Abstract
Subsequent malignant neoplasms (SMNs) are associated with significant morbidity and are a major cause of premature mortality among cancer survivors. Several large studies have demonstrated a strong association between the radiation and/or chemotherapy used to treat the primary cancer and the risk of developing SMNs. However, for any given therapeutic exposure, the risk of developing an SMN varies between individuals. Genomic variation can potentially modify the association between therapeutic exposures and SMN risk, and can possibly explain the observed inter-individual variability. This article provides a brief overview of the current knowledge regarding the role of genomic variation in the development of therapy-related SMNs. This article also discusses the methodological challenges in undertaking an endeavor to develop a deeper understanding of the molecular underpinnings of therapy-related SMNs, such as, an appropriate study design, identification of an adequately sized study population together with a reliable plan for collecting and maintaining high quality DNA, clinical validation of the phenotype, and selection of an appropriate approach or platform for genotyping. Understanding the modifiers of risk of treatment-related SMNs is critical to developing targeted intervention strategies and optimizing risk-based health care of cancer survivors.
The number of cancer survivors in the U.S. has tripled since 1971, and is growing by 2% each year.1 There is a clear recognition of long-term morbidity in cancer survivors; the incidence of severe or life-threatening chronic health conditions exceeds 40% several years from diagnosis.2, 3 One of the most serious treatment-related adverse events is the development of histologically distinct new cancers or subsequent malignant neoplasms (SMNs) – a major cause of premature death.4, 5 Two types of SMNs are recognized, based on well-defined associations with specific therapeutic exposures: i) therapy-related myelodysplasia or acute myeloid leukemia (t-MDS/AML) associated with alkylating agents and topoisomerase II inhibitors; and ii) radiation-related solid SMNs. SMNs account of 18% of all incident adult-onset cancers, surpassing de novo breast, lung and prostate.6 The incidence of SMNs exceeds 20% at 30 years after diagnosis of childhood cancer, representing a 4 to 6-fold increased risk of developing a new malignancy for cancer survivors, compared to the general population.7, 8 The magnitude of association between specific therapeutic exposures and SMNs are moderate to large (OR: 3.1 to 15.9)7, with a clear dose-response relation adding further biological credibility to that association9–11. Despite the unambiguous relation between therapeutic exposures and SMNs, there exists a wide variation in individual susceptibility – a topic that has not been explored comprehensively.
Mutations in high-penetrance genes (e.g., Li-Fraumeni syndrome 12–14, RB (retinoblastoma)15–17, NF1 (neurofibromatosis)18, PTCH1 (Gorlin or nevoid basal cell carcinoma syndrome)19, 20, WT1 (Wilms’ tumor)21 and ATM (ataxia telangiectasia)22, 23) could possibly modify the association between therapeutic exposures and SMNs. Many of the genes associated with familial cancer syndromes are responsible for mediating cellular response to DNA damage (e.g., ATM, BRCA) induced by genotoxic insults such as radiation and chemotherapy. Cancer survivors who carry a deleterious high-penetrance mutation are likely to be at increased risk for additional primary cancers (reviewed in6). For example, NF1 patients with a primary neoplasm are at increased risk of SMNs. The incidence of SMN is 11% among NF1 patients with primary neoplasms; the risk was 75% among NF1 patients treated for a primary embryonal cancer.24 In another case series of NF1 individuals, the risk of SMNs after exposure to radiation was reported to be 3-fold higher when compared with the risk among those not exposed to radiation.18 Furthermore, several studies of Ataxia telangiectasia families have demonstrated that heterozygosity for A-T causing mutations in ATM is associated with breast cancer risk.25, 26 Cancer survivors carrying these genetic variants should be followed closely for the development of therapy-related SMNs.
However, the low frequency of these mutations in the general population27 suggests the attributable risk to the development of SMNs is likely very small. The interindividual variability in risk of therapy-related SMNs is more likely related to common polymorphisms in low-penetrance genes that regulate drug metabolism/ disposition or, those responsible for DNA repair. Between 20% and 95% of the variability in cytotoxic drug disposition can possibly be explained by genetic variation28, and polymorphisms in genes involved in drug metabolism/ disposition contribute to disease-free survival and drug toxicity29. Several studies have demonstrate the role played by variation in DNA repair in susceptibility to de novo cancer;30–33 using the same argument, variation in DNA repair could possibly modify SMN risk among cancer patients exposed to DNA-damaging agents, such as radiation and chemotherapy. Finally, it is conceivable that gene-environment (therapeutic exposure) interactions could magnify functional impact of the polymorphisms.
Drug Metabolism and Disposition
Metabolism of genotoxic agents occurs in two phases. Phase I involves activation of substrates into highly reactive electrophilic intermediates that can damage DNA – a reaction principally performed by the cytochrome p450 (CYP) family of enzymes. The xenobiotic substrates of CYP proteins include cyclophosphamide, ifosfamide, thiotepa, doxorubicin, and dacarbazine. Phase II enzymes function to inactivate genotoxic substrates. The more commonly examined phase II proteins comprise the glutathione S-transferase (GST) and NAD(P)H:quinone oxidoreductase-1 (NQO1). GSTs detoxify doxorubicin, lomustine, busulfan, chlorambucil, cisplatin, cyclophosphamide, melphalan, etc. NQO1 uses the cofactors NADH and NADPH to catalyze the electron reduction of its substrates, produces less reactive hydroquinones, and therefore prevents generation of reactive oxygen species and free radicals which may subsequently lead to oxidative damage of cellular components. The balance between the two sets of enzymes is critical to the cellular response to xenobiotics; e.g., high activity of phase I enzyme and low activity of a phase II enzyme can result in DNA damage from the excess of harmful substrates. Polymorphisms in drug metabolizing genes are very common in the population; many are functionally significant, and may contribute to the risk of SMNs.
P-glycoprotein (encoded by MDR1) traps hydrophobic drugs in the plasma membrane of cells and effluxes them using an ATP-dependent process; many chemotherapeutic drugs are substrates of this protein. A number of functional polymorphisms exist in the MDR1 gene and could play a role in the development of SMNs.
DNA repair
DNA repair mechanisms protect somatic cells from mutations in tumor suppressor genes and oncogenes that can lead to cancer initiation and progression. Small differences in an individual’s DNA repair capacity may be magnified in conjunction with exposure to chemotherapy or radiotherapy. An individual’s DNA repair capacity appears to be genetically determined.34 A number of DNA repair genes contain polymorphic variants, resulting in large inter-individual variations in DNA repair capacity. In fact, one tenth of the general population is known to have a reduced capacity to repair DNA damage.35 Thus, individuals with altered DNA repair mechanisms are likely susceptible to the development of genetic instability that drives the process of carcinogenesis as it relates to SMNs.
Mismatch repair (MMR) functions to correct mismatched DNA base pairs that arise as a result of misincorporation errors that have escaped polymerase proofreading during DNA replication. Defects in the MMR pathway result in genetic instability or a mutator phenotype, manifested by an elevated rate of spontaneous mutations characterized as multiple replication errors in simple repetitive DNA sequences (microsatellites) – functionally identified as microsatellite instability (MSI).
Double-Strand Breaks (DSBs) in DNA as a consequence of chemotherapy or radiation, lead to loss of genetic material, resulting in chromosomal aberrations. Cellular pathways available to repair DSBs include homologous recombination (HR), non-homologous end-joining (NHEJ), and single-strand annealing. HR uses the second, intact copy of the chromosome as a template to copy the information lost at the DSB site on the damaged chromosome – a high-fidelity process. NHEJ pathway joins broken DNA ends containing very little homology. This process is not always precise and can result in small regions of non-template nucleotides around the site of the DNA break.
Base Excision Repair (BER) pathway corrects individually damaged bases occurring as a result of ionizing radiation. The XRCC1 protein plays a central role in the BER pathway, by acting as a scaffold and recruiting other DNA repair proteins.
Nucleotide Excision Repair (NER) removes structurally unrelated bulky damage induced by radiation and chemotherapy. The NER pathway is linked to transcription, and components of the pathway comprise the basal transcription factor IIH complex (TFIIH), which is required for transcription initiation by RNA polymerase II.
There are two approaches to the study of genetic variation in SMNs: (1) candidate gene studies based on the selection of a limited number of biologically relevant genes/ pathways; and (2) genome-wide association studies (GWAS) using DNA arrays capable of detecting a million or more SNPs. The candidate gene approach is guided by a specific hypothesis whereas the agnostic nature of the genome-wide approach is necessary for comprehensive discovery analysis (i.e., ability to study action/ interaction of many genes and discover/ identify new genes) – a distinct advantage over candidate gene approach. However, unless the magnitude of genotype/phenotype association is anticipated to be large, a GWAS approach requires a large sample size to account for false discovery, and can become an expensive (and sometimes logistically difficult) endeavor. In addition, there is the need for a replication cohort so that the genes identified in the discovery set can be validated in the test set.
The studies presented here (and summarized in Table 1 and Table 2) include single gene studies, where there was ample pre-clinical (in vitro and/or in vivo) data that provided a compelling rationale for examination of the association in a single gene setting; candidate gene studies utilizing curated sets of genes with biological plausibility; those studies using a GWAS approach with successful validation of the findings in independent cohorts or extension of the findings with some functional data. A paucity of scientifically/methodologically robust studies in the extant literature highlights issue that there are methodological challenges to conducting such studies; these are summarized at the end of the review. Nonetheless, an understanding of the etiopathogenetic pathways that lead to SMNs is critical to developing targeted prevention/ intervention strategies, and optimizing risk-based care of survivors.
Table 1.
Role of Genetic Susceptibility in the Development of therapy-related myelodysplasia/acute myeloid leukemia
| Study | Candidate Genes | Study design | Replication study | Results |
|---|---|---|---|---|
| Candidate Gene Studies | ||||
| Genes responsible for drug metabolism and disposition | ||||
| Felix et al*, 199850 | CYP3A4 |
Case control Cases (n=30): Childhood primary cancers Controls (de novo AML: n=99): childhood cancers |
No | 21% of de novo leukemias with MLL gene translocations and 0% of 22 t- MDS/AMLs with MLL gene translocations carried the CYP3A4-V (P=0.016). Individuals with CYP3A4-W genotype may be at increased risk for t-MDS/AML. |
| Rund et al**, 200551 | Drug metabolism and disposition genes |
Case-control Cases (n=96): Childhood and adult-onset primary cancers Controls: healthy Israeli population |
No | A protective effect of CYP3A4 1nB genotype against development of t-MDS/AML Reduced expression of MSH2 and MLH1 in 6/10 patients with MSI as compared to 0/5 of patients without MSI. Genetic predisposition as well as epigenetic events contributed to the etiology of t-AML/MDS in this study. |
| Blanco*, 200252 | CYP3A5*3, CYP3A4*1B, NQO1609 C→T |
Case Control Cases (n=53): Childhood ALL patients with t-MDS/AML Controls (n=224): Childhood ALL with no t-MDS/AML |
No | No differences between the control and t-MDS/AML group in the incidence of homozygous CYP3A5*3 genotypes: 82.0% vs. 85.4% in whites (P 0.4), 6.5% vs. 12.5% in blacks (P 0.5), and 69.6% vs. 75.0% in Hispanics (P 0.7). Data do not support an association between common CYP3A4, NQO1 or CYP3A5 polymorphisms and the risk of t-MDS/AML in children treated for ALL. |
| Bolufer*, 200753 | CYP1A1*2A(T6235C), CYP2E1*5B (C-1019T), CYP3A4*1B (A-290G), del(GSTT1), del(GSTM1), NQO1*2(C609T), MTHFR(C677T), TYMS |
Case-Control Cases (n=78): Childhood and adult-onset primary cancers Healthy Controls (n=458): children and adults |
No | Polymorphism profile consisting of CYP1A1*2A, del(GSTT1) and NQO1*2 strongly modified the risk of t-MDS/AML. The absence of all three polymorphisms decreased the risk of t-MDS/AML 18-fold (OR = 0.05, 95% CI = 0.005–0.63, P= 0.02), whereas the presence of only NQO1*2 or all three polymorphisms enhanced the risk of t-MDS/AML (OR = 2.1, 95% CI = 1.1–4.0, P = 0.03 and OR=18.4, 95% CI = 1.6–212.8, P = 0.02 respectively). |
| Larson et al**, 199954 | NQO1 |
Case control Cases (n=56): Childhood and adult-onset primary cancers Controls (de novo myeloid leukemia=48): Childhood and adult-onset primary cancers |
No | Frequency of an inactivating polymorphism in NQO1 appears to be increased among those with t-MDS/AML. |
| Naoe et al***, 200055 | NQO1, GSTM1, GSTT1, CYP3A4 |
Case control Cases (n=58): Adult-onset primary cancers Cancer controls (de novo AML: n=411): Adult-onset primary cancers Healthy controls (n=150): adults |
No | Homozygous Ser/Ser genotype of NQO1 at codon 187, causing loss of function, was more frequent in patients with t-MDS/AML (24.1%) than in those with de novo AML (15.6%), and control (10.6%), P= 0.002. |
| Allan et al**, 200156 | GSTM1, GSTT1, GSTP1 |
Case control Cases (n=89): Childhood and adult-onset primary cancers Cancer Controls (de novo AML: n=420): Childhood and adult-onset primary cancers Healthy controls (n=1022): Children and adults |
No | Individuals with at least one GSTP1 codon 105 Val allele were significantly over-represented in t-MDS/AML cases compared with de novo AML cases (OR=1.8, 95%CI, 1.1–2.9). Also, relative to de novo AML, the GSTP1 codon 105 allele occurred more often among t-MDS/AML patients with prior exposure to chemotherapy (OR=2.7, 95%CI, 1.4–5.1), particularly among those with prior exposure to known GSTP1 substrates (OR=4.3, 95%CI, 1.4–13.2). |
| Woo, et al*. 200057 | GSTM1, GSTT1 |
Cohort study Cohort of children with ALL (n=302); 57 developed t-MDS/AML |
No | The study did not identify any associations between GSTM1 or GSTT1 null genotypes and t-MDS/AML |
| DNA Repair | ||||
| Ellis et al***, 200862 |
MDM2 SNP309 TtP53 codon 72 |
Case-control (healthy controls) Cases (discovery set: n=80; replication set: n=91): Adult-onset primary cancers Cancer Controls (de novo AML: n=721): adult-onset Healthy controls (n=2392): adults |
yes | Neither polymorphism alone influenced the risk of t-MDS/AML; however an interactive effect was detected such that MDM2 TT TP53 Arg/Arg double homozygotes, and individuals carrying both a MDM2 G allele and a TP53 Pro allele, were at increased risk of t-MDS/AML (OR=2.04, 95%CI, 1.20–3.48, Pinteraction=0.009) |
| Worrillow et al**, 200365 |
hMSH2 –6exon 13 Evaluation of MSI |
Case control Cases (n=91): Childhood and adult-onset primary cancers Cancer Controls (de novo AML: n=420): Childhood and adult-onset primary cancers Healthy controls (n=837): Children and adults |
Verification by direct sequencing | The variant (C) hMSH2 allele was significantly overrepresented in t-MDS/AML cases that had previously been treated with alkylating agents, compared with controls (OR=4.0, 95%CI 1.4–11.4); 38% of the patients were MSI positive. |
| Worrillow et al**, 200864 | MLH1 (rs1800734) |
Case control Cases (n=133): Childhood and adult-onset primary cancers Cancer Controls (de novo AML: n=420): Childhood and adult-onset primary cancers Healthy controls (n=1177): Children and adults |
No | Carrier frequency of MLH1 -93 variant was higher in patients who developed t-MDS/AML after alkylating agents exposure for HL, compared to patients without alkylating agent exposure. MLH1 -93 variant allele was also over- represented in t-MDS/AML cases compared with de novo AML cases and was associated with increased risk of t-MDS/AML (OR=5.3, 95%CI, 1.4–20.2) in patients exposed to alkylating agents |
| Jawad et al***, 200660 | HLX; RAD51 |
Case control Cases (n=42): Adult-onset primary cancers Cancer Controls (de novo AML: n=166): Adult-onset primary cancers Healthy controls (n=189): adults |
No | Variant HLX1 allele significantly increased risk of t-MDS/AML (OR=3.4, 95%CI, 1.7–6.8). Polymorphism in RAD51 (135G/C-5′ UTR) also increased risk of t- MDS/AML. Combined analysis revealed a synergistic 9.5-fold increase (95%CI, 2.22–40.64) in risk. |
| Allan et al**, 200461 | XPD |
Case-control Cases (n=91): Childhood and adult-onset primary cancers Cancer Controls (de novo AML: n=420): Childhood and adult-onset primary cancers Healthy controls (n=729): Children and adults |
No | Homozygosity for the XPD codon 751 glutamine variant was associated with a significantly increased risk of developing t-MDS/AML (OR=2.2 for Gln/Gln vs. Lys/Ly; 95% CI. 1.0–4.7). |
| DNA repair and drug metabolizing genes | ||||
| Seedhouse et al***, 200258 | XRCC1, XRCC3, XPD, NQO1 |
Case control Cases (n=34): adult-onset primary cancers Cancer controls (de novo AML: n=134): Adult-onset cancer survivors Healthy controls (n=178): adults |
No | Presence of at least one XRCC1 399Gln allele indicated a protective effect for the allele in controls compared with patients with t-MDS/AML (OR=0.4, 95%CI, 0.2–0.9) |
| Seedhouse et al***, 200459 |
RAD51, XRCC3 GSTM1 |
Case control Cases (n=51): adult-onset primary cancers Cancer controls (de novo AML: n=216): Adult-onset cancer survivors Healthy controls (n=186): adults |
No | When both variant RAD51-135C and XRCC3-241Met alleles are present, the risk of t-MDS/AML is increased (OR=8.1, 95% CI, 2.2–29.7). The risk of t- MDS/AML is further increased (OR=15.3, 95% CI, 1.8–127.3) among patients with GSTM1 deletion. |
| Guillem et al***, 200767 | NQO1, GSTP1, XRCC1. XRCC3, NBS1, ERCC5, XPC, MTHFR |
Case-control Cases (n=81): Adult-onset primary cancers Healthy controls (n=64): adults |
No | Significant association between the MTHFR haplotype (SNPs 677 and 1298) and the risk of developing t-MDS/AML in breast cancer patient group (p=0.02) and cyclophosphamide-treated hematological disease group (p=0.005). Risk haplotypes were different for each case, corresponding to the 677T1298A haplotype after breast cancer treatment and the 677C1298C haplotype after hematological malignancy. |
| Ding***, 201268 | CYP1A1, CYP3A4, GSTM1, GSTT1, GSTP1, NQO1, MTHFR, TP53, MDM2, MLH1, MSH2, XPD, RAD51, XRCC1, XRCC2, XRCC3, XRCC4, LAMC2, NMAT2 |
Case-control Cases (n=49): adult-onset primary cancers Cancer controls (n=49): Adult-onset cancer survivors (matched on primary diagnosis) |
correlated with gene expression | A synergistic effect between TP53 (P72R) and MTHFR. (C677T and 11298C) (p=0.0035). Expression of both TP53 and MTHFR was significantly lower in cases compared with controls, supporting their role in t-MDS/AML development. |
| Genome-wide Association Studies | ||||
| Knight et al**., 200969 | GWAS |
Case-control Cases (Discover: n=80; replication: n=70): Childhood and adult-onset primary cancers Healthy controls (discover: n=150; replication: n=95): Children and adults |
Yes | Among patients with acquired abnormalities of chromosomes 5 or 7; 3 SNPs (rs1394384 [OR=0.3, 95% CI, 0.2–0.6], rs1381392 [OR=2.1, 95%CI, 1.3–3.4], and rs1199098 [OR=0.5, 95% CI, 0.3–0.8]) were associated with t-MDS/AML; |
The study populations included primarily children
The study populations included primarily adults, although some children were also included
The study populations included primarily adults
Table 2.
Role of Genetic Susceptibility in the Development of therapy-related solid subsequent malignant neoplasms
| Study | GWAS vs. Candidate gene | Study design | Replication study | Results |
|---|---|---|---|---|
| Any solid | ||||
| Best et al****, 201170 | SMN GWAS |
Case-control Cases (discovery: n=100; replication: n=62): childhood and adult-onset primary cancers Cancer controls (discovery: n=89; replication: n=71): childhood and adult-onset cancer survivors (matched on primary diagnosis) |
Yes | Two variants at chromosome 6q21 (rs4946728 [OR=11.4, 95%CI, 3.2–40.3]; rs1040411 [OR=6.6, 95%CI, 3.2–13.5]) were associated with SMNs in childhood HL survivors. The variants comprise a risk locus associated with decreased basal expression of PRDM1 and impaired induction of PRDM1 protein after radiation exposure. |
| Mertens et al*, 200493 |
Candidate gene GSTM1, GSTT1, XRCC1 |
Cohort study Cohort (n=650): Childhood HL survivors of which 178 patients were diagnosed with SMNs |
No | Individuals lacking GSTM1 were at increased risk of any SMN (OR=1.5, 95%CI, 1.0–2.3). |
| Radiation-related breast cancer | ||||
| Bernstein et al***, 201023 |
Candidate gene Full mutation screen of ATM gene |
Case control Cases (n=708): breast cancer patients with contralateral breast cancer (adult-onset) Controls (n=1397): U/L breast cancer (adult-onset) |
No | Among women who carried a rare, deleterious ATM missense variant, radiation exposure was associated with a statistically significantly higher risk of contralateral breast cancer compared with unexposed women who carried the wild-type genotype (0.01–0.99 Gy: RR=2.8, 95%CI, 1.2–6.5; ≥1 Gy: RR=3.3, 95%CI, 1.4–8.0) or compared with unexposed women who carried the same deleterious missense variant (0.010.99 Gy: RR=5.3, 95%CI: 1.6–17.3; ≥1.0 Gy: RR=5.8, 95% CI: 1.8–19.0, Ptrend=0.04) |
| Brooks et al***, 201294 |
Candidate gene CHEK2, MRE11A, MDC1, NBN, RAD50, TP53BP1 |
Case-control Cases (n=649): breast cancer patients with contralateral breast cancer (adult-onset) Controls (n=1284): U/L breast cancer (adult-onset) |
No | Carriers of a RAD50 haplotype exposed to ≥1 gray had an increased risk of contralateral breast cancer compared with unexposed carriers (RR=4.3, 95% CI, 1.9–9.6) with an excess relative risk per Gray=2.1 (95% CI, 0.6–5.3) |
| Radiation-associated meningioma | ||||
| Hosking et al***, 201174 | Family-based association test (FBAT) |
Family-based association study 15 families segregating radiation-associated meningioma using high-density SNP arrays (adults) |
No | The strongest haplotype associations were attained at 18q21.1 (p=7.5 × 10−5), 18q21.31 (p=2.8 × 10−5) and 10q21.3 (p=1.6 × 10−4). The 18q21.1 and 10q21.3 associations provide support for variation in PIAS2, KATNAL2, TCEB3C, TCEB3CL, CTNNA3 genes as risk factors for radiation- associated meningioma. |
| Head and Neck Cancer | ||||
| Gal et al*, 200585 |
Candidate gene XRCC1 Arg399Gln, XRCC3 Thr241Met, XPD Lys751Gln, MGMT Leu84Phe, Val143Ile |
Cohort study Cohort (n=246): 18–65 yo with oral cancer; 88 developed a head and neck SMN |
No | A significant increased risk of SMNs (all sites combined; upper airway digestive tract sites; head and neck squamous cell carcinomas) was observed among XRCC3 241 Met allele homozygotes (HR=2.65–3.44, p<0.02). |
| Jin et al***, 201386 |
Candidate gene P53, p73, p14ARF, MDM2, MDM4 |
Cohort study Cohort (n=1283): Adults with squamous cell carcinoma of the head and neck diagnosed between 1995 and 2007; of these 120 developed an SMN |
No | SMN risk increased with increasing number of risk genotypes (Ptrend<0.0001). Compared with the low risk group (0–3 combined risk genotypes), both the medium-risk (4–5 combined risk genotypes) and high-risk (6–9 combined risk genotypes) groups had significantly increased risk of SMNs (HR: 1.6, 95% CI: 1.0–2.6; HR: 3.0, 95%CI, 1.8–5.0, respectively) |
| Zhang, et al***. 2012,87 |
Candidate gene p53 codon 72 and p73 G4C14-to-A4T14 |
Cohort study Cohort (n=1269)of adults with squamous cell carcinoma of the head and neck; of these 109 developed an SMN |
No | Patients with p53 WP + PP and p73 GC/GC genotypes had an increased risk of SMNs compared with the corresponding p53 WW and p73 GC/T + AT/AT genotypes. After combining the 2 polymorphisms, a significantly increased risk of SMN was observed in the high risk group (p53 P carriers and p73 GC/GC) compared with the low risk group (p53 WW and p73 AT carriers). |
| Radiation-associated papillary carcinoma of thyroid gland | ||||
| Akulevich*, 200990 |
Candidate gene ATM, XRCC1, TP53, XRCC3, MTF1 |
Case-control Cases (n=123): Childhood primary cancers Healthy controls (596): Children |
No | In the analyses of ATM/TP53 (rs1801516/rs664677/rs609429/rs1042522) combinations, the GG/TC/CG/GC genotype was associated with radiation-induced papillary thyroid carcinoma (OR=2.1, 95% CI, 1.2–3.8) |
The study populations included primarily children
The study populations included primarily adults, although some children were also included
The study populations included primarily adults
The study population includes an equal mix of children and adults
Role of genetic susceptibility in therapy-related myelodysplasia/ acute myeloid leukemia
t-MDS/AML has been reported after conventional treatment of Hodgkin lymphoma (HL), non-Hodgkin lymphoma (NHL), acute lymphoblastic leukemia (ALL), sarcomas, and breast, ovarian and testicular cancers36–42, and after autologous hematopoietic cell transplantation (aHCT) for HL or NHL, where it is the major cause of non-relapse mortality43–48. The cumulative incidence of t-MDS/AML ranges from 2% at 15 years after conventional therapy37 to 8.6% at 6 years after autologous HCT43. Two types are recognized by the WHO classification: alkylating agent-related type, and topoisomerase II inhibitor-related type.49 Alkylating agents associated with t-MDS/AML include cyclophosphamide, ifosfamide, mechlorethamine, melphalan, busulfan, nitrosureas, chlorambucil, dacarbazine, and platinum compounds. Mutagenicity is related to the ability of alkylating agents to form crosslinks and/or transfer alkyl groups to form DNA monoadducts. Alkylation results in inaccurate base pairing during replication and single- and double-strand breaks in the double helix as the alkylated bases are repaired. Alkylating-agent associated t-MDS/AML is associated with abnormalities involving chromosomes 5 (−5/del[5q]) and 7 (−7/del[7q]). DNA topoisomerase II (topo II) inhibitors include antitumor antibiotics (doxorubicin, daunorubicin mitoxantrone) and epipodophyllotoxins (etoposide and teniposide). Therapy-related leukemias associated with topo II inhibitors are characterized by chromosomal rearrangements involving chromosome 11q23, as well as a variety of balanced translocations (t(8;21), t(15;17), or t(9;22) and others).
Drug Metabolism and risk of t-MDS/AML
A limited number of studied have described the association between genes responsible for drug-metabolizing enzymes and the risk of t-MDS/AML. Two studies found variant allele G of CYP3A4 1B(A290G) to be under-represented in patients with t-MDS/AML when compared with those with de novo AML or healthy individuals50, 51 – while two others found no association52, 53. A polymorphism of NQO1 gene that results in an amino acid change (Pro to Ser), located in codon 187, produces the complete loss of enzyme activity in homozygous subjects (Ser/Ser) and has been associated with an increased risk of alkylating agent-induced t-MDS/AML.54, 55 Inheritance of the GSTP1 (GST pi) valine allele in codon 105 was associated with an increased risk of t-MDS/AML, particularly in those patients who have been treated with chemotherapeutic drugs that are substrates of GSTP1 and not among t-MDS/AML patients with exposure to radiation alone.56 On the other hand, GSTM1 or GSTT1 null genotypes were found not be associated with t-MDS/AML.57
DNA damage and repair
XRCC1, XRCC3 and XPD are polymorphic genes belonging to the major DNA repair pathways. XRCC1 is involved in BER and repair of single strand breaks. The XRCC3 protein functions in the homologous DNA DSB repair pathway and directly interacts with and stabilizes Rad51. The XPD protein is involved in the NER pathway, and functions to remove bulky damaged adducts from DNA. The presence of at least one XRCC1 399Gln allele indicated a protective effect for the allele in controls compared with patients with t-MDS/AML.58 RAD51 and XRCC3 are involved in the repair of DNA by the HR pathway, and the two genes play a critical role in genomic stability. Rad51 protein binds to DNA and promotes homologous pairing. Xrcc3 protein stabilizes Rad51 – and both are part of a complex consisting of Xrcc2, Xrcc3, Rad51B, Rad51C, and Rad51D. Polymorphisms have been identified in both the RAD51 (RAD51-G135C) and XRCC3 (XRCC3-Thr241Met) genes, and t-MDS/AML risk was found to be significantly increased when both variant RAD51-135C and XRCC3-241Met alleles are present.59 These results suggest that DNA double-strand breaks and their repair are important in the pathogenesis of t-MDS/AML.
Studies of radiation-induced t-MDS/AML in mice suggest that the number of target stem cells is a risk factor, and the HLX1 homeobox gene, which is important for hematopoietic development, could be a candidate gene. A combined analysis of RAD51 and HLX1 variant alleles demonstrated a synergistic 9.5-fold increase in the risk of t-MDS/AML.60
ERCC2 encodes a DNA helicase integral to nucleotide excision DNA repair, and a common functional variant at codon 751 (rs13181) defines a low-penetrance risk allele for t-MDS/AML.61 An association between the ERCC2 variant and t-MDS/AML (with alterations in chromosomes 5/7), possibly indicates that the protein encoded by ERCC2 plays a role in the repair of alkylating agent-induced DNA damage.
Genetic variation in the p53 pathway has been hypothesized to affect t-MDS/AML risk, and the association between t-MDS/AML and common functional p53-pathway variants (MDM2 SNP309 and TP53 codon 72 polymorphism) has been examined.62 Although neither polymorphism alone influenced the risk, MDM2 and TP53 variants interacted to modulate responses to genotoxic therapy.62 This interactive effect was observed primarily among patients previously treated with alkylating agents.
Methylating agents such as procarbazine are commonly used to treat HL and are associated with an increased risk of t-MDS/AML.63 Cytotoxicity of methylating agents is mediated primarily by the DNA MMR system. Loss of MLH1, a major component of DNA MMR, results in persistence of mutagenized cells that are at high risk of malignant transformation. A common polymorphism at position −93 (rs1800734) in the core promoter of MLH1 was overrepresented among patients who developed t-MDS/AML after methylating chemotherapy for HL, compared to patients who did not receive methylating therapy.64 Furthermore, the variant (C) hMSH2 allele was found to be significantly overrepresented in t-MDS/AML cases that had previously been treated with O6-guanine alkylating agents, including cyclophosphamide and procarbazine, implicating this allele in conferring a nondisabling DNA mismatch repair defect and predisposing the patienst to the development of t-MDS/AML.65
Impact of antimetabolite drugs and DNA synthesis/ repair on t-MDS/AML risk
Methylene tetrahydrofolate reductase (Mthfr) enzyme plays a role in DNA synthesis/ repair by directing 5,10-methylene tetrahydrofolate toward methionine synthesis. The negative effect on DNA synthesis/ repair induces chromosomal aberrations in the hematopoietic precursor cells. Polymorphisms of MTHFR (C677T and A1298C) are known to be associated with decreased Mthfr activity.66 An association between the MTHFR haplotype and risk of t-MDS/AML among patients with breast cancer or hematological malignancies exposed to alkylating agents or topoisomerase II inhibitors have been.67 Furthermore, a synergistic effect between TP53 and MTHFR has been reported.68 Expression of both TP53 and MTHFR was significantly lower in cases compared to controls, supporting their role in t-MDS/AML development.
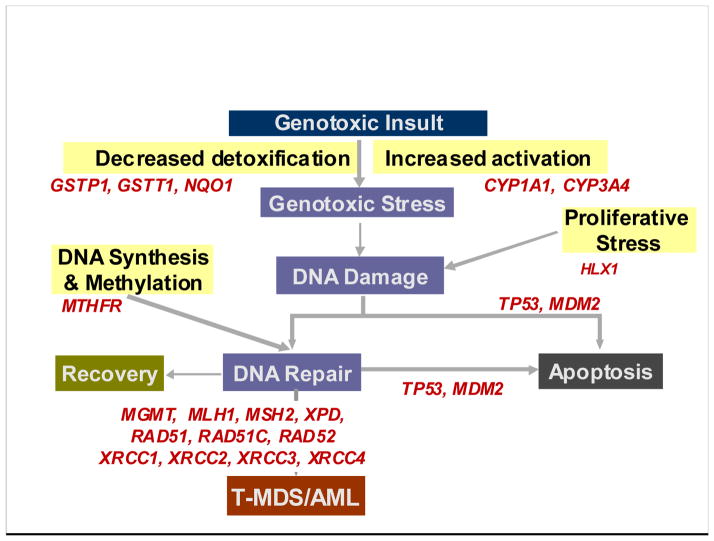
The proposed model (Figure 1) integrates findings from the studies described above to help explain the pathogenesis of t-MDS/AML. Thus, high activity of phase I enzyme (CYP3A4) and low activity of a phase II enzyme (GSTP1 and NQO1) can result in DNA damage from the excess of harmful substrates. The damaged DNA undergoes imperfect repair in the face of impaired repair (XRCC1, XRCC3, RAD51, ERCC2, MLH1) and/ or impaired apoptosis (TP53, and MDM2). Impaired hematopoiesis (HLX1) in the face of increased proliferative stress increases the risk of chromosomal aberrations. Reduced MTHFR activity is associated with chromosomal aberrations during DNA repair. When combined with higher TP53 activity, it would normally result in efficient clearance of damaged cells through apoptosis. However, when combined with less efficient TP53, it would result in accumulation of progenitor cells with chromosomal damage and increased the risk of t-MDS/AML. On the other hand, with normal MTHFR activity to support DNA repair, allele variants of TP53 do not impact t-MDS/AML development, since efficient DNA repair would maximize DNA recovery and minimize the risk of chromosomal aberrations. As shown in Table 1, while these findings provide biological plausibility to the pathogenesis of t-MDS/AML, very few studies have incorporated all the candidate genes in a single study that is large enough to have sufficient power to overcome issues related multiple comparisons. The number of cases included in these studies ranged from 30 to 133. Furthermore, the studies did not take into account the cytogenetic or morphologic heterogeneity of t-MDS/AML. Other major issues stem from the use of healthy (non-cancer) controls (67% of the studies) or use of patients with de novo AML (55% of the studies) as controls. The concerns related to the use of healthy controls or cancer controls consisting of patients with de novo myeloid malignancies are discussed later in the article. Finally, none of the studies have utilized a validation/ replication population to confirm their findings, nor have they extended their findings by conducting functional studies.
Figure 1.
Role of genetic variation in risk of therapy-related leukemia – a proposed model
Genome-wide association studies
Using a case-control study design, 3 SNPs (rs1394384, rs1381392, and rs1199098) were found to be associated with t-MDS/AML with chromosome 5/7 abnormalities.69 The findings were confirmed in an independent replication cohort. rs1394384 is intronic to ACCN1, a gene encoding an amiloride-sensitive cation channel that is a member of the degenerin/ epithelial sodium channel; rs1199098 is in LD with IPMK, which encodes a multikinase that positively regulates the prosurvival AKT kinase and may modulate Wnt/beta-catenin signaling; rs1381392 is not near any known genes, miRNAs, or regulatory elements, although it lies in a region recurrently deleted in lung cancer.
Role of genetic susceptibility in therapy-related solid SMNs
Therapy-related solid SMNs demonstrate a strong relation with ionizing radiation. The risk of solid SMNs is highest when the exposure occurs at a younger age, increases with the total dose of radiation, and with increasing follow-up after radiation.7 Some of the well-established radiation-related solid SMNs include breast cancer, thyroid cancer, brain tumors, sarcomas, and basal cell carcinomas (BCC).7, 39 A GWAS identified two variants at chromosome 6q21 associated with solid SMNs.70 The variants comprise a risk locus associated with decreased basal expression of PRDM1 and impaired induction of the PRDM1 protein after radiation exposure. These data implicate PRDM1 in the etiology of radiation-induced SMNs. The role of genomic variants in the risk of specific solid SMNs is described below.
Breast Cancer
Ionizing radiation is an established breast carcinogen. Breast cancer is the most common solid SMN after HL, largely due to chest radiation for treatment of HL. The risk of radiation-related breast cancer among female survivors of childhood cancer ranges from 25- to 55-fold that of the general population.71 For female HL patients treated with chest radiation at less than 16 years of age, the cumulative incidence of breast cancer approaches 20% by age 45 years.38 The latency after chest radiation ranges from 8 to 10 years, and the risk of breast cancer increases in a linear fashion with radiation dose with an estimated relative risk of 6.4 at a dose of 20 Gy and 11.8 at a dose of 40 Gy.10 Breast cancer risk is attenuated among women who also received radiation doses of 5 Gy or greater to the ovaries, reflecting the important role of hormonal stimulation on radiation-induced breast cancer.10
The ATM gene is a key regulator of cellular responses to the DNA damage induced by ionizing radiation. Women who carry rare deleterious ATM missense variants and who are exposed to radiation may have an elevated risk of developing contralateral breast cancer.23 However, the rarity of these deleterious missense variants (<1%) implies that ATM mutations could account for only a small portion of radiation-related breast cancers.
Meningioma
Meningiomas develop after cranial radiation used to treat histologically distinct brain tumors or used for management of central nervous system (CNS) disease among ALL or NHL patients.72 There is a linear relation between risk of meningioma and radiation dose.9 While ionizing radiation is an established risk factor for meningioma, a very small fraction of irradiated individuals develop this tumor, suggesting the role for genetic susceptibility. The SNP rs4968451, which maps to intron 4 of the gene that encodes breast cancer susceptibility gene 1 – interacting protein 1, has been shown to associated with an increased risk of developing meningioma.73 Given that approximately 28% of the European population are carriers of at-risk genotypes for rs4968451, the variant is likely to make a substantial contribution to the development of meningioma. Another study used the family-based association test program, and showed that haplotype associations were attained at 18q21.1, 18q21.31 and 10q21.3, providing support for a variation in PIAS2, KATNAL2, TCEB3C, TCEB3CL and CTNNA3 genes as risk factors for radiation-associated meningioma.74 These findings suggest that genetic susceptibility to radiation-associated meningioma is likely mediated through the co-inheritance of multiple risk alleles.
Melanoma
Survivors of childhood cancer are at increased risk of melanoma.75, 76 Melanoma is also reported in HCT recipients.77 Radiotherapy may contribute to an increased risk of melanoma, but only at very high doses of low linear energy transfer radiation.78 Certain variants of MC1R, CDKN2A, MTAP and PLAS2G6 genes are associated with an increased risk of de novo melanoma.79–82 The excess risk of melanoma following retinoblastoma83 is probably due to common etiological factors between these two tumor types: the retinoblastoma protein (pRB) is phosphorylated by CDK4 and CDK6, the two target kinases of CDKN2A. Sunlight exposure increases risk of melanoma. Sunlight also potentiates cutaneous synthesis of vitamin D, which can inhibit melanoma cell growth and promote apoptosis. Vitamin D effects are mediated through the vitamin D receptor (VDR). The risk of multiple primary melanoma has been shown to be increased in people who have the BsmI variant of VDR.84 These findings suggest complex interacting pathways that interact with the environment to increase the risk of melanoma.
Upper aerodigestive tract (UADT) neoplasms
Polymorphisms in genes involved in DNA repair pathways were examined for their association with the development of SMNs of the UADT in patients previously diagnosed with head and neck squamous cell cancers. An increased risk of SMNs (all sites combined, as well as for UADT sites and for head and neck squamous cell cancers) was observed among XRCC3 241Met allele homozygotes.85 Because of their important roles in mediating the stabilization and expression of p53, high-risk genotypes of polymorphisms in p53-related genes (p53, p73, p14ARF, MDM2 and MDM4) were examined for their role in increased risk of SMNs after index squamous cell carcinoma of the head and neck. Each p53-related polymorphism had a moderate effect on increasing SMN risk; the risk increased with increasing number of risk genotypes. Compared with the low-risk group (0–3 combined risk genotypes), both the medium risk (4–5 combined risk genotypes) and high-risk (6–9 combined risk genotypes) groups had significantly increased SMN risk. These findings suggest that combined risk genotypes of p53-related genes may jointly modify SMN risk.86 Similar associations were observed in a study that tested the hypothesis that structural and biochemical similarities between p53 and p73 proteins would result in higher risk of SMN after index squamous cell carcinoma of the head and neck among individuals who carry high-risk genotypes of p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms (individually or in combination).87
Thyroid cancer
Thyroid cancer is observed after neck radiation for HL, ALL, brain tumors and after total body irradiation (TBI) for HCT.7, 39 A linear dose-response relation between thyroid cancer and radiation is observed up to 20 Gy, with a decline in the odds ratio at higher doses, demonstrating evidence for a cell kill effect.88, 89 The ATM G5557A and XRCC1 Arg399Gln polymorphisms (DNA damage response genes), is shown to be associated with a decreased risk of papillary thyroid cancer. TP53 Arg72Pro is associated with increased risk of radiogenic papillary thyroid cancer. In the analyses of ATM/TP53 (rs1801516/rs664677/rs609429/rs1042522) combinations, the GG/TC/CG/GC genotype is associated with radiation-induced papillary thyroid cancer. The results indicate that polymorphisms of DNA damage response genes may be potential risk modifiers of ionizing radiation-induced papillary thyroid cancer.90 Significant associations have also been reported for rs1801516 in ATM and rs1867277 in the promoter region of FOXE1, suggesting that thyroid morphogenesis pathway, in addition to DNA double-strand break repair pathway are involved in the etiology of papillary thyroid cancer risk.91 Telomere shortening is observed in response to ionizing radiation exposure. An inverse relation between telomere content and radiation-related thyroid cancer has been observed,92 suggesting that shorter telomeres (resulting in genomic instability) may contribute to thyroid cancer in childhood cancer survivors.
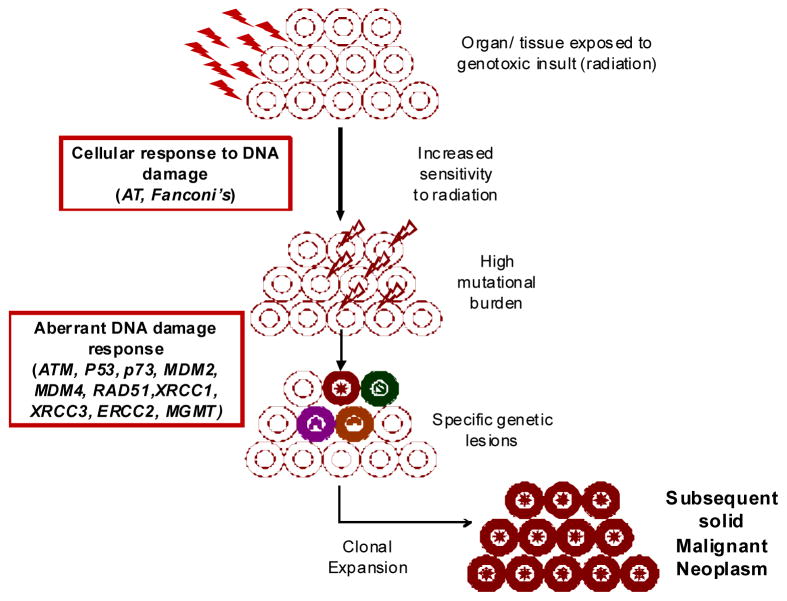
The proposed model (Figure 2) integrates findings from the studies described above to help explain the pathogenesis of radiation-related solid malignancies. Thus, radiation exposure to an un-involved organ (e.g., breast, thyroid, brain, etc.) results in DNA damage, which in turn, initiates cellular responses to the DNA damage. Aberrant DNA damage response results in an increase in mutational burden. Inability to repair the DNA damage results in the development of specific genetic lesions. Finally, clonal expansion of cells carrying specific genetic lesions results in the development of solid SMNs. These studies are summarized in Table 2, demonstrating that the vast majority of these studies are limited in scope and size. The vast majority of the studies are focused on specific DNA repair genes and have demonstrated the role for deleterious ATM missense variant and RAD50 haplotype (contralateral radiation-related breast cancer), XRCC3 241 Met allele homozygotes, P53, p73, p14ARF, MDM2, MDM4 (smoking-related head and neck cancer), and ATM and TP53 (radiation-related papillary carcinoma of the thyroid gland).
Figure 2.
Role of genetic variation in risk of solid subsequent malignant neoplasms – a proposed model
Methodological Issues
In order to develop a deeper understanding of the molecular underpinnings of therapy-related SMNs, careful attention needs to be devoted to study design, sample size, a precise definition of the phenotypes, and high-quality DNA. Consideration should be given to survival bias when designing prevalent case-control studies, especially where the endpoint is associated with high early lethality. Study design must include rigorous power estimations to determine the number of subjects necessary to meet statistical objectives. An efficient and cost-effective methodology is the use of a nested case-control study design, especially when the samples have been banked on the entire cohort and a comprehensive longitudinal follow-up of the cohort has resulted in a near-complete ascertainment of the outcome of interest. Finally, use of appropriate comparison groups is critical. Several studies have either used healthy individuals or individuals with histologically identical de novo cancer as comparison groups (e.g., de novo AML as a reference group for t-MDS/AML). This strategy could be problematic because of the possibility of shared genotoxic insults (e.g., benzene), driving the association towards null. The ideal comparison group should consist of individuals identical to the cases with respect to primary cancer but who do not develop the outcome of interest. It is also important to ensure that the controls have been followed for at least as long as the cases from the time of diagnosis and preferably for a longer duration. This is done to ensure that the “controls” have had ample opportunity to develop the outcome of interest.
Summary and Future Directions
This review provides examples of the modifying influence of genetic variation on treatment-related SMN risk. Most of these studies have examined a limited number of polymorphisms in small heterogeneous samples, contributing to largely inconclusive results. Functional redundancy often results in the availability of more than one gene product to detoxify the same substrate or repair the same damage type. Hence, a variant in one gene may have minimal consequences, whereas the combination of variants in two or more genes could have more serious consequences resulting in the emergence of a malignant phenotype. Furthermore, previous studies have often generally failed to systematically examine gene-therapy interactions, because of the absence of detailed therapeutic exposure data coupled with small sample sizes. There remains a critical need to replicate these findings in large independent cohorts before these findings can be incorporated into the clinical management of the patients. The discovery of functional genetic variants associated with key outcomes will have significant implications for future research aimed at improving risk assessment. Identification of new and informative genetic markers have utility for developing objective pre-therapy risk assessment and patient counseling, and serving as rational tools for clinical management and treatment planning. The ultimate goal is to identify those at highest risk, such that targeted prevention and intervention strategies can be instituted.
Acknowledgments
Supported in part by NCI grant U10 CA98543 (P Adamson); U24 55727 (LL Robison); R01 CA 139633 (S Bhatia); and The Leukemia and Lymphoma Society: 6093-08 (S Bhatia).
References
- 1.Anonymous. Cancer survivors: living longer, and now, better. Lancet. 2004;364:2153–4. doi: 10.1016/S0140-6736(04)17601-0. [DOI] [PubMed] [Google Scholar]
- 2.Oeffinger KC, Mertens AC, Sklar CA, et al. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355(15):1572–82. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- 3.Sun C-L, Francisco F, Kawashima T, et al. Burden of Long-Term Morbidity after Hematopoietic Cell Transplantation: A Report from the Bone Marrow Transplant Survivor Study (BMTSS) Blood. 2007;110:832. [Google Scholar]
- 4.Bhatia S, Robison LL, Francisco L, et al. Late mortality in survivors of autologous hematopoietic cell transplantation: report from the Bone Marrow Transplant Survivor Study. Blood. 2005;105:4215–22. doi: 10.1182/blood-2005-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mertens AC, Liu Q, Neglia JP, et al. Cause-specific late mortality among 5-year survivors of childhood cancer: The childhood cancer survivor study. Journal of the National Cancer Institute. 2008;100(19):1368–79. doi: 10.1093/jnci/djn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis LB, Demark Wahnefried W, Allan JM, Ng AK. Aetiology, genetics and prevention of secondary neoplasms in adult cancer survivors. Nat Rev Clin Oncol. 2013;10:289–310. doi: 10.1038/nrclinonc.2013.41. [DOI] [PubMed] [Google Scholar]
- 7.Friedman DL, Whitton J, Leisenring W, et al. Subsequent neoplasms in 5-year survivors of childhood cancer: the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2010;102(14):1083–95. doi: 10.1093/jnci/djq238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reulen RC, Frobisher C, Winter DL, et al. Long-term risks of subsequent primary neoplasms among survivors of childhood cancer. JAMA. 2011;305(22):2311–9. doi: 10.1001/jama.2011.747. [DOI] [PubMed] [Google Scholar]
- 9.Neglia JP, Robison LL, Stovall M, et al. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2006;98(21):1528–37. doi: 10.1093/jnci/djj411. [DOI] [PubMed] [Google Scholar]
- 10.Inskip PD, Robison LL, Stovall M, et al. Radiation dose and breast cancer risk in the childhood cancer survivor study. J Clin Oncol. 2009;27(24):3901–7. doi: 10.1200/JCO.2008.20.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatti P, Veiga LH, Ronckers CM, et al. Risk of second primary thyroid cancer after radiotherapy for a childhood cancer in a large cohort study: an update from the childhood cancer survivor study. Radiat Res. 2010;174(6):741–52. doi: 10.1667/RR2240.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limacher JM, Frebourg T, Natarajan-Ame S, Bergerat JP. Two metachronous tumors in the radiotherapy fields of a patient with Li-Fraumeni syndrome. Int J Cancer. 2001;96:238–42. doi: 10.1002/ijc.1021. [DOI] [PubMed] [Google Scholar]
- 13.Birch JM. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20:4621–28. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 14.Talwalkar SS, et al. Myelodysplastic syndromes arising in patients with germline TP53 mutation and Li-Fraumeni syndrome. Arch Pathol Lab Med. 2010;134:1010–15. doi: 10.5858/2009-0015-OA.1. [DOI] [PubMed] [Google Scholar]
- 15.Wong FL, Boice JD, Jr, Abramson DH, et al. Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA. 1997;278:1262–7. doi: 10.1001/jama.278.15.1262. [DOI] [PubMed] [Google Scholar]
- 16.Kleinerman RA, Tucker MA, Tarone RE, et al. Risk of new cancers after radiotherapy in long-term survivors of retinoblastoma: An extended follow-up. J Clin Oncol. 2005;23:2272–79. doi: 10.1200/JCO.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 17.Draper GJ, Sanders BM, Kingston JE. Second primary neoplasms in patients with retinoblastoma. Br J Cancer. 1986;53:661–71. doi: 10.1038/bjc.1986.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharif S. Second primary tumors in neurofibromatosis 1 patients treated for optic glioma: substantial risks after radiotherapy. J Clin Oncol. 2006;24:2570–75. doi: 10.1200/JCO.2005.03.8349. [DOI] [PubMed] [Google Scholar]
- 19.Evans DG, Farndon PA, Burnell LD, Gattamaneni HR, Birch JM. The incidence of Gorlin syndrome in 173 consecutive cases of medulloblastoma. Br J Cancer. 1991;64:959–61. doi: 10.1038/bjc.1991.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein AM, Yuen J, Tucker MA. Second cancers after medulloblastoma: population-based results from the United States and Sweden. Cancer Causes Control. 1997;8:865–71. doi: 10.1023/a:1018464328836. [DOI] [PubMed] [Google Scholar]
- 21.Breslow NE, Lange JM, Friedman DL, et al. Secondary malignant neoplasms after Wilms tumor: an international collaborative study. Int J Cancer. 2010;127(3):657–66. doi: 10.1002/ijc.25067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swift M, Morrell D, Massey RB, Chase CL. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325:1831–6. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein JL, et al. Radiation exposure, the ATM gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102:475–83. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maris JM, Wiersma SR, Mahgoub N, et al. Monosomy 7 myelodysplastic syndrome and other second malignant neoplasms in children with neurofibromatosis type 1. Cancer. 1997;79:1438–46. [PubMed] [Google Scholar]
- 25.Swift M, Morrell D, Massey RB, Chase CL. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325:1831–36. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 26.Olsen JH, Hahnemann JM, Borresen-Dale AL, et al. Breast and other cancers in 1445 blood relatives of 75 Nordic patients with ataxia telangiectasia. Br J Cancer. 2005;93:260–5. doi: 10.1038/sj.bjc.6602658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan JM. Genetic susceptibility to radiogenic cancer in humans. Health Phys. 2008;95:677–86. doi: 10.1097/01.HP.0000326339.06405.ea. [DOI] [PubMed] [Google Scholar]
- 28.Kalow W, Ozdemir V, Tang BK, Tothfalusi L, Endrenyi L. The science of pharmacological variability: an essay. Clin Pharmacol Ther. 1999;66:445–7. doi: 10.1016/S0009-9236(99)70006-8. [DOI] [PubMed] [Google Scholar]
- 29.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–49. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 30.Berwick M, Vineis P. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst. 2000;92:874–97. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
- 31.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–30. [PubMed] [Google Scholar]
- 32.Bhatti P, Struewing JP, Alexander BH, et al. Polymorphisms in DNA repair genes, ionizing radiation exposure and risk of breast cancer in U.S. Radiologic technologists. Int J Cancer. 2008;122:177–82. doi: 10.1002/ijc.23066. [DOI] [PubMed] [Google Scholar]
- 33.Rajaraman P, Bhatti P, Doody MM, et al. Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. Int J Cancer. 2008;123:2713–6. doi: 10.1002/ijc.23779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins A, Harrington V. Repair of oxidative DNA damage: assessing its contribution to cancer prevention. Mutagenesis. 2002;17(6):489–93. doi: 10.1093/mutage/17.6.489. [DOI] [PubMed] [Google Scholar]
- 35.Rai R, Peng G, Li K, Lin SY. DNA damage response: the players, the network and the role in tumor suppression. Cancer Genomics Proteomics. 2007;4:99– 106. [PubMed] [Google Scholar]
- 36.Bhatia S, Krailo MD, Chen Z, et al. Therapy-related myelodysplasia and acute myeloid leukemia after Ewing sarcoma and primitive neuroectodermal tumor of bone: a report from the Children’s Oncology Group. Blood. 2007;109:46–51. doi: 10.1182/blood-2006-01-023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia S, Robison LL, Oberlin O, et al. Breast cancer and other second neoplasms after childhood Hodgkin’s disease. N Engl J Med. 1996;334(12):745–51. doi: 10.1056/NEJM199603213341201. [DOI] [PubMed] [Google Scholar]
- 38.Bhatia S, Yasui Y, Robison LL, et al. High risk of subsequent neoplasms continues with extended follow-up of childhood Hodgkin’s disease: report from the Late Effects Study Group. J Clin Oncol. 2003;21(23):4386–94. doi: 10.1200/JCO.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 39.Bhatia S, Sklar C. Second cancers in survivors of childhood cancer. Nat Rev Cancer. 2002;2(2):124–32. doi: 10.1038/nrc722. [DOI] [PubMed] [Google Scholar]
- 40.Bhatia S, Sather HN, Pabustan OB, Trigg ME, Gaynon PS, Robison LL. Low incidence of second neoplasms among children diagnosed with acute lymphoblastic leukemia after 1983. Blood. 2002;99(12):4257–64. doi: 10.1182/blood.v99.12.4257. [DOI] [PubMed] [Google Scholar]
- 41.Travis LB, Andersson M, Gospodarowicz M, et al. Treatment-associated leukemia following testicular cancer. J Natl Cancer Inst. 2000;92:1165–71. doi: 10.1093/jnci/92.14.1165. [DOI] [PubMed] [Google Scholar]
- 42.Travis LB, Holowaty EJ, Bergfeldt K, et al. Risk of leukemia after platinum-based chemotherapy for ovarian cancer. N Engl J Med. 1999;340:351–7. doi: 10.1056/NEJM199902043400504. [DOI] [PubMed] [Google Scholar]
- 43.Krishnan A, Bhatia S, et al. Predictors of Therapy-Related Leukemia and Myelodysplasia following Autologous Transplantation for Lymphoma: an Assessment of Risk Factors. Blood. 2000;95:1588–93. [PubMed] [Google Scholar]
- 44.Howe R, Micallef IN, Inwards DJ, et al. Secondary myelodysplastic syndrome and acute myelogenous leukemia are significant complications following autologous stem cell transplantation for lymphoma. Bone Marrow Transplant. 2003;32:317–24. doi: 10.1038/sj.bmt.1704124. [DOI] [PubMed] [Google Scholar]
- 45.Laughlin MJ, McGaughey DS, Crews JR, et al. Secondary myelodysplasia and acute leukemia in breast cancer patients after autologous bone marrow transplant. J Clin Oncol. 1998;16:1008–12. doi: 10.1200/JCO.1998.16.3.1008. [DOI] [PubMed] [Google Scholar]
- 46.Stone RM. Myelodysplastic syndrome after autologous transplantation for lymphoma: the price of progress. Blood. 1994;83:3437–40. [PubMed] [Google Scholar]
- 47.Govindarajan R, Jagannath S, Flick JT, et al. Preceding standard therapy is the likely cause of MDS after autotransplants for multiple myeloma. Br J Haematol. 1996;95:349–53. doi: 10.1046/j.1365-2141.1996.d01-1891.x. [DOI] [PubMed] [Google Scholar]
- 48.Bhatia S, Ramsay NK, Steinbuch M, et al. Malignant neoplasms following bone marrow transplantation. Blood. 1996;87(9):3633–9. [PubMed] [Google Scholar]
- 49.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 50.Felix CA, Walker AH, Lange BJ, et al. Association of CYP3A4 genotype with treatment-related leukemia. Proc Natl Acad Sci U S A. 1998;95(22):13176–81. doi: 10.1073/pnas.95.22.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rund D, Krichevsky S, Bar-Cohen S, et al. Therapy-related leukemia: clinical characteristics and analysis of new molecular risk factors in 96 adult patients. Leukemia. 2005;19:1919–28. doi: 10.1038/sj.leu.2403947. [DOI] [PubMed] [Google Scholar]
- 52.Blanco JG, Edick MJ, Hancock ML, et al. Genetic polymorphisms in CYP3A5, CYP3A4 and NQO1 in children who developed therapy-related myeloid malignancies. Pharmacogenetics. 2002;12:605–11. doi: 10.1097/00008571-200211000-00004. [DOI] [PubMed] [Google Scholar]
- 53.Bolufer P, Collado M, Barragan E, et al. Profile of polymorphisms of drug metabolizing enzymes and the risk of therapy-related leukaemia. Br J Haematol. 2007;136:590–96. doi: 10.1111/j.1365-2141.2006.06469.x. [DOI] [PubMed] [Google Scholar]
- 54.Larson RA, Wang Y, Banerjee M, et al. Prevalence of the inactivating 609C-->T polymorphism in the NAD(P)H:quinone oxidoreductase (NQO1) gene in patients with primary and therapy-related myeloid leukemia. Blood. 1999;94:803–07. [PubMed] [Google Scholar]
- 55.Naoe T, Takeyama K, Yokozawa T, et al. Analysis of genetic polymorphism in NQO1, GST-M1, GST-T1, and CYP3A4 in 469 Japanese patients with therapy-related leukemia/myelodysplastci syndrome and de novo acute myeloid leukemia. Clin Cancer Res. 2000;6:4091–5. [PubMed] [Google Scholar]
- 56.Allan JM, Wild CP, Rollingson S, et al. Polymorphism in glutathione S-transferase P1 is associated with susceptibility to chemotherapy-induced leukemia. PNAS. 2001;98(20):11592–97. doi: 10.1073/pnas.191211198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo MH, Shuster JJ, Chen CL, et al. Gluthathione S-transferase genotypes in children who develop treatment-related acute myeloid malignancies. Leukemia. 2000;14(232–7) doi: 10.1038/sj.leu.2401660. [DOI] [PubMed] [Google Scholar]
- 58.Seedhouse C, Bainton R, Lewis M, Harding A, Russell N, Das-Gupta E. The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood. 2002;100(10):3761–6. doi: 10.1182/blood-2002-04-1152. [DOI] [PubMed] [Google Scholar]
- 59.Seedhouse C, Faulkner R, Ashraf N, Das-Gupta E, Russell N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clin Cancer Res. 2004;10(8):2675–80. doi: 10.1158/1078-0432.ccr-03-0372. [DOI] [PubMed] [Google Scholar]
- 60.Jawad M, Seedhouse CH, Russell N, Plumb M. Polymorphisms in human homeobox HLX1 and DNA repair RAD51 genes increase the risk of therapy-related acute myeloid leukemia. Blood. 2006;108:3916–18. doi: 10.1182/blood-2006-05-022921. [DOI] [PubMed] [Google Scholar]
- 61.Allan JM, Smith AG, Wheatley K, et al. Genetic variation in XPD predicts treatment outcome and risk of acute myeloid leukemia following chemotherapy. Blood. 2004;104:3872–7. doi: 10.1182/blood-2004-06-2161. [DOI] [PubMed] [Google Scholar]
- 62.Ellis NA, Huo D, Yildiz O, et al. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112(3):741–9. doi: 10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Josting A, Wiedenmann S, Franklin J, et al. Secondary myeloid leukemia and myelodysplastic syndromes in patients treated fpr Hodgkin’s disease: a report from the German Hodgkin’s lymphoma Study Group. J Clin Oncol. 2003;21:3440–6. doi: 10.1200/JCO.2003.07.160. [DOI] [PubMed] [Google Scholar]
- 64.Worrillow LJ, Smith AG, Scott K, et al. Polymorphic MLH1 and risk of cancer after methylating chemotherapy for Hodgkin lymphoma. J Med Genet. 2008;45:142–6. doi: 10.1136/jmg.2007.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Worrillow LJ, Travis LB, Smith AG, et al. An intron splice acceptor polymorphism in hMSH2 and risk of leukemia after treatment with chemotherapeutic alkylating agents. Clin Cancer Res. 2003;9(8):3012–20. [PubMed] [Google Scholar]
- 66.Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169–72. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 67.Guillem VM, Collado M, Terol MJ, et al. Role of MTHFR (677, 1298) haplotype in the risk of developing secondary leukemia after treatment of breast cancer and hematological malignancies. Leukemia. 2007;21:1413– 22. doi: 10.1038/sj.leu.2404709. [DOI] [PubMed] [Google Scholar]
- 68.Ding Y, Sun C-L, Li L, et al. Genetic susceptibility to therapy-related leukemia after Hodgkin lymphoma or non-Hodgkin lymphoma: role of drug metabolism, apoptosis, and DNA repair. BCJ. 2012;2:e58. doi: 10.1038/bcj.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knight JA, Skol AD, Shinde A, et al. Genome-wide association study to identify novel loci associated with therapy-related myeloid leukemia susceptibility. Blood. 2009;113(22):5575–82. doi: 10.1182/blood-2008-10-183244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Best T, Li DL, Skol AD, et al. Variants at 6q21 implicate PRDM1 in the etiology of therapy-induced second malignancies after Hodgkin’s lymphoma. Nature Medicine. 2011;17(8):941–43. doi: 10.1038/nm.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Henderson TO, Amsterdam A, Bhatia S, et al. Systematic review: surveillance for breast cancer in women treated with chest radiation for childhood, adolescent, or young adult cancer. Ann Intern Med. 2010;152(7):444–55. W144–54. doi: 10.1059/0003-4819-152-7-201004060-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowers DC, Nathan PC, Constine L, et al. Subsequent neoplasms of the CNS among survivors of childhood cancer: a systematic review. Lancet Oncology. 2013;14(8):E321–E28. doi: 10.1016/S1470-2045(13)70107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bethke L, Murray A, Webb E, et al. Comprehensive Analysis of DNA Repair Gene Variants and Risk of Meningioma. J Natl Cancer Inst. 2008;100:270– 76. doi: 10.1093/jnci/djn004. [DOI] [PubMed] [Google Scholar]
- 74.Hosking FJ, Feldman D, Bruchim R, et al. Search for inherited susceptibility to radiation-associated meningioma by genomewide SNP linkage disequilibrium mapping. British Journal of Cancer. 2011;104:1049–54. doi: 10.1038/bjc.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pappo AS, Armstrong GT, Liu W, et al. Melanoma as a subsequent neoplasm in adult survivors of childhood cancer: A report from the childhood cancer survivor study. Pediatric Blood & Cancer. 2013;60(3):461–66. doi: 10.1002/pbc.24266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Braam KI, Overbeek A, Kaspers GJ, et al. Malignant melanoma as second malignant neoplasm in long-term childhood cancer survivors: a systematic review. Pediatr Blood Cancer. 2012;58(5):665–74. doi: 10.1002/pbc.24023. [DOI] [PubMed] [Google Scholar]
- 77.Socie G, Curtis RE, Deeg HJ, et al. New malignant diseases after allogeneic marrow transplantation for childhood acute leukemia. Journal of Clinical Oncology. 2000;18(2):348–57. doi: 10.1200/JCO.2000.18.2.348. [DOI] [PubMed] [Google Scholar]
- 78.Guerin S, Dupuy A, Anderson H, et al. Radiation dose as a risk factor for malignant melanoma following childhood cancer. Eur J Cancer. 2003;39(16):2379–86. doi: 10.1016/s0959-8049(03)00663-4. [DOI] [PubMed] [Google Scholar]
- 79.Soufir N, Avril MF, Chompret A, et al. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet. 1998;7(2):209–16. doi: 10.1093/hmg/7.2.209. [DOI] [PubMed] [Google Scholar]
- 80.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–5. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falchi M, Bataille V, Hayward NK, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nature Genetics. 2009;41(8):915–U76. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tucker MA. Melanoma epidemiology. Hematol Oncol Clin North Am. 2009;23(3):383–95. vii. doi: 10.1016/j.hoc.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eng C, Li FP, Abramson DH, et al. Mortality from second tumors among long-term survivors of retinoblastoma. J Natl Cancer Inst. 1993;85(14):1121–8. doi: 10.1093/jnci/85.14.1121. [DOI] [PubMed] [Google Scholar]
- 84.Mandelcorn-Monson R, Marrett L, Kricker A, et al. Sun exposure, vitamin D receptor polymorphisms FokI and BsmI and risk of multiple primary melanoma. Cancer Epidemiology. 2011;35:e105–e10. doi: 10.1016/j.canep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gal TJ, Huang W-Y, Chen C, Hayes RB, Schwartz SM. DNA Repair Gene Polymorphisms and Risk of Second Primary Neoplasms and Mortality in Oral Cancer Patients. Laryngoscope. 2005;115:2221–31. doi: 10.1097/01.mlg.0000183736.96004.f7. [DOI] [PubMed] [Google Scholar]
- 86.Jin L, Sturgis EM, Zhang Y, et al. Genetic variants in p53-related genes confer susceptibility to second primary malignancy in patients with index squamous cell carcinoma of head and neck. Carcinogenesis. 2013;34:1551–57. doi: 10.1093/carcin/bgt096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, Sturgis EM, Huang Z, Zafereo ME, Wei Q, Li G. Genetic Variants of the P53 and P73 Genes Jointly Increase Risk of Second Primary Malignancies in Patients After Index Squamous Cell Carcinoma of the Head and Neck. Cancer. 2012;118:485–92. doi: 10.1002/cncr.26222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sigurdson AJ, Ronckers CM, Mertens AC, et al. Primary thyroid cancer after a first tumour in childhood (the Childhood Cancer Survivor Study): a nested case-control study. Lancet. 2005;365(9476):2014–23. doi: 10.1016/S0140-6736(05)66695-0. [DOI] [PubMed] [Google Scholar]
- 89.Ronckers CM, Sigurdson AJ, Stovall M, et al. Thyroid cancer in childhood cancer survivors: a detailed evaluation of radiation dose response and its modifiers. Radiat Res. 2006;166(4):618–28. doi: 10.1667/RR3605.1. [DOI] [PubMed] [Google Scholar]
- 90.Akulevich NM, Saenko VA, Rogounovitch TI, et al. Polymorphisms of DNA damage response genes in radiation-related and sporadic papillary thyroid carcinoma. Endocrine-Related Cancer. 2009;16:491–503. doi: 10.1677/ERC-08-0336. [DOI] [PubMed] [Google Scholar]
- 91.Damiola F, Byrnes G, Moissonnier M, et al. Contribution of ATM and FOXE1 (TTF2) to risk of papillary thyroid carcinoma in Belarusian children exposed to radiation. Int J Cancer. 2014;134:1659–68. doi: 10.1002/ijc.28483. [DOI] [PubMed] [Google Scholar]
- 92.Gramatges MM, Liu Q, Yasui Y, et al. Telomere content and risk of second malignant neoplasm in survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. Clin Cancer Res. 2014;20(4):904–11. doi: 10.1158/1078-0432.CCR-13-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mertens AC, Mitby PA, Radloff G, et al. XRCC1 and glutathione-S-transferase gene polymorphisms and susceptibility to radiotherapy-related malignancies in survivors of Hodgkin disease. Cancer. 2004;101:1463–72. doi: 10.1002/cncr.20520. [DOI] [PubMed] [Google Scholar]
- 94.Brooks JD, Teraoka SN, Reiner AS, et al. Variants in Activators and downstream targets of ATM, radiation exposure, and contralateral breast cancer risk on the WECARE study. Hum Mutat. 2012;33:158–64. doi: 10.1002/humu.21604. [DOI] [PMC free article] [PubMed] [Google Scholar]