Abstract
Changes in DNA methylation and subsequent changes in gene expression regulation are the hallmarks of age- and tissue-dependent epigenetic drift and plasticity resulting from the combinatorial integration of genetic determinants and environmental cues. To determine whether perinatal lead exposure caused persistent DNA methylation changes in target tissues, we exposed mouse dams to 0, 3 or 30 ppm of lead acetate in drinking water for a period extending from 2 months prior to mating, through gestation, until weaning of pups at postnatal day-21, and analyzed whole-genome DNA methylation in brain cortex and hippocampus of 2-month old exposed and unexposed progeny. Lead exposure resulted in hypermethylation of three differentially methylated regions in the hippocampus of females, but not males. These regions mapped to Rn4.5s, Sfi1, and Rn45s loci in mouse chromosomes 2, 11 and 17, respectively. At a conservative fdr<0.001, 1,623 additional CpG sites were differentially methylated in female hippocampus, corresponding to 117 unique genes. Sixty of these genes were tested for mRNA expression and showed a trend towards negative correlation between mRNA expression and methylation in exposed females but not males. No statistically significant methylome changes were detected in male hippocampus or in cortex of either sex. We conclude that exposure to lead during embryonic life, a time when the organism is most sensitive to environmental cues, appears to have a sex- and tissue-specific effect on DNA methylation that may produce pathological or physiological deviations from the epigenetic plasticity operative in unexposed mice.
Keywords: DNA methylation, Brain, Gene expression, Lead, Heavy metals
INTRODUCTION
Environmental signals enable organisms to react and adapt to changing living conditions. More than a filter that selects potential phenotypic variations, the environment is itself the source of the variation through cues that enable the developing organism to increase its fitness in that particular environment (West-Eberhard 2005). Because embryonic life is a time when the organism is most sensitive to environmental signals (Yamazaki et al. 2003), this phenotypic plasticity is particularly critical during development. Developmental plasticity, however, is not always adaptive and often gives rise to maladaptive pathophysiological consequences either in the embryo or in later adult life, as is the case with the responses to lead exposure. There is good agreement that the most important cognitive, behavioral and psychiatric health effects of lead exposure are manifest long after exposure has ceased (Wright et al. 2008; Yuan et al. 2006), suggestive of either a genetic (mutational) or an epigenetic component. However, the causes of the long-term morbidity associated with prenatal and early postnatal exposure to lead are poorly understood. The variability in genetic or epigenetic factors as exacerbating or protective agents of human neurodevelopmental morbidity has not been adequately examined in relationship to early exposure to lead. Studies linking attention deficits, aggressive and disruptive behavior, and poor self-regulation have shown that early exposure to lead results in an increased likelihood of engaging in antisocial behavior in later life (Dietrich et al. 2001; Needleman et al. 1996; Needleman et al. 2002; Wright et al. 2008). Current debate centers on the identification of the developmental periods during which the organism is most vulnerable to the effects of lead and on the exposure level and duration that produce adverse effects. Risk factors and biomarkers are needed to identify individuals at high risk for lead-associated maldevelopment.
In humans, early life exposure to lead can produce persistent alterations in the brain structure of adults, including loss of gray matter in the cortex (Brubaker et al. 2009; Cecil et al. 2008), changes in myelin structure in white matter (Brubaker et al. 2009), and low level of activation in brain areas associated with language function, such as left frontal cortex and left middle temporal gyrus (Yuan et al. 2006). Additionally, mice exposed to lead in utero have also neurochemistry alterations in the hippocampus, including increment of myoinositol/creatine (Ins/Cr) and glutamine (Gln) (Lindquist, unpublished). Recently, gestational lead exposure in Wistar rats was shown to reduce the number of pyramidal cells in the hippocampus (Baranowska-Bosiacka et al. 2013). In addition, differentiation of embryonic stem cells into glutamatergic neurons in the presence of lead caused alterations in the expression of glutamate receptor subunits Grin1, Grin2D, Grik5, Gria4, and Grm6 that were also observed in hippocampus and cortex of mice gestationally exposed to this metal (Sanchez-Martin et al. 2013). Using primary rat hippocampal cultures, lead was found to negatively modify important neuronal pathways implicated in synaptic plasticity, such as learning, memory, and cell survival (Guilarte and McGlothan 2003; Neal et al. 2011). These in vivo and in vitro findings suggest that cortex and hippocampus are the key target tissues of lead toxicity in the brain.
Heavy metals such as lead elicit environmental signals that modulate epigenetic mechanisms often associated with regulation of gene expression, of which DNA methylation at CpG sites is the most common (Rountree et al. 2001). Expression and activity of DNA methyltransferases (DNMTs) is highly regulated in the central nervous system (CNS) (Feng et al. 2005). Important genes triggered during memory formation and synaptic plasticity, such as Reelin and brain-derived neurotrophic factor (BDNF), show dramatic changes in promoter methylation when DNMT activity is inhibited in hippocampus of young adult mice (Levenson et al. 2006), leading to the hypothesis that DNMT activity may be crucial to regulate brain function. Consistent with this hypothesis, 700-day old mice exposed during gestation to lead showed changes in the induction or repression of 150 genes that correlated with their DNA methylation profiles (Dosunmu et al. 2012). Strikingly, Macaca fascicularis monkeys exposed to lead during infancy showed epigenetic changes twenty-three years later that caused reduced levels of total DNA methylation, DNA methyltransferases-1 and -3A, methyl CpG binding protein-2, and modified histone marks critical for the regulation of gene expression. As a result of these changes, the aging brains of these monkeys showed elevated expression of Alzheimer diseaserelated genes, including β-amyloid precursor protein (APP) and β-site APP cleaving enzyme 1 (BACE1), as well as an increase of total amyloid plaques in the cortex (Bihaqi et al. 2011; Wu et al. 2008).
We used DNA methylation analyses to determine whether prenatal to early postnatal exposure to lead acetate were associated with persistent DNA methylation changes in the brain tissues of exposed mice. Our results show that, at a time point when blood lead levels of perinatally exposed and unexposed adult mice are undistinguishable from each other, there is a highly significant change in DNA methylation in the specific brain regions of the exposed mice, with a trend to be negatively correlated to gene expression levels. The effect is sex- and tissue-dependent, with females showing greater hypermethylation than males, and more so in hippocampus than cortex. Exposure to lead during embryonic life appears to have a sex- and tissue-specific effect that may produce pathological or physiological deviations from the epigenetic plasticity operative in unexposed mice. Further analyses to correlate DNA methylation and regulatory gene expression changes will be crucial to understand the mechanisms of lead neurotoxicity.
MATERIALS AND METHODS
Animals and lead exposure
C57BL/6 mice (Charles River) were housed in the Vivarium at the Cincinnati Children’s Hospital Medical Center under standard conditions (10 h light/14 h dark) and given ad libitum access to food and water. The Animal Care and Use Committees of the Cincinnati Children’s Hospital Medical Center and the University of Cincinnati approved all experimental procedures conducted with these mice. Mice were treated humanely and with regard for alleviation of suffering. Female mice were given drinking water containing 0, 3 (low dose), or 30 ppm (high dose) lead acetate (Sigma Aldrich, St. Louis, Mo.), approximately 60- and 600-fold, respectively the current action level, for a minimum of 2 months prior to mating to stabilize their circulating lead levels and were maintained on this water through weaning. Male breeding mice were exposed to leaded water only while they were with the females. Pups were weaned at postnatal day 21 and were put on normal drinking water for the duration of the experiment. Drinking patterns and water consumption showed no appreciable differences between the groups. One male and one female from each of four litters were evaluated at post-natal day 60 (± 1 day) at a time when blood was collected for blood lead analysis and brain regions were collected for DNA methylation analyses. Blood and tissue samples were stored at −80° C until analyzed.
Tissue collection and total DNA isolation
Brain cortex and hippocampus were collected by dissection using a rodent brain slicer matrix (Zivic Instruments, Pittsburgh, PA). Total DNA from the two brain areas was isolated using a DNeasy blood and tissue kit from Qiagen following the manufacturer’s protocols. The incubation time for lysis was 60 and 150 minutes for cortex and hippocampus respectively.
Global methyl-seq analysis
To prepare the sequencing library we used the SureSelect Methyl-Seq Target Enrichment System for mouse kit (Agilent, Santa Clara, CA). Following Agilent recommended conditions, we used 2–3 µg of high quality mouse genomic DNA sheared in a Covaris S2 focused-ultrasonicator (Covaris, Woburn, MA) to a size of 150–200 bp, as validated by 2100 Bioanalyzer (Agilent). The DNA fragments were end-repaired, 3’-end adenylated and ligated to the methylated adaptor. The size of the ligated libraries was validated in the Bioanalyzer, followed by hybridization with biotin-labeled RNA-baits to capture the regions where methylation impacts gene regulation, including CpG islands, CpG island shores, undermethylated regions, promoters, and differentially methylated regions. After hybridization, the libraries were captured with streptavidin beads, bisulfite-modified with the EZ DNA Methylation-Gold kit (Zymo, Irvine, CA), and enriched by 8 cycles of PCR. Individually amplified libraries were labeled with unique indices by 6 cycles of PCR and purified and size-selected using AMPure XP beads (Beckman Coulter, Indianapolis IN). The quality and quantity of the libraries were assessed by the Bioanalyzer High Sensitivity DNA assay. To accurately quantify the library concentration for clustering generation, the libraries were diluted 1:106 in a buffer containing 10 mM Tris-HCl, pH 8.0 and 0.05% Tween 20, and analyzed by qPCR using a Kapa Library Quantification kit (Kapabiosystem, Woburn, MA) in the ABI's 9700HT Fast Real-Time PCR System (Lifetech, Grand Island, NY).
Cluster generation and HiSeq sequencing
Equal amounts of four uniquely indexed libraries were pooled to fill one lane of the flow cell for clustering in the cBot system (Illumina, San Diego, CA). The pooled libraries at a final concentration of 15 pM were clustered onto a flow cell using Illumina’s TruSeq PE Cluster kit v3, and sequenced for paired-end 2×100 cycles using TruSeq SBS kit v3 on Illumina HiSeq system according to the manufacturer’s recommended protocol. All library preparation, clustering and sequencing steps were performed by the Genomics, Epigenomics and Sequencing Core of the University of Cincinnati.
Statistical differential methylation analysis
Methyl-seq data was demultiplexed and converted to fastq files using Illumina’s CASAVA 1.8. The sequencing quality was assessed by FastQC software (Andrews 2014). Paired-end sequence was aligned to UCSC mm10 mouse genome using Bismark (Krueger and Andrews 2011) with default parameters. Bismark was then used to extract methylation status from each read. This resulted in counts of methylated and un-methylated reads at each CpG site in every sample. In order to identify differentially methylated CpG sites caused by lead exposure, the male and female lead-exposed samples were separately compared to control samples of the same sex. The beta-binomial test from ibb R package (Pham et al. 2010) was used to test for differential methylation on the counts. Basically, the count of methylated C of a CpG site was distributed according to a binomial distribution with success probability p in n total reads of the site, and p was modeled as a random variable from a beta distribution. Since the differential methylation analysis was performed on raw counts, no filtering on coverage was performed. A Manhattan plot was used to visualize the distribution of differentially methylated CpG sites on the genome. Significant differentially methylated CpG sites were selected with FDR adjusted p-values of <0.001, <0.01, or <0.1. Two adjacent significant CpG sites were combined into one significant region if they were less than 1000 bp apart from each other. The significant region was extended until there were no more significant CpG sites within 1000 bp from the current region. The closest genes of significant sites and regions were identified using Bioconductor package VariantAnnotation (Obenchain et al. 2014).
Reverse transcription and real-time PCR
Total RNA was isolated from the tissue samples by using the RNeasy Mini kit (Qiagen). Reverse transcription was performed using random hexamer primers and SuperScript III transcriptase (Invitrogen) as indicated previously (Wang et al. 2010). Real-time qPCR was performed to quantify the expression levels of different genes which were normalized to Gapdh mRNA. Supplemental Table S1 shows a list of primers used for each gene. Raw data was analyzed using the 2ΔΔCt method and it is shown as the log2 of the normalized level.
RESULTS
Blood lead levels in PND60 mice
We exposed dams to 0, 3, or 30 ppm lead acetate in drinking water for a minimum of 2 months prior to mating and maintained these conditions through weaning, while male breeding mice were exposed to leaded water only while they were with the females. Pups were weaned at post-natal day 21 and were given drinking water without lead for the duration of the experiment. At PND60, the time when they were subjected to DNA methylation analyses, we found no significant difference in blood lead values among the three groups of pups. Blood lead levels in pups (n=16 per group) of control, 3 ppm- and 30 ppm-exposed dams were 1.2 ± 1.1, 0.9 ± 0.8 and 1.3 ± 3.1 µg/dl, well below the current action level of 5 µg/dl, in good agreement with values observed in unexposed mice fed a normal diet (Ercal et al. 1996; Iavicoli et al. 2003) and consistent with prior observations by others (Virgolini et al. 2008; Widzowski et al. 1994).
CpG site coverage
Bismark alignment was performed after bisulfite sequencing to analyze the coverage of CpG data reads obtained. The number of CpG sites covered was higher in cortex than in hippocampus samples, ranging between 1.5 million for males and 1.2 million for females in both groups of lead exposure. In the female hippocampus, sequencing covered slightly more than 0.8 million CpG sites for mice exposed to 3 ppm and 1.5 million CpG sites when exposed to 30 ppm. In the male hippocampus, the coverage was 0.8 million CpG sites for both lead doses (Supplemental Table S2).
Comparison of DMR methylation levels in exposed and unexposed mice
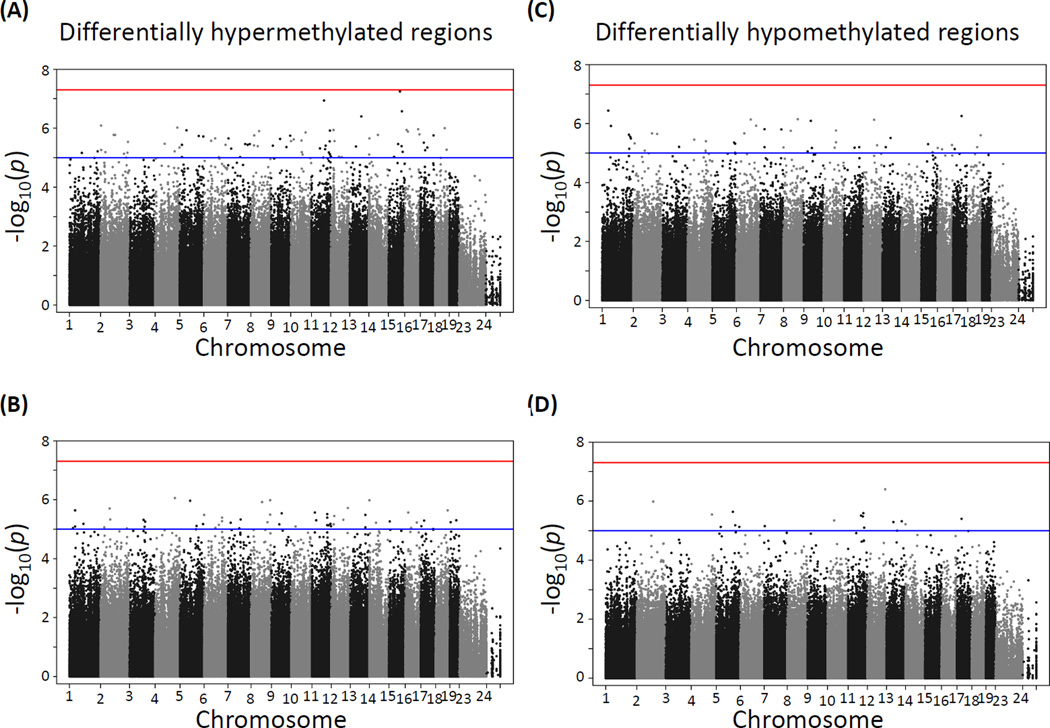
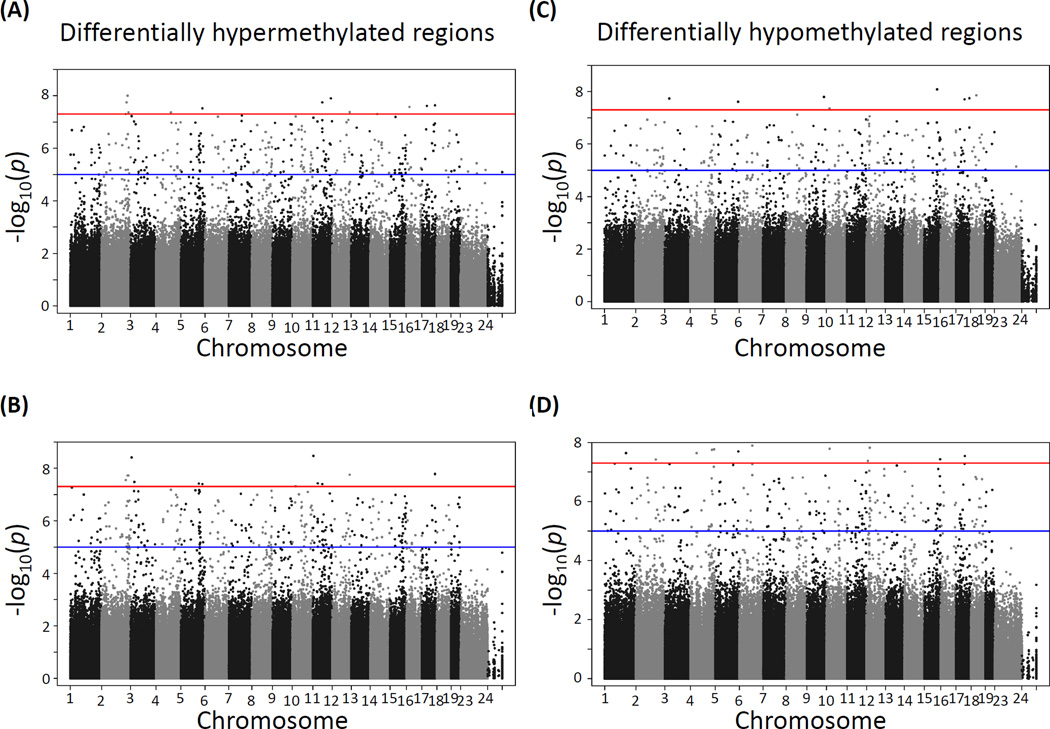
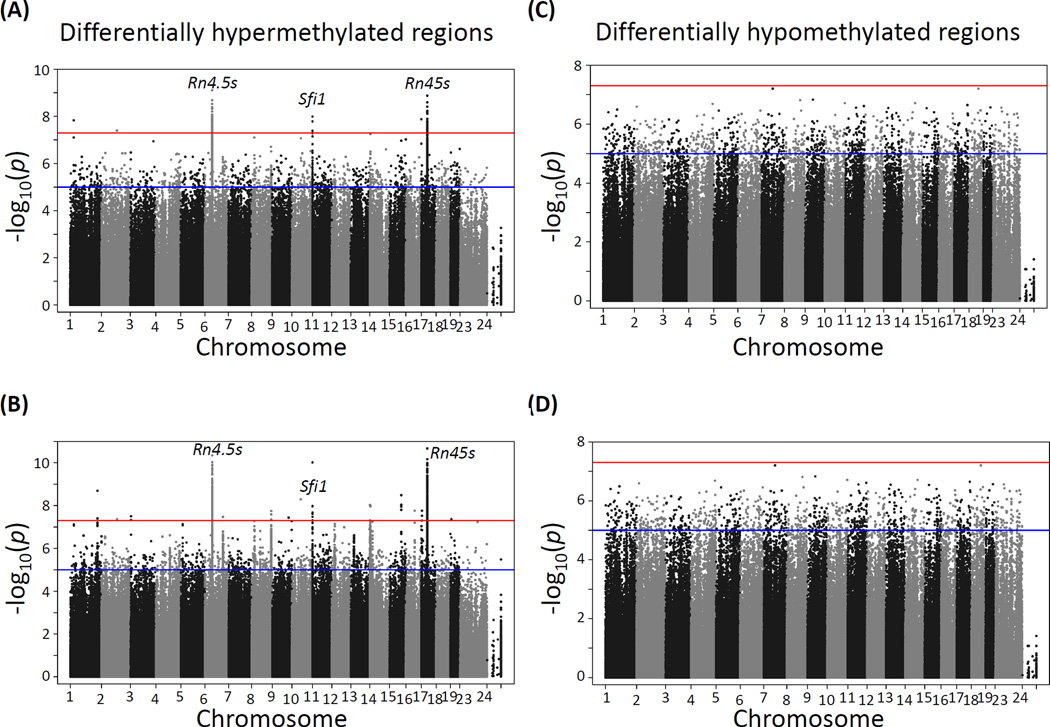
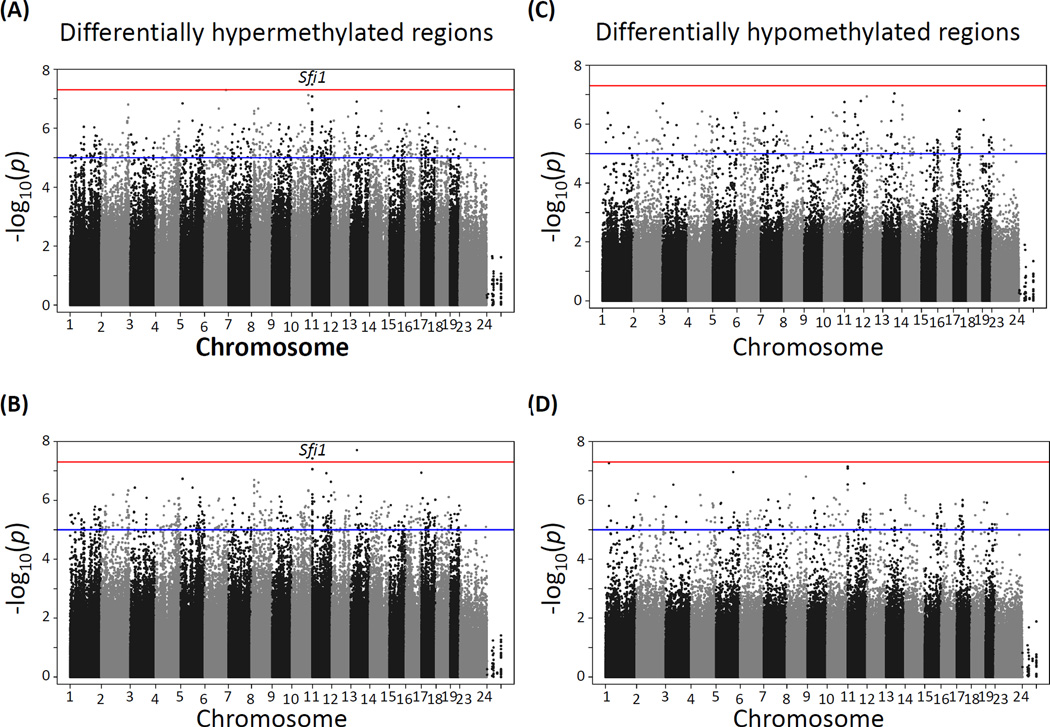
CpG site methylation in cortex and hippocampus at various lead doses relative to the unexposed control groups showed a large number of differentially methylated sites at p-values between 1×10−5 and 5×10−8 and below, the later corresponding to the Bonferroni correction for an approximate one million multiple tests at p≤0.05. The distribution of differentially methylated sites appears to be fairly homogeneous across all chromosomes, except for the X chromosome, which as expected is significantly hypomethylated in males compared to females (see comparison of male vs female data for cortex in Supplemental Fig. S1). Using p≤5×10−8 as the threshold for statistical significance, no significant hyper- or hypomethylated sites were detected in the hippocampus (Fig. 1) or cortex (Fig. 2) of male mice exposed to either 3 (panels A and C) or 30 (panels B and D) ppm lead doses. In contrast, three prominent hypermethylation regions were found in the hippocampus of females exposed to both 3 ppm (Fig. 3A) and 30 ppm doses (Fig. 3B). These three DMRs mapped to Rn4.5s in chromosome 2, Sfi1 in chromosome 11, and Rn45s in chromosome 17; of the three sites, Sfi1 was also hypermethylated in cortex of female mice exposed to both 3 and 30 ppm of lead, albeit at a lower significance level (Fig. 4A and 4B). In contrast to the hypermethylation results, no lead dose induced local DNA hypomethylation (Figs. 1–4, panels C and D).
Figure 1. Manhattan plot of −log10 p-values of differential methylation across the mouse genome induced by lead acetate in the hippocampus of male mice.
(A) and (B): differentially hypermethylated regions in mice exposed to 3 ppm (A) and 30 ppm (B) of lead. (C) and (D): differentially hypomethylated regions in mice were exposed to 3 ppm (C) and 30 ppm (D) of lead.
Figure 2. Manhattan plot of −log10 p-values of differential methylation across the mouse genome induced by lead acetate in the cortex of male mice.
(A) and (B): differentially hypermethylated regions in mice exposed to 3 ppm (A) and 30 ppm (B) of lead. (C) and (D): differentially hypomethylated regions in mice were exposed to 3 ppm (C) and 30 ppm (D) of lead.
Figure 3. Manhattan plot of −log10 p-values of differential methylation across the mouse genome induced by lead acetate in the hippocampus of female mice.
(A) and (B): differentially hypermethylated regions in mice exposed to 3 ppm (A) and 30 ppm (B) of lead. (C) and (D): differentially hypomethylated regions in mice were exposed to 3 ppm (C) and 30 ppm (D) of lead.
Figure 4. Manhattan plot of −log10 p-values of differential methylation across the mouse genome induced by lead acetate in the cortex of female mice.
(A) and (B): differentially hypermethylated regions in mice exposed to 3 ppm (A) and 30 ppm (B) of lead. (C) and (D): differentially hypomethylated regions in mice were exposed to 3 ppm (C) and 30 ppm (D) of lead.
In addition to the three prominent regions described above, a number of hyper- and hypomethylated sites were evident in the Manhattan plots in Figs. 1–4 with p-values comprised between the 1×10−5 and 5×10−8 thresholds indicated earlier. We set three different false discovery rate (fdr) values to categorize the significance of these sites and determine whether specific regions of the genome showed statistically significant changes in methylation patterns due to lead exposure. For this analysis, we considered that a group of contiguous sites plus 500 bp at either side constituted a genome region associated with its corresponding gene locus. At fdr≤ 0.1, most of the female genome showed methylation changes after exposure to 3 or 30 ppm of lead, with greater than 160,000 CpG sites, 60,000 genome regions and 16,000 genes differentially methylated in hippocampus. No significant number of sites were affected in the hippocampus of exposed males (Table 1). At this cut-off level, cortices in both sexes showed significant differential methylation, 10-times higher in females than in males (Supplemental Table S3).
Table 1.
Number of significant CpG sites, genome regions and genes affected by Pb exposure at different fdr cut-offs
| FDR | CpG Sites | Genome Regions | Genes |
|---|---|---|---|
| 0.001 | 1623 | 222 | 117 |
| 0.01 | 20129 | 12435 | 6180 |
| 0.1 | 159619 | 66539 | 16141 |
At fdr≤0.01, we found 20,129 differentially methylated sites in female hippocampus, corresponding to 12,435 genome regions encompassing 6,180 genes (Table 1). At this cut-off value, neither the hippocampus of males nor the cortex of females exposed to lead presented significant sites with changes in the methylation pattern, but we found112 hypermethylated sites in the cortex of males exposed to 3 ppm and 181when exposed to 30 ppm (Supplemental Table S3).
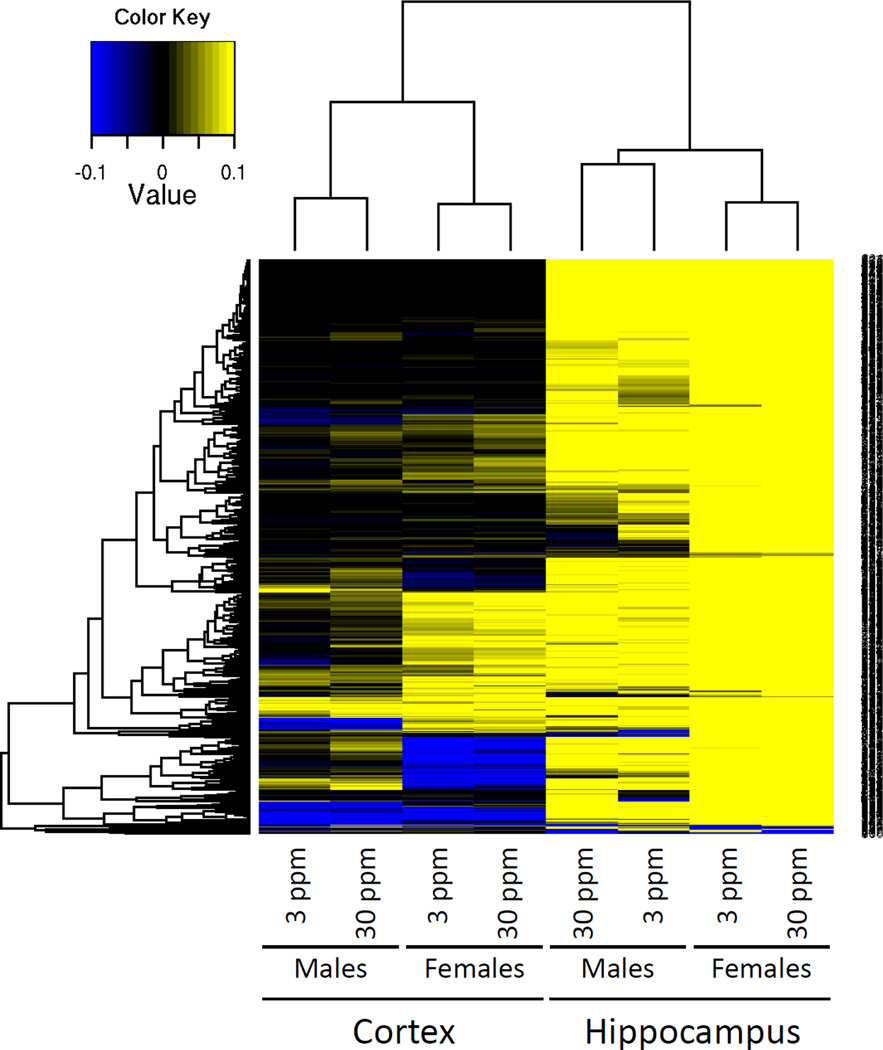
At fdr≤0.001, we found 1,623 differentially methylated CpG sites in the hippocampus of exposed females only, and no significant sites in cortex of either sex. Of these 1,623 CpG sites, 322 corresponded to females exposed to 3 ppm and 1,537 (with some overlap with the previous dose) to 30 ppm exposure, encompassing 222 regions of the genome and affecting 117 unique genes (Table 1 and Supplemental Tables S3 and S4). Interestingly, most of the 1,623 significant CpG sites observed in the hippocampus at this cut-off were hypermethylated after lead exposure with little evidence of hypomethylation (Fig. 5 and Supplemental Table S4). In addition, data from male and female differential methylation clustered together for tissue and treatment dose, but separately for sex (Fig. 5), indicating that perinatal lead exposure induces differential methylation in male and female brain tissues, albeit at very different levels.
Figure 5. Heat map of significant CpG sites differentially methylated at a fdr≤0.001.
The values shown for the females correspond to the normalized β-values calculated from the number of methylated counts divided by the number of methylated plus unmethylated counts for each one of the the1,623 CpG sites in the females. The male β-values are the corresponding values in the males.
Gene expression analysis by real-time PCR
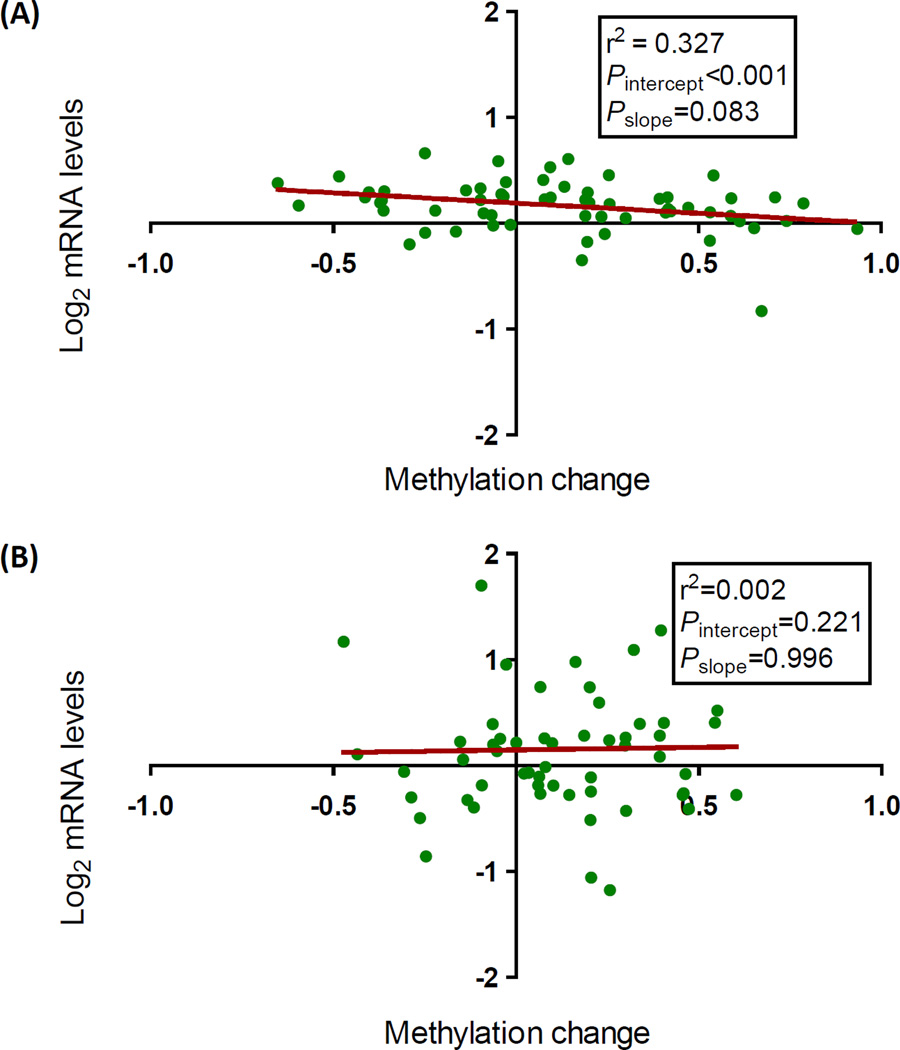
We used real-time PCR to analyze the expression of 60 of the 117 differentially methylated unique genes at fdr≤0.001 in the hippocampus of male and female mice exposed to 0 and 3 ppm lead. The selected genes had differentially methylated CpG sites with p<0.05 for females exposed to both lead doses, and the sites were located between −10 and +10 kb from the transcription start site (Supplemental Table S4). Expression of these genes in lead exposed mice, whether male or female, was very similar to their expression in control mice when the genes were analyzed individually (Supplemental Fig. S2); however, there was a trend towards a significant negative correlation between methylation change and mRNA level (pslope=0.083; r2=0.327) in the hippocampus of female mice exposed to 3 ppm lead (Fig. 6A) and not in the males (pslope=0.996; r2=0.002) (Fig. 6B). These data suggest that lead not only induces hypermethylation in the female hippocampus, but also that it may silence the expression of genes involved in brain function as a consequence of epigenetic modification.
Figure 6. Analysis of the correlation between mRNA levels and methylation change in hippocampus of females (A) and males (B) exposed to 3 ppm of lead.
The log2 mRNA levels determined by RT-PCR of the 60 genes tested (see Supplemental Fig. S2) is shown as a function of the corresponding methylation changes (Supplemental Table S4). Negative methylation change values indicate hypomethylation and positive, hypermethylation.
DISCUSSION
In this study we show that in utero and lactational lead exposure induces persistent DNA methylation changes in hippocampus and cortex, two brain tissues that are lead targets in mice. The majority of changes result from DNA hypermethylation and are highly sex dependent, with female mice substantially more affected than males. Of the two brain tissues, the hippocampus had significantly higher levels of differentially methylated CpG sites than the cortex. At a restrictive p-value≤5×10−8, only CpG sites in the Sfi1 gene were hypermethylated in the female cortex at both exposure levels. In contrast, exposure led to high levels of hypermethylation in the hippocampus of females, that affected sites in three loci, Rn4.5s, Sfi1, and Rn45s, mapping to chromosomes 2, 11, and 17, respectively, showing a striking degree of DNA hypermethylation. At a conservative fdr≤0.001, we found 1,623 differentially methylated CpG sites in the hippocampus of exposed females, the majority (>90%) showing hypermethylation. These sites defined 222 regions of the genome, corresponding to an additional 117 unique genes. After analyzing the expression of 60 from this group, we found a trend towards a significant negative correlation between expression and methylation change in exposed female mice, but not males. This analysis is inherently limited for it examines only a single time point of expression and does not take into consideration the timing of expression of the genes tested. These genes are distributed throughout the genome and do not appear to be related through either regulatory or functional connections, even though the methylation changes occur reproducibly in multiple mice at the same locations.
Early life exposure to lead may have toxic effects in the developing brain. Lead exposure during early childhood has been linked to deficits in cognitive functions and IQ, behavioral effects, and attention deficit hyperactivity disorder (Bellinger et al. 1994; Chen et al. 2007; Froehlich et al. 2009), suggesting that lead neurotoxicity may result from the alteration of mechanisms like DNA methylation that regulate transcriptional pathways contributing to synapse function, neurogenesis and ultimately, expression of memory-related genes. Lead has recently been proposed to act in a locus-specific way on the epigenome, depending on the genomic features in which affected CpG sites are located (Faulk et al. 2013; Faulk et al. 2014). Our current results are in conceptual agreement with this hypothesis.
Lead exposure alters the expression of genes involved in DNA methylation, such as methyl-cytosine-phosphate-guanine binding protein-2 (MeCP2) and DNA methyltransferases-1 and -3A (Bihaqi et al. 2011; Schneider et al. 2013; Wu et al. 2008). Exposure to lead from gestational day 13 to PND-20 resulted in significant changes of gene expression in cortical regions of mouse brain, at 20 and 700 days of age; these changes were correlated with changes in DNA methylation profile and repressed many genes normally up-regulated during normal aging, suggesting that early life exposure to lead disturbs developmental processes in the brain and compromises its ability to cope with adult challenges (Dosunmu et al. 2012). Hypermethylation of genes involved in neurogenesis signaling pathways has also been found in neuronal precursor cells derived from human embryonic stem cells chronically exposed to lead. These cells exhibit shorter neurites and less branching, as well as a significant decrease in the expression levels of the neural marker genes PAX6 and MSI1(Senut et al. 2014).
Methylation and expression show a strong sex-dependence, with changes more evident in females than in males. Similar sex-specific effects have been observed as the consequence of maternal separation, which caused repression of BDNF expression in hippocampus of C57BL/6J female mice but not males, and hypermethylation in males but not females (Kundakovic et al. 2013). Brain differences between males and females are a common phenomenon, since sexual differentiation in the brain takes place during a perinatal sensitive window as a result of gonadal hormone-induced developmental organization (Auger and Auger 2013; Chung and Auger 2013; Menger et al. 2010). Sex differences in gene expression patterns and in the regulation of genes coding for DNA methylases have been observed in hippocampus and frontal cortex of rats exposed to lead (Schneider et al. 2011; Schneider et al. 2012b), and shown to be differentially expressed depending on the developmental timing of the exposure (Schneider et al. 2012a). The effect of sex in the regulation of the genome and epigenome is largely unexplored. Sex influences genomic methylation status although the mechanisms underlying this effect are unknown (Liu et al. 2010). Possibly, interactions between sex hormones and lead exposure during development make the female hippocampus more susceptible to differentially methylation. In contrast to our results, observations in humans, have shown that early exposure to lead causes neuropsychological alterations in mid-adolescent males not, so much in females (Ris et al. 2004). These epigenetic sex differences observed in mice may be independent consequences of exposure, underlining the complexity of the sexspecific response to early life adversities that can affect the epigenetic regulation of gene expression in males and females.
Rn4.5s codes for a 98-nucleotide nuclear RNA with unknown function that is transcribed by RNA polymerase III (Gogolevskaya et al. 2010) and Rn45s codes for the RNA precursor to 18S, 5.8S and 28S rRNA (Grozdanov et al. 2003); both of them show changes in methylation in the hippocampus of females exposed to 3 ppm of lead (Supplemental Figs. S3 and S4). Although neither of these two genes has been linked to metal toxicity or lead neurotoxicity previously, their hypermethylation by lead exposure may compromise ribosome structure or overall protein synthesis capacity in the hippocampus. Changes in expression of Sfi1 have been observed in a genetic mouse model of neurodevelopmental disorder, being up-regulated in young brain and down-regulated in older brain (Trent et al. 2014). This gene has two regions that are differentially methylated in the hippocampus of females exposed to 3 ppm of lead (Supplemental Fig. S5) but its expression is not changed in either sex (Supplemental Fig. S2) possibly because the gene is already highly methylated and changes in these two regions are not enough to alter the expression levels.
The promoter of the Dynlt1b (dynein light chain Tctex-type 1B) gene is hypermethylated in the hippocampus of females exposed to 3 and 30 ppm of lead (Supplemental Fig. S6). In the dentate gyrus of the hippocampus, new neurons continue to be generated throughout life from progenitor cells at the subgranular zone (Eriksson et al. 1998; Kornack and Rakic 1999). These newborn neurons serve not only to maintain the pool of neurons, but also to build memory (Ernst et al. 2014; Nakashiba et al. 2012). Dynlt1b transcription is high in these cells (Dedesma et al. 2006), and acts as a regulator for the genesis of neurons (Gauthier-Fisher et al. 2009; Tseng et al. 2010) and possibly for neurite outgrowth and axon formation as well (Sachdev et al. 2007).
Although the expression level is not changed at PND60 after early life exposure to lead (Supplemental Fig. S2), it is plausible that prenatal and early postnatal exposure to lead may inhibit the formation of new neurons, decrease the total pool of neurons, and eventually compromise memory formation. Although the use of lead has been reduced in the last few decades, exposure is still an important concern due to its non-biodegradable nature and its ubiquitous presence, which poses a potential health risk as a result of the increased sensitivity of children to lead toxicity (Markowitz 2000). During the gestational period, lead crosses the placenta and blood-brain barrier reaching the developing fetal brain (Hu et al. 1998). It is becoming increasingly evident that early-life exposure to lead may produce enduring changes in the epigenetic mechanisms that regulate gene expression in the brain, contributing to pathological and physiological outcomes.
Supplementary Material
HIGHLIGHTS.
Perinatal lead exposure caused persistent DNA methylation changes in brain
Lead exposure caused hypermethylation in the hippocampus of females, but not males
1,623 CpG sites, corresponding to 117 unique genes, were differentially methylated
60 of these genes showed negative correlation between mRNA expression and methylation
Developmental lead exposure has sex- and tissue-specific effects on DNA methylation
ACKNOWLEDGMENTS
We thank Ying Xia, Andrew Vonhandorf, Chia-I Ko, Hisaka Kurita, Vinicius Carreira, Qin Wang and Yunxia Fan for critically reading the manuscript and providing helpful criticisms. This work was supported by NIH grants R21 ES020048 and the Center for Environmental Genetics P30 ES06096.
REFERENCES
- Andrews S. FastQC: A quality control tool for high throughput sequence data. 2014 http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Auger CJ, Auger AP. Permanent and plastic epigenesis in neuroendocrine systems. Front Neuroendocrinol. 2013;34(3):190–197. doi: 10.1016/j.yfrne.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Baranowska-Bosiacka I, Struzynska L, Gutowska I, Machalinska A, Kolasa A, Klos P, Czapski GA, Kurzawski M, Prokopowicz A, Marchlewicz M, Safranow K, Machalinski B, Wiszniewska B, Chlubek D. Perinatal exposure to lead induces morphological, ultrastructural and molecular alterations in the hippocampus. Toxicology. 2013;303:187–200. doi: 10.1016/j.tox.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Bellinger D, Leviton A, Allred E, Rabinowitz M. Pre- and postnatal lead exposure and behavior problems in school-aged children. Environ. Res. 1994;66(1):12–30. doi: 10.1006/enrs.1994.1041. [DOI] [PubMed] [Google Scholar]
- Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer's disease. J. Alzheimers. Dis. 2011;27(4):819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- Brubaker CJ, Schmithorst VJ, Haynes EN, Dietrich KN, Egelhoff JC, Lindquist DM, Lanphear BP, Cecil KM. Altered myelination and axonal integrity in adults with childhood lead exposure: a diffusion tensor imaging study. Neurotoxicology. 2009;30(6):867–875. doi: 10.1016/j.neuro.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil KM, Brubaker CJ, Adler CM, Dietrich KN, Altaye M, Egelhoff JC, Wessel S, Elangovan I, Hornung R, Jarvis K, Lanphear BP. Decreased brain volume in adults with childhood lead exposure. PLoS. Med. 2008;5(5):e112. doi: 10.1371/journal.pmed.0050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Cai B, Dietrich KN, Radcliffe J, Rogan WJ. Lead exposure, IQ, and behavior in urban 5- to 7-year-olds: does lead affect behavior only by lowering IQ? Pediatrics. 2007;119(3):e650–e658. doi: 10.1542/peds.2006-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WC, Auger AP. Gender differences in neurodevelopment and epigenetics. Pflugers Arch. 2013;465(5):573–584. doi: 10.1007/s00424-013-1258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedesma C, Chuang JZ, Alfinito PD, Sung CH. Dynein light chain Tctex-1 identifies neural progenitors in adult brain. J. Comp Neurol. 2006;496(6):773–786. doi: 10.1002/cne.20958. [DOI] [PubMed] [Google Scholar]
- Dietrich KN, Ris MD, Succop PA, Berger OG, Bornschein RL. Early exposure to lead and juvenile delinquency. Neurotoxicol. Teratol. 2001;23:511–518. doi: 10.1016/s0892-0362(01)00184-2. [DOI] [PubMed] [Google Scholar]
- Dosunmu R, Alashwal H, Zawia NH. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech. Ageing Dev. 2012;133(6):435–443. doi: 10.1016/j.mad.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercal N, Treeratphan P, Hammond TC, Matthews RH, Grannemann NH, Spitz DR. In vivo indices of oxidative stress in lead-exposed C57BL/6 mice are reduced by treatment with meso-2,3-dimercaptosuccinic acid or N- acetylcysteine. Free Rad. Biol. Med. 1996;21:157–161. doi: 10.1016/0891-5849(96)00020-2. [DOI] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat. Med. 1998;4(11):1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, Frisen J. Neurogenesis in the striatum of the adult human brain. Cell. 2014;156(5):1072–1083. doi: 10.1016/j.cell.2014.01.044. [DOI] [PubMed] [Google Scholar]
- Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics. 2013;5(5):487–500. doi: 10.2217/epi.13.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Liu K, Barks A, Goodrich JM, Dolinoy DC. Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics. 2014;9(7):934–941. doi: 10.4161/epi.29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79(6):734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Froehlich TE, Lanphear BP, Auinger P, Hornung R, Epstein JN, Braun J, Kahn RS. Association of tobacco and lead exposures with attention-deficit/hyperactivity disorder. Pediatrics. 2009;124(6):e1054–e1063. doi: 10.1542/peds.2009-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Fisher A, Lin DC, Greeve M, Kaplan DR, Rottapel R, Miller FD. Lfc and Tctex-1 regulate the genesis of neurons from cortical precursor cells. Nat. Neurosci. 2009;12(6):735–744. doi: 10.1038/nn.2339. [DOI] [PubMed] [Google Scholar]
- Gogolevskaya IK, Veniaminova NA, Kramerov DA. Nucleotide sequences of B1 SINE and 4.5S(I) RNA support a close relationship of zokors to blind mole rats (Spalacinae) and bamboo rats (Rhizomyinae) Gene. 2010;460(1–2):30–38. doi: 10.1016/j.gene.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Grozdanov P, Georgiev O, Karagyozov L. Complete sequence of the 45-kb mouse ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 2003;82(6):637–643. doi: 10.1016/s0888-7543(03)00199-x. [DOI] [PubMed] [Google Scholar]
- Guilarte TR, McGlothan JL. Selective decrease in NR1 subunit splice variant mRNA in the hippocampus of Pb2+exposed rats: implications for synaptic targeting and cell surface expression of NMDAR complexes. Brain Res. Mol. Brain Res. 2003;113(1–2):37–43. doi: 10.1016/s0169-328x(03)00083-4. [DOI] [PubMed] [Google Scholar]
- Hu H, Rabinowitz M, Smith D. Bone lead as a biological marker in epidemiologic studies of chronic toxicity: conceptual paradigms. Environ. Health Perspect. 1998;106(1):1–8. doi: 10.1289/ehp.981061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavicoli I, Carelli G, Stanek EJ, Castellino N, Calabrese EJ. Effects of low doses of dietary lead on red blood cell production in male and female mice. Toxicol. Lett. 2003;137(3):193–199. doi: 10.1016/s0378-4274(02)00404-6. [DOI] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Continuation of neurogenesis in the hippocampus of the adult macaque monkey. Proc. Natl. Acad. Sci. U. S. A. 1999;96(10):5768–5773. doi: 10.1073/pnas.96.10.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Front Psychiatry. 2013;4:78. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006;281(23):15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS. One. 2010;5(4):e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz M. Lead poisoning. Pediatr. Rev. 2000;21(10):327–335. doi: 10.1542/pir.21-10-327. [DOI] [PubMed] [Google Scholar]
- Menger Y, Bettscheider M, Murgatroyd C, Spengler D. Sex differences in brain epigenetics. Epigenomics. 2010;2(6):807–821. doi: 10.2217/epi.10.60. [DOI] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez BV, Chittajallu R, Iwamoto KS, McBain CJ, Fanselow MS, Tonegawa S. Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell. 2012;149(1):188–201. doi: 10.1016/j.cell.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal AP, Worley PF, Guilarte TR. Lead exposure during synaptogenesis alters NMDA receptor targeting via NMDA receptor inhibition. Neurotoxicology. 2011;32(2):281–289. doi: 10.1016/j.neuro.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman HL, McFarland C, Ness RB, Fienberg SE, Tobin MJ. Bone lead levels in adjudicated delinquents. A case control study. Neurotoxicol. Teratol. 2002;24(6):711–717. doi: 10.1016/s0892-0362(02)00269-6. [DOI] [PubMed] [Google Scholar]
- Needleman HL, Riess JA, Tobin MJ, Biesecker GE, Greenhouse JB. Bone lead levels and delinquent behavior. JAMA. 1996;275(5):363–369. [PubMed] [Google Scholar]
- Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, Morgan M. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014;30(14):2076–2078. doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TV, Piersma SR, Warmoes M, Jimenez CR. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 2010;26(3):363–369. doi: 10.1093/bioinformatics/btp677. [DOI] [PubMed] [Google Scholar]
- Ris MD, Dietrich KN, Succop PA, Berger OG, Bornschein RL. Early exposure to lead and neuropsychological outcome in adolescence. J. Int. Neuropsychol. Soc. 2004;10(2):261–270. doi: 10.1017/S1355617704102154. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20(24):3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- Sachdev P, Menon S, Kastner DB, Chuang JZ, Yeh TY, Conde C, Caceres A, Sung CH, Sakmar TP. G protein beta gamma subunit interaction with the dynein light-chain component Tctex-1 regulates neurite outgrowth. EMBO J. 2007;26(11):2621–2632. doi: 10.1038/sj.emboj.7601716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin FJ, Fan Y, Lindquist DM, Xia Y, Puga A. Lead induces similar gene expression changes in brains of gestationally exposed adult mice and in neurons differentiated from mouse embryonic stem cells. PLoS. One. 2013;8(11):e80558. doi: 10.1371/journal.pone.0080558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Anderson DW, Sonnenahalli H, Vadigepalli R. Sex-based differences in gene expression in hippocampus following postnatal lead exposure. Toxicol. Appl. Pharmacol. 2011;256(2):179–190. doi: 10.1016/j.taap.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Anderson DW, Talsania K, Mettil W, Vadigepalli R. Effects of developmental lead exposure on the hippocampal transcriptome: influences of sex, developmental period, and lead exposure level. Toxicol. Sci. 2012a;129(1):108–125. doi: 10.1093/toxsci/kfs189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Kidd SK, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol. Lett. 2013;217(1):75–81. doi: 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Mettil W, Anderson DW. Differential effect of postnatal lead exposure on gene expression in the hippocampus and frontal cortex. J. Mol. Neurosci. 2012b;47(1):76–88. doi: 10.1007/s12031-011-9686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut MC, Sen A, Cingolani P, Shaik A, Land SJ, Ruden DM. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicol. Sci. 2014;139(1):142–161. doi: 10.1093/toxsci/kfu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent S, Fry JP, Ojarikre OA, Davies W. Altered brain gene expression but not steroid biochemistry in a genetic mouse model of neurodevelopmental disorder. Mol. Autism. 2014;5(1):21. doi: 10.1186/2040-2392-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Gruzdeva N, Li A, Chuang JZ, Sung CH. Identification of the Tctex-1 regulatory element that directs expression to neural stem/progenitor cells in developing and adult brain. J. Comp Neurol. 2010;518(16):3327–3342. doi: 10.1002/cne.22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgolini MB, Rossi-George A, Weston D, Cory-Slechta DA. Influence of low level maternal Pb exposure and prenatal stress on offspring stress challenge responsivity. Neurotoxicology. 2008;29(6):928–939. doi: 10.1016/j.neuro.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan Y, Puga A. Dioxin Exposure Disrupts the Differentiation of Mouse Embryonic Stem Cells into Cardiomyocytes. Toxicol. Sci. 2010;115:225–237. doi: 10.1093/toxsci/kfq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and the origin of species differences. Proc. Natl. Acad. Sci. U. S. A. 2005;102(Suppl 1):6543–6549. doi: 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widzowski DV, Finkelstein JN, Pokora MJ, Cory-Slechta DA. Time course of postnatal lead-induced changes in dopamine receptors and their relationship to changes in dopamine sensitivity. Neurotoxicology. 1994;15(4):853–865. [PubMed] [Google Scholar]
- Wright JP, Dietrich KN, Ris MD, Hornung RW, Wessel SD, Lanphear BP, Ho M, Rae MN. Association of prenatal and childhood blood lead concentrations with criminal arrests in early adulthood. PLoS. Med. 2008;5(5):e101. doi: 10.1371/journal.pmed.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Brock B, Cox DP, Cardozo-Pelaez F, McPherson CA, Harry J, Rice DC, Maloney B, Chen D, Lahiri DK, Zawia NH. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J. Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Mann MR, Lee SS, Marh J, McCarrey JR, Yanagimachi R, Bartolomei MS. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc. Natl. Acad. Sci. U. S. A. 2003;100(21):12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Holland SK, Cecil KM, Dietrich KN, Wessel SD, Altaye M, Hornung RW, Ris MD, Egelhoff JC, Lanphear BP. The impact of early childhood lead exposure on brain organization: a functional magnetic resonance imaging study of language function. Pediatrics. 2006;118(3):971–977. doi: 10.1542/peds.2006-0467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.