Introduction
A key challenge in neuroscience and, in particular, neuroimaging, is to move beyond identification of regional activations toward the characterization of functional circuits underpinning perception, cognition, behavior, and consciousness. Granger causality (G-causality) analysis provides a powerful method for achieving this, by identifying directed functional (“causal”) interactions from time-series data. G-causality implements a statistical, predictive notion of causality whereby causes precede, and help predict, their effects. It is defined in both the time and frequency domains, and it allows for the conditioning out of common causal influences. In this paper we explain the theoretical basis and computational implementation of G-causality analysis in neuroimaging and, more broadly, in neurophysiology, noting both its exciting potential and the assumptions that govern its application and interpretation.
Concepts of brain connectivity are becoming increasingly prevalent as neuroscientists seek to unravel the detailed circuitry underlying perception, cognition, and behavior. Efforts to characterize structural connectivity—the brain's “connectome”—through anatomical methods are now complemented by an intense focus on “functional” and “effective” connectivity, through statistical analysis of neural signals. The distinction between functional and effective connectivity is crucial when implementing and interpreting these analyses, but it remains sometimes misunderstood. In a nutshell, functional connectivity aims at describing the statistical dependencies between two or more variables, where these dependencies can be undirected (as in correlation or coherence) or directed (as in G-causality and “transfer entropy;” Granger, 1969; Schreiber, 2000). Being descriptions of data, functional connectivity analyses make minimal assumptions about the underlying (physical) mechanisms. In contrast, effective connectivity analyses aim to find the simplest possible circuit diagram explaining observed responses (Friston et al., 2013) and work in general by comparing how well distinct mechanistic models perform in accounting for observed data. In neuroscience this approach is exemplified by dynamic causal modeling (DCM; Friston et al., 2003), which applies a Bayesian framework to assess model performance. The distinction between effective and functional connectivity clarifies why methods like G-causality and DCM are complementary rather than competitive—they make different assumptions, and they permit different interpretations (Friston et al., 2013). Here, we provide a brief introduction to G-causality, explaining its theoretical basis, computational strategy, and the possibilities and limitations that arise in neuroscience and neuroimaging applications.
G-causality analysis
G-causality is based on the simple idea that causes both precede and help predict their effects. This idea can be traced to at least Norbert Wiener and was operationalized by the econometrician Clive Granger (Granger, 1969) in terms of linear vector autoregressive (VAR) models of stochastic time-series data, with important generalizations later provided by John Geweke (Geweke, 1982). VAR models are simple mathematical models in which the value of a variable at a particular time is modeled as a (linear) weighted sum of its own past (usually over a number of discrete time-steps), and of the past of a set of other variables. Each variable is a vector stochastic process representing a time series. Fitting a VAR model amounts to finding the optimal weights so that estimation errors are minimized; many standard techniques exist for doing this. In this setting, G-causality says that a variable X “G-causes” another variable Y if the past of X contains information that helps predict the future of Y, over and above the information already in the past of Y itself (and in the past of other “conditioning” variables Z). In general terms, when this condition is satisfied one can say that there is “information flow” from X to Y. This is justified because G-causality is an approximation to transfer entropy (the approximation is exact for Gaussian variables), which is a directed version of Shannon's mutual information, which in turn is a very general way of characterizing the statistical dependency or shared information between two variables (Barnett et al., 2009).
In practice, the computational strategy for implementing G-causality analysis (GCA) rests on estimating and comparing two VAR models, given a set of time-series data. Assume we have 3 variables: X, Y, and Z, and are interested in measuring information flow from X to Y. First, a “full” VAR model is estimated jointly for all the variables. This leads to a particular prediction/estimation error for each variable within the set. A second “reduced” VAR model is then estimated, which omits the potential cause (X, in the example above). This leads to a second set of prediction errors for each remaining variable. If the prediction error for Y is significantly smaller for the full regression (including X), as compared with the reduced regression (excluding X), then we say that X G-causes Y, conditioned on Z. (Note that we also have, from the same models, the G-causality from X to Z conditioned on Y). Technically, the magnitude of the G-causality is given by the ratio of the variance of the prediction-error terms for the reduced and full regressions. If the data are Gaussian (i.e., normally distributed) the equivalence with transfer entropy (Barnett et al., 2009) means that G-causality magnitudes can be interpreted in terms of information-theoretic bits-per-unit-time.
Since G-causality rests on comparisons of model error, it only makes sense in situations where variables can be modeled as having random variations—i.e., as stochastic. Another important assumption, at least standardly, is that the data are (weakly) stationary, which means that the means and variances of the variables are stable over time. Given these assumptions, G-causality has several attractive properties (Geweke, 1982; Barnett and Seth, 2014). First, it is easy to compute since there are several standard algorithms for optimal estimation of (linear) VAR models (e.g., ordinary least squares, Levinson–Wiggs–Robinson equations). Second, since VAR models are very general, there is no need to make a priori assumptions about the underlying physical mechanisms, other than that they generate data suitable for VAR modeling. Third, the G-causality statistic follows (asymptotically) a χ2 distribution, so for large sample sizes one can perform statistical significance testing without recourse to permutation or bootstrapping methods. Fourth, VAR models can be easily transformed into the frequency domain, allowing spectral (“band-limited”) estimations of G-causality (though here surrogate methods are needed to test statistical significance). Finally, G-causality is invariant under rescaling of variables, meaning that it is unaffected by overall signal strength. Altogether, G-causality offers a simple yet powerful means for characterizing information flow within a network of variables, in both time and frequency domains, while making minimal assumptions about the underlying generative mechanisms. We note, but do not discuss here, that G-causality is closely related to other data-driven methods including partial directed coherence and the directed transfer function (Florin et al., 2011). Nonparametric implementations of G-causality are also possible (Dhamala et al., 2008).
Although GCA, like most statistical analyses, usually requires adaptation to each specific application, a general approach can be identified. This approach is based on the freely available multivariate Granger causality analysis (MVGC) MATLAB (MathWorks) toolbox, which was designed for applications in neuroscience (Barnett and Seth, 2014). The process starts with the recording of time-series data, either during steady-state conditions or while a task is being performed. These data are then checked for stationarity via standard techniques. If the data are initially nonstationary, a range of preprocessing steps can be considered. Drifts and slow fluctuations can be removed by detrending (linear or piecewise), differencing, and/or high-pass filtering. [Filters should be chosen to have a low model order, to minimize difficulties in VAR model fitting (Barnett and Seth, 2014).] Oscillatory nonstationary features like electrical line noise can be removed by notch filtering or other methods. The data can also be windowed into shorter, possibly more stationary, epochs: given sufficient data, this method can shed light on time-varying G-causality (Ding et al., 2000). Finally, analysis can focus on limited data segments where stationarity applies. Whereas in most neuroscience contexts some combination of these steps will suffice, if nonstationarities persist, other more sophisticated approaches will be needed (Hesse et al., 2003; also see below, Interpretations, limitations, and emerging directions).
Next, full and reduced VAR models are estimated (note that the MVGC toolbox is able to extract full and reduced VAR model coefficients from a single full model, avoiding a source of bias). This process requires estimation of model order—i.e., the number of past observations (time-steps) to be included in the VAR models. Excessively high (low) model orders will lead to overfitting (underfitting) of the data. Again, standard techniques exist for estimating an optimal model order, which are based on balancing model complexity (broadly, number of parameters) against error. It is worth noting that model order estimation is constrained by the data, not by the underlying physical mechanism. Thus, data obtained with a low sampling rate (e.g., fMRI data) will typically be associated with low model orders (1 or 2 at most). Similarly, fast-sampled data (e.g., EEG) may require prior downsampling to achieve an appropriate balance between model complexity and the time span covered by a VAR model (e.g., a model order of 25 at 250 Hz covers a time scale of 100 ms). Finally, G-causality values can be computed and checked for statistical significance, either by comparison against standard analytic distributions (time domain) or via surrogate statistics (frequency domain).
The interpretation of G-causality naturally rests on the experimental design. In general, GCA is better suited to revealing differences in directed functional connectivity between experimental conditions than to characterizing these connectivity patterns per se. Another powerful approach is to correlate trial-by-trial differences in behavioral variables (e.g., response times) with G-causality values (Wen et al., 2012). Given the need for stationarity, steady-state or prestimulus periods may be more suitable for GCA than complex time-varying induced or evoked signals.
G-causality in neuroimaging and neurophysiology
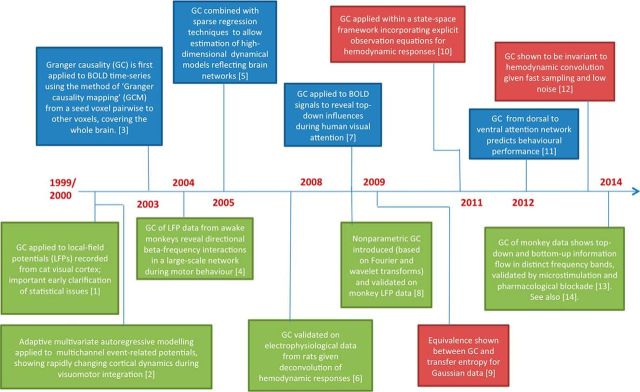
A popular setting for GCA in neuroscience is in functional neuroimaging, where there is an increasing appetite to move beyond the identification of regional activations toward the characterization of functional circuits (Friston et al., 2013). The common neuroimaging modalities, fMRI, MEG/EEG, and electrocorticography (ECoG), each offer distinctive opportunities and limitations. GCA of intracranially recorded electrophysiological data like local field potentials and intracranial EEG (iEEG) follows the same principles as for EEG and MEG, but without reference to the indirect nature of “sensor space” data. A selection of landmark empirical and theoretical developments in GCA for neuroscience and neuroimaging is given in Figure 1.
Figure 1.
Selected landmarks in the development and application of G-causality for neuroscience and neuroimaging. Blue boxes refer to fMRI studies and green boxes to electrophysiological studies; red boxes show theoretical/modeling advances. The embedded references are as follows: [1], Bernasconi and König (1999); [2], Ding et al., (2000); [3], Goebel et al. (2003); [4], Brovelli et al. (2004); [5], Valdés-Sosa et al. (2005); [6], David et al. (2008); [7], Wang et al. (2008); [8], Dhamala et al. (2008); [9], Barnett et al. (2009); [10], Ryali et al. (2011); [11], Wen et al. (2012); [12], Seth et al. (2013); [13], van Kerkoerle et al. (2014); [14], Bastos et al. (2015).
G-causality of magneto/electrophysiological data
Data obtained from electrical or magnetic recordings of continuous neural activity are well suited to GCA in virtue of having high temporal resolution. For steady-state data the primary consideration is to ensure stationarity. As mentioned, this may require detrending, differencing, windowing, and/or filtering. Importantly, as explained below, (temporal) filtering should be used sparingly and only as needed for stationarity purposes. For event-related or induced data, nonstationarity is likely to be a common issue which can be tackled either by (1) a “vertical” regression in which VAR models are estimated for very short windows across trials, rather than for each trial separately, or (2) removing the ensemble average ERP. The former assumes that each trial is an independent realization of the same underlying stochastic process; the latter assumes minimal intertrial variation in the ERP (Wang et al., 2008). For analyses in sensor space it can be useful to apply a spatial filter (e.g., a surface Laplacian) to spatially decorrelate the data and ameliorate the effects of volume conduction (especially relevant for EEG; Cohen, 2014).
GCA of electrophysiological data offers the important advantage of spectral analysis; that is, G-causal influences can be tied to specific frequency bands (Geweke, 1982). This is very useful when testing neurophysiological hypotheses that attribute specific functional roles for different neural oscillations. For example, recent applications to monkey electrophysiological data have revealed “top-down” G-causality influences in the alpha and beta ranges, and “bottom-up” influences in the gamma range, consistent with popular predictive processing frameworks (Bressler and Richter, 2014; van Kerkoerle et al., 2014; Bastos et al., 2015). It is critical that analyses like these are conducted using band-limited G-causality directly in the frequency domain (Geweke, 1982; Barnett and Seth, 2014), rather than prefiltering the data into the desired frequency bands and applying time-domain G-causality. The reason is that, perhaps surprisingly, G-causality is theoretically invariant to (invertible) filtering operations (Barnett and Seth, 2011). In practice, GCA of (temporally) filtered data will produce results different from those for GCA of unfiltered data; however, the differences will reflect sub-optimalities in VAR model fitting of filtered data and not the desired band-limited G-causalities. Note that integrating band-limited G-causality across all frequencies (up to the Nyquist frequency) should recover the time-domain G-causality. GCA of related signals such as ECoG and iEEG data follows the same principles, although nonstationarity issues may manifest differently. Application to spike-train data derived from single-unit recordings requires a different approach recognizing the point-process nature of such data (Kim et al., 2011).
G-causality of fMRI data
G-causality analysis of fMRI data has been highly controversial (David et al., 2008; Roebroeck et al., 2011), and a detailed treatment is beyond the current scope. Although fMRI furnishes time-series data with high spatial precision, the indirect relationship between the fMRI BOLD signal and the underlying neural mechanisms pose important challenges for GCA beyond those already faced by standard regional or correlational analyses. Two features are especially problematic: (1) the sampling rate (repetition time or TR) for fMRI data is usually very low, of the order of seconds, which is much slower than the millisecond timescale of the underlying neuronal responses; and (2) fMRI responses reflect convolution with an HRF, which imposes long delays (in terms of “time-to-peak”) with respect to neural activity and, even worse, may have significant inter-regional variability (Handwerker et al., 2012). A typical worrisome scenario might be one in which region X causally influences region Y at the neuronal level, but the HRF for X has a longer time-to-peak than the HRF for Y. One might suspect that GCA of BOLD signals from X and Y would lead to the incorrect inference that Y G-causes X as a result of the confounding hemodynamic delays. However, this suspicion does not hold up under scrutiny. In fact, because the HRF acts as a (slow, moving-average) filter, the invariance to filtering implies that G-causality is invariant to HRF variability, a result now established both theoretically and in detailed modeling (Seth et al., 2013). Unfortunately, for this invariance to apply in practice, the TR must be of the order of the neuronal delays, which is not currently feasible, although recent developments in ultrafast sequences are promising (Feinberg and Setsompop, 2013). Also, full invariance requires that the HRF act as an invertible filter, which may not always be the case. Even if HRFs do not vary among regions, GCA of heavily downsampled and convolved data can still lead to inflated false negatives and false positives (Seth et al., 2013). Other simulations have indicated that a monotonic relationship is preserved between G-causality at the neural level and G-causality in simulated BOLD signals under a broad range of convolution parameters and sampling rates, at least in bivariate situations (Wen et al., 2013). In view of these results, although GCA is invariant to HRF variability given sufficiently fast sampling and low measurement noise, current applications of G-causality to fMRI should be treated cautiously and require carefully chosen experimental paradigms (e.g., comparison between conditions wherein HRFs can be assumed to remain unchanged).
Interpretations, limitations, and emerging directions
Connectivity analyses can be arranged along a scale from model-free to model-based. Model-free approaches like mutual information and transfer entropy impose the fewest assumptions on the data but—because of this—are challenging to estimate given limited data. At the other extreme, methods like dynamic causal modeling (DCM) specify detailed state-space models that describe how dynamics are coupled at the level of underlying mechanisms (the “state” equation), and how these dynamics are manifest in observable data (the “observation” equation). By jointly inverting candidate models and finding the most likely (among a set of competitors), DCM can address claims about physical-causal mechanisms. GCA occupies a useful middle ground between fully model-free and highly model-dependent approaches, where a generic dynamical model (the VAR process) is combined with a liberal structural model (i.e., no assumptions are made about the underlying structural connectivity). Unlike DCM, GCA can be applied directly to any time-series to obtain a measure of coupling among empirically sampled neuronal systems. Additionally, because fewer parameters need estimating than for a comparable DCM, GCA can typically be applied to larger networks, which is an increasingly important consideration in neuroscience settings. One useful way to summarize the relationship between GCA and DCM is that the former can furnish “data-driven” hypotheses for subsequent testing by explicitly specified DCMs. Another is to recall that GCA and DCM ask and answer fundamentally different questions, so that choosing one or the other (or both) depends on whether one is interested in describing the data in terms of information flow (GCA) or exposing the underlying physical-causal mechanism (DCM).
A limitation of standard GCA is that it only models linear interactions. However, the equivalence with transfer entropy for Gaussian variables means that linear VAR modeling is guaranteed to capture all the relevant variance for Gaussian data, and most of it for approximately Gaussian data. Even substantially nonlinear interactions that unfold over a small number of observations can sometimes be approximated by a (linear) VAR model with a large model order (Anderson, 1971). Care should be taken if the degree of nonlinearity differs across conditions in a study, as this could confound GCA. Finally, the conceptual framework of GCA can easily be extended to incorporate more sophisticated dynamical models. An attractive opportunity is to analyze G-causality in the framework of VARMA models, which augment standard VAR models with a moving average (MA) component. This could be very useful since VARMA models behave sensibly under both downsampling and additive noise (Solo, 2007). An alternative approach is to place GCA in a state-space framework by augmenting standard VAR models with explicit observation equations. This may be valuable in contexts like fMRI where detailed models linking neural (state) dynamics to observed (BOLD) responses are available (Ryali et al., 2011).
Conclusions
GCA provides a powerful and generic statistical tool for characterizing directed functional interactions from time-series data, that finds natural application in neuroscience and neuroimaging. Used carefully, it has the potential to shed distinctive light on the functional circuits underlying perception, cognition, and behavior, in a variety of experimental settings and using a range of data acquisition methods. Used poorly, like most statistical methods, it has the potential to obscure rather than illuminate. Care is needed not only in ensuring that the data and analysis process respect the necessary assumptions (mainly stationarity), but also in interpretation. Most importantly, G-causality does not make direct claims about underlying physical-causal mechanisms. Testing these claims instead requires statistical models which explicitly incorporate generative models linking such mechanisms to observable data, and which typically fall in the domain of effective connectivity. G-causality is fundamentally about describing observed data in terms of directed functional interactions or information flow. For this reason, some authors prefer terms like “Granger prediction” which avoid mention of “causality” (Cohen, 2014).
Although the principles governing the application and interpretation of GCA are not difficult to assimilate, they are not yet part of a standard conceptual toolkit for neuroscience. It may therefore be tempting to set GCA and its cousins aside, as perhaps a passing fashion. In our view this would be a mistake because neuroscience is moving, inevitably if unevenly, toward characterizing functional circuits. GCA, rooted firmly in information theory and calling on a mature approach to statistical inference, will be increasingly useful as the challenges of its application in specific neuroscience settings are addressed. We look forward to a scenario in which neuroscientists are readily able to select the connectivity analysis method best suited for their specific question, and for the data they have at hand. Several freely available software toolboxes can now facilitate the application of G-causality (Cui et al., 2008; Barnett and Seth, 2014). Of these, the MVGC toolbox (Barnett and Seth, 2014) most closely reflects the principles outlined in this article.
Footnotes
Editor's Note: Toolboxes are intended to briefly highlight and evaluate an emerging approach or a resource that is becoming widely used in neuroscience. For more information, see http://www.jneurosci.org/misc/itoa.shtml.
This work is supported by a donation from the Dr. Mortimer and Theresa Sackler Foundation via the Sackler Centre for Consciousness Science (www.sussex.ac.uk/sackler). A.B.B. is additionally supported by Engineering and Physical Sciences Research Council Grant EP/L005131/1 and A.K.S. by ERC FP7 project CEEDS (FP7-ICT-258749).
References
- Anderson TW. The statistical analysis of time series. New York: Wiley; 1971. [Google Scholar]
- Barnett L, Seth AK. Behaviour of Granger causality under filtering: theoretical invariance and practical application. J Neurosci Methods. 2011;201:404–419. doi: 10.1016/j.jneumeth.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Barnett L, Seth AK. The MVGC multivariate Granger causality toolbox: a new approach to Granger-causal inference. J Neurosci Methods. 2014;223:50–68. doi: 10.1016/j.jneumeth.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Barnett L, Barrett AB, Seth AK. Granger causality and transfer entropy are equivalent for Gaussian variables. Phys Rev Lett. 2009;103:238701. doi: 10.1103/PhysRevLett.103.238701. [DOI] [PubMed] [Google Scholar]
- Bastos AM, Vezoli J, Bosman CA, Schoffelen JM, Oostenveld R, Dowdall JR, De Weerd P, Kennedy H, Fries P. Visual areas exert feedforward and feedback influences through distinct frequency channels. Neuron. 2015;85:390–401. doi: 10.1016/j.neuron.2014.12.018. [DOI] [PubMed] [Google Scholar]
- Bernasconi C, König P. On the directionality of cortical interactions studied by structural analysis of electrophysiological recordings. Biol Cybern. 1999;81:199–210. doi: 10.1007/s004220050556. [DOI] [PubMed] [Google Scholar]
- Bressler SL, Richter CG. Interareal oscillatory synchronization in top-down neocortical processing. Curr Opin Neurobiol. 2014;31C:62–66. doi: 10.1016/j.conb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Brovelli A, Ding M, Ledberg A, Chen Y, Nakamura R, Bressler SL. Beta oscillations in a large-scale sensorimotor cortical network: directional influences revealed by Granger causality. Proc Natl Acad Sci U S A. 2004;101:9849–9854. doi: 10.1073/pnas.0308538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MX. Analyzing neural time series data. Cambridge, MA: MIT; 2014. [Google Scholar]
- Cui J, Xu L, Bressler SL, Ding M, Liang H. BSMART: a Matlab/C toolbox for analysis of multichannel neural time series. Neural Netwk. 2008;21:1094–1104. doi: 10.1016/j.neunet.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O, Guillemain I, Saillet S, Reyt S, Deransart C, Segebarth C, Depaulis A. Identifying neural drivers with functional MRI: an electrophysiological validation. PLoS Biol. 2008;6:2683–2697. doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamala M, Rangarajan G, Ding M. Analyzing information flow in brain networks with nonparametric Granger causality. Neuroimage. 2008;41:354–362. doi: 10.1016/j.neuroimage.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Bressler SL, Yang W, Liang H. Short-window spectral analysis of cortical event-related potentials by adaptive multivariate autoregressive modeling: data prepocessing, model validation, and variability assessment. Biol Cybern. 2000;83:35–45. doi: 10.1007/s004229900137. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Setsompop K. Ultra-fast MRI of the human brain with simultaneous multi-slice imaging. J Magn Reson. 2013;229:90–100. doi: 10.1016/j.jmr.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin E, Gross J, Pfeifer J, Fink GR, Timmermann L. Reliability of multivariate causality measures for neural data. J Neurosci Methods. 2011;198:344–358. doi: 10.1016/j.jneumeth.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273–1302. doi: 10.1016/S1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Friston K, Moran R, Seth AK. Analysing connectivity with Granger causality and dynamic causal modelling. Curr Opin Neurobiol. 2013;23:172–178. doi: 10.1016/j.conb.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geweke J. Measurement of linear dependence and feedback between multiple time series. J Am Statistical Assoc. 1982;77:304–313. doi: 10.1080/01621459.1982.10477803. [DOI] [Google Scholar]
- Goebel R, Roebroeck A, Kim DS, Formisano E. Investigating directed cortical interactions in time-resolved fMRI data using vector autoregressive modeling and Granger causality mapping. Magn Reson Imaging. 2003;21:1251–1261. doi: 10.1016/j.mri.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424–438. doi: 10.2307/1912791. [DOI] [Google Scholar]
- Handwerker DA, Gonzalez-Castillo J, D'Esposito M, Bandettini PA. The continuing challenge of understanding and modeling hemodynamic variation in fMRI. Neuroimage. 2012;62:1017–1023. doi: 10.1016/j.neuroimage.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse W, Möller E, Arnold M, Schack B. The use of time-variant EEG Granger causality for inspecting directed interdependencies of neural assemblies. J Neurosci Methods. 2003;124:27–44. doi: 10.1016/S0165-0270(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Kim S, Putrino D, Ghosh S, Brown EN. A Granger causality measure for point process models of ensemble neural spiking activity. PLoS Comput Biol. 2011;7:e1001110. doi: 10.1371/journal.pcbi.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. The identification of interacting networks in the brain using fMRI: model selection, causality and deconvolution. Neuroimage. 2011;58:296–302. doi: 10.1016/j.neuroimage.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Ryali S, Supekar K, Chen T, Menon V. Multivariate dynamical systems models for estimating causal interactions in fMRI. Neuroimage. 2011;54:807–823. doi: 10.1016/j.neuroimage.2010.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T. Measuring information transfer. Phys Rev Lett. 2000;85:461–464. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- Seth AK, Chorley P, Barnett LC. Granger causality analysis of fMRI BOLD signals is invariant to hemodynamic convolution but not downsampling. Neuroimage. 2013;65:540–555. doi: 10.1016/j.neuroimage.2012.09.049. [DOI] [PubMed] [Google Scholar]
- Solo V. On causality I: sampling and noise. Proceedings of the 46th IEEE Conference on Decision and Control; December, 12–14, 2007; New Orleans, LA. 2007. pp. 3634–3639. [Google Scholar]
- Valdés-Sosa PA, Sánchez-Bornot JM, Lage-Castellanos A, Vega-Hernández M, Bosch-Bayard J, Melie-García L, Canales-Rodríguez E. Estimating brain functional connectivity with sparse multivariate autoregression. Philos Trans R Soc Lond B Biol Sci. 2005;360:969–981. doi: 10.1098/rstb.2005.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkoerle T, Self MW, Dagnino B, Gariel-Mathis MA, Poort J, van der Togt C, Roelfsema PR. Alpha and gamma oscillations characterize feedback and feedforward processing in monkey visual cortex. Proc Natl Acad Sci U S A. 2014;111:14332–14341. doi: 10.1073/pnas.1402773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen Y, Ding M. Estimating Granger causality after stimulus onset: a cautionary note. Neuroimage. 2008;41:767–776. doi: 10.1016/j.neuroimage.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Yao L, Liu Y, Ding M. Causal interactions in attention networks predict behavioral performance. J Neurosci. 2012;32:1284–1292. doi: 10.1523/JNEUROSCI.2817-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Rangarajan G, Ding M. Is Granger causality a viable technique for analyzing fMRI data? PLoS One. 2013;8:e67428. doi: 10.1371/journal.pone.0067428. [DOI] [PMC free article] [PubMed] [Google Scholar]