Abstract
In bacteria, animals, fungi, and many single celled eukaryotes, division is initiated by the formation of a ring of cytoskeletal protein at the nascent division site. In bacteria, the tubulin-like GTPase FtsZ serves as the foundation for the cytokinetic ring. A conserved feature of FtsZ is an intrinsically disordered peptide known as the C-terminal linker. Chimeric experiments suggest the linker acts as a flexible boom allowing FtsZ to associate with the membrane through a conserved C-terminal domain and also modulates interactions both between FtsZ subunits and between FtsZ and modulatory proteins in the cytoplasm.
Mechanistically, cytokinesis exhibits an extraordinary degree of conservation across all domains of life. In bacteria, animals, fungi, and many single celled eukaryotes, division is initiated by the formation of a ring of cytoskeletal protein at the nascent division site. This ring serves as a framework for assembly of the division machinery and constricts at the leading edge of the closing septum during cytokinesis. In animals and fungi this ring is composed of the ATPases actin and myosin.
In bacteria, the essential GTPase FtsZ serves as the foundation for the cytokinetic ring. A distant relative of eukaryotic tubulin, FtsZ forms single stranded polymers in vitro that resemble the 13 protofilaments that run the length of microtubules. Like tubulin, FtsZ binds GTP as a monomer but the active site for GTP hydrolysis is formed at the interface between the two subunits. In vitro, GTP-binding promotes FtsZ polymerization, which further promotes GTP hydrolysis. Although it appears static in conventional micrographs, fluorescence recovery after photo bleaching (FRAP) indicates that the FtsZ ring is highly dynamic with subunit turnover on the order of seconds [1].
In contrast to tubulin, FtsZ does not form structures as highly ordered as a microtubule under standard in vitro assembly conditions. Single stranded polymers, however, do interact laterally in vitro, forming parallel bundles and sheets [2,3]. Super-resolution microscopy and electron cryotomography (ECT) support a model in which single-stranded FtsZ polymers also interact laterally in vivo to form a loosely associated wreath-like structure that varies in thickness around its circumference[4-8].
The single-stranded nature of FtsZ filaments and the apparently cooperative nature of its assembly in vitro, has for years presented a perplexing paradox. Multi-stranded filaments, such as actin and microtubules, undergo cooperative assembly [9-11]. Cooperative filaments assemble in two stages: an energetically unfavorable nucleation phase and an ensuing elongation phase. The affinity of subunits for the filament end is higher than the affinity of a subunit for itself due to the interaction between an adding subunit and multiple other subunit interfaces already at the filament end. Critical concentration and a concentration dependent lag phase are defining features of cooperative assembly. Cooperative filaments display a critical concentration (Cc), a minimum concentration of subunits below which polymerization will not occur. When the subunit concentration rises above the Cc, all additional protein assembles into filaments. At steady state, the equilibrium concentration of monomers will equal the critical concentration. Additionally, cooperative assembly features polymers undergoing a concentration-dependent lag phase during which the nucleus, or smallest species for which elongation is preferred over disassembly, is formed.
Quantitative calorimetric and spectroscopic methods indicate that FtsZ assembly is cooperative [12-14]. FtsZ polymers display a critical concentration for assembly and exhibit the characteristic lag phase seen in many other cytoskeletal filaments. At the same time, the observation that FtsZ forms stable single-stranded filaments in vitro, is ad odds with standard models for cooperative assembly in which the adding subunit interacts with multiple subunits on the growing polymer. Single stranded polymer formation is more consistent with an isodesmic assembly model in which the affinity for dimer formation is the same as polymer elongation. This discrepancy between in vitro experimental data and FtsZ polymer structure have rendered the existing kinetic models of actin or tubulin assembly insufficient for modeling FtsZ assembly kinetics [15]. To overcome these limitations, numerous models have been employed to describe how FtsZ assembles cooperatively yet primarily appears as single-stranded filaments [15,16]. These models predict that subunits undergo conformational changes that facilitate a high affinity active state favoring polymerization. However, detailed molecular and structural evidence for such a high affinity state remains elusive.
The FtsZ C-terminal linker: a model intrinsically disordered region (IDR)
Structurally FtsZ consists of four functional domains: a globular tubulin-like core, an intrinsically disordered C-terminal “linker” (CTL), a highly conserved ~11 residue region known as the C-terminal tail or grappling hook peptide (CTT or GHP), and short, variable set of residues termed the C-terminal variable region or CTV (Figure 1). The negatively charged core encompasses FtsZ’s GTP binding site and the coordinating T7 “loop.” The interface between the nucleotide binding site and the T7 loop in dimers and higher order polymers forms the active site for GTP hydrolysis. The CTT or GHP is the site of interaction between FtsZ and a host of modulatory proteins including the conserved membrane associated protein FtsA that serves to anchor FtsZ to the plasma membrane. Finally, depending on its charge, the CTV can play a significant role in mediating lateral interactions between FtsZ polymers in vitro and in the integrity of the FtsZ ring in vivo.
Figure 1. Structure of B. subtilis FtsZ.
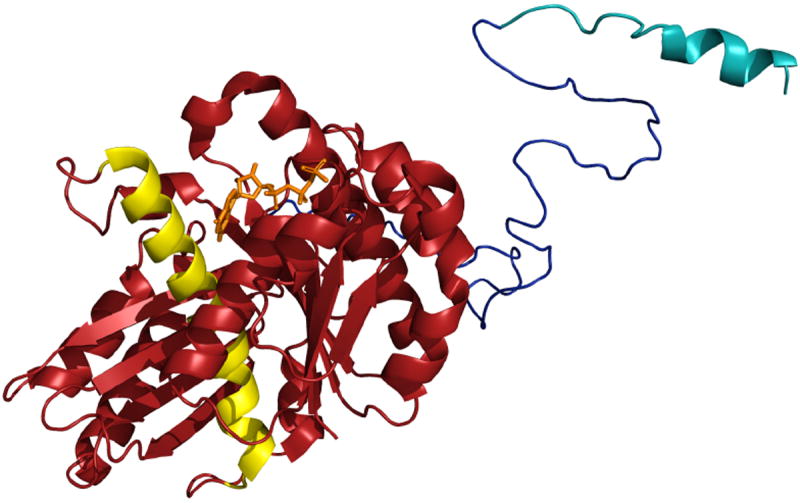
The N-terminal core (maroon) is depicted as bound to GDP [from PDB entry 2RHO [61]]. The flexible C-terminal linker (red) is depicted here as an unstructured peptide. The conserved C-terminal tail (CTT), in blue is depicted as a short α helix based on the structure of the E. coli CTT in complex with ZipA [62].
The CTL, the focus of this review, has received scant attention until recently. The sequence of the CTL is highly variable and like most archetypal IDPs a singular structure cannot be resolved from crystals of FtsZ that contain the CTL. Experimental data support a model in which the CTL serves as a flexible tether between FtsZ and the plasma membrane [17,18]. The importance of the intrinsically disordered CTL to the function of FtsZ has been established unequivocally in recent investigations.
Consistent with its intrinsically disordered nature being important for function, the primary sequences of CTLs are largely fungible as determinants of FtsZ activity. Not only is the linker poorly conserved at the sequence level across species, the CTL of both E. coli and B. subtilis FtsZ can be replaced by non-homologous linkers from other FtsZs and intrinsically disordered regions (IDR) from unrelated proteins without obvious deleterious effects on FtsZ assembly or function[17,18]. CTLs of varying lengths are also tolerated, up to a point. Replacing the ~50 residue native linker with one that is approximately twice as long (~100 residues) has only a modest deleterious impact on FtsZ ring formation in B. subtilis and E. coli. However, increasing linker length further (over 95 residues in E. coli and 100 residues in B. subtilis) completely disrupts FtsZ function in vivo, leading to a lethal block in division[17,18]. Reducing linker length by 50% (from ~50 to ~25 residues for B. subtilis FtsZ) has little effect on FtsZ assembly, whereas replacing E. coli’s linker with a significantly shorter (≤24 residues) IDP from alpha adducin results in a non-functional FtsZ that is unable to complement the heat sensitivity of a conditional ftsZ allele, ftsZ84 [17,18].
The net charge of CTLs has little impact in vivo, although it can interfere with lateral interactions between FtsZ polymers in vitro. Chimeric E. coli FtsZ’s CTLs with charges ranging from +3 to -7 all complement the heat sensitivity of conditional ftsZ84 mutants [17]. While modest changes in the charge pattern of the CTL (e.g. swapping the native B. subtilis CTL with the CTL from E. coli FtsZ) have a minor impact on lateral interactions between FtsZ protofilaments in vitro, it does not meaningfully impair FtsZ assembly or function in vivo. [18]. This impact is likely due to reductions in charge shielding between the linker and the negatively charged core domains in adjacent single stranded FtsZ polymers, as similar effects have been observed when CTV charge is altered [2]. Changes in lateral interaction potential have been implicated in the failure of intrinsically disordered peptides that included an 11-residue sequence from alpha-adducin (QQREKTRWLNS) to complement the heat sensitivity of the ftsZ84ts allele [17]. Dubbed the ‘black sequence’, these 11 residues appeared to strongly promote lateral interactions between polymers of E. coli FtsZ, resulting in paired protofilaments that presumably interfere with FtsZ’s ability to assemble into functional rings in vivo.
In contrast to primary sequence or charge, CTL flexibility is critical for function. B. subtilis FtsZ filaments missing the entire CTL (FtsZΔCTL) display significantly slower assembly kinetics than wild-type filaments, and form irregular, branched filaments [18] (Figure 2). FtsZΔCTL is unable to form protofilaments. Taken together with a decreased GTP turnover rate (~2.5 fold lower than wild type FtsZ) and increased critical concentration for GTP hydrolysis, the data point toward reduced subunit-subunit interactions in the absence of the CTL. Conversely, replacing the CTL with more structurally rigid alpha helices significantly impairs FtsZ filament formation and GTPase activity, implying this chimeric FtsZ lacks conformational flexibility in the N-terminal core locking the subunits in an inactive conformation (Figure 2).
Figure 2. A flexible and disordered C-terminal linker is essential for FtsZ assembly and cell division.
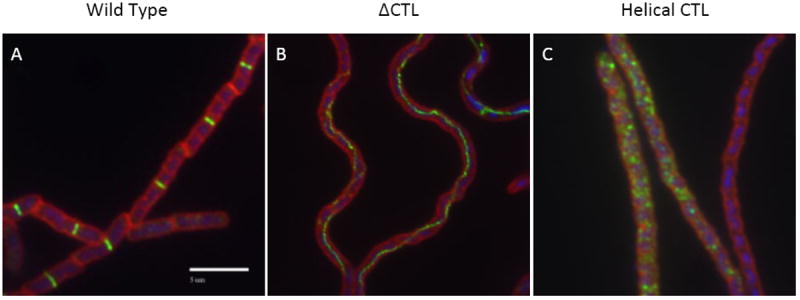
Immunofluorescence micrographs of B. subtilis cells expressing (A) wild type FtsZ (B) a mutant FtsZ missing the CTL but retaining the grappling hook peptide and C-terminal variable region (FtsZ ΔCTL), and (C) a chimeric FtsZ in which the CTL has been replaced with an alpha helical domain of the same size from human beta-catenin (FtsZ CTLH). FtsZ is green DNA is blue and cell wall is red in these images. Wild type FtsZ forms ordered ring-like structures that traverse the width of the cell. (Cells adhere to the slide lengthwise such that the FtsZ rings appear as bands in the micrographs). Neither FtsZ ΔCTL or FtsZ CTLH are able to support division as evidenced by the lack of distinct FtsZ rings and the filamentous nature of the cells. Deletion of the CTL appears to fundamentally change the architecture of FtsZ assembly leading to the formation of filaments or cords of FtsZ polymers that traverse the length of the filaments. FtsZ CTLH is also defective in ring formation forming randomly positioned puncta suggestive of aggregation. Bar = 5μm.
Together these findings suggest that a flexible CTL is critical for the efficient transition of subunits into an active conformation needed for polymerization and cooperative assembly of FtsZ filaments. A role for a flexible CTL in subunit-subunit interaction supports computational results and structural data suggesting that the active FtsZ subunit goes through significant reorientation in the N-terminal core that positions the T7 loop in the nucleotide binding pocket of the adjacent subunit [19-21]. Further structural and biochemical studies will be needed to shed light on specifically how the CTL is able to communicate structurally with the globular core. A highly charged disordered C-terminal tail has recently been shown to coordinate the cooperative assembly of PhuZ, a tubulin homolog from bacteriophage 201φ2-1, into a three-stranded filament [22]. It is possible that the FtsZ CTL, in conjunction with the highly conserved GHP peptide, plays a similar role in the cooperative assembly of single stranded FtsZ polymers.
FtsZ as a generator of constrictive force
Beyond serving as a framework for assembly of the division machinery, a growing body of evidence suggests that FtsZ also serves to generate at least a portion of the force required for constriction of the septum during cytokinesis. In contrast to animal cells in which interaction between a motor protein, myosin, and actin generates the force required for bacterial division, FtsZ does not appear to be associated with any obvious motor proteins. In fact, a recent search of the bacterial database at NCBI revealed no obvious homologues of eukaryotic motor protein in the over 4700 bacterial genomes sequenced to date.
The initial discovery that FtsZ constricts at the leading edge of the invaginating septum in a dividing E. coli cell implicated FtsZ in this process [23]. Coupled with lack of candidate motor proteins, the ability of FtsZ to self-assemble into filaments in the presence of GTP, and its status as the only cell division protein conserved across bacteria and archaea, has led to what has been described as the “Z-centric hypothesis” in which FtsZ not only serves as the framework for assembly of the division machinery, but is also the primary generator of the constrictive force necessary for cytokinesis[24].
Liposome experiments conducted by the Erickson group, suggest that under at least some conditions, FtsZ alone and more convincingly in combination with another highly conserved protein, the membrane anchored ATPase FtsA, is sufficient to drive membrane invagination [25,26]. A YFP tagged FtsZ fusion in which the GHP motif of FtsZ has been replaced with an amphipathic helix to ensure association between FtsZ and the membrane, is sufficient to support assembly of FtsZ rings around the interior circumference of tubular liposomes. In the context of these liposomes, the fluorescently tagged FtsZ (NAME) was able to assemble into rings that appeared to constrict slightly in the presence of GTP[25], suggesting a role for nucleotide hydrolysis in FtsZ force generation. More recently, however, the same group determined that constriction can occur in the absence of GTP, although constrictions are more transient [27].
Additional studies provide strong evidence that FtsA and FtsZ together form a dynamically self-organizing system able to adapt to the rapidly progressing division septum. Experiments in which native E. coli FtsZ and FtsA are added to unilamellar liposomes provide the most convincing demonstration of FtsZ’s force generating potential. Under these conditions, FtsZ forms a ring, which constricts in the presence of both GTP and ATP to form two daughter “cells” [26]. A recent in vitro reconstitution study by Loose and Mitchison revealed that FtsZ is capable of forming large, dynamic structures on lipid bilayers driven by interaction with FtsA and the addition of both GTP and ATP [28]. Together, these experiments suggest that FtsA plays a much more active role in division than previously thought and certainly well beyond its role as a membrane “anchor” for FtsZ.
At the same time, the specific mechanics of FtsZ ring constriction remain elusive. A plethora of models exist for force generation by FtsZ filaments [e.g. [29-34]. ECT and single molecule imaging, specifically photoactivatable light microscopy (PALM), support a model in which short FtsZ protofilaments, anchored to the membrane via interactions with FtsA, form a loose, wreathlike structure around the inner circumference of the cell [4-6]. This type of organization is reminiscent of the sliding filament mechanism of force generation exhibited by actin-myosin filaments in muscle fibers. In this model, FtsZ protofilaments, anchored at different points in the membrane and held together via electrostatic interactions, would slide across one another, bringing their anchor points closer together. On the scale of the entire FtsZ ring, this type of action would lead to membrane “gathering” and constriction. Consistent with this idea, structured illumination microscopy (SIM) suggests the presence of higher concentrations of FtsZ at specific points along the circumference of the cytokinetic ring [8]. The role of these concentrated regions of FtsZ (called “nodes”) is not clear. One, as yet untested, possibility, is that these nodes serve as anchors between sliding filament arrays. At the same time however, in the absence of motor proteins, the forces necessary to drive filament sliding are not readily apparent.
Alternatively, it has been suggested that the intrinsic bend of FtsZ protofilaments after nucleotide hydrolysis might result in generation of contractile force [35-37]. Such models follow closely with the ‘iterative pinching’ model proposed based on the discontinuous ECT images of the Z ring in C. crescentus [6]. Driven by continual dynamic polymerization and depolymerization of FtsZ, short filaments formed on the membrane bend and subsequently generate an inward force over many cycles. Importantly, such a model is consistent with the liposome experiments from the Erickson lab showing that FtsZ is sufficient to bend membranes and recent SIM data revealing the Z ring is undergoing continual redistribution around the cell circumference during a cell cycle. Whether the strength of the bending force is sufficient for constriction remains a topic of intense debate [e.g. [29,38-41].
Models that invoke nucleotide hydrolysis-induced bending of filaments must be reconciled with experimental evidence for cell division in GTP-hydrolysis-deficient FtsZ mutants. An FtsZ mutant in the “synergy loop” (D212G) is GTPase-dead but still supports cell division [42]. GTP hydrolysis clearly is not necessary for constriction. However, Hsin et al., show even the slightly bent FtsZ-GTP filaments can generate ~10 pN of force, still sufficient to carry out division [29,33]. In these GTPase deficient mutants, other cellular factors might contribute to force generation as well. A recent model also shows curvature of filaments can drive formation of either a circular or helical Z ring, as straighter filaments generate helices or spirals[31]. This model perhaps explains structures observed in different ftsZ mutants [43,44].
A role for the CTL in force generation
Even assuming the filaments are capable of sliding, in the absence of a motor protein, specifically how force is generated is not clear. One possibility, suggested by Erickson and colleagues[17], is that the CTL itself functions as an entropic spring maintaining contacts between protofilaments and the membrane via interactions between the GHP and FtsA, as well another membrane associated protein, ZipA [17]. In this model, sliding or bending FtsZ filaments put tension on the CTL springs, which in turn pulls the membrane in towards the cytokinetic ring. Gardner et al estimate that the force necessary to stretch the CTL to half its length (~8.5 nm) requires a force of 9.3 pN. This type of force, exerted at multiple sites around the FtsZ ring would be sufficient to initiate invagination of the septum, if not complete it. The proposed model is consistent with data indicating that linker length is an important determinant of function, although at the same time the range of tolerated lengths is quite large; linkers between 25 and 100 residues were phenotypically indistinguishable in B. subtilis [17,18]. Additionally, substituting the CTL with a stiff polypeptide, such as a portion of the α-helical domain from beta-catenin, not only interferes with division but also interferes with FtsZ ring assembly, making it difficult to assess the specific requirement for flexibility in constriction.
Genetic and cytological data strongly argues against a model where FtsZ generated force is sufficient to drive cytokinesis. Inhibiting the activity of the dozen or so well characterized cell division proteins known to function downstream of FtsZ assembly in E. coli or B. subtilis, results in filamentous cells with regularly spaced FtsZ rings. Although these FtsZ rings likely remain dynamic with regard to subunit turnover, they are unable to support division, maintaining what appears to be a constant diameter over many generations until the cells finally senesce.
Instead of FtsZ doing all the work of constriction, we propose a model in which the initial constriction of the FtsZ ring serves as a physical trigger for cytokinesis (Figure 3). In this model the force generated by bending protofilaments and a constricting Z ring result in mechanical stress sufficient to alter the direction of cell envelope synthesis from lateral to septal. This type of model is reminiscent of the model of Sun and Jiang [45] and is consistent with both cytological data from conditional E. coli mutants suggesting division is an all or nothing phenomenon. A model in which a minor FtsZ-driven constriction triggers cross wall biosynthesis is supported by electron micrographs of the Gram-positive bacterium Bacillus subtilis in which small nubs of cell wall are visible at the poles of sporulating cells that previously harbored unused polar FtsZ rings [46-48].
Figure 3. The flexible FtsZ linker as a transducer of constriction force.
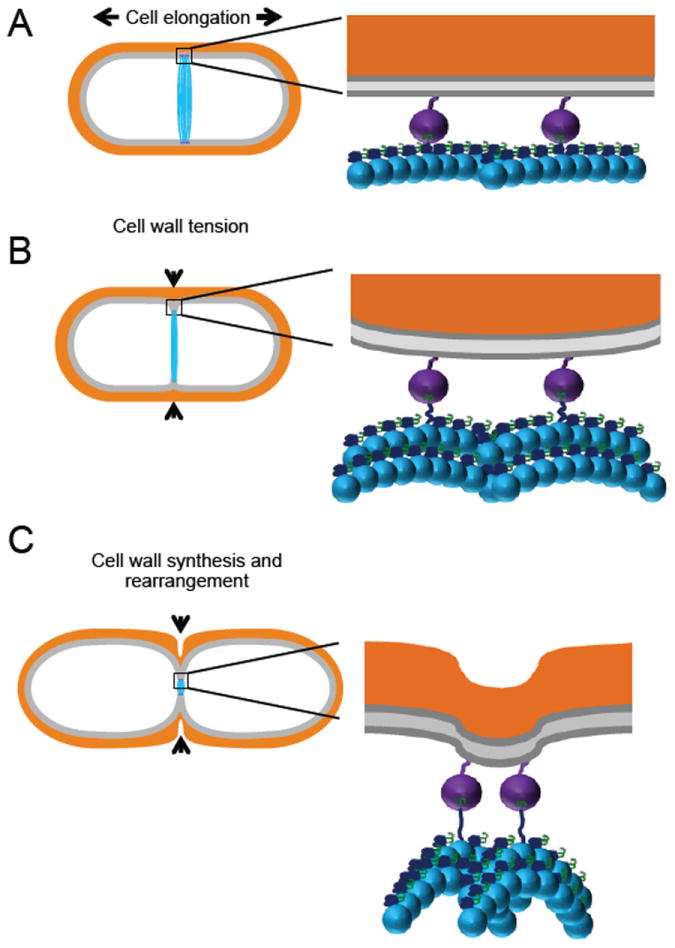
A. As cell division begins the Z ring (blue) anchored by FtsA (purple) localizes to midcell. Growth of the cell wall is directed outward. Inset: FtsZ filaments (blue) are anchored to the membrane through the interaction between its grappling hook peptide (GHP; green) and FtsA (purple). After initial filament formation and tethering to the membrane FtsZ filaments remain primarily straight in their GTP-bound form. B. Initial constriction of the Z ring created inward tension on the cell wall. Inset: Curved FtsZ filaments induced after GTP hydrolysis bend in away from the membrane. The force from bending filaments causes the CTL (dark blue) of subunits anchored to FtsA to extend and deform the membrane inward. C. Continued Z-ring constriction and invagination of the membrane serves as a signal for the cell-wall synthesis machinery to form of the leading division septum. Inset: FtsZ filaments continue to bend further as more subunits inhabit their GDP-bound state. The flexible CTL extends taught and further pulls on the membrane leading to re-arrangement of the cell wall to follow. Membrane = gray and cell wall = orange.
In this model, the key step is deformation of the plasma membrane at the initiation of division. Based on the computational data that estimates the constriction force required for cell division is on the order of ~8 pN [40] and that a FtsZ filament can generate 10-30 pN of force through bending due to GTP hydrolysis [33,37], the question remains how does the bending force act upon the membrane? Since the filament does not interact directly with the membrane but is anchored to the membrane through another protein like FtsA, this suggests that the force must be transduced through another mechanism.
We would suggest that the initial constrictive force is transmitted through the CTL itself. Subunits at the end of a filament bend inwards away from the membrane, due to the intrinsic properties associated with GTP hydrolysis. In response, the linker, because it is flexible, extends to keep the filament anchored to the membrane. In effect, the linker pulls the membrane inward as the filament continues to bend and the linker approaches its contour length. Filament ends have been estimated to be approximately 20 nm from the membrane when bent in an intermediate conformation and their center anchored at the membrane [16]. With a contour length of ~17 nm, the FtsZ linker should be almost fully extended in this conformation and further bending would lead to distortion of the membrane. Support for such a model comes from observations that an FtsZ construct in which the CTL is approximately five times longer than wild type forms Z rings but cannot support cell division [18]. Such a long linker is unable to be extended to its contour length and transduce any force to the membrane even when FtsZ filaments are fully bent, resulting in assembled but impotent cytokinetic rings.
An opportunity to connect sequence-disorder relationships to function
Computational analysis suggests that the FtsZ CTL is an archetypal example of an intrinsically disordered peptide or IDP. Systematic investigations have enabled the establishment of a predictive diagram-of-states for inferring the relationship between sequence features and conformational properties of IDPs (Figure 4) [49-57]. The diagram-of-states is based on composition specific parameters, such as the net charge per residue (NCPR) and the fraction of charged residues (FCR), as inputs to make inferences regarding the predominant conformational states – globules, coils, or chimeras of globules and coils – for a specific IDP sequence. The classification of IDPs into distinct conformational classes applies to sequences that are at least thirty residues long, have low overall hydrophobicity, and low proline contents.
Figure 4. Diagram-of-states for IDPs.
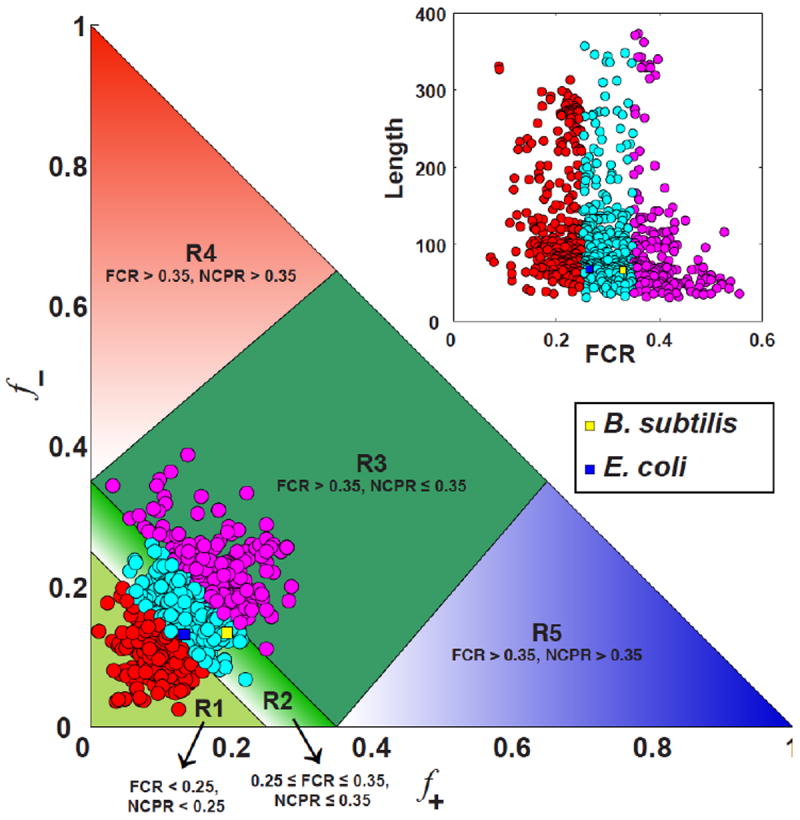
Sequences with R1 are predicted to form globules those with R2 will be chimeras of globules and coils whereas the sequences within R3 will form classic random coils if the charges are well mixed within the linear sequence (low κ), and hairpins if the charges are segregated within the linear sequence (high κ). The conformational properties are expected to vary continuously as a function of FCR. We annotated the diagram-of-states using information gleaned from the analysis of CTL sequences from different FtsZ proteins of 1,209 distinct bacteria. The inset shows a scatter plot of all 1,209 sequences in the two-parameter space of FCR and CTL sequence length. The color-coding used in this scatter plot corresponds to the annotation of the diagram-of-states.
Polar tracts (NCPR ≈ 0 and FCR ≈ 0) and weak polyelectrolytes / polyampholytes (NCPR ≤ 0.25 and FCR ≤ 0.25) are predicted to form heterogeneous distributions of globules that minimize their interface with aqueous solvents [49-52,58,59]. Polyelectrolytic sequences for which NCPR > 0.3 and FCR > 0.35 should form swollen random coil conformations that maximize chain-solvent interactions and minimize intra-chain electrostatic repulsions [52]. Sequences of polyampholytic IDPs include both types of charged residues and the sequences of CTLs are best described as either weak or strong polyampholytes based on their FCR values.
Recent work has shown that the conformational properties of polyampholytes are governed by the linear sequence distribution of opposite charges [56]. The mixing versus segregation of oppositely charged residues is calculated using a parameter κ. For a given IDP sequence, f+ and f− denote the fraction of positive and negatively charged residues, respectively. These fractions are calculated using the intrinsic pka values of amino acids and essentially tallies the fraction of Lys / Arg (f+) and Asp / Glu (f−) residues within a sequence. The overall charge asymmetry is defined as [60]. For each sequence variant we calculate κ by partitioning the sequence into n overlapping windows of size g. For each g-residue window we calculate , which is the charge asymmetry for blob i in the sequence of interest. Using this we quantify the mean squared deviation from σ as . Different sequence variants will have different values of δ, and the maximal value δmax for a given amino acid composition is used to define such that 0 ≤ κ ≤ 1. We calculate κ using two values for the window size, g = 5 and 6, and the final κ for a given sequence variant is an average of the two values.
Low values of κ are realized for well-mixed sequence variants and κ → 1 if oppositely charged residues are segregated in the linear sequence. If opposite charges are well-mixed within the linear sequence, then electrostatic repulsions and attractions counterbalance each other and the result is random coil like conformations. Conversely, if opposite charges are well segregated in the linear sequence, then IDPs form semi-compact hairpin-like conformations. Sequences of intermediate compositional biases (NCPR < 0.25 and 0.25 < FCR < 0.35) form a fuzzy boundary between the sequences of low and high FCR.
What do quantitative sequence-disorder relationships say about the CTL of FtsZ? Figure 4 shows our annotation of the predictive diagram-of-states using 1,209 different sequences for FtsZ CTLs. Of these 1,209 sequences, 34% fall in region R1, 47% in region R2, and 19% correspond to region R3 (Figure 5). The inset to Figure 4 depicts the properties of sequences drawn from regions R1, R2, and R3 in the space of FCR and CTL length. As shown in panel A of Figure 5, longer CTL sequences generally have lower FCR values, whereas sequences with higher FCR values tend to be shorter in length. This observation highlights the fact that inasmuch as length and FCR are covarying quantities, one must proceed with caution in interpreting the effects of changes to CTL length on FtsZ function, both in cytokinesis and force generation. Simple models that rely on worm-like chain model descriptions for the CTL are likely to be misleading.
Figure 5. Properties of FtsZ CTL sequences.
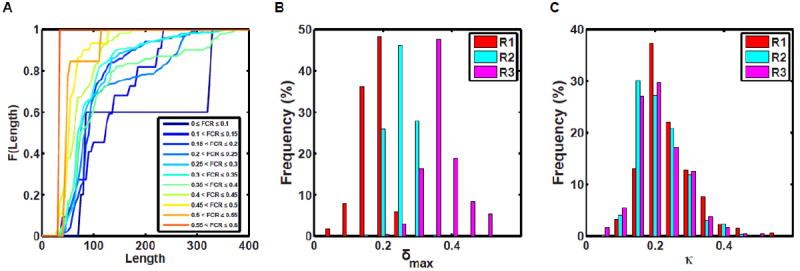
(A) Cumulative distribution functions (CDFs) quantifying the probability that a CTL sequence corresponding to a specific FCR interval will have a chain length less than or equal to a specific value. The CDFs corresponding to sequences with lower FCR values are shown using the cooler colors whereas CDFs corresponding to higher FCR values are shown using hotter colors. The overall message of this analysis is that CTL sequences with higher FCR values tend to be shorter (< 100 residues) than those with lower FCR values. (B) Histograms quantifying the frequencies with which CTL sequences corresponding to different values of δmax are realized in different regions of the diagram-of-states. The lower the δmax values the fewer the number of sequences one can generate with distinct conformational properties. Therefore, δmax quantifies the overall designability for a give amino acid composition and this designability increases with FCR and is highest for sequences drawn from R3 and is lowest for sequences drawn from R1. (C) Histograms quantifying the distribution of κ values for CTL sequences drawn from different regions of the diagram-of-states.
The linear patterning of oppositely charged residues within the CTL sequences are likely to also be relevant for FtsZ functions, for the evolution of these sequences, and for the inferred robustness / fungible nature of CTLs. For a given amino acid composition, one can generate numerous distinct sequences that span the range of κ values. The number of distinct sequences one can generate for a given composition will be set by the FCR value, which in turn determines δmax. Panel B in Figure 5 shows the distribution of δmax values for CTL sequences drawn from different regions of the diagram-of-states shown in Figure 4. We see a clear trend of δmax shifting toward higher values as FCR increases, which is in line with the expectation that a wider range of sequences with distinct charge patterns can be generated as FCR increases. Panel C in Figure 5 shows the distribution of κ values for CTL sequences drawn from regions R1, R2, and R3 of the diagram-of-states shown in Figure 5. Each of the distributions is peaked around a κ value of 0.2 and the tails of these distributions suggest a selection against sequences of very low κ values or κ values greater than 0.4.
The sequence characteristics of CTLs imply two testable predictions: First, we expect there to be a covariation between FCR and chain length, whereby the CTL length increases as FCR decreases and vice versa. This covariation should conserve coarse grain properties such as the overall size and shape distributions of disordered CTLs and this could explain the ability to interchange CTLs between different bacterial sequences. Secondly, for a given amino acid composition, we expect that changes to κ, achieved through sequence design, should change the overall conformational properties thus allowing us to test the impact of κ and hence the impact of sequence-disorder relationships of CTLs on bacterial cytokinesis mediated by FtsZ. Specifically, the distribution of κ values suggests that CTL sequences with κ values below 0.1 and above 0.45 are likely to yield functionally impaired FtsZs.
In the context of the overall model that we propose for the critical role of the CTL in constriction, we expect that CTL sequences that result from de novo design will lead to deleterious consequences for FtsZ functions if one of the following were true: (i) The parent CTL sequence belongs to region R1, and the de novo designed sequence conserves FCR and significantly alters the CTL length. (ii) The properties of the designed sequence are fundamentally altered by changes to the amino acid composition that lead to a significant decrease or increase of FCR, which would imply a change in conformational class vis-à-vis the diagram-of-states. (iii) For a parent sequence derived from R2 or R3, if we were to leverage the intrinsic designability afforded by the amino acid composition to deploy CTL designs with κ values that lie well outside the range of observed values. If the κ becomes too high, the CTL should either become unresponsive or occlude longitudinal interactions because it is locked up in strong intra-chain electrostatic interactions, whereas CTL variants with very low κ are likely to either be readily degraded or exceed a putative flexibility threshold, which could impair lateral associations and / or force generation.
It is clear that the CTL plays an essential if underappreciated role in FtsZ assembly and bacterial cell division. Taking advantage of computational modeling, improved biophysical characterization of IDPs, and cytological, biochemical and genetic analysis we are confident that ongoing and future work will help us elucidate the connection between sequence-disorder relationships of CTLs and their connection to FtsZ function. Success in such an endeavor will, in all likelihood, provide a template for leveraging the growing body of sequence-disorder studies to understand the role of specific sequence-disorder relationships in IDP functions for a wide range of systems.
Acknowledgments
Work in the Levin laboratory is funded in part by a National Institutes of Health Public Health Service Grant GM64671 to PAL. RVP acknowledges support from the National Science Foundation through grant MCB-1121867
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–81. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buske P, Levin PA. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem. 2012 doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–54. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang S-C, Hatem C, et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89:1099–120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS ONE. 2010;5:e12680–16. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilhofer M, Jensen GJ. The bacterial cytoskeleton: more than twisted filaments. Current Opinion in Cell Biology. 2013;25:125–33. doi: 10.1016/j.ceb.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss MP, Liew ATF, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM Super Resolution Microscopy Reveals a Bead-Like Arrangement for FtsZ and the Division Machinery: Implications for Triggering Cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oosawa F, Kasai M. A theory of linear and helical aggregations of macromolecules. J Mol Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 10.Wegner A, Engel J. Kinetics of the cooperative association of actin to actin filaments. Biophys Chem. 1975;3:215–25. doi: 10.1016/0301-4622(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 11.Gaskin F, Cantor CR, Shelanski ML. Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. J Mol Biol. 1974;89:737–55. doi: 10.1016/0022-2836(74)90048-5. [DOI] [PubMed] [Google Scholar]
- 12.Caplan MR, Erickson HP. Apparent cooperative assembly of the bacterial cell division protein FtsZ demonstrated by isothermal titration calorimetry. J Biol Chem. 2003;278:13784–8. doi: 10.1074/jbc.M300860200. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Bjornson K, Redick SD, Erickson HP. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J. 2005;88:505–14. doi: 10.1529/biophysj.104.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Erickson HP. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem. 2005;280:22549–54. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miraldi E, Thomas PJ, Romberg L. Allosteric Models for Cooperative Polymerization of Linear Polymers. Biophys J. 2008 doi: 10.1529/biophysj.107.126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–28. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner KAJA, Moore DA, Erickson HP. The C-terminal Linker of Escherichia coli FtsZ Functions as an Intrinsically Disordered Peptide. Mol Microbiol. 2013 doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buske P, Levin PA. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol. 2013 doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martín-Galiano AJ, Buey RM, Cabezas M, Andreu JM. Mapping flexibility and the assembly switch of cell division protein FtsZ by computational and mutational approaches. J Biol Chem. 2010;285:22554–65. doi: 10.1074/jbc.M110.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsui T, Han X, Yu J, Yao M, Tanaka I. Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop. J Biol Chem. 2014;289:3501–9. doi: 10.1074/jbc.M113.514901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Erickson HP. Conformational changes of FtsZ reported by tryptophan mutants. Biochemistry. 2011;50:4675–84. doi: 10.1021/bi200106d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehr EA, Kraemer JA, Erb ML, Coker JKC, Montabana EA, Pogliano J, et al. The structure and assembly mechanism of a novel three-stranded tubulin filament that centers phage DNA. Structure. 2014;22:539–48. doi: 10.1016/j.str.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–4. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 24.Erickson HP. FtsZ, a tubulin homologue in prokaryote cell division. Trends in Cell Biology. 1997;7:362–7. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- 25.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–4. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USa. 2013 doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osawa M, Erickson HP. Inside-out Z rings--constriction with and without GTP hydrolysis. Mol Microbiol. 2011;81:571–9. doi: 10.1111/j.1365-2958.2011.07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allard JF, Cytrynbaum EN. Force generation by a dynamic Z-ring in Escherichia coli cell division. Proc Natl Acad Sci USa. 2009;106:145–50. doi: 10.1073/pnas.0808657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews SS, Arkin AP. A mechanical explanation for cytoskeletal rings and helices in bacteria. Biophys J. 2007;93:1872–84. doi: 10.1529/biophysj.106.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer-Friedrich E, Friedrich BM, Gov NS. FtsZ rings and helices: physical mechanisms for the dynamic alignment of biopolymers in rod-shaped bacteria. Phys Biol. 2012;9:016009. doi: 10.1088/1478-3975/9/1/016009. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh B, Sain A. Force generation in bacteria without nucleotide-dependent bending of cytoskeletal filaments. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:051924. doi: 10.1103/PhysRevE.83.051924. [DOI] [PubMed] [Google Scholar]
- 33.Hsin J, Gopinathan A, Huang KC. Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. Proc Natl Acad Sci USa. 2012;109:9432–7. doi: 10.1073/pnas.1120761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shlomovitz R, Gov NS. Membrane-mediated interactions drive the condensation and coalescence of FtsZ rings. Phys Biol. 2009;6:046017. doi: 10.1088/1478-3975/6/4/046017. [DOI] [PubMed] [Google Scholar]
- 35.Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–70. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H, Si F, Margolin W, Sun SX. Mechanical control of bacterial cell shape. Biophys J. 2011;101:327–35. doi: 10.1016/j.bpj.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Hsin J, Zhao L, Cheng Y, Shang W, Huang KC, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–5. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol. 2008;18:235–44. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 39.Huecas S, Llorca O, Boskovic J, Martin-Benito J, Valpuesta JM, Andreu JM. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys J. 2008;94:1796–806. doi: 10.1529/biophysj.107.115493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan G, Wolgemuth CW, Sun SX. Z-ring force and cell shape during division in rod-like bacteria. P Natl Acad Sci USA. 2007;104:16110–5. doi: 10.1073/pnas.0702925104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner DJ, Portman I, Dafforn TR, Rodger A, Roper DI, Smith CJ, et al. The mechanics of FtsZ fibers. Biophys J. 2012;102:731–8. doi: 10.1016/j.bpj.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee A, Saez C, Lutkenhaus J. Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J Bacteriol. 2001;183:7190–7. doi: 10.1128/JB.183.24.7190-7197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Addinall SG, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–72. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monahan LG, Robinson A, Harry EJ. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol Microbiol. 2009;74:1004–17. doi: 10.1111/j.1365-2958.2009.06914.x. [DOI] [PubMed] [Google Scholar]
- 45.Sun SX, Jiang H. Physics of bacterial morphogenesis. Microbiol Mol Biol Rev. 2011;75:543–65. doi: 10.1128/MMBR.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Gene Dev. 1996;10:478–88. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 47.Ryter A, Schaeffer P, Ionesco H. Classification cytologique, par leur stade de blocage des mutants de sporulation de Bacillus subtilis Marburg [Cytologic classification by their blockage stage of sporulation mutants of Bacillus subtilis Marburg] Ann Inst Pasteur (Paris) 1966;110:305–15. [PubMed] [Google Scholar]
- 48.Ryter A. Etude morphologique de la sporulation deBacillus subtilis. Annales De l’Institut Pasteur. 1964;108:40–60. [PubMed] [Google Scholar]
- 49.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci USa. 2006;103:16764–9. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitalis A, Wang X, Pappu RV. Quantitative Characterization of Intrinsic Disorder in Polyglutamine: Insights from Analysis Based on Polymer Theories. Biophysical Journal. 2007;93:1923–37. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran HT, Mao A, Pappu RV. Role of Backbone Solvent Interactions in Determining Conformational Equilibria of Intrinsically Disordered Proteins. J Am Chem Soc. 2008;130:7380–92. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 52.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USa. 2010;107:8183–8. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das RK, Mittal A, Pappu RV. How is functional specificity achieved through disordered regions of proteins? Bioessays. 2012;35:17–22. doi: 10.1002/bies.201200115. [DOI] [PubMed] [Google Scholar]
- 54.Das RK, Crick SL, Pappu RV. N-Terminal Segments Modulate the α-Helical Propensities of the Intrinsically Disordered Basic Regions of bZIP Proteins. J Mol Biol. 2012;416:287–99. doi: 10.1016/j.jmb.2011.12.043. [DOI] [PubMed] [Google Scholar]
- 55.Das RK, Mao AH, Pappu RV Science Signaling ∣ Science/AAAS. Science Signaling. 2012 doi: 10.1126/scisignal.2003091. [DOI] [PubMed] [Google Scholar]
- 56.Mao AH, Lyle N, Pappu RV. Describing sequence–ensemble relationships for intrinsically disordered proteins. Biochem J. 2012;449:307–18. doi: 10.1042/BJ20121346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babu MM, Kriwacki RW, Pappu RV. Versatility from protein disorder. Science. 2012 doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 58.Halfmann R, Alberti S, Krishnan R, Lyle N, O’Donnell CW, King OD, et al. Opposing Effects of Glutamine and Asparagine Govern Prion Formation by Intrinsically Disordered Proteins. Molecular Cell. 2011;43:72–84. doi: 10.1016/j.molcel.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins. 2005;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- 60.Dobrynin AV, Rubinstein M. Flory Theory of a Polyampholyte Chain. J Phys II France. 1995;5:677–95. [Google Scholar]
- 61.Raymond A, Lovell S, Lorimer D, Walchli J, Mixon M, Wallace E, et al. Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer. BMC Biotechnol. 2009;9:37. doi: 10.1186/1472-6750-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, et al. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–91. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]


