Summary
The bacterial spore can rapidly convert from a dormant to a fully active cell. Here we study this remarkable cellular transition in Bacillus subtilis and reveal the identity of the newly synthesized proteins throughout spore revival. Our analysis uncovers a highly ordered developmental program that correlates with the spore morphological changes and reveals the spatial and temporal molecular events fundamental to reconstruct a cell. As opposed to current knowledge, we found that translation takes place during the earliest revival event, termed germination, a process hitherto considered to occur without the need for any macromolecule synthesis. Furthermore, we demonstrate that translation is required for execution of germination and relies on the bona fide translational factors RpmE and Tig. Our study sheds light on the spore revival process and on the vital building blocks underlying cellular awakening, thereby paving the way for designing new antimicrobial agents to eradicate spore-forming pathogens.
Graphical Abstract
Highlights
-
•
We reveal the identity of the newly synthesized proteins throughout spore revival
-
•
We define the timeline of molecular events occurring during spore revival
-
•
Protein synthesis occurs during germination and is essential for its execution
-
•
RpmE and Tig are required for protein synthesis during germination
Sinai et al. describe the spatial and temporal molecular events occurring throughout the remarkable awakening process of a dormant bacterial spore and show that, in contrast to current knowledge, protein synthesis takes place during the earliest revival event, termed germination, and is required for its execution.
Introduction
The Gram-positive bacterium Bacillus subtilis (B. subtilis) belongs to a unique type of bacteria, capable of altering between modes of growth and sporulation. The process of sporulation is induced by nutrient deprivation and is initiated by the formation of an asymmetrically positioned septum. This polar septum divides the cell into a small forespore compartment, which ultimately becomes a spore, and a larger mother cell, which nurtures the developing spore. Subsequently, the forespore is engulfed by the mother cell, and a thick layer of peptidoglycan, termed the cortex, as well as inner and outer shells of proteinaceous coats, is deposited around the spore (Stragier and Losick, 1996). These protective layers confer unique mechanical properties, acting as a shield between the spore and its surroundings, enabling it to face extremes of heat, radiation, and chemical assault for long periods of time. Consequently, spore-forming bacteria, including dangerous pathogens such as Clostridium difficile (C. difficile) and Bacillus anthracis (B. anthracis), are highly resistant to antibacterial treatments and difficult to eradicate (Driks, 2002; Setlow, 2003). Sporulation culminates when the mother-cell lyses and the mature spore is discharged. The common notion is that from this moment on the spore is dormant and metabolically inert (Stragier and Losick, 1996). However, we have recently shown that entering dormancy lasts a few days, during which the spore RNA content is actively modified, changing according to spore age and temperature of incubation (Segev et al., 2012).
Once nutrients become available, dormancy ceases, and the spore rapidly resumes a vegetative life form (Setlow, 2003; Stragier and Losick, 1996). This revival process is classically divided into two major consecutive phases: (1) germination, during which the spore undergoes rehydration, release of dipicolinic acid (DPA), cortex hydrolysis, and coat disassembly (this phase is accompanied by transition from a phase-bright spore to a phase-dark cell, as manifested by light microscopy) and (2) outgrowth, in which the spore activates the synthesis of macromolecules to become a vegetative cell and emerges from the disintegrating spore shells (Moir, 2006; Santo and Doi, 1974; Setlow, 2003). Previously, we identified an intermediate phase, designated the “ripening period,” taking place early during outgrowth prior to cell elongation, in which no morphological change is evident. We found that this period is dedicated to molecular reorganization and demonstrated that it varies in length according to the initial molecular resources contained within the spore (Segev et al., 2013). We refer here to the entire process of resuming growth, which includes germination, ripening, and outgrowth, as “spore revival.”
Spore germination is triggered upon exposure to an array of molecules, including amino acids, sugars, and cell wall muropeptides that bind to receptors in the inner spore membrane. These ligand-receptor interactions activate downstream signaling events, directing the spore to revive (Moir, 2006; Setlow, 2003; Shah et al., 2008). In B. subtilis, the GerA receptor induces germination in response to l-alanine, while the GerB and GerK receptors collaborate to trigger germination in response to a mixture of aspargine, glucose, fructose, and potassium ions (AGFK) (Setlow, 2003). Currently held views consider germination to occur without the need for any macromolecule synthesis (Moir, 2006; Setlow, 2003, 2013; Steinberg et al., 1965; Vinter, 1970), while the subsequent stages are characterized by massive production of RNA and proteins. A detailed investigation of the reviving spore proteome, utilizing 2D gel analysis, revealed the process to be highly ordered, with specific classes of proteins produced in a temporal manner (Hecker et al., 1984; Torriani and Levinthal, 1967); however, the identity of these proteins was not determined. A more recent study monitoring the expression profile of mRNA throughout spore revival corroborated the existence of a complex developmental program, involving the temporal expression of at least 30% of the B. subtilis genome (Keijser et al., 2007). Here, we present a comprehensive time resolution analysis describing the identity of proteins synthesized in the course of spore revival. Through our investigation, we uncover how a dormant cell restores life. Furthermore, we provide evidence that, in contrast to current thinking, protein synthesis occurs during germination and is essential for its execution.
Results
Determining the Temporal Landscape of the Newly Synthesized Proteins during Spore Revival
The remarkable transition from a dormant spore to a fully active cell offers a unique opportunity to follow the molecular events required to build a cell. To explore this transition, we attempted to determine the proteomic landscape of a reviving spore throughout germination, ripening, and outgrowth. To differentiate the newly synthesized proteins during revival from the pre-existing spore protein pool, we employed the BONCAT (BioOrthogonal Non-Canonical Amino-acid Tagging) protein tagging technique (Figure 1A; see Experimental Procedures) (Dieterich et al., 2007). BONCAT allows the specific labeling and identification of newly translated proteins due to incorporation of an azide-bearing artificial amino acid termed azidohomoalanine (AHA), which is a substitute for methionine.
Figure 1.
Identifying the Newly Synthesized Proteins in the Course of Spore Revival
(A) Schematic description of the BONCAT strategy (Dieterich et al., 2007) for labeling, detection, and identification of the newly synthesized proteins tagged with AHA during spore revival. The general flow of the procedure (left) and detailed illustration of specific steps (right) are shown. Spores of LS5 (ΔmetE) strain are incubated in revival medium containing AHA (red hexagons), and the newly synthesized proteins (red curved lines) are separated from the existing spore proteins (blue curved lines). Spore proteins are extracted and tagged with TAP tag (green circles) using click chemistry (1, right). Next, newly produced proteins are isolated using avidin beads and identified by mass spectrometry (2, right). Scale bar represents 1 μm.
(B) Spores of LS5 (ΔmetE) strain were incubated in revival medium in which methionine was replaced by AHA. The sequence of events during revival is shown as captured by phase contrast images: germination is visualized as the loss of spore brightness (red line), the ripening period in which no morphological change is evident (orange line), and outgrowth is characterized by increasing cell length (green line). The average length of the reviving spores is presented below. Scale bar represents 1 μm.
(C) Spores of LS5 (ΔmetE) strain were incubated in revival medium and optical density (OD600) was measured at the indicated time points. Data are presented as a fraction of the initial OD600 of the phase-bright spores. Decreasing OD600 signifies spore germination, and increasing OD600 indicates spore outgrowth (Moir and Smith, 1990).
(D) Dot blot analysis of protein samples (in duplicates) collected throughout revival of LS5 (ΔmetE) spores at the indicated time points. Samples were diluted (1:100 and 1:200) and spotted on a membrane that was subsequently probed with anti-biotin antibodies. The obtained signal was compared with known amounts of biotinylated BSA.
Since B. subtilis is capable of synthesizing methionine, we constructed an auxotrophic strain to enhance AHA uptake by deleting the metE gene. Cells were induced to sporulate in the presence of methionine, and the resulting spores were purified. Spores were then subjected to revival in minimal medium containing the germination triggering molecules AGFK and l-alanine, supplemented with amino acids excluding methionine, which was replaced by AHA. The auxotrophic B. subtilis spores revived normally in the presence of AHA, likely due to sufficient amounts of endogenous methionine contained within spores (Setlow, 1988). As can be seen, germination, determined as the transition from a phase-bright spore to a phase-dark cell (Figure 1B), was accompanied by a drop in OD600 and took place from 0 to 15 min (Figure 1C). The subsequent ripening period, in which no morphological changes were evident, extended from 15 to 60 min (Figures 1B and 1C), and finally outgrowth, characterized by increase in cell length and OD600, occurred between 60 and 150 min (Figures 1B and 1C). Proteins were extracted from dormant spores (t = 0) and at 30 min intervals during revival until the first vegetative division took place at t = 150 min (Figure 1B). The extracted protein samples were then incubated with an alkyne-bearing biotin-Flag tag (TAP tag) to covalently label the newly synthesized proteins harboring AHA (Figure 1A). The amount of tagged proteins was estimated using dot blot analysis with antibody against biotin. Synthesis of labeled proteins was readily detected at t = 30 min, increasing gradually with time (Figure 1D). To enrich for newly synthesized proteins, samples were incubated with NeutrAvidin beads, and the captured proteins were cleaved with trypsin and identified by mass spectrometry (Figure 1A). At least two valid peptides per locus or one peptide containing an AHA-derived modification served as the minimal requirement to classify a detected protein as translated during the AHA-labeling step (Dieterich et al., 2007).
Utilizing this approach, a total of 653 proteins were identified as translated during the transition from dormant state to vegetative growth (Table S1 available online). Notably, 217 newly synthesized proteins were detected as early as 30 min into revival (Table S1). The synthesis of these proteins was largely dependent on de novo transcription, since when spores were subjected to revival in the presence of the two transcriptional inhibitors rifampicin and actinomycin D, only 12 proteins were detected at the same time point (Table S2). Additional 158 newly identified proteins were monitored at t = 60 min, when spores were at the ripening phase. At t = 90 min, when outgrowth was evident, 176 new proteins were detected. Finally, about 50 proteins were added to the pool at each subsequent time point during the transition to vegetative growth (Table S1).
To analyze the awakening pattern of molecular processes upon revival, we categorized the identified proteins into distinct groups according to their molecular function, using various B. subtilis genomic databases (Subtiwiki, GenoList, Uniport) (Table S1). A global view of this transition is presented in Figure 2. The activated cellular processes display different patterns, with several having all protein components detected at the first time point (e.g., glycolysis, ATP synthesis), while others exhibit a gradual awakening pattern (e.g., utilization of amino acids, rRNA and tRNA processing). Further, some pathways, in which all components were initially dormant, displayed synchronized synthesis at a given time point (e.g., fatty acid and teichoic acid biosyntheses). To corroborate our BONCAT-based data, representative proteins from different categories were fused to GFP, and their production during revival was followed. Indeed, fluorescence from GFP precisely coincided with the time of protein detection by BONCAT (Figure 3).
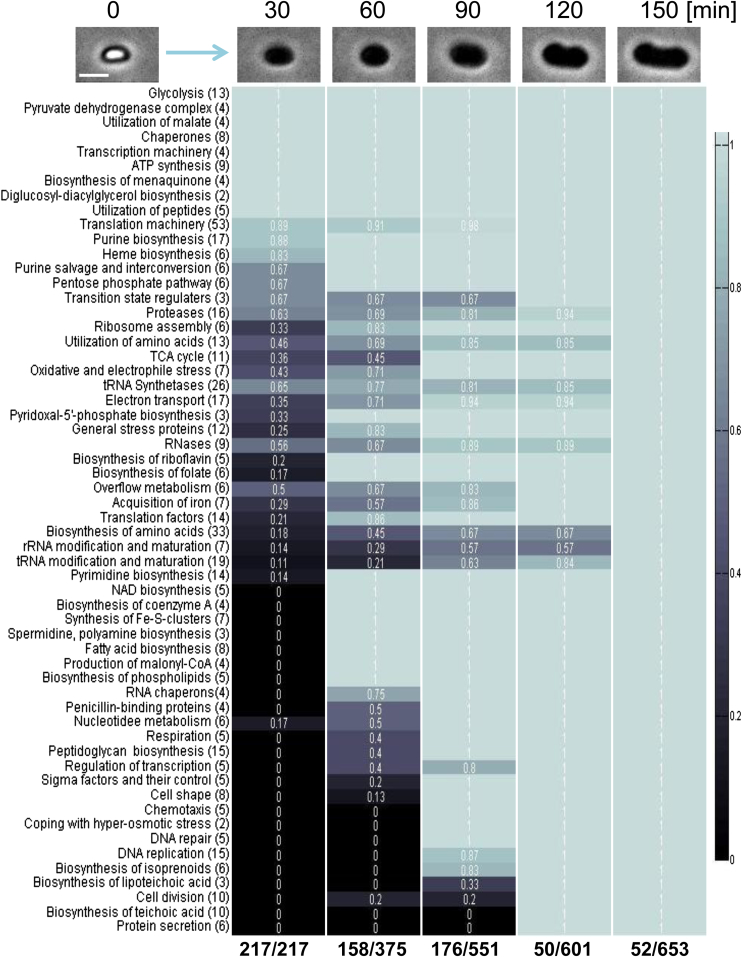
Figure 2.
The Temporal Landscape of the Newly Synthesized Proteins during Spore Revival
Spores of LS5 (ΔmetE) strain were incubated in revival medium in which methionine was replaced by AHA. Samples were collected at the indicated time points and processed as described in Figure 1A. The newly synthesized proteins were categorized into groups representing different molecular processes. The total number of proteins related to each group is indicated in parenthesis (data extracted from Table S1). The colors (see colors bar) and numbers (in white) indicate the relative fraction of proteins that was detected at a specific time point out of the total identified proteins for each group throughout revival. The numbers below summarize the amount of newly detected proteins out of the total detected proteins at each time point. Shown above are corresponding phase contrast images of a single reviving spore. Scale bar represents 1 μm.
See also Figure S1.
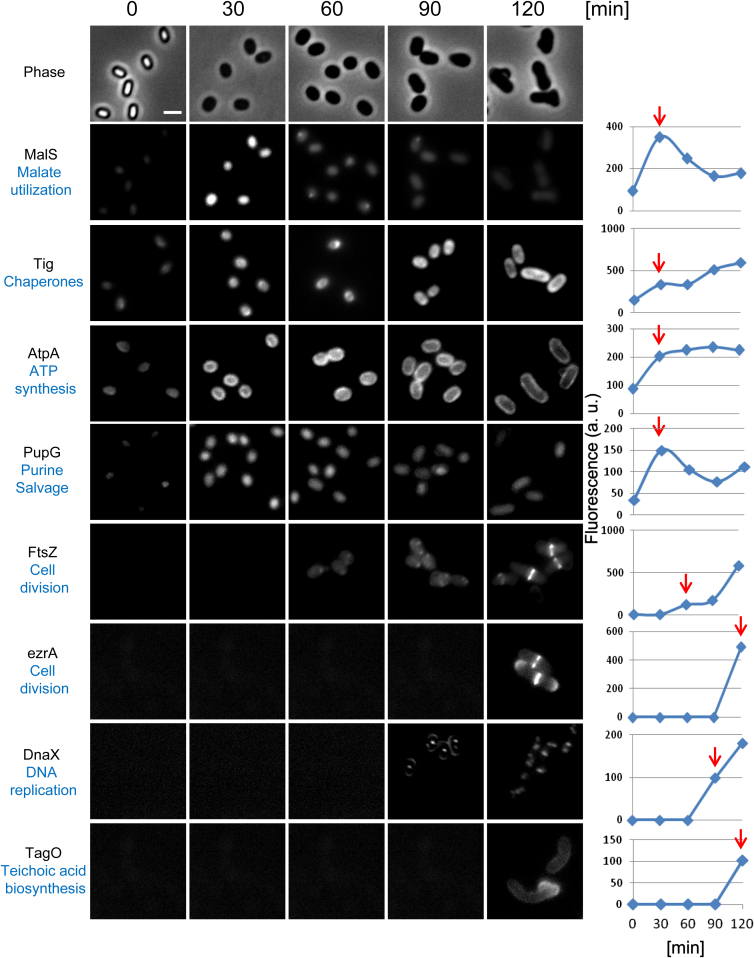
Figure 3.
Monitoring Fluorescence of Representative Proteins Fused to GFP throughout Spore Revival
Representative proteins from the indicated functional groups (derived from Table S1 and Figure 2) were fused to GFP and monitored for time of production during spore revival using fluorescence microscopy. GFP fluorescence images acquired at the indicated time points are shown. Upper panels show phase-contrast images of a strain harboring malS-gfp. For each strain, a representative experiment of three independent biological repeats is shown. Quantification of the fluorescence signal in arbitrary units (a.u.) is shown on the right. Fluorescence from at least 300 cells from three different fields was measured and averaged for each time point. The intensity of a WT (PY79) strain, lacking the gfp gene, was subtracted from the net average fluorescence intensity. Red arrows indicate the earliest time point at which a representative protein was identified by BONCAT. Scale bar represents 1 μm.
The Early Spore Revival Proteome
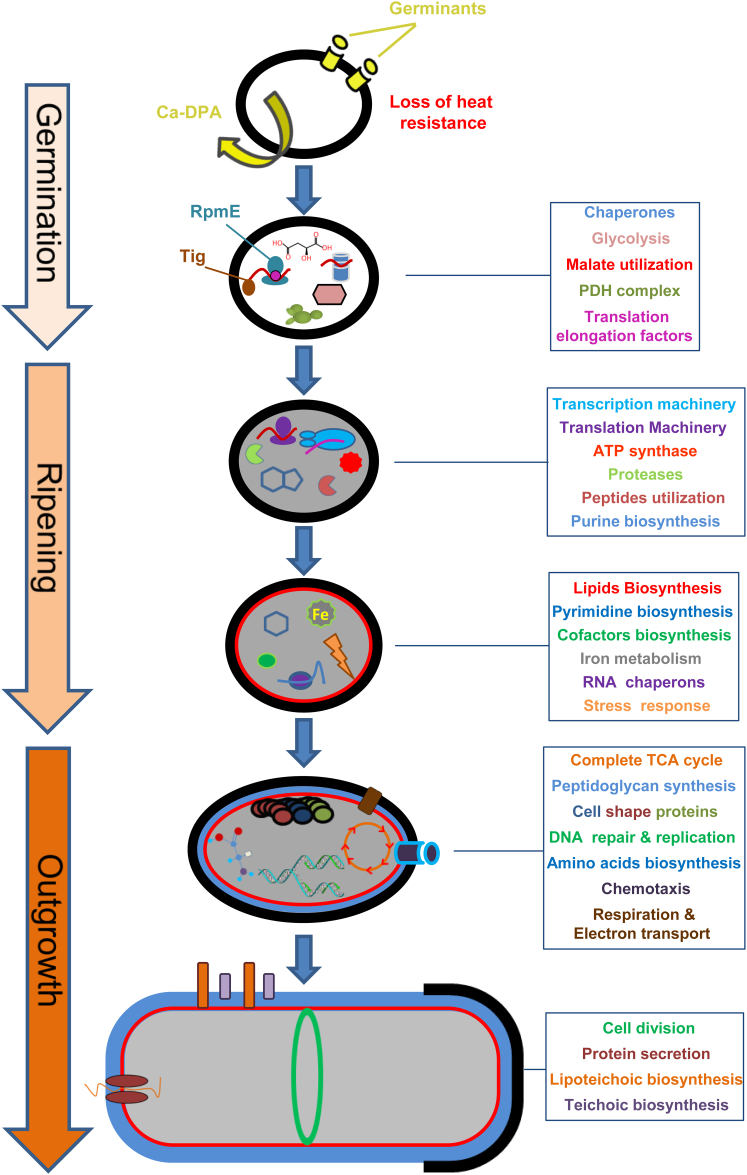
During the first 30 min of the revival process, the spore undergoes germination (after 15 min, approximately 80% of the spores complete germination) and enters the ripening period (Figures 1B and 1C). During these initial stages, 217 proteins were detected, which corresponds to 33% of the spore revival proteome, demonstrating rapid acquisition of metabolism. Accordingly, the core components of the transcriptional and translational machineries were produced along with a group of chaperones required for proper protein folding. Energy generation machineries, including all ATP synthesis, glycolysis, and pyruvate dehydrogenase (PDH) components, were readily detected at this time, providing fuel to the ongoing revival process (Figures 2 and 3; Table S1).
Notably, the metabolic pathway components required for malate utilization, including all four malic enzymes (MaeA, MalS, MleA, and YtsJ) that convert malate into pyruvate through oxidative decarboxylation (Meyer and Stülke, 2013), were produced at this early stage (Figures 2 and 3; Table S1). Malate, which is a preferred carbon source for B. subtilis (Meyer and Stülke, 2013), was not supplemented into the medium, implying that the spore stores malate as a key carbon source to energize revival. Consistent with this view, spores compared with vegetative cells were found to harbor large amounts of malate (Figure S1A). Furthermore, a strain deleted of all four genes encoding malic enzymes showed no detectable growth defect but displayed an extended ripening period in comparison to reviving WT spores (Figures S1B–S1D). Additionally, spores of the quadro mutant contained malate levels similar to those of WT spores (Figure S1A), indicating that the observed ripening defect is due to deficiency in malate utilization.
The Progression of the Ripening Period
No morphological change was evident 60 min into spore revival, indicating that the spore was still at the ripening period, although a substantial group of 158 additional proteins was produced at this time point (Figure 2; Table S1). The pentose phosphate pathway was among the most prominent fully activated processes. This pathway is required to generate pentoses and NADPH; the former is essential for nucleotide biogenesis, whereas the latter is required for fatty acid biosynthesis (Zamboni et al., 2004). In line with this finding, processes related to lipid biosynthesis were fully activated after 60 min, including fatty acid and phospholipid formation and production of malonyl-CoA, indicating that membrane construction was initiated (Figure 2; Table S1).
A manifested activation was also observed for the pyrimidine biosynthesis pathway, in which all 14 proteins involved where detected at t = 60 min. Notably, most of the purine biosynthesis proteins were already detected 30 min into revival (Figure 2; Table S1). These observations are consistent with the premise that the pur genes are induced in response to glucose contained in the revival medium, whereas the pyr genes are known to be controlled by pyrimidine availability (Blencke et al., 2003). This proteomic profile supports a model whereby RNA transcription is initially based on the spore pre-existing nucleotide reservoir, and only during ripening newly generated nucleotides are utilized. This model is in accord with the observation of Setlow and Kornberg (1970a) that de novo nucleotide biosynthesis initiates early during revival and relies on re-establishment of translation, as well as with our earlier finding that during ripening the spore is engaged in synthesizing large amounts of rRNA required to build up new ribosomes (Segev et al., 2013).
Along with membrane and nucleotide biosynthesis, several pathways related to cofactor production were induced at this time, including proteins involved in NAD, riboflavin, pyridoxal-phosphate, folate and coenzyme A production, which are required for numerous enzymatic activities. In parallel, general and oxidative stress proteins were highly induced (Figure 2; Table S1). This activation might be associated with the hydration of the spore core and activation of metabolism, events likely to generate oxidative stress (Ibarra et al., 2008).
From Outgrowth into Vegetative Growth
At 90 min into revival, spore size was increased (Figures 1B and 1C), indicating the initiation of outgrowth. Accordingly, processes related to cell wall synthesis and cell elongation were fully activated, as evidenced by the production of proteins involved in peptidoglycan (PG) biosynthesis and cell shape determination (Figure 2; Table S1). Furthermore, the main energy producing cellular pathways, tricarboxylic acid cycle (TCA) and respiration, were entirely induced to facilitate spore growth (Figure 2; Table S1). At this point, the spore was capable of generating the complete set of building molecules, essential for cell outgrowth and elongation. In light of this increased metabolic activity, the spore produced DNA metabolic proteins required to repair the damaged genome and to carry out replication in preparation for the upcoming division (Figures 2 and 3; Table S1).
A more significant increase in size of the reviving spore was apparent at t = 120 min, with the spore accomplishing the conversion into a rod shape vegetative cell by t = 150 min (Figure 1B). Approximately 100 proteins were added to the revival proteome during these later phases, with the majority being components associated with ongoing processes. Nevertheless, three major pathways, including cell division, biosynthesis of teichoic and lipoteichoic acids, and protein secretion, were largely activated only at t = 120 min (Figures 2 and 3; Table S1). Interestingly, the assembly of the division machinery correlated with the observed morphological changes. A total of 10 bona fide cell division proteins were identified during the course of spore revival. The two initial division components, FtsZ and FtsA, were detected as early as 60 min post-induction of revival, while the others were first identified only at t = 120 min (Figure 2; Table S1). Observing the dynamics of FtsZ-GFP during revival revealed detectable fluorescence at t = 60 min; however, Z-ring formation occurred only at t = 120 min (Figure 3). Consistently, EzrA, one of the late cell division proteins, was produced and localized to the division site of a reviving spore only at t = 120 min (Figure 3). Thus, it seems that the temporal production of cell division components contributes to the proper assembly of the division ring. A similar gradual activation pattern was observed for components required for cell wall construction. Notably, the PG synthesizing enzymes were produced at t = 90 min, early during outgrowth, while the teichoic and lipoteichoic acids biosynthesis enzymes were produced later on (Figures 2 and 3; Table S1). This sequential pattern of cell wall production suggests that PG construction precedes teichoic and lipoteichoic acid synthesis.
Protein Synthesis Is Carried Out during Germination and Required for the Process to Occur
The first 30 min of revival are composed of two phases: germination, taking place approximately at the first 15 min, and early ripening (Figure 1C). We next zoomed into this time period to investigate the long-outstanding question of whether protein synthesis takes place during germination. It has been shown previously that Bacilli spores are capable of undergoing germination in the presence of RNA and protein synthesis inhibitors (Steinberg et al., 1965; Vinter, 1970). However, some doubts have been raised concerning these observations, as drugs may not be capable of penetrating the spore protective shells at this early stage (Sussman and Douthit, 1973; Tisdale and DeBusk, 1972). To elucidate this issue, we incubated spores with antibiotics that inhibit translation and are soluble in ethanol, which was shown to increase spore permeability (Tanimoto et al., 1996). Remarkably, when spores were incubated with either lincomycin or tetracycline and then induced to germinate with l-alanine, approximately 80%–90% of the spore population was clearly inhibited from germinating, as manifested by their phase bright appearance (Figure 4A; Table S3). Moreover, incubation with both antibiotics blocked germination in more than 99% of the spores (Table S3), indicating that translation is required for the process. Further analysis of the antibiotic treated spores revealed that upon induction of germination, they released DPA at kinetics similar to that of untreated spores and lost their heat resistance (Figure 4B; Table S4). Thus, the antibiotic-treated spores initiated germination, but were most likely blocked prior to cortex hydrolysis as they still remained phase bright (Moir and Smith, 1990).
Figure 4.
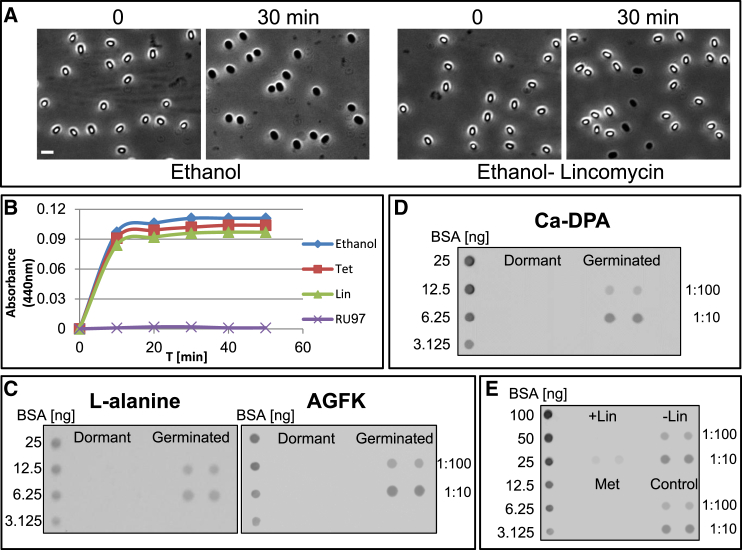
Protein Synthesis Is Carried Out during Germination and Required for the Process to Occur
(A) PY79 (WT) spores were incubated at 37°C for 2 hr in ethanol with or without lincomycin (250 μg/ml). Next, spores were washed with phosphate buffer and incubated with l-alanine for 30 min. Shown are phase-contrast images of spores incubated with (right) or without (left) lincomycin, before (t = 0) and after (t = 30 min) l-alanine addition. Scale bar represents 1 μm.
(B) PY79 (WT) spores were incubated at 37°C for 2 hr in ethanol with or without lincomycin (250 μg/ml) or tetracycline (250 μg/ml). Next, spores were washed with phosphate buffer and incubated with l-alanine. The DPA release to the medium was determined as a measurement of OD440. The nongerminating strain RU97 (ΔgerAA) was used as a control.
(C and D) Spores of LS5 (ΔmetE) strain were induced to germinate with l-alanine (C, left), AGFK (C, right) for 30 min or with Ca-DPA (D) for 60 min in the presence of AHA. Shown are dot blot analyses of protein samples (in duplicates) that were collected before (t = 0, dormant) and after (t = 30, germinated) germination induction. Samples were diluted (1:100 and 1:10) and spotted on a membrane that was subsequently probed with antibiotin antibodies. The obtained signal was compared with known amounts of biotinylated BSA.
(E) Spores of LS5 (ΔmetE) strain were incubated at 37°C for 2 hr in ethanol with (+Lin) or without (–Lin) lincomycin (250 μg/ml) or in double-distilled water (DDW) (control). Next, spores were washed with phosphate buffer and incubated with l-alanine and AHA for 30 min. As a negative control, spores incubated in DDW were also germinated in the presence of methionine instead of AHA (Met). Shown is a dot blot analysis conducted as in (C and D).
We next utilized the BONCAT technique to substantiate that proteins are produced during germination. We took advantage of the observation that dormant spores undergo germination without the ability to execute outgrowth when the germinant factors l-alanine or AGFK are introduced in the absence of additional nutrients (Setlow, 2003). Accordingly, spores were incubated with l-alanine or AGFK in the presence of AHA and the absence of other nutrients. Proteins were extracted, incubated with TAP tag, and dot-blot analysis conducted. Synthesis of labeled proteins was evident for both l-alanine and AGFK germinating spores (Figure 4C). Repeating this assay with the non-nutrient germinant Ca-DPA (Setlow, 2003) corroborated that proteins are being synthesized during germination without the need for external nutrients (Figure 4D). Furthermore, protein synthesis was almost completely abolished in the presence of lincomycin (Figure 4E).
Labeled proteins synthesized during germination were then identified by mass spectrometry. A total of 116 newly synthesized proteins were monitored upon AGFK addition, while 32 and 27 proteins were identified when l-alanine and Ca-DPA were utilized as germinant factors, respectively (Table S5). Analysis of the synthesized proteins revealed that the majority of the proteins detected following l-alanine or Ca-DPA addition were identical (24 of the identified proteins) and included in the list of proteins monitored upon AGFK supplementation (Table S5). The proteins monitored in all assays were part of the revival proteome at t = 30 min (Tables S1 and S5). A comparison of the obtained proteomes when categorized into functional groups provided a higher resolution of the earliest events occurring during revival (Table S6). Ca-DPA and l-alanine, which are relatively minimal sources that can trigger spore germination, activated synthesis of proteins categorized into glycolysis, PDH complex, malate utilization, translation elongation, chaperones, and oxidative stress (Table S6). On the other hand, the combination of AGFK triggering molecules supported the synthesis of components belonging to additional pathways, including transcription and translation machineries, electron transport, RNA degradation, and proteolysis (Table S6).
We conclude that protein synthesis is carried out during germination and necessary for its completion. Furthermore, the small group of proteins synthesized in response to Ca-DPA and l-alanine may exclusively represent the germination proteome.
The Translational Factors RpmE and Tig Play a Key Role in Spore Germination
A detailed investigation of the proteins produced in response to l-alanine revealed four newly synthesized translational factors RplU, RplP, RpmE, and Tig (Table S5). RplU, RplP, and RpmE are ribosomal components constituting the 50S subunit (Akanuma et al., 2012), whereas Tig is a ribosome-associated chaperone, which interacts directly with emerging nascent polypeptides (Merz et al., 2008). We reasoned that these factors might be involved in translation of proteins during germination. Since rpmE and tig genes are nonessential for vegetative growth (Akanuma et al., 2012; Reyes and Yoshikawa, 2002), we constructed strains deleted for these genes and followed the ability of mutant spores to revive. Remarkably, both ΔrpmE and Δtig mutant spores exhibited an apparent germination deficiency, as evidenced by their delay in converting from a phase-bright to a phase-dark state in comparison to the WT spores (Figures 5A and 5B). Moreover, spores harboring deletion of both factors displayed a more severe germination defect (Figures 5A and 5B). The ability of these spores to eventually germinate suggests that additional translational factors, such as the essential components RplU and RplP (Akanuma et al., 2012), mediate this developmental transition. Closer investigation of the germinating mutant spores revealed that they released DPA and turned heat sensitive similarly to WT spores (Figure 5C; Table S4). Thus, like the antibiotic treated spores, the mutant spores initiated germination but failed to efficiently progress throughout the process. Analysis of the mutant strains during vegetative growth revealed that the Δtig strain grew similarly to WT, while the ΔrpmE strain exhibited only a slight growth perturbation, suggesting that both genes acquired unique germination properties (Figure S2A). Importantly, ΔrpmE and Δtig mutant strains sporulated similarly to WT cells (Figure S2D), and their germination defect was rescued by ectopic insertion of the corresponding WT alleles (Figure 5B). Since RpmE and Tig are bona fide translational factors, they could affect the global protein level within spores. It was therefore plausible that the defective germination displayed by the mutant strains was due to decreased amounts of germination receptors in spores. To rule out this possibility, we determined the levels of GerAA, GerAC, GerBC, GerKA, and SpoVAD in mutant and WT spores by western blot analysis (Ramirez-Peralta et al., 2012). The levels of all the investigated proteins were similar in WT and in the mutant spores (Figures S2B and S2C). In addition, the mutant spores showed lysozyme resistance levels similar to WT spores (Figure S2E), indicating that their coat is intact. Taken together, these results imply that the observed defect of the mutant strains is not due to a general deficiency in translation during sporulation but is specific to germination.
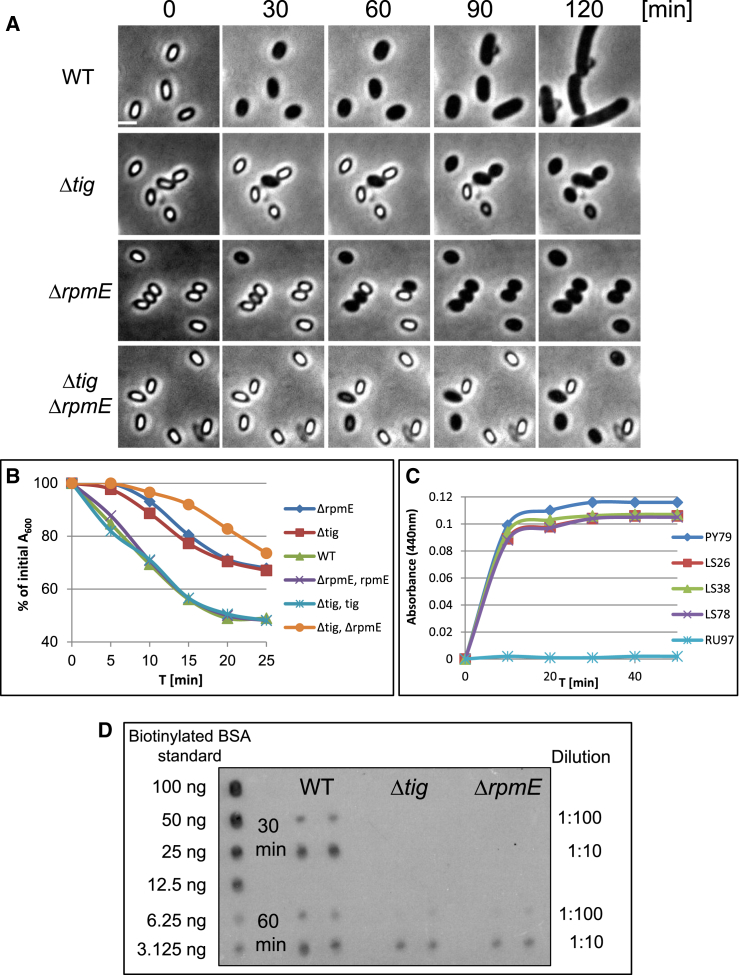
Figure 5.
The Translational Factors RpmE and Tig Play a Key Role in Spore Germination
(A) Spores of PY79 (WT), LS38 (Δtig), LS26 (ΔrpmE), and LS78 (Δtig ΔrpmE) strains were incubated in LB medium supplemented with l-alanine (4 mM) and monitored by time lapse microscopy. Shown are phase contrast images from a representative experiment out of three independent biological repeats. Scale bar represents 1 μm.
(B) Spores of PY79 (WT), LS38 (Δtig), LS26 (ΔrpmE), LS78 (Δtig, ΔrpmE), LS101 (Δtig, amyE::tig), and LS100 (ΔrpmE, amyE::rpmE) strains were incubated with AGFK+ l-alanine, and optical density (OD600) was measured at the indicated time points. Data are presented as a fraction of the initial OD600 of the phase-bright spores. Decreasing OD600 signifies spore germination (Moir and Smith, 1990).
(C) Spores of PY79 (WT), LS38 (Δtig), LS26 (ΔrpmE), LS78 (Δtig, ΔrpmE), and RU97 (ΔgerAA) strains were incubated with l-alanine to trigger germination. DPA release to the medium was determined as a measurement of OD440.
(D) Spores of LS5 (ΔmetE) (WT), LS82 (ΔmetE, Δtig), and LS83 (ΔmetE, ΔrpmE) strains were induced to germinate with l-alanine in the presence of AHA for 60 min. Shown is a dot blot analysis of protein samples (in duplicates) collected 30 and 60 min after germination induction. Samples were diluted (1:100 and 1:10) and spotted on a membrane that was subsequently probed with antibiotin antibodies. The obtained signal was compared with known amounts of biotinylated bovine serum albumin.
See also Figure S2.
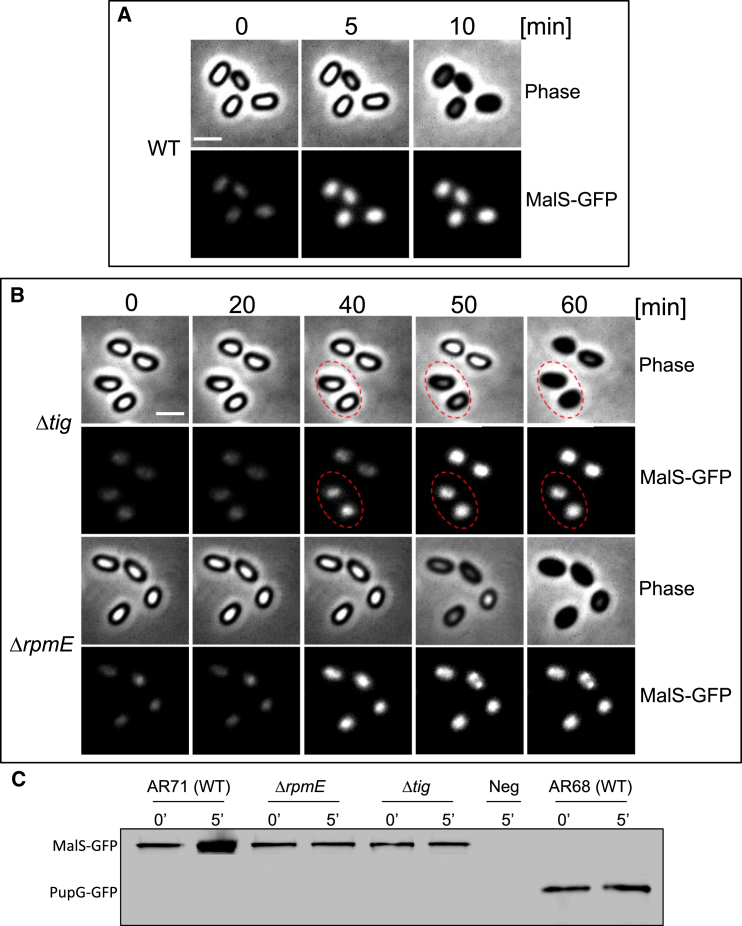
To substantiate that RpmE and Tig are required for protein production during germination, the BONCAT system was used to follow protein synthesis of ΔrpmE and Δtig mutant spores during this phase. To this end, spores of the different strains were incubated only with l-alanine and AHA, and proteins were extracted from spores at 30 and 60 min post-germination induction and subjected to tagging with TAP tag followed by dot blot analysis. No detectable protein synthesis took place in the ΔrpmE and Δtig mutants at t = 30 min, whereas protein synthesis was clearly evident in WT spores (Figure 5D). After 60 min, relatively small amounts of newly synthesized proteins were detected in the ΔrpmE and Δtig mutants, approximately 10-fold less than those found in WT spores (Figure 5D).
Finally, we wished to observe protein synthesis during germination in real time. To achieve this goal, we investigated the production of MalS-GFP fusion by time lapse microscopy, as MalS is one of the earliest proteins produced in germinating spores (Table S5). Remarkably, WT spores harboring MalS-GFP fusion exhibited a dramatic increase in the GFP signal as early as 5 min after germination induction by l-alanine. Evidently, under these conditions GFP folding was faster than previous estimates (Tsien, 1998). At this time, spores were clearly residing in their phase-bright state, indicating that germination was still in progress (Figure 6A). As a control, we monitored the production of PupG-GFP that was detected in spores (Figure 3), but was excluded from the germination proteome (Table S5). Consistent with our data, fluorescence from PupG-GFP did not increase during germination (Figure S3A). Furthermore, the increase in fluorescence from MalS-GFP was considerably delayed in ΔrpmE and Δtig mutant spores, detectable only 40 min after germination induction (Figure 6B). Of note, mutant spores that exhibited an increase in fluorescence progressed through germination and completed the process at the subsequent time point (Figure 6B, circles). These results were further corroborated by western blot analysis (Figures 6C and S3B).
Figure 6.
Real-Time Protein Synthesis during Spore Germination
(A and B) Spores of AR71 (WT) (A) and LS80 (Δtig) and LS81 (ΔrpmE) (B) harboring malS-gfp were incubated with l-alanine and followed by time lapse microscopy. Shown are phase contrast (upper) and fluorescence (lower) images taken at the indicated time points. Dashed circles in (B) highlight spores in which increase in MalS-GFP fluorescence is followed by germination. For each strain, images were scaled to the same intensity range. The intensity of a WT (PY79) strain, lacking the gfp gene, was subtracted from the net average fluorescence intensity. A representative experiment out of three independent biological repeats is shown. Scale bars represent 1 μm.
(C) Spores of AR71 (WT, malS-gfp), LS81 (Δtig, malS-gfp), LS82 (ΔrpmE, malS-gfp), and AR68 (wild-type, pupG-gfp) strains were induced to germinate with l-alanine, and proteins were extracted before (t = 0) and after (t = 5 min) l-alanine addition. Equal amounts of protein extracts were subjected to western blot analysis. Membrane was probed with antibody against GFP. Proteins extracted from PY79 spores lacking gfp (t = 5 min) were used as a negative control (Neg).
We surmise that protein synthesis during germination considerably relies on the translational factors RpmE and Tig, which are produced early during the process.
Discussion
The fascinating awakening of the bacterial spore provides an exceptional opportunity to delineate the spatial and temporal molecular events vital to construct a fully functional cell. Here we succeeded in defining the timeline of the spore revival proteome. The key cellular processes turned on at each time point in the course of spore revival are summarized in Figure 7. Our investigation revealed a highly orchestrated developmental process that correlates with the spore morphological changes. Surprisingly, we found that protein synthesis is necessary to facilitate germination, a process traditionally considered to occur without the requirement for any macromolecule synthesis (Moir, 2006; Setlow, 2003, 2013; Steinberg et al., 1965; Vinter, 1970).
Figure 7.
The Molecular Timeline of a Reviving Bacterial Spore
The temporal activation of the indicated cellular processes, as determined by our findings, was assigned to the morphological changes occurring during spore revival. The awakening cellular processes are indicated in the square boxes and their color corresponds to the illustrated cellular processes.
Germination of Bacilli spores was found to occur in the presence of RNA and protein synthesis inhibitors (Steinberg et al., 1965; Vinter, 1970). However, here we show that by increasing spore permeability we were able to introduce protein synthesis inhibitors into spores, subsequently impeding germination. Furthermore, antibiotic-treated spores initiated germination, as indicated by DPA release, but seem staled prior to cortex hydrolysis. The DPA release indicates that at least partial core rehydration occurred, allowing resumption of protein synthesis among other enzymatic activities. Accordingly, ATP synthesis from spore endogenous sources was shown to increase sharply 5 min post-germination induction and to follow DPA release (Setlow et al., 2001; Setlow and Kornberg, 1970b). In line with this view, the metabolic enzymes required for glycolysis and malate utilization were produced during germination. Interestingly, Bacilli spores were found to contain high levels of phosphenolglycerate (PGA) (Nelson and Kornberg, 1970), which could serve as an immediately accessible substrate for the glycolytic enzymes to produce energy. In a similar manner, we found that the spore harbors significant amounts of malate that can serve as a “ready to use” resource for the malic enzymes to energize revival. The observation that protein synthesis takes place during germination induced by the non-nutrient germinant Ca-DPA implies that the spore harbors a pool of amino acids required to facilitate the process. Indeed, it has been shown that the abundant small acid-soluble spore proteins, which comprise 10%–20% of the spore protein pool, are degraded in the first minutes of spore revival, providing an instant source of amino acids for early protein synthesis (Setlow, 1988). Taken together, our findings support previous studies showing that the spore reservoir contains all elements needed for initial protein synthesis, including amino acids, ribosomes, and energy sources (Kieras et al., 1978; Setlow and Kornberg, 1970a, 1970b; Setlow and Primus, 1975).
It still remains elusive how the ribosomes are rapidly activated during germination. Our data indicate that, at least in part, this transition is facilitated by translation components, such as RpmE and Tig, that associate with the spore ribosomes. The contribution of the existing spore proteome is therefore crucial to initiate translational events. Characterization of the dormant spore proteome revealed the existence of 154 proteins of which the majority (110) are spore specific proteins or proteins with unknown function (Kuwana et al., 2002). However, only a few ribosomal components were monitored, suggesting that a more comprehensive investigation of the dormant spore proteome is required.
Our study defines the molecular events taking place during the ripening period. We have previously demonstrated that rRNA assembly is a key event occurring throughout ripening (Segev et al., 2013), and here we show that the entire array of ribosomal protein components is produced during this phase. In a complementary fashion, we found that the transcription machinery components, along with enzymes required for nucleotide biosynthesis, are produced at this stage. Notably, 12 metabolites have been defined as basic precursors for synthesis of all cellular constituents (Rokem et al., 2007). Our study suggests that spore outgrowth is initiated only after all the metabolic pathways producing these components are awakened. Accordingly, the glycolytic enzymes and the PDH complex are produced during germination, generating seven of the defined essential components: glucose-6P, fructose-6P, glyceraldehyde-3P, 3P-glycerate, phosphoenolpyruvate, pyruvate, and acetyl-CoA. Subsequently, the pentose phosphate pathway enzymes are translated at the onset of the ripening period, generating erytrose-4P and ribose-5P. Finally, the complete set of the TCA cycle enzymes is synthesized at the initiation of outgrowth, producing the remaining essential components oxaloacetate, 2-oxoglutarate, and succinyl-CoA. Thus, once the reviving spore has acquired all essential building blocks, cell size starts increasing.
Studies that use 2D gel analysis to characterize the reviving spore proteome of Streptomyces coelicolor (S. coelicolor) identified approximately 150 proteins as produced in the course of 5.5 hr of revival (Strakova et al., 2013a, 2013b). Among the earliest proteins identified were chaperones and protein modification enzymes. Our data indicate that proteins with comparable functions are produced during germination in B. subtilis, suggesting that these proteins play a common role in the spore awakening program of distinct bacterial species. Curiously, the investigation of spore revival in S. coelicolor revealed little correlation between transcription and translation (Strakova et al., 2013b). Comparing our proteome timeline to the transcriptional profile of reviving B. subtilis spores (Keijser et al., 2007) also indicated a partial correlation. For instance, processes that were shown by BONCAT and light microscopy to awaken during outgrowth, such as DNA replication, cell division, and cell wall biosynthesis, seem to be transcribed during the ripening period. Thus, translational regulation could play a role in the spore revival program.
The impermeability of the spore at the time of protein synthesis initiation should be taken into consideration when designing drugs against spore forming pathogens such as B. anthracis, which causes anthrax, and Bacillus cereus, which causes food borne disease (Driks, 2002; Setlow, 2003). Our identification of RpmE and Tig as key factors for protein synthesis during germination may provide new therapeutic targets and pave the way for development of more effective drugs. In line with this possibility, Tig was found to be among the seven proteins identified to be synthesized during revival of B. anthracis spores (Huang et al., 2004). RpmE and Tig are not specific to spore formers and are highly conserved among both Gram-positive and -negative bacteria. However, their dispensability for vegetative growth in B. subtilis suggests that they might acquire unique properties in spore formers, yet to be elucidated.
Experimental Procedures
Strains and General Methods
B. subtilis strains are derivatives of PY79 and are listed in Table S7. Plasmids construction is described in Supplemental Experimental Procedures. Sporulation was induced at 37°C by suspending cells in Schaeffer’s liquid medium (Difco Sporulation Medium [DSM]) (Harwood and Cutting, 1990), and mature spores were purified as described in Supplemental Experimental Procedures. Purified spores were heat activated (80°C, 30 min) prior to germination and revival experiments. Unless indicated differently, spore revival was induced at 37°C by suspending spores in revival medium composed of S7 minimal medium (Vasantha and Freese, 1980) supplemented with AGFK (2.5 mM l-aspargine, 5 mg/ml d-glucose, 5 mg/ml d-fructose, 50 mM KCl), 0.01 M alanine, and all additional amino acids in concentrations suitable for B. subtilis (Harwood and Cutting, 1990). For BONCAT experiments, AHA (200 mg/l; Anaspec) was used instead of methionine. For germination induction by l-alanine and AGFK, heat-activated spores were incubated for 30 min, a time in which 99% of the WT spores completed germination. For germination induced by Ca2+-DPA, heat-activated spores were incubated in 60 mM Ca2+-DPA (pH 8.3) for 60 min at room temperature.
BONCAT Spore Revival Experiments
Cultures of 100 ml of reviving spores were centrifuged and washed with PBS ×1. Pellets were resuspended in PBS ×1 supplemented with protease inhibitors (Thermo, 78439), lysed using FastPrep (MP) (6.5, 60 s, ×3), and centrifuged (5 min, 14,000 RPM). Under these conditions, less than 0.01% of the spore population remained intact. Supernatants containing a mixed population of AHA-labeled newly synthesized proteins and unlabeled pre-existing proteins were collected. The enrichment for newly translated proteins was performed using BONCAT, as previously described (Dieterich et al., 2007), with the following modifications. For tagging of AHA-labeled proteins, samples were incubated overnight at 4°C with triazole ligand (0.25 mM; Sigma), alkyne-bearing biotin-flag tag (0.063 mM; Genscript), and CuBr (2 mM in DMSO; Sigma). Trypsin-digested samples were analyzed by LC-MS/MS on LTQ-Orbitrap (Thermo). Each presented BONCAT dataset is based on three independent biological repeats. The proteins displayed in Tables S1, S2, and S5 were detected in all repeats. In each experiment, at least 35% of the proteins were identified based on AHA containing peptides, and the majority of the proteins (>95%) were identified based on at least three different peptides. In each experiment, a methionine sample was added as a control. The number of identified proteins in the methionine control samples was always below 1% of the proteins identified in the parallel AHA samples and included mainly abundant spore specific proteins. These proteins were excluded from the analysis.
Additional experimental procedures, including light microscopy, spores purification, l-malate determination, DPA measurements, determination of the levels of spore germination proteins, western blot analysis, and spore resistance to lysozyme treatment, are described in the Supplemental Experimental Procedures.
Acknowledgments
We are grateful to members of the Ben-Yehuda laboratory and G. Bachrach, I. Goldberg, J.S. Rokem, I. Rosenshine, and A. Taraboulos (Hebrew University), R. Losick (Harvard University), and A. Rouvinski (Pastuer Institute) for valuable discussions and comments on the manuscript. We thank P. Setlow (UConn) for generously providing antibodies and K.M. Devine (Trinity College) and A. Driks (Loyola University) for strains. We thank the Smoler Proteomic Center at the Department of Biology, Technion, for mass spectrometry services. This work was supported by the European Research Council (ERC) Starting Grant (209130) and ERC Advance Grant (339984) awarded to S.B.-Y.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
The proteomic timeline of reviving LS5 (ΔmetE) spores as determined by BONCAT. The analysis is based on three independent biological repeats. Only proteins detected in all repeats are displayed.
The proteome of germinating LS5 (ΔmetE) spores incubated with l-alanine, AGFK, or Ca-DPA, as determined by BONCAT. The analysis is based on three independent biological repeats. Only proteins detected in all repeats are displayed.
References
- Akanuma G., Nanamiya H., Natori Y., Yano K., Suzuki S., Omata S., Ishizuka M., Sekine Y., Kawamura F. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J. Bacteriol. 2012;194:6282–6291. doi: 10.1128/JB.01544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencke H.M., Homuth G., Ludwig H., Mäder U., Hecker M., Stülke J. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 2003;5:133–149. doi: 10.1016/s1096-7176(03)00009-0. [DOI] [PubMed] [Google Scholar]
- Dieterich D.C., Lee J.J., Link A.J., Graumann J., Tirrell D.A., Schuman E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- Driks A. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 2002;10:251–254. doi: 10.1016/s0966-842x(02)02373-9. [DOI] [PubMed] [Google Scholar]
- Harwood C.R., Cutting S.M. Wiley; Chichester, New York: 1990. Molecular Biological Methods for Bacillus. [Google Scholar]
- Hecker M., Wachlin G., Dunger A.M., Mach F. Protein-synthesis during outgrowth of Bacillus subtilis spores. A two-dimensional gel-electrophoresis study. FEMS Microbiol. Lett. 1984;25:57–60. [Google Scholar]
- Huang C.M., Foster K.W., DeSilva T.S., Van Kampen K.R., Elmets C.A., Tang D.C. Identification of Bacillus anthracis proteins associated with germination and early outgrowth by proteomic profiling of anthrax spores. Proteomics. 2004;4:2653–2661. doi: 10.1002/pmic.200400831. [DOI] [PubMed] [Google Scholar]
- Ibarra J.R., Orozco A.D., Rojas J.A., López K., Setlow P., Yasbin R.E., Pedraza-Reyes M. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J. Bacteriol. 2008;190:2031–2038. doi: 10.1128/JB.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser B.J.F., Ter Beek A., Rauwerda H., Schuren F., Montijn R., van der Spek H., Brul S. Analysis of temporal gene expression during Bacillus subtilis spore germination and outgrowth. J. Bacteriol. 2007;189:3624–3634. doi: 10.1128/JB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieras R.M., Preston R.A., Douthit H.A. Isolation of stable ribosomal subunits from spores of Bacillus cereus. J. Bacteriol. 1978;136:209–218. doi: 10.1128/jb.136.1.209-218.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana R., Kasahara Y., Fujibayashi M., Takamatsu H., Ogasawara N., Watabe K. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology. 2002;148:3971–3982. doi: 10.1099/00221287-148-12-3971. [DOI] [PubMed] [Google Scholar]
- Merz F., Boehringer D., Schaffitzel C., Preissler S., Hoffmann A., Maier T., Rutkowska A., Lozza J., Ban N., Bukau B., Deuerling E. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J. 2008;27:1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F.M., Stülke J. Malate metabolism in Bacillus subtilis: distinct roles for three classes of malate-oxidizing enzymes. FEMS Microbiol. Lett. 2013;339:17–22. doi: 10.1111/1574-6968.12041. [DOI] [PubMed] [Google Scholar]
- Moir A. How do spores germinate? J. Appl. Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Moir A., Smith D.A. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- Nelson D.L., Kornberg A. Biochemical studies of bacterial sporulation and germination. XIX. Phosphate metabolism during sporulation. J. Biol. Chem. 1970;245:1137–1145. [PubMed] [Google Scholar]
- Ramirez-Peralta A., Zhang P., Li Y.Q., Setlow P. Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl. Environ. Microbiol. 2012;78:2689–2697. doi: 10.1128/AEM.07908-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes D.Y., Yoshikawa H. DnaK chaperone machine and trigger factor are only partially required for normal growth of Bacillus subtilis. Biosci. Biotechnol. Biochem. 2002;66:1583–1586. doi: 10.1271/bbb.66.1583. [DOI] [PubMed] [Google Scholar]
- Rokem J.S., Lantz A.E., Nielsen J. Systems biology of antibiotic production by microorganisms. Nat. Prod. Rep. 2007;24:1262–1287. doi: 10.1039/b617765b. [DOI] [PubMed] [Google Scholar]
- Santo L.Y., Doi R.H. Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J. Bacteriol. 1974;120:475–481. doi: 10.1128/jb.120.1.475-481.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E., Smith Y., Ben-Yehuda S. RNA dynamics in aging bacterial spores. Cell. 2012;148:139–149. doi: 10.1016/j.cell.2011.11.059. [DOI] [PubMed] [Google Scholar]
- Segev E., Rosenberg A., Mamou G., Sinai L., Ben-Yehuda S. Molecular kinetics of reviving bacterial spores. J. Bacteriol. 2013;195:1875–1882. doi: 10.1128/JB.00093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spore germination. Curr. Opin. Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Setlow P. Summer meeting 201—when the sleepers wake: the germination of spores of Bacillus species. J. Appl. Microbiol. 2013;115:1251–1268. doi: 10.1111/jam.12343. [DOI] [PubMed] [Google Scholar]
- Setlow P., Kornberg A. Biochemical studies of bacterial sporulation and germination. 23. Nucleotide metabolism during spore germination. J. Biol. Chem. 1970;245:3645–3652. [PubMed] [Google Scholar]
- Setlow P., Kornberg A. Biochemical studies of bacterial sporulation and germination. XXII. Energy metabolism in early stages of germination of Bacillus megaterium spores. J. Biol. Chem. 1970;245:3637–3644. [PubMed] [Google Scholar]
- Setlow P., Primus G. Protein metabolism during germination of Bacillus megaterium spores. I. Protein synthesis and amino acid metabolism. J. Biol. Chem. 1975;250:623–630. [PubMed] [Google Scholar]
- Setlow B., Melly E., Setlow P. Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J. Bacteriol. 2001;183:4894–4899. doi: 10.1128/JB.183.16.4894-4899.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I.M., Laaberki M.H., Popham D.L., Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg W., Halvorso H., Keynan A., Weinberg E. Timing of protein synthesis during germination and outgrowth of spores of Bacillus cereus strain T. Nature. 1965;208:710–712. [Google Scholar]
- Stragier P., Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 1996;30 doi: 10.1146/annurev.genet.30.1.297. 297–241. [DOI] [PubMed] [Google Scholar]
- Strakova E., Bobek J., Zikova A., Rehulka P., Benada O., Rehulkova H., Kofronova O., Vohradsky J. Systems insight into the spore germination of Streptomyces coelicolor. J. Proteome Res. 2013;12:525–536. doi: 10.1021/pr300980v. [DOI] [PubMed] [Google Scholar]
- Strakova E., Bobek J., Zikova A., Vohradsky J. Global features of gene expression on the proteome and transcriptome levels in S. coelicolor during germination. PLoS ONE. 2013;8:e72842. doi: 10.1371/journal.pone.0072842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman A.S., Douthit H.A. Dormancy in microbial spores. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1973;24:311–352. [Google Scholar]
- Tanimoto Y., Ichikawa Y., Yasuda Y., Tochikubo K. Permeability of dormant spores of Bacillus subtilis to gramicidin S. FEMS Microbiol. Lett. 1996;136:151–156. doi: 10.1111/j.1574-6968.1996.tb08041.x. [DOI] [PubMed] [Google Scholar]
- Tisdale J.H., DeBusk A.G. Permeability problems encountered when treating conidia of neurospora crassa with RNA synthesis inhibitors. Biochem. Biophys. Res. Commun. 1972;48:816–822. doi: 10.1016/0006-291x(72)90680-8. [DOI] [PubMed] [Google Scholar]
- Torriani A., Levinthal C. Ordered synthesis of proteins during outgrowth of spores of Bacillus cereus. J. Bacteriol. 1967;94:176–183. doi: 10.1128/jb.94.1.176-183.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Vasantha N., Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J. Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinter V. Symposium on bacterial spores: V. Germination and outgrowth: effect of inhibitors. J. Appl. Bacteriol. 1970;33:50–59. doi: 10.1111/j.1365-2672.1970.tb05233.x. [DOI] [PubMed] [Google Scholar]
- Zamboni N., Fischer E., Laudert D., Aymerich S., Hohmann H.P., Sauer U. The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J. Bacteriol. 2004;186:4528–4534. doi: 10.1128/JB.186.14.4528-4534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The proteomic timeline of reviving LS5 (ΔmetE) spores as determined by BONCAT. The analysis is based on three independent biological repeats. Only proteins detected in all repeats are displayed.
The proteome of germinating LS5 (ΔmetE) spores incubated with l-alanine, AGFK, or Ca-DPA, as determined by BONCAT. The analysis is based on three independent biological repeats. Only proteins detected in all repeats are displayed.