Abstract
Gold nanoparticles functionalized with oligonucleotides that bear a cholesterol group are used as gene sensors. The hydrophobic molecule is buried inside the nanostructure but when the complementary RNA sequence is present the structure unfolds exposing the cholesterol group to the water. This rearrangement leads to the aggregation of the nanostructures.
Modified gold nanoparticles (AuNPs) have multiple applications in biology or medicine such as delivery systems of biological active materials or as focal enhancers of photothermal therapy in cancer cells.1 Among the most remarkable applications is the use of AuNPs as sensors of different materials,2 such as metals,3 cells,4 proteins,5 nucleic acids6 or the pH level.7 There have been reported AuNP sensors based on electrochemistry,8 as well as, electroluminescence9 or surface enhanced Raman scattering (SERS).10
Most of AuNPs sensors utilize colour changes of the solution, which can be modulated by the aggregation of the nanoparticles. When AuNPs are in colloidal dispersion the colour of the solution is reddish, changing to bluish upon their aggregation.
The selectivity of the sensor is achieved by modifying AuNPs with structures, such as antibodies,11 peptides,12 nucleic acids13 or small molecules,14 that are able to recognize corresponding targets. In the presence of the matching ligand, the AuNP sensor aggregates, resulting in the observed colour change.
Other sensors are based on the fluorescence quenching properties of AuNPs. In this case, the fluorescence of organic dyes is modulated by the proximity to the nanoparticle. The interaction with the target induces a conformational change of the structure enabling the detection of a fluorescent signal. This strategy has been employed in the detection of DNA sequences where AuNPs are modified with fluorescent Molecular Beacons.15 Due to the hairpin structure of the Molecular Beacon the fluorescent molecule is close to the nanoparticle in the absence of the target sequence. In the presence of the complementary DNA or RNA strand the Molecular Beacon undergoes a structural change placing the fluorescent molecule away from the nanoparticle unquenching the fluorescent signal.
Herein we report a gene sensor using AuNPs modified with Molecular Beacon-like structures that bear a cholesterol derivative. In the absence of the target sequence the cholesterol molecules are buried inside the nanostructure, stabilizing the nanostructure in aqueous media. In the presence of the target sequence the hairpin unfolds exposing the cholesterol units to the water molecules (Scheme 1). Hydrophobic molecules have been employed to induce the aggregation of lipid particles,16 but in this case we have used a cholesterol derivative to build a gene sensor using AuNPs modified with oligonucleotides.
Scheme 1.
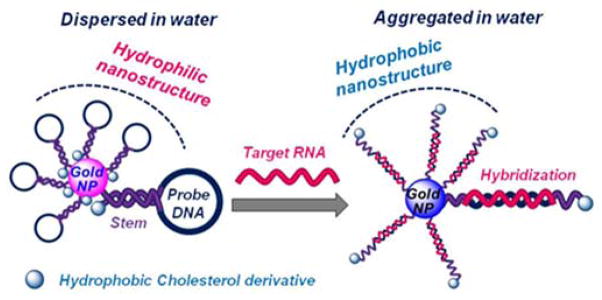
Schematic representation of the AuNP sensor with hydrophobic Molecular Beacons.
We are able to show a significant change in the solubility of our nanoparticles when the cholesterol moiety is exposed to the water molecules. This alteration can be seen by naked eye since the particles precipitate in the presence of the target sequence, reducing the colour intensity of the solution. Related gene sensors based on the aggregation of oligonucleotides covalently attached to AuNPs require the use of two sets of modified AuNPs to promote the aggregation.17 In our case, only one system is needed.
To evaluate this approach we have selected three target genes in which specific single-point mutations are involved in different diseases: GNAQ, PDE6B and KRAS. GNAQ mutations are present in over 50% of uveal melanomas,18 mutant PDE6B is a gene responsible for retinitis pigmentosa (RP) and KRAS is frequently mutant in pancreatic cancer, among other malignant diseases.19
AuNPs were prepared following the established procedure developed by Turkevich, which yields nanoparticles of 13 nm size in aqueous solution.20 Then, the nanoparticles were functionalized with modified oligonucleotides to yield the final sensor. Oligonucleotides used were designed to target the selected genes and were prepared with two modifications, 1) a cholesterol derivative at the 5′-end and 2) a dithiolane derivative at the 3′-end. The cholesterol group is used to modulate the hydrophobic properties of the nanoparticle in the presence of the target gene. The dithiolane moiety allows the functionalization of AuNPs due to the high affinity of sulfur to gold. This group has several advantages compared with the standard thiol groups previously used in the preparation of modified oligonucleotides. First, deprotection of dithiolane group is not requiered and oligonucleotides can be used after standard synthesis and deprotection. Second, binding is fast and modified AuNPs are more stable compared to AuNPs bearing thiol modified oligonucleotides. We have prepared a modified solid support to introduce this group at the 3′-end of oligonucleotides (ESI†).
The required oligonucleotides were prepared in a MerMade4 DNA synthesizer using the modified CPG, and commercial phosphoramidites, including the cholesterol molecule. The oligonucleotide strands were purified by polyacrylamide gel electrophoresis and characterized by MALDI-TOF. These oligonucleotides were incubated with AuNPs overnight and modified nanostructures were purified by iterative centrifugation. Modified AuNPs were obtained as a well dispersed and very stable solution in water. Particles can be stored at −20 over months without loss of their properties.
Next, modified AuNPs were tested for their ability to detect the complementary oligonucleotide. A solution of the modified gold nanoparticles was incubated with different oligonucleotides: 1) the corresponding target, 2) a scramble and 3) a mismatched sequence. Results were compared to evaluate the selectivity of modified AuNPs for recognizing the target oligonucleotide.
Optimization of the experimental conditions by adjusting the amount of NaCl as well as the incubation time allowed us to detect single-point mutations at 32 nM in the case of KRAS and at 3.2 nM in the case of PDE6B and GNAQ (ESI†). The best results were obtained with the sensors designed to detect GNAQ. In this case, two different sets of sensors were able to detect the desired mutation. One set was designed to detect the wild-type sequence (GQ-WT) and another to detect the mutated one (GQ-MUT).
In our experiments we did not observe a change in color from reddish to bluish, as we expected from reported sensing systems that involve gold nanoparticles. However, we noticed a decrease of the saturation of the reddish color and the formation of insoluble larger particles (ESI†). The precipitate is induced in the presence of the target oligonucleotide, which opens the hairpin structure exposing the cholesterol moiety to the water. To evaluate the role of the cholesterol group in the AuNP sensor a derivative without the cholesterol moiety was prepared. Incubation of this structure with its target sequence did not yield changes in absorbance (ESI†), highlighting the key role of the cholesterol moiety in the sensor.
Our aim for the next round of experiments was to detect mutations in total mRNA extracts from different cell lines. Due to the excellent results obtained with the GNAQ sensor we focused on this gene and we used the following cell lines: OMM1.3, a uveal melanoma cell line bearing GNAQ mutation (626A>C; Q209P), C918 a uveal melanoma cell line expressing wild type GNAQ and RNA extracts from mouse liver that do not contain the GNAQ gene.
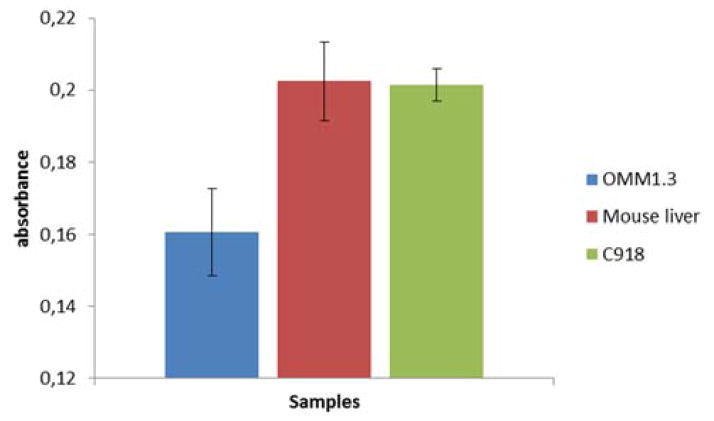
After the incubation of AuNPs designed to detect wild type GNAQ with total mRNA extracts of OMM1.3, C918 and mouse liver we were able to observe differences in absorbance indicating the detection of the wild type sequence. In Figure 1 a drop in absorbance is observed when the wild type sequence is present. The decrease in absorbance is less intense for extracts that contain mutant GNAQ.
Figure 1.
Detection of wt gene using modified AuNPs and total mRNA extracts (5.5 ng/μL) of different cell lines. C918 expresses wild type GNAQ, OMM1.3 expresses mutated GNAQ and mRNA from mouse liver that do not contain the GNAQ gene. The nanoparticles are designed to detect the wt GNAQ gene present in the C918 cell line. The drop in absorbance in the blue bar is due to the presence of the wt GNAQ gene. Values obtained as an average of three experiments.
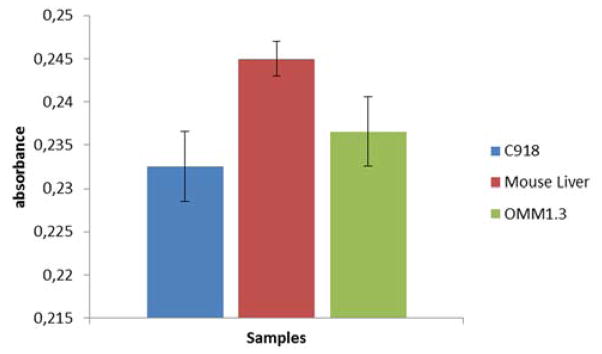
Nanoparticles designed to detect the single-point mutation in GNAQ showed even better results. Absorbance only dropped when cell extracts from the mutant OMM1.3 cells were used (Figure. 2). In this case, the mutation can be detected even in the presence of a large amount of the wild-type gene (ESI†).
Figure 2.
Detection of mutated gene using modified AuNPs and total mRNA extracts (5.5 ng/μL) of different cell lines. C918 expresses wild type GNAQ, OMM1.3 expresses mutated GNAQ and mRNA from mouse liver that do not contain the GNAQ gene. The nanoparticles are designed to detect the mutated GNAQ gene present in the OMM1.3 cell line. The drop in absorbance in the blue bar is due to the presence of the mutated GNAQ gene. Values obtained as an average of three experiments.
The detection of the mutant sequence could be observed with the naked eye. As shown in Figure 3, the color of the solution in tube A containing extracts from GNAQ mutant cells is less intense compared to tube B and C that hold extracts that do not contain GNAQ and wild type GNAQ respectively. In addition, a higher amount of aggregates could be detected in tube A.
Figure 3.
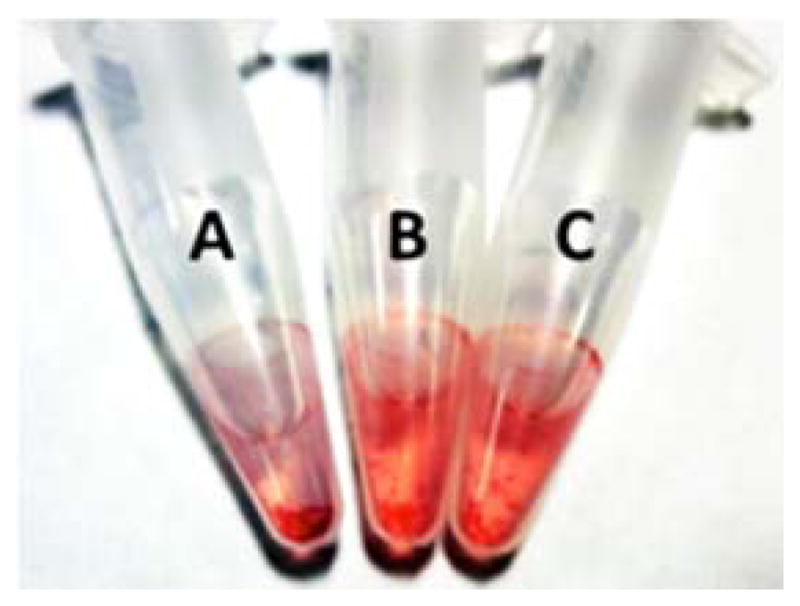
The AuNP sensor for mutant GNAQ allows the detection of the mutation with the naked eye. A: Extract from OMM1.3 cell line where the mutated gene is expressed. B: Extract from mouse liver where the gene is not expressed. C: Extracts from C918 cell line where the wild type gene is expressed. The total mRNA amount employed was (5.5 ng/μL).
Conclusions
In summary, we present herein a new sensor for the detection of specific DNA and RNA sequences, which does not require the use of any additional equipment to confirm the presence of a gene. The read-out of the detection can be done with the naked eye due to changes in the intensity and saturation in the colour of the solution. We want to highlight that the sensor is able to detect single-point-mutations at concentrations as low as 3.2 nM of the target sequence. Furthermore, the sensor was able to detect specific mutations in total mRNA cell extracts at low concentration without the need of prior amplification.
Supplementary Material
Acknowledgments
The authors are grateful to Timothy Dattels for his generous support. The authors further thank Boris Bastian for providing the cell lines used in this study. This work has been supported by NIH (1K08CA155035-01A1), the Dermatology Foundation, the American Skin Association, the American Cancer Association, the Max Kade Foundation, the Spanish Ministry of Economy and competitiveness (grant SAF2010-15440) and IMDEA Nanociencia. This work has further been supported by the Vereinfür Dermatologie Krankenanstalt Rudolfstiftung and by a grant from Family Honzak, Vienna.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/c000000x/
Notes and references
- 1.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew Chem. 2010;122:3352–3366. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; b) Saha K, Agasti SS, Kim C, Li X, Rotello VM. Chem Rev (Washington, DC, U S) 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JS, Han MS, Mirkin CA. Angew Chem. 2007;119:4171–4174. [Google Scholar]; Angew Chem, Int Ed. 2007;46:4093–2768. [Google Scholar]
- 4.Lu W, Arumugam SR, Senapati D, Singh AK, Arbneshi T, Khan SA, Yu H, Ray PC. ACS Nano. 2010;4:1739–1749. doi: 10.1021/nn901742q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu M, Jia C, Huang Y, Lou X, Yao S, Jin Q, Zhao J, Xiang J. Analyst. 2010;135:327–31. doi: 10.1039/b916629g. [DOI] [PubMed] [Google Scholar]
- 6.a) Mirkin MC, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]; b) Xia F, Zuo XL, Yang RQ, Xiao Y, Kang D, Vallee-Belisle A, Gong X, Yuen JD, Hsu BBY, Heeger AJ, Plaxco KW. Proc Natl Acad Sci USA. 2010;107:10837–10841. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu XY, Cheng F, Liu Y, Li WG, Chen Y, Pan H, Liu HJ. J Mater Chem. 2010;20:278–284. [Google Scholar]
- 8.Mani V, Chikkaveeraiah BV, Patel V, Gutkind ḰJS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jie GF, Liu P, Zhang SS. Chem Commun. 2010;46:1323–1325. doi: 10.1039/b919186k. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Puebla R, Liz-Marzán LM. Small. 2010;6:604–610. doi: 10.1002/smll.200901820. [DOI] [PubMed] [Google Scholar]
- 11.Lou S, Ye J, Li K, Wu A. Analyst. 2012;137:1174–1181. doi: 10.1039/c2an15844b. [DOI] [PubMed] [Google Scholar]
- 12.Hosta-Rigau L, Olmedo I, Arbiol J, Cruz LJ, Kogan MJ, Albericio F. Bioconjugate Chem. 2010;21:1070–1078. doi: 10.1021/bc1000164. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Wu Z, Zhou G, He Z, Zhou X, Shen A, Hu J. Chem Commun (Cambridge, UK) 2012;48:3164–3166. doi: 10.1039/c2cc16741g. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Tian Y, Cui Y, Liu W, Ma W, Jiang X. J Am Chem Soc. 2010;132:12349–12356. doi: 10.1021/ja1028843. [DOI] [PubMed] [Google Scholar]
- 15.a) Tu Y, Wu P, Zhang H, Cai C. Chem Commun. 2012;48:10718–10720. doi: 10.1039/c2cc35564g. [DOI] [PubMed] [Google Scholar]; b) Qiao G, Gao Y, Li N, Yu Z, Zhuo L, Tang B. Chem--Eur J. 2011;17:11210–11215. doi: 10.1002/chem.201100658. [DOI] [PubMed] [Google Scholar]
- 16.Lovell JF, Jin H, Ng KK, Zheng G. Angew Chem. 2010;122:8089–8091. doi: 10.1002/anie.201003846. [DOI] [PubMed] [Google Scholar]; Angew Chem, Int Ed. 2010;49:7917–7919. doi: 10.1002/anie.201003846. [DOI] [PubMed] [Google Scholar]
- 17.a) Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]; b) Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 18.Wu X, Zhu M, Fletcher JA, Giobbie-Hurder A, Hodi FS. PloS one. 2012;7:e29622. doi: 10.1371/journal.pone.0029622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colicelli J. Sci STKE. 2004:re13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turkevich J, Stevenson PC, Hillier J. Discuss Faraday Soc. 1951;11:55–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.