikingly, we found that the pseudogene-expression profile can accurately classify these two histological subtypes (RF, AUC score = 0.944, SVM, AUC score = 0.962, LR, AUC score = 0.892, Fig. 3c). Moreover, the best-performing algorithm, LR, achieved a high AUC of 0.922 on the independent test set (Fig. 3d), with accuracy = 0.90, positive predictive value = 0.80, and negative predictive value = 0.93. The predictive power of pseudogene expression is comparable to those achieved by the mRNA expression profiles, suggesting that both pseudogene and mRNA expression can classify the UCEC subtypes independently (Supplementary Fig. 6). These results indicate that pseudogene expression can effectively capture clinically relevant information and may provide an independent approach to validate the classification of tumor subtypes.
Figure 3. The predictive power of pseudogene expression in classification of UCEC subtypes.
(a) The UCEC dataset (n = 306) was split into training (n = 223) and test (n = 83) sets. (b) Schematic representation of feature selection and classifiers building through five-fold cross-validation within the training set. (c) The ROC curves of the three classifiers based on the cross-validation within the training set. (d) The ROC curve from applying the best-performing classifier (LR) built from the whole training set to the test set. (RF: random forest, SVM: support vector machine, LR: logistic regression.)
Assessment of pseudogene expression tumor subtypes
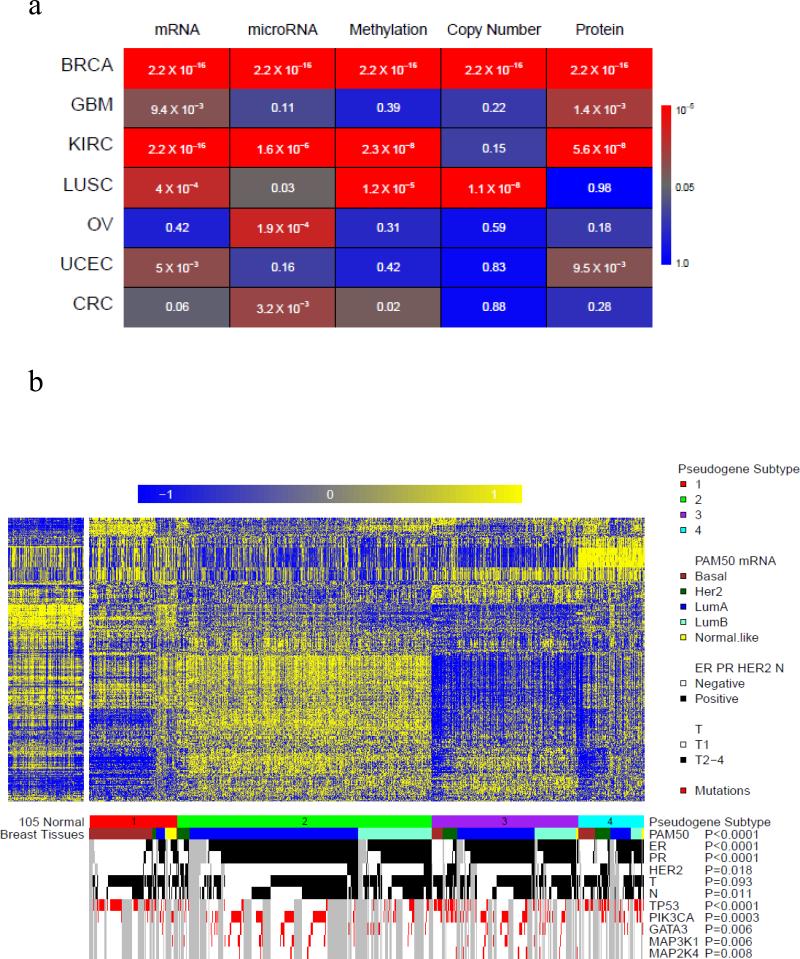
Cancer is a complex disease involving multiple layers of aberrations that cannot be sufficiently captured by any single type of molecular data. In recent years, various “omic” data, such as mRNA expression, microRNA expression, DNA methylation, somatic copy number alteration, and protein expression, have been widely used to classify tumor samples into different molecular subtypes13-19. The integrative analysis across these molecular subtypes, especially through the efforts in TCGA, often provide crucial insights into pathobiology and help stratify patients for predicting prognosis and selecting effective treatment. To complement the supervised analysis in the above section, we next performed unsupervised analyses and explored the biomedical relevance of tumor subtypes based on pseudogene-expression profiles. For each cancer type, we selected the pseudogenes with the most variable expression (500 for each cancer in the paired-end group and 100 for each cancer in the single-end group, respectively) and used non-negative matrix factorization (NMF)29 to classify tumor samples into subtypes (clusters). Strikingly, in multiple cancer types, we observed that subtypes based on pseudogene expression had high concordance with other molecular subtypes (Fig. 4a, chi-squared tests).
Figure 4. Correlations of pseudogene expression subtypes with other tumor subtypes.
(a) Concordance between pseudogene expression subtypes and molecular subtypes defined by other genomic data in seven TCGA cancer types. Pseudogene-expression subtypes were defined based on the expression of 500 or 100 pseudogenes with the most variable patterns through unsupervised analysis using non-negative matrix factorization (NMF)29. The colors indicate the statistical significance of the chi-squared tests for assessing the concordance between the pseudogene-expression subtypes and other molecular subtypes. (b) Concordance between pseudogene expression subtypes and other subtypes in BRCA. Pseudogene expression: subtype 1, red (n = 144); subtype 2, green (n = 390); and subtype 3, purple (n = 303). PAM50 subtypes: basal-like (brown), HER2-enriched (dark green), luminal A (blue), luminal B (aquamarine), and normal-like (yellow). The status of ER, PR, HER2 or N is marked in black (positive) and white (negative); T status is marked in black (T2-T4) and white (T1). Mutations of TP53, PIK3CA, GATA3, MAP3K1, and MAP2K4 are marked in red. Correlations were assessed by chi-squared tests.
Here, we present breast cancer as an example (Fig. 4b). Based on the NMF consensus clustering, 837 BRCA samples can be classified into three distinct subtypes: subtype 1 (n = 144), subtype 2 (n = 390), and subtype 3 (n = 303) (Fig. 4b, Supplementary Data 2). These pseudogene subtypes show high concordance with the well-established PAM50 molecular subtypes25 and the status of ER/PR/HER2 markers (chi-squared test, Fig. 4b). Subtype 1 is significantly enriched for basal-like samples, containing 70 of 139 basal-like samples; subtype 2 is enriched for luminal A and luminal B samples that 382 of 390 samples are these two subtypes; subtype 3 is enriched for Her2 samples, containing 50 of 67 HER2 samples. The pseudogene expression subtypes also correlate with the mutation status of key cancer genes13: subtype 1 shows a depletion of GATA3 mutations; and subtype 2 has many samples of TP53 mutations. These results strongly indicate that pseudogene expression represents a novel and relevant dimension for investigating cancer-related molecular mechanisms; and integrating it with other molecular data related analysis may help characterize the molecular basis of tumorigenesis in a more comprehensive way.
Prognostic power of pseudogene expression in kidney cancer
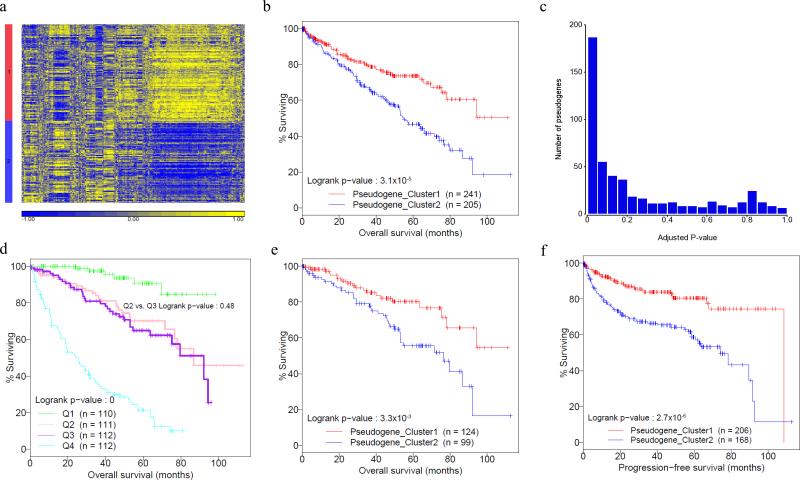
To study the potential clinical value of pseudogene-expression, we examined whether the pseudogene subtypes correlate with clinical outcomes in KIRC. Currently, neither prognostic nor predictive markers are recommended for clinical use by the College of American Pathologists. Based on the 500 pseudogenes with the most variable expression, we were able to classify 446 KIRC samples into two distinct subtypes (Fig. 5a, Supplementary Data 3). Tumor samples in subtype 1 convey a much better patient prognosis (n = 234, survival time of 75.8 ± 3.7 months) than those in subtype 2 (n = 212, survival time of 63.1 ± 3.7 months) (Fig. 5b, log-rank test P = 0.019). To assess whether individual pseudogenes can confer prognostic power given clinical variables, for each pseudogene, we built the full multivariate Cox model, consisting of both clinical variables and the pseudogene expression. We observed an enrichment of pseudogenes (115 out of 500) with a statistically significant P-value (FDR < 0.05) (Fig. 5c, Supplementary Data 4). Noteworthy, among the 115 pseudogenes, only 19 (16.5%) showed relatively high expression correlations (Spearman correlation ≥ 0.5) with their WT genes, suggesting that the predictive power of pseudogene expression is largely independent of the corresponding WT genes.
Figure 5. Prognostic value of pseudogene expression in KIRC.
(a) KIRC subtypes are classified based on the expression of 500 pseudogenes with the most variable patterns through unsupervised analysis using non-negative matrix factorization (NMF, n = 446). (b) Kaplan-Meier plot showing correlations of the two pseudogene expression subtypes with overall survival (log-rank test P = 0.019). Red denotes pseudogene expression subtype 1 (n = 241); blue denotes pseudogene-expression subtype 2 (n = 205). (c) P-value distribution of individual pseudogene expressions in multivariate Cox proportional hazards model containing clinical variables. (d) Kaplan-Meier plot of the four risk groups defined by clinical variables in terms of overall survival, and the two middle risk groups cannot be separated (Q2 [n = 111] vs. Q3 [n =112], log-rank test P = 0.48). (e) Kaplan-Meier plot showing that the two pseudogene expression subtypes can effectively separate the samples in the two medium risk groups in terms of overall survival (Q2 [n = 113] vs. Q3 [n = 110], log-rank test P = 9.6×10−3).
To further assess the clinical utility of the observed pseudogene-expression subtypes, we classified the KIRC samples into four risk quartiles based on the risk scores (in terms of overall survival) calculated from the multivariate Cox model, employing only clinical variables: low risk group (Q1, n = 110), low-medium risk group (Q2, n = 111), medium-high risk group (Q3, n = 112), and high risk group (Q4, n = 112) (Methods, Supplementary Data 3). Although the survival curves of these four risk groups are significantly separated (Fig. 5d, log-rank test P = 0), the clinical variables actually fail to separate the two medium risk groups (Fig. 5d, Q2 vs. Q3, log-rank test P = 0.48). In contrast, the samples in these two groups can be well separated based on the pseudogene-expression subtypes (Fig. 5e, log-rank test P = 9.6×10−3). For comparison, we performed the same analysis on the two medium-risk groups (Q2 and Q3) using the subtypes defined by mRNA and microRNA expression (obtained from TCGA KIRC Analysis Working Group15) or other molecular data (obtained from TCGA Pan-Cancer Analysis Working Group) and observed no significant correlations with overall survival (log-rank test, mRNA expression, P = 0.84; microRNA expression, P = 0.13; DNA methylation, P = 0.44; somatic copy number alteration, P = 0.77; and protein expression, P = 0.14). The robust results in the above survival data analyses underscore the potential prognostic value of pseudogene expression in KIRC.
Although they do not generate functional protein products, pseudogenes may act as regulatory RNAs and affect the expression of coding genes through multiple mechanisms5. To gain some mechanistic insight into how expressed pseudogenes contribute to the observed KIRC pseudogene-expression subtypes, we performed a systematic analysis (Supplementary Fig. 9a and Supplementary Data 5). Among 102 expressed pseudogenes without a clear WT cognate gene, 44 pseudogenes showed a significant differential expression between the two subtypes (t-test, corrected P < 0.05, fold change > 1.5), with potential function as lncRNAs5. For those pseudogenes with a WT cognate gene, 93 pairs of pseudogenes and their WT genes showed a significant differential expression between the two subtypes (t-test, corrected P < 0.05). Among them, 64 showed strong positive correlations (Rs ≥ 0.3), suggesting that they may regulate their WT counterparts through competing for shared regulatory RNAs5,30; while 4 showed strong negative correlations with their WT cognate genes (Rs ≤ -0.3), suggesting that they may function as antisense transcripts to inhibit the WT-gene expression. Further analyses on independent, strand-specific RNA-seq data would provide more insights into these mechanisms. Among the WT cognate genes with strong positive correlations with their pseudogenes, we noticed that the survival correlations of individual WT genes with prognostic value match the survival pattern of the pseudogene-expression subtypes: WT genes with better prognosis (potentially tumor suppressors, hazard ratio < 1) show higher expression levels in subtype 1 (the better survival group) and the genes with worse prognosis (potentially oncogenes, hazard ratio > 1) show higher expression levels in subtype 2 (the worse survival group, Supplementary Fig. 8b). Finally, we examined the classic miRNA decoy model as proposed in Poliseno et al. (2010)9 and identified 38 such candidates (Methods and Supplementary Data 5). One candidate of interest is the potential regulation of a putative tumor suppressor α-catenin (CTNNA1) by the pseudogene PGOHUM00000257111 through competition for up to 9 shared miRNA regulators (Supplementary Data 5). Indeed, the expression levels of PGOHUM00000257111 were significantly higher in cluster 1 (t-test P = 1.48×10-7), which may lead to the elevated levels of CTNNA1 in subtype 1 (Supplementary Fig. 8b) and therefore better survival.
Discussion
Recently, pseudogenes have emerged as new players in tumor biology5,10. However, a crucial question remains unclear: does pseudogene expression, as a whole, represent a biologically meaningful dimension that can characterize tumor heterogeneity and provide clinical applications? Here, we performed a pan-cancer analysis of pseudogene expression for what is, to our knowledge, the largest number of cancer patient samples (~3,000) in one such analysis. Utilizing TCGA patient cohorts with a sufficient sample size, we show the predictive power of pseudogene expression in classifying established tumor types and the high concordance of tumor subtypes based on pseudogene expression with other molecular subtypes as well as clinically established biomarkers (such as ER and PR status in breast cancer). It should be emphasized that a large number of tumor-lineage-specific pseudogenes identified through between-disease comparisons10 do not imply our findings through the within-disease analyses. Because many tumor-lineage or cancer-specific pseudogenes could arise from tissue-related rather than tumorigenesis-related effects, they may or may not have the power to differentiate tumor subtypes.
Strikingly, our analysis reveals an unexpected prognostic power of pseudogene expression in kidney cancer: pseudogene-expression subtypes not only correlate with patient survival but also confer additional prognostic powers for a group of patients whose survival times cannot be well predicted based on conventional clinical variables. This finding implies a novel prognostic strategy that incorporates both the risk scores defined by the clinical-variable model and the tumor subtypes revealed by pseudogene expression (subtype 1 and subtype 2): among medium-risk patients, patients of subtype 2 may benefit from earlier, more aggressive therapies. Interestingly, although the tumor subtypes defined by other molecular data (e.g., mRNA and miRNA) show high concordance with the pseudogene-expression subtypes based on the whole patient cohort, they do not confer additional prognostic power based on the medium-risk patient subset. These aggregate results provide a strong rationale for further investigation of the clinical utility of pseudogene expression, which has been understudied in the field. Since TCGA patient samples were collected for the purpose of comprehensive molecular profiling and were collected from different institutions, this practice might introduce some bias. In addition, the resulting clinical annotation of patient samples and related records may not be as rigorous and complete as those obtained from standard clinical trials. Therefore, further efforts should be made to validate the clinical utility of pseudogene expression in a more formal clinical setting (e.g., clinical trials).
While our study primarily focused on the biomedical significance and clinical relevance of pseudogene expression as a whole (i.e., the subtypes that collectively represent the information of many pseudogenes), an intriguing question is how individual pseudogenes are functionally involved in tumorigenesis. This is a challenging but exciting topic since pseudogenes may affect their WT coding genes or unrelated genes through multiple mechanisms such as microRNA decoys and antisense transcripts. From a systems biology point of view, the informative behavior of pseudogenes may originate from a role such as “regulator.” Our preliminary analysis here revealed some candidates of potential interest. Further efforts are required to elucidate how these pseudogenes functionally contribute to tumor initiation and development and how they are regulated through the complex gene regulatory network.
Methods
Pseudogene expression quantification
We downloaded RNA-seq BAM files of 2,808 samples (only primary tumor samples) in seven TCGA cancer types and their related normal tissue samples (if available) from UCSC Cancer Genomics Hub on January, 2013 (CGHub, https://cghub.ucsc.edu/). TCGA BAM files were generated based on Mapsplice2 algorithm32 for alignment against the hg19 reference genome using default parameters. We further filtered the reads mapped with multiple locations in BAM files. To perform a comprehensive survey of pseudogenes, we obtained the genomic information of 16,892 human pseudogenes through combining the latest pseudogene annotations from the Yale Pseudogene Database (build 73)11 and the GENCODE Pseudogene Resource (version 18)2. We further filtered those pseudogenes that overlapped with any known coding genes. To address the potential cross-mapping issue, we calculated the alignability score12 for each pseudogene exon. Alignability provides a measure of how often the sequence at a given location will align within the whole genome (up to 2 mismatches). For each 75-mer window, an alignability score S was defined as 1/(number of matches found in the genome): S = 1 means one match in the genome, S = 0.5 for two matches in the genome, and so on12. To count mapped reads for a pseudogene, we only retained those exons with an average alignability score S ≥ 0.95 to ensure mapping accuracy; and quantified pseudogene expression as RPKM20. The pseudogenes with detectable expression were defined as those with an average RPKM ≥ 1 across all samples in each cancer type, as used in the literature21,22. We then log-transformed the RPKM values for further analysis. We used Spearman rank correlations to assess the coexpression patterns between pseudogenes and their WT cognate genes. The pseudogene expression data have been deposited into Synapse (https://www.synapse.org/) with ID syn1732077.
Supervised analysis of expressed pseudogenes
To identify tumor-lineage-specific/cancer-specific pseudogenes, or those differentially expressed among established molecular or histological subtypes, we used analysis of variance (ANOVA) or student t-test to detect the statistical difference between two or more groups. To correct for multiple comparisons, we used the Bonferroni method, with a corrected P-value cutoff of 0.05.
To assess the predictive power of pseudogene expression for two UCEC histological subtypes (endometrioid vs. serous), we divided the samples into training and test sets according to the institutions where the samples were collected. Adapted from Yuan et al. 33, we applied three well-established machine learning algorithms (random forest [RF]27, support vector machine [SVM]28 and logistic regression [LR]) to predict the subtype (as a binary variable) using the log-transformed expression levels of pseudogenes (or mRNA) as candidate features. We evaluated the performance of classifiers through 5-fold cross validation within the training set. In detail, we randomly divided the training set into five equal portions; then, during each of the five iterations, we first applied the least absolute shrinkage and selector operator (LASSO34) as the feature selection method on 4/5 of the training data and trained the classifiers (1000 trees for RF, radial kernel for SVM, other parameters set by default) with the selected features. Next, we applied the trained classifiers to the remaining 1/5 of the training data for prediction. The predictions from all five iterations were then combined and compared with the truth, based on which a ROC curve was drawn35 and the AUC score, were calculated accordingly. Finally, we performed feature selection (Supplementary Data 6) and built the classifier from the whole training set using the best-performing algorithm (with the highest AUC) identified through the cross-validation, and applied it to the test set in order to independently validate the predictive power.
Analysis of tumor subtypes revealed by pseudogene expression
To classify tumor subtypes based on pseudogene expression, for each cancer type, we selected the 500 pseudogenes with the most variable expression pattern, used NMF to classify the tumor samples into clusters29, and then used the cophenetic correlation to select the optimized clusters. To perform an objective assessment, we obtained independently defined molecular subtypes by other genomic data from TCGA marker papers13-19 whenever possible; and if not, then from TCGA Pan-Cancer Analysis Working Group (through a similar NMF-based unsupervised analysis) (syn1688309 for microRNA expression, syn1701558 for DNA methylation, and syn1682511 for mRNA expression, copy number variation, and protein expression [Reverse phase protein array]36). All related subtype classifications and method details are publically available at Synapse37. To assess correlations among the subtypes, we used the chi-squared test or Fisher's exact test, as applicable, and considered P < 0.05 to be statistically significant.
KIRC patient survival analysis
We obtained the clinical information associated with the KIRC samples, including the patient's overall survival time, age, and the tumor grade, and stage from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). We compiled progression-free survival (PFS) data based on TCGA clinical follow-up records. In this study, we defined PFS as the interval from the date of treatment to the date of an event (disease progression or recurrence, or new tumor diagnosis), or the date of last follow-up or decease if none of the events listed above occurred before that date. We used a log-rank test to examine whether the subtypes significantly correlated with patient survival, and a multivariate Cox proportional hazards model to assess whether the subtype provided additional prognostic power, given the clinical variables; to correct for multiple comparisons, we used the Benjamini-Hochberg method38, with an adjusted FDR cutoff of 0.05. To calculate the risk score for patients, we first built a Cox proportional hazard model by fitting the clinical variables (i.e., patient age, cancer stage and grade) with the censored survival data, and then plugged the original clinical variables back into the obtained model (i.e., the regression function) to calculate the linear predictor or the risk score for each patient. Patients were classified into quartiles grouped by the risk scores (which are essentially continuous values). To display the difference between groups, we used Kaplan-Meier plots, presenting the average survival time as the means ± s.e.m., for which we estimated the mean survival time as the area under the survival curve39.
Mechanistic analysis of pseudogene driven regulation
We downloaded KIRC mRNA expression and miRNA expression from Synapse (syn300013), and used ANOVA (Bonferroni corrected P < 0.05) to identify differentially expressed pseudogenes or mRNAs among the subtypes. We used Spearman rank correlations to assess the expression patterns between a pseudogene and its WT cognate genes: Rs ≥ 0.3 (or ≤ -0.3) were considered as strong positive (or negative) correlation. To identify candidates for the miRNA-decoy model, we obtained the predicted conserved target sites from MicroRNA.org40 and used the following criteria: (i) the expression levels of a pseudogene and its WT cognate genes were strongly positively correlated (Rs ≥ 0.3); (ii) its WT cognate gene showed a significant negative correlation with the miRNA of interest (FDR < 0.05) and contained predicted target sites in its 3’ UTR; and (iii) the pseudogene showed a significant negative correlation with the expression of the same miRNA (FDR < 0.05).
Supplementary Material
Acknowledgements
We gratefully acknowledge contributions from the TCGA Research Network and its TCGA Pan-Cancer Analysis Working Group (contributing consortium members are listed in Supplementary Note 1). The TCGA Pan-Cancer Analysis Working Group is coordinated by J.M. Stuart, C. Sander and I. Shmulevich. This study was supported by the National Institutes of Health (CA143883 and CA016672 to H.L.); NIH/UTMDACC Uterine SPORE Career Development Award and the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine to H.L. We thank MD Anderson high performance computing core facility for computing resources and LeeAnn Chastain for editorial assistance.
Footnotes
Author Contribution
H.L. conceived of and supervised the project. L.H. and H.L. designed and performed the research. Y.Yuan performed survival and subtype prediction analysis. S.Z., Y. Yang., J.L., M.E., L.D., Y.X., R.W. contributed to the data analysis. L.H., Y.Yuan and H.L. wrote the manuscript with input from all other authors.
References
- 1.Balakirev ES, Ayala FJ. Pseudogenes: are they “junk” or functional DNA? Annu. Rev. Genet. 2003;37:123–51. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 2.Pei B, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li WH, Gojobori T, Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–9. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- 4.Pink RC, et al. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–8. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poliseno L. Pseudogenes: newly discovered players in human cancer. Sci. Signal. 2012;5:re5. doi: 10.1126/scisignal.2002858. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Cantz T, et al. Absence of OCT4 expression in somatic tumor cell lines. Stem cells. 2008;26:692–7. doi: 10.1634/stemcells.2007-0657. [DOI] [PubMed] [Google Scholar]
- 9.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalyana-Sundaram S, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149:1622–34. doi: 10.1016/j.cell.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karro JE, et al. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 2007;35:D55–60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derrien T, et al. Fast computation and applications of genome mappability. PLoS One. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Cancer Genome Atlas Research Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Gan QA, et al. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiita AP, et al. Global cellular response to chemotherapy-induced apoptosis. Elife. 2013;2:e01236. doi: 10.7554/eLife.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lax SF, Kurman RJ. A dualistic model for endometrial carcinogenesis based on immunohistochemical and molecular genetic analyses. Verh Dtsch Ges Pathol. 1997;81:228–32. [PubMed] [Google Scholar]
- 24.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8:261–71. doi: 10.1038/nrclinonc.2010.216. [DOI] [PubMed] [Google Scholar]
- 27.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 28.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 29.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 32.Wang K, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan Y, Xu Y, Xu J, Ball RL, Liang H. Predicting the lethal phenotype of the knockout mouse by integrating comprehensive genomic data. Bioinformatics. 2012;28:1246–1252. doi: 10.1093/bioinformatics/bts120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996:267–288. [Google Scholar]
- 35.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 36.Li J, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–7. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Omberg L, et al. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nat Genet. 2013;45:1121–1126. doi: 10.1038/ng.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 39.Chen D, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 2012;18:1511–7. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.