Abstract
The initial cloning of receptor protein tyrosine phosphatases (RPTPs) was met with excitement because of their hypothesized function in counterbalancing receptor tyrosine kinase signaling. In recent years, members of a subfamily of RPTPs with homophilic cell-cell adhesion capabilities, known as the R2B subfamily, have been shown to have functions beyond that of counteracting tyrosine kinase activity, by independently influencing cell signaling in their own right and by regulating cell adhesion. The R2B subfamily is composed of four members: PTPmu (PTPRM), PTPrho (PTPRT), PTPkappa (PTPRK), and PCP-2 (PTPRU). The effects of this small subfamily of RPTPs is far reaching, influencing several developmental processes and cancer. In fact, R2B RPTPs are predicted to be tumor suppressors and are among the most frequently mutated protein tyrosine phosphatases (PTPs) in cancer. Confounding these conclusions are more recent studies suggesting that proteolysis of the full-length R2B RPTPs result in oncogenic extracellular and intracellular protein fragments. This review discusses the current knowledge of the role of R2B RPTPs in development and cancer, with special detail given to the mechanisms and implications that proteolysis has on R2B RPTP function. We also touch upon the concept of exploiting R2B proteolysis to develop cancer imaging tools, and consider the effects of R2B proteolysis on axon guidance, perineural invasion and collective cell migration.
Keywords: Proteolysis, Perineural invasion, Axon guidance, Collective cell migration, RPTP, Receptor PTP
1. Introduction to the R2B subfamily of receptor-like tyrosine phosphatases
Protein tyrosine phosphatases are identified based on the presence of a stretch of approximately 280 amino acids with ten conserved motifs [1]. Within the group of the 37 classical tyrosine phosphatases known today, 16 are non-transmembrane PTPs, such as PTP1B. 21 are RPTPs that are further divided into eight subfamilies based on their extracellular domain structure [2] or sequence similarity of the first phosphatase domain [3]. The type IIb family of RPTPs (R2B) is of interest due to the presence of cell adhesion molecule-like domains in their extracellular region. R2Bs have an N-terminal MAM (Meprin, A5 (neuropilin), PTP mu) domain, an immunoglobulin (Ig) domain and four fibronectin III (FNIII) repeats in their extracellular segment [4]. The combination of Ig domains and FNIII repeats are found in the cell-cell adhesion molecules (CAMs) NCAM and L1. Given their structural similarity to CAMs, R2B RPTPs were predicted to function in regulating cell adhesion and altering tyrosine phosphorylation levels in cells in response to adhesion [4, 5].
R2B phosphatases have a longer juxtamembrane domain than other RPTPs. This domain shares 20% identity with the cytoplasmic domain of classical cadherins [5, 6], which suggested that in addition to functioning as cell adhesion molecules in their own right, R2Bs might also regulate this major group of CAMs. R2Bs contain two cytoplasmic tyrosine phosphatase domains (D1 and D2), of which only the membrane proximal phosphatase domain (D1) is catalytically active [7]. The second phosphatase domain, along with other regions of the cytoplasmic domain, likely regulates tyrosine phosphatase activity of the first PTP domain [1, 8]. R2B phosphatases are only found in vertebrates, suggesting they evolved to control processes unique to higher organisms [9].
2. Mechanisms of mediating cell-cell adhesion
2.1 Homophilic cell-cell adhesion
The first R2B identified was PTPμ (also known as PTPmu, PTPRM and RPTPM) [10], followed by PTPκ (PTPkappa and PTPRK) [11], PCP-2 (PTPpi, PTPpsi, PTPλ, hPTP-J, PTPRO and PTPRU) [12], and PTPρ (PTPrho and PTPRT) [13]. Because of the presence of the CAM-like extracellular domains, R2B PTPs were evaluated for their ability to mediate cell-cell adhesion. Expression of PTPμ [14, 15], PTPκ [16], and PTPρ [17] in non-adherent cells induces cell-cell aggregation by binding to an identical protein on the adjacent cell’s surface, confirming that these proteins do function as CAMs that mediate homophilic cell-cell adhesion. While extracellular domain fragments of PCP-2 can mediate homophilic binding [12], PCP-2 is unable to mediate homophilic cell-cell adhesion when expressed in non-adherent cells [18]. The R2B RPTPs do not bind heterophilically to one another [17–19], and no other heterophilic-binding partners of R2B RPTPs have been identified.
Based on studies of PTPμ, the MAM, Ig and first two FN III repeats are required for mediating homophilic cell-cell adhesion [19–25]. Cell and in vitro binding studies demonstrated that the MAM domain is responsible for sorting distinct R2B subfamily members from each other to maintain strictly homophilic cell-cell adhesions [19], while the Ig domain promotes direct homophilic binding even in vitro [20]. Crystallographic studies of PTPμ suggest that the MAM and Ig domain of one PTPμ molecule binds to the first and second FN III domain of a second PTPμ molecule in trans to mediate cell-cell adhesion [23]. Differences in the peripheral areas of the homophilic dimer interface are also hypothesized to account for the binding specificity of R2B RPTPs [23].
The MAM and Ig domain of PCP-2 swapped into a chimeric PTPρ protein are sufficient to make non-adhesive cells adhesive, while also conferring a unique adhesion molecule identity to the PCP-2-swapped-PTPρ-chimera, as these chimeric cells sort away from wild-type PTPρ-expressing cells [18]. Yet the entire extracellular domain of PCP-2 swapped into an otherwise wild-type PTPρ protein does not mediate adhesion [18]. Evaluation of exposed surface charges of PCP-2 revealed a more positive electrostratic potential on the backside of the first and second FN III repeats of PCP-2 compared to other R2Bs [18]. Comparison of the amino acid residues present in PCP-2 with those identified as being essential for homophilic binding of PTPμ by Aricescu et al. [18] demonstrate a few minor sequence differences between PCP-2 and PTPμ. It is not known whether those minor differences in the FN III domains are responsible for the inability of PCP-2 to mediate cell-cell adhesion, although it is clear that the MAM and Ig domain of PCP-2 do retain some adhesive capability.
2.2 Cadherin-based adhesion
In addition to sharing sequence homology with the cytoplasmic domains of classical cadherins [5, 6], R2B RPTPs localize to sites of adherens junctions [26–28]. PTPμ and PCP-2 also regulate cadherin-based adhesion [29, 30], and PTPκ stabilizes E-cadherin at adherens junctions [31]. PTPμ expression and tyrosine phosphatase activity are required for a process analogous to axon extension of neurons called neurite outgrowth on cadherin substrates [32]. R2B RPTPs interact with classical cadherins, including E-cadherin, N-cadherin, R-cadherin and VE-cadherin [26, 33–35]. Classical cadherins regulate cell-cell adhesion and the actin cytoskeleton via the catenin proteins. Catenin family members include α, β, δ, γ and p120. R2B RPTPs interact with a number of catenins and, in some cases, have been shown to dephosphorylate catenins to regulate adherens junctions. For instance, PTPκ interacts with β-catenin and plakoglobin/γ-catenin [27], dephosphorylates β-catenin [27, 36] and regulates the localization of β-catenin in cells [31]. β-catenin is also a substrate of PCP-2 [37]. PTPρ interacts with α-actinin, α-, β-, and γ-catenin, p120-catenin and N-,E-, and VE-cadherins [34]. PTPμ binds E-, N-, R- and VE-cadherin complexes that contain α, β, γ, and p120 catenin [26, 33, 35, 38]. p120 catenin and E-cadherin are PTPρ and PTPμ substrates [26, 33, 34, 38]. PIPKIγ90 is an additional PTPμ substrate that, when dephosphorylated by PTPμ, inhibits integrin mediated cell-matrix adhesion and promotes cell-cell adhesion at adherens junctions [39].
While R2B RPTPs clearly have a function in maintaining the structure of adherens junctions by regulating the phosphorylation state of cadherins and catenins, they are also hypothesized to have a more structural role in adherens junctions. Electron micrographs demonstrate that changing the length of the extracellular domain of PTPμ changes the distance between two cell membranes [23], suggesting that PTPμ functions as a rigid “spacer-clamp” to structurally reinforce adherens junctions [23, 24]. Therefore, R2B RPTPs regulate cell-cell adhesion at adherens junctions via two general mechanisms: through homophilic trans interactions to act as “spacer-clamps” and via cis interactions with the cadherin-catenin complex to stabilize adherens junctions by regulating tyrosine phosphorylation of cadherins and catenins.
3. Role of R2Bs in Development
R2B RPTPs are expressed in a variety of tissues during development [4]. While each R2B has a unique expression pattern in terms of timing and tissue location during development, every R2B RPTP is expressed in the nascent nervous system. Various techniques have been employed to assess the function of individual R2B RPTPs, from in vitro axonal migration studies, known as neurite outgrowth assays, to zebrafish morpholino experiments. These studies show that R2B RPTPs have indispensable functions in somitogenesis, and neural, endothelial cell, and immune system development (Table 1).
Table 1.
R2B RPTPs in Development
| Developmental function | Site/Cells involved | R2B involved | Reference |
|---|---|---|---|
| Nervous System | |||
| Retinal lamination | Retina | PTPμ | [41] |
| Neurite outgrowth | Retinal ganglion cells | PTPμ | [32,40,42] |
| Cerebellar granule cells | PTPκ | [49] | |
| Synapse formation | Hippocampal neurons | PTPρ | [54] |
| Dendritic arborization | Hippocampal neurons | PTPρ | [55] |
| Regulation of neuronal survival or differentiation | Ventral tegmental area and substantia nigra | PCP-2 | [56] |
| Other | |||
| T-cell development | CD4+ T cells | PTPκ | [57,58,60] |
| Somitogenesis | First and second somites | PCP-2 | [61,62] |
| Bone marrow development | Megakaryocytes | PCP-2 | [63] |
3.1 R2Bs in Neural Development
3.1.1 PTPμ
PTPμ is expressed in the developing chick retina and tectum [32, 40, 41]. In ex vivo retinal organ cultures, PTPμ was shown to regulate the formation of neuronal layers within the retina, a process known as lamination [41]. PTPμ regulates neurite outgrowth of retinal ganglion cells, the large projection neurons of the retina that connect the light input signal generated in the retina to the higher processing centers of the brain [32, 40]. Tyrosine phosphatase activity of PTPμ is necessary for its regulation of neurite outgrowth [42]. Neurite outgrowth on a purified PTPμ substrate requires signaling via PLCγ1, PKCδ, the Rho GTPases Cdc42 and Rac1, and IQGAP1 [43–48].
3.1.2 PTPκ
Despite its extensive expression in the nervous system, a definitive role for PTPκ during neural development has yet to be demonstrated. PTPκ regulates in vitro neurite outgrowth of embryonic cerebellar granule cells, via MEK1 and Grb2 signaling mechanisms [49]. A β-galactosidase knock-in mouse with β-gal inserted into the phosphatase domain of PTPκ, thereby rendering it inactive [50], resulted in viable mice, demonstrating that PTPκ phosphatase activity is not necessary during development [50]. This may be due to redundancy with other family members. The function of adhesion mediated by the extracellular domain of PTPκ during development remains unknown.
3.1.3 PTPρ
PTPρ is an intriguing R2B RPTP from a developmental standpoint because it is found in a region of the human genome known as human accelerated region 9 (HAR 9), one of 49 regions of our genome with a rapid rate of divergence from chimpanzee [51]. PTPρ was thought to be exclusively expressed in the brain [13, 52, 53], although recent studies have demonstrated its expression elsewhere in the adult animal (see section 4.1). In the nervous system, PTPρ interacts with neuroligin and neurexins to promote synapse formation in cultured hippocampal neurons, functioning as a trans homophilic adhesion molecule with itself while reinforcing the neurexin-neuroligin interaction [54]. Phosphorylation of PTPρ by Fyn reduced the interaction of PTPρ with itself in trans and with neuroligin and instead promoted increased PTPρ-PTPρ cis interactions, resulting in attenuated synapse formation [54]. PTPρ also regulates dendritic arborization in hippocampal neurons, by dephosphorylating and thereby regulating the activity of the Rac1 GAP activating protein called breakpoint cluster region protein (BCR) [55].
3.1.4 PCP-2
PCP-2 also regulates the development of the nervous system. The development and maintenance of meso-diencephalic dopamine neurons (mdDA) of the substantia nigra and ventral tegmental area of the midbrain depends on the activity of the transcription factors Nurr1 and Pitx3. PCP-2 is downstream of both of these transcription factors and its expression is lost in Nurr −/− mice, which lose most mdDA neurons late in development [56]. This suggests that PCP-2 expression may be necessary for mdDA cell generation and survival. Given that PCP-2 does not mediate cell-cell adhesion [18], the mechanism of how PCP-2 regulates mdDA cell survival is unlikely to be due to the direct regulation of mdDA cell-cell adhesion, but instead might be via the regulation of cadherin based adhesion or the modulation of PCP-2 signaling.
3.2 Role of R2Bs in the development of other tissues
Besides their role in the development of the nervous system, R2B PTPs also regulate other important developmental processes, including somitogenesis and T-cell and megakaryocyte development. PTPκ, for instance, regulates the differentiation of CD4+ T cells in the Long-Evans Cinnamon (LEC) strain of rat [57, 58]. Deletion of PTPκ generates T-helper cell immunodeficiency in the LEC rats, while transduction of PTPκ into LEC bone marrow cells was sufficient to replenish half of the missing CD4+ T cells in the thymus of the LEC rats [59]. Furthermore, CD4+ T cell development was reduced in bone marrow-derived stem cells transduced with the dominant negative form of PTPκ [60]. Both a PTPκ dominant negative construct and short-hairpin RNA directed against PTPκ reduced c-raf, MEK, and ERK 1/2 phosphorylation in T cell lines, suggesting that PTPκ expression positively regulates the c-Raf/MEK/Erk 1/2 signaling pathways required for CD4+ T cell development [60].
PCP-2 is expressed in the first somites of the chick during development [61]. Morpholino knock-down experiments demonstrate that PCP-2 regulates oscillatory genes necessary for body segment formation [62]. Notch-delta signaling is required for segment formation via the regulation of the repressive transcription factors her1 and her7. Notably, PCP-2 morpholino-injected embryos resemble delta morpholino-injected embryos, with defects in segment polarity followed by death 2–3 days post-fertilization [62]. Expression of her1, her7, and deltaC are disrupted in PCP-2 morpholino-injected embryos [62]. In her1 morpholino-injected embryos, her7 transcript levels are reduced and her1 transcript levels are increased, due to hypothesized transcript stabilization by the morpholino [62]. In PCP-2 morpholino injected embryos, her7 transcripts are further reduced. When PCP-2 and her1 morpholinos are co-injected, her1 transcript levels are reduced, demonstrating that PCP-2 is capable of regulating her1 and her7 transcripts independently, either upstream of or in parallel with the Notch/delta system in presomitic mesoderm pattern formation [62].
PCP-2 also regulates the proliferation of megakaryocyte cells in vitro [63]. The growth factor stem cell factor induces hematopoietic progenitor cell differentiation via binding to and activating the c-Kit receptor. PCP-2 was found to constitutively interact with the c-Kit receptor, and PCP-2 was both tyrosine phosphorylated and proteolytically cleaved in response to stem cell factor stimulation. The addition of PCP-2 antisense oligonucleotides to primary bone marrow cells significantly reduced the number of megakaryocyte colonies, demonstrating the role of PCP-2 in megakaryocyte proliferation and/or differentiation [63].
4. Alteration of R2B RPTPs in Cancer
R2B RPTPs have been classified as both tumor suppressors and oncogenes, depending on the specific R2B, the type of cancer, and the changes observed in the gene or proteins [64]. Furthermore, genetic, epigenetic, transcriptional and post-translational changes in R2B RPTPs have been observed in cancer, in some cases all four events have been described for one RPTP [64, 65]. The known alterations of R2B RPTPs in human cancers are outlined in Table 2.
Table 2.
R2B RPTPs in Cancer
| Cancer type | R2B involved | Reference |
|---|---|---|
| Glioblastoma | PTPμ | [75,95] |
| Malignant Glioma Population – including samples from patients with oligodendroglioma, astrocytoma, anaplastic oligodendroglioma, anaplastic astrocytoma and glioblastoma | PTPκ | [76] |
| Head and Neck Squamous Cell Carcinoma | PTPρ | [67] |
| PTPμ | [67] | |
| PTPκ | [67] | |
| Esophageal | PTPρ | [65] |
| Lung | PTPρ | [65] |
| Breast | PTPμ | [74] |
| PTPκ | [74] | |
| Pancreatic | PTPκ | [79] |
| Stomach | PTPρ | [65] |
| Bladder | PTPρ | [65] |
| Colorectal | PTPρ | [66] |
| PTPμ | [65,68] | |
| PTPκ | [81] | |
| PCP-2 | [65] | |
| Endometrial | PTPρ | [65] |
| PTPμ | [65] | |
| PTPκ | [65] | |
| PCP-2 | [65] | |
| Melanoma | PTPκ | [80,86] |
| PCP-2 | [86] | |
| Central Nervous System Lymphoma | PTPκ | [77] |
| Primary Intraocular lymphoma | PTPκ | [78] |
| Hodgkin Lymphoma | PTPκ | [83] |
4.1 PTPρ
A large body of data exists that demonstrates that PTPρ is a frequently disrupted PTP in cancer. PTPρ functions as a tumor suppressor in colorectal cancer cells and is the most frequently mutated RPTP gene in colorectal cancer [66], in head and neck squamous cell carcinoma (HNSCC), and in cancer in general [65, 67]. In addition to genetic mutations of the PTPRT coding sequence, alterations in promoter methylation status of the PTPRT gene have been noted in colorectal cancer [68]. microRNAs targeting PTPρ mRNA have been observed following hepatitis virus B infection [69] and in side population cells (that are similar to cancer stem cells) from the MCF7 breast cancer cell line [70]. These microRNAs disrupt PTPρ function in cancer cells by reducing the amount of wild-type PTPρ protein.
Many of the PTPρ mutations identified result in frameshift or nonsense mutations in either the extracellular adhesive domain or in its catalytic domains [66]. Expression of full-length wild-type PTPρ suppresses colorectal and HNSCC cell growth in vitro, whereas expression of a mutant version of PTPρ promotes cell growth [66, 67]. Recombinant D1 or D2 domains designed with the PTPρ mutations observed in cancer have reduced activity to the 6,8-difluoro-4-methylumbelliferyl phosphate substrate compared to control [66]. These PTPρ mutations yield a destabilized D1 domain that compromises phosphatase activity [71]. Mutations observed in the MAM, Ig and FN III repeats of PTPρ are also loss of function, as they all disrupt the ability of PTPρ to mediate cell-cell adhesion [17, 72].
Downstream effects of PTPρ D1 domain mutations have begun to be elucidated. PTPρ D1 domain mutations impact both STAT3 [67] and paxillin activity [73]. A catalytic domain mutation of PTPρ increases the phosphorylation state of the PTPρ substrate STAT3, resulting in STAT3 hyperactivity [67]. Paxillin is another PTPρ substrate [73]. PTPρ engineered with cancer-occurring D1 mutations cannot dephosphorylate paxillin on tyrosine 88 (Y88), while PTPρ knock-out mice show higher levels of paxillin Y88 phosphorylation [73]. PTPρ knock-out mice were observed to be more susceptible to colon cancer induction while also displaying higher levels of colonic paxillin Y88 phosphorylation [73]. The PTPρ mutations observed in cancer demonstrate the multiplicity of effects that RPTP mutations can have on both adhesion and catalytic activity and the downstream effects that these mutations have on STAT3 and paxillin signaling pathways.
4.2 PTPμ
Full-length PTPμ functions as a tumor suppressor in breast cancer and glioma [74, 75]. The PTPRM promoter is hypermethylated in sporadic colorectal cancer [68], and the PTPRM gene is frequently mutated in HNSCC [67]. Functionally, reduced levels of PTPμ mRNA in breast cancer patients are associated with poor prognosis and shorter disease free survival time [74]. Similarly, less full-length PTPμ protein is observed in high-grade brain tumor glioblastoma multiforme (GBM) than in lower grade astrocytomas or in noncancerous brain tissue [75]. Concomitant with the reduction of PTPμ protein and/or transcript levels is an increase in tumor cell invasiveness, motility and cell growth [74, 75]. The identity of some of the downstream signaling defects predicted to occur in cancer cells following PTPμ disruption are known. For example, in glioma cells, PLCγ1 activity is downstream of PTPμ signaling and PLCγ1 activity is required for mediating cancer cell migration in the absence of PTPμ protein [45]. In breast cancer cells, ERK and JNK activity are required to promote migration when PTPμ expression is reduced [74].
4.3 PTPκ
Loss of heterozygosity in the PTPRK coding region on chromosome 6 is associated with malignant glioma [76], central nervous system lymphomas [77], primary intraocular lymphoma [78], and pancreatic cancers [79]. The PTPRK gene is mutated in HNSCC [67], and a mutated PTPκ was identified as a melanoma antigen on CD4+ T-cells [80]. PTPRK is also found to be part of gene fusions with R-spondin family members, agonists of the Wnt/β-catenin signaling pathway, in 10% of colorectal cancer [81]. PTPRK was identified in a mouse transposon study of colorectal cancer as one of the ten most likely causative mutations observed in cancer patients [82]. Both Epstein-Barr Virus infection of Hodgkin lymphoma cells and the androgen receptor-targeted microRNA MiR-133b induce the loss of PTPκ mRNA and protein expression [83, 84]. Low levels of PTPκ transcripts are observed in breast cancer tissue [85] and in 20% of melanoma cells and tissue biopsies [86]. The consensus is that PTPκ is a tumor suppressor, as reduced PTPκ expression is correlated with shorter disease free-survival times in breast cancer [85], and loss of heterozygosity of the PTPRK region of chromosome 6 is associated with shorter patient survival with CNS lymphoma and primary ocular lymphoma [77, 78]. In at least one instance, however, PTPκ has a hypothesized oncogenic function as an inhibitor of apoptosis [87].
U-87 MG glioma and Me1 melanoma cells expressing exogenous full-length PTPκ have reduced levels of proliferation and migration [31, 88], as expected for a tumor suppressor. In agreement with this hypothesized tumor suppressor role, expression of mutant versions of PTPκ found in glioma cells in the loss of heterozygosity in U-87 MG cells [77] promote cell growth, motility and invasion [88]. Likewise, in non-Epstein-Barr Virus infected Hodgkin lymphoma cells, loss of PTPκ by si-RNA increases the survival and proliferation of the cells [83]. Knock-down of PTPκ protein in breast cancer cells using ribozymes also increases cell proliferation, matrix-adhesion and invasiveness [85]. PTPκ mediates these cellular responses by regulating EGFR and β-catenin activity in glioma and melanoma cells [88]. Specifically, PTPκ sequesters β-catenin at adherens junctions to limit the transcription and translation of cyclin D1 and c-myc by nuclear β-catenin [31]. In Hodgkin lymphoma, Epstein-Barr Virus infection reduces PTPκ protein expression by inactivating the transforming growth factor β - Smad2 – pathway that induces PTPκ transcription [83, 89].
4.4 PCP-2
PCP-2 also has hypothesized functions as a tumor suppressor. PCP-2 mRNA transcripts are expressed in normal melanocytes but are reduced in 30% of melanoma cell lines and present in only one out of nine melanoma biopsies [86]. PTPκ mRNA is also reduced in melanoma cell lines and biopsies [86, 90]. Occasionally both PTPκ and PCP-2 mRNA levels are reduced in the same tissue or cell samples [86, 90]. Expression of full length PCP-2 in SW480 colon cancer cells reduces their ability to migrate and proliferate, likely by promoting E-cadherin-based adhesion at adherens junctions while reducing β-catenin’s nuclear signaling [30]. In summary, R2B RPTPs all have proposed tumor suppressor functions. More recently, however, additional post-translational alterations observed in the R2B subfamily have added complexity to this straightforward tumor suppressor story.
5. Proteolysis
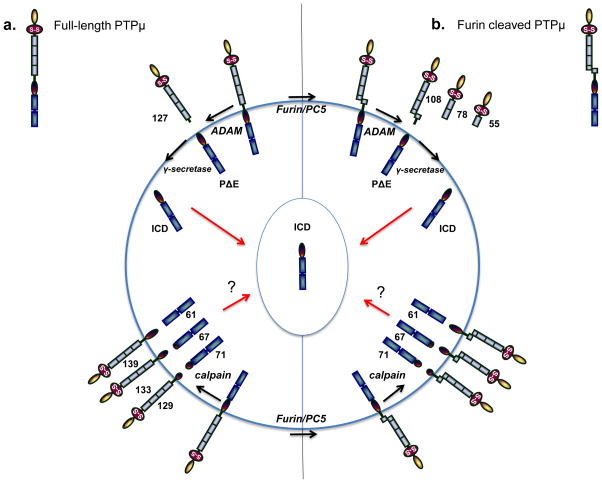
Post-translational changes to R2B RPTPs occur preferentially in cancer tissue. In particular, R2B RPTPs are included in the list of cell-cell CAMs preferentially proteolyzed in cancer [91, 92]. The modification of homophilic cell-cell CAMs by proteolysis is proposed to be one mechanism of promoting loss of contact inhibition of proliferation and migration in cancer cells [92]. Unlike their corresponding full-length isoforms, proteolytically cleaved fragments of R2B RPTPs have oncogenic properties [93, 94]. In the case of PTPμ, extracellular and intracellular fragments of PTPμ preferentially accumulate in GBM tissue and cells [93, 95]. At the cell surface in “normal” immortalized and in unstimulated GBM cancer cells, PTPμ exists in two forms, either a full-length 200 kDa form or as two non-covalently associated fragments of 100 kDa each, the E (or extracellular) subunit and the P (phosphatase) subunit generated by furin [93, 94] or proprotein convertase 5 (PC5) cleavage [96]. Following stimulation with a calcium ionophore, which mimics growth factor stimulation, cell-surface PTPμ is cleaved by an ADAM protease (Figure 1; [94]). Depending on which form of cell-surface PTPμ is used as the substrate (ie. either the full-length protein or the protein composed of two non-covalently associated fragments), ADAM activity generates shed extracellular fragments of PTPμ of 119 and 100 kDa in “normal” immortalized cells, and of 127 and 108 kDa in GBM tumor cells [94] (Figure 1). The membrane-tethered PTPμ fragment left after ADAM cleavage, termed PΔE, is then proteolyzed by γ-secretase in the plasma membrane to generate a membrane-free cytoplasmic fragment of PTPμ, the intracellular domain fragment (ICD) [36, 93]. ICD is capable of translocating into the nucleus, where it is hypothesized to act on a different set of substrates than at the plasma membrane [93] (Figure 1). In GBM tumor cells, calpain proteolyzes PTPμ in its cytoplasmic domain to generate three additional membrane-free fragments of 71, 67 and 61 kDa with three complementary transmembrane fragments of 139, 133, and 129 kDa, all three of which have intact extracellular cell-cell adhesion domains [94] (Figure 1). Calpain cleavage of PTPμ has the effect of dissociating adhesion from phosphatase activity. Additional unidentified serine proteases cleave PTPμ in GBM cells to yield smaller extracellular fragments with MWs of 78 and 55 kDa [94] (Figure 1).
Figure 1.
PTPμ proteolysis in cancer cells as a model for R2B proteolysis. PTPμ, PTPκ, and PCP-2 have all been shown to be proteolyzed in cancer cells, and may follow the same steps as has been observed for PTPμ proteolysis. The molecular weights shown are for PTPμ. PTPμ exists at the cell surface in two full-length forms: either the non-furin processed full-length PTPμ protein or the furin-processed form that consists of two non-covalently associated PTPμ subunits (furin cleaved PTPμ). In (a), ADAM protease cuts full-length PTPμ in the extracellular domain to yield a shed 127 kDa fragment and a membrane-tethered PΔE fragment. The PΔE fragment is then proteolyzed by the γ-secretase complex to release the ICD fragment from the plasma membrane. The ICD is capable of translocating into the cell nucleus. PTPμ is proteolyzed differently in cancer cells in response to ionomycin stimulation. Full-length PTPμ is also cleaved by calpain inside of the cell, to yield three membrane-free fragments of 71, 67, and 61 kDa and three integral membrane forms of PTPμ with intact extracellular domains of 129, 133, and 139 kDa. In (b), furin cleaves PTPμ in the golgi network to yield two non-covalently associated subunits at the plasma membrane. Furin-processed forms of PTPμ are also cleaved in the extracellular domain by ADAM to yield shed fragments of 108 kDa. The membrane-tethered fragment is then cleaved by the γ-secretase complex to yield the ICD fragment. In cancer cells, calpain cleavage of furin processed PTPμ yields membrane-free cytoplasmic fragments of 71, 67, and 61 kDa, as is observed for full-length PTPμ (a). Three integral membrane forms of PTPμ with intact extracellular domains are also generated in response to calpain cleavage. PTPκ is proteolyzed by a furin-like protease, an ADAM, and the γ-secretase complex, just like PTPμ.
Biologically, the absence of full-length PTPμ [75] and the presence of PTPμ fragments contribute to cell survival and migration [93]. Tyrosine phosphatase activity of the cytoplasmic fragments of PTPμ promote cancer cell migration, as use of a wedge inhibitor that blocks tyrosine phosphatase function reduces cell migration in GBM cells [93]. Reduction of full-length PTPμ and PTPμ fragments in GBM tumor models with short hairpin RNA reduces GBM cell growth in vitro and blocks tumor formation in flank and intracranial tumor models [97]. The shed and membrane associated extracellular fragments of PTPμ have different lengths but intact adhesive domains. The function of individual cancer-generated extracellular fragments of PTPμ remains unknown, even though collectively we know they contribute to tumor growth and migration. Given the marked accumulation of shed PTPμ fragments in the tumor microenvironment, as demonstrated immunohistochemically [95] and using novel imaging agents (see section 5.1 below), it is very likely that these fragments have unique biological functions in cancer.
Like PTPμ, full-length PTPκ is proteolyzed by furin to generate two cleaved fragments, the E-(110 kDa) and P-subunits (100 kDa), which remain associated at the plasma membrane [11]. At high cell density, PTPκ is cleaved by ADAM 10 to yield a shed extracellular fragment (MW 120 kDa) and an 80 kDa transmembrane fragment (PΔE [36]). An additional cytoplasmic fragment of approximately 70 kDa, termed phosphatase intracellular portion (PIC), was identified when the proteosome was inhibited. PIC is generated by presenilin-1 activity in the gamma secretase complex, which cuts PΔE in the transmembrane domain, thereby releasing PIC from the membrane and facilitating its translocation to the nucleus [36]. And while PIC and PTPκ are both able to dephosphorylate β-catenin, they have opposing effects on TCF-mediate transcription: full-length PTPκ decreases TCF-mediated transcription, whereas PIC increases it [36]. Therefore, the localization of the different forms of PTPκ yields opposite cellular effects.
Interestingly, over-expression of N-acetylglucosaminyl transferase V (GnT-V) in WiDr colon cancer cells results in preferential glycosylation and shedding of a 110 kDa PTPκ extracellular fragment [98]. Shedding of PTPκ in the WiDr cells is regulated by the secreted proprotein convertase 5 (PC5A) [99]. It remains to be investigated whether PC5A/furin cleavage of PTPκ yields two membrane associated fragments, as suggested by Anders and colleagues [36], or whether PC5A/furin cleavage results in the shedding of the extracellular segment, as suggested by Kim et al. [99]. Glycosylation and shedding of PTPκ correlated with increased migration of the colon cancer cells, again suggesting that the proteolyzed isoforms of PTPκ are oncogenic [98].
Proteolytic fragments of PCP-2 have also been observed [12, 63]. Extracellular proteolytic fragments of 115 and 70 kDa were isolated from mouse brain extracts and from 293 cells over-expressing PCP-2 isoforms [12]. Stimulation of Cos-7 cells with stem cell factor results in the phosphorylation of PCP-2 and the generation of phosphorylated and proteolyzed membrane-spanning PCP-2 fragments of 100 and 80 kDa with some parts of the extracellular domain remaining [63]. In addition, a non-phosphorylated 73 kDa intracellular PCP-2 fragment is generated following stem cell factor stimulation [63]. Proteolysis is generalizable to the entire R2B subfamily, as PTPρ has been observed to be cleaved by furin during neural development [54]. The stimulus for proteolysis varies between the R2B members, suggesting that proteolysis may occur in different contexts, including cancer, development [54], and in response to high cell density [36], growth factor stimulation [63], and calcium influx [94].
5.1 Exploitation of proteolysis to generate molecular imaging probes for tumors
GBM is a particularly invasive/dispersive tumor type making the actual location of the tumor cells difficult to image. Yet the best method of promoting GBM patient survival is thorough surgical removal of the tumor tissue, which is exacerbated by this imaging challenge and the need to limit the removal of healthy tissue [100, 101]. To be effective GBM imaging agents, tumor-specific probes need to first cross the blood brain barrier and then produce signals to specifically identify invading cells at the tumor edge to facilitate surgical removal. To address this, molecular imaging agents are being developed that specifically bind to tumor cells and can be imaged using fluorescent surgical microscopes [102]. Given that R2Bs are homophilic binding proteins and that their shed extracellular fragments are observed in cancer cells and tissues, imaging probes designed to homophilically bind to the shed R2B fragments are being developed for use as cancer diagnostics, as reviewed in [91].
Within the last few years, substantial progress has been made in demonstrating the utility of probes generated to bind to PTPμ in GBM. Extracellular fragments of PTPμ are present at high levels at the tumor edge [95, 103]. PTPμ probes generated to the MAM or Ig domain of PTPμ, termed SBK probes, robustly and specifically label GBM tumor and edge tissue sections [95]. When labeled with a fluorescent dye, the SBK probes recognize tumor flank models of GBM within minutes [95]. Importantly, the SBK probes cross the blood brain barrier to label intracranial GBM tumors [95]. Intracranial SBK2 probe labeling is observed 25 minutes after injection to accommodate for clearance from the brain [95]. The SBK2 probe labels greater than 99% of all CNS-1 GBM tumor cells in a xenograft tumor of CNS-1 cells in athymic nude mice [103]. The probe also labels cells that dispersed several millimeters away from the main tumor, and the level of probe fluorescence is often brighter in the region of tumor dispersal than in the main tumor [103]. This is remarkable because dispersed cell labeling is a major disadvantage of the best currently available GBM fluorescent probe 5-Aminolevulinic acid (5-ALA) [104, 105]. When conjugated to a gadolinium chelate, the SBK2 probe completely and rapidly labels LN-229 GBM tumors with higher contrast enhancement than the generic MRI contrast enhancement agent, Gadoteridol (ProHance) [106].
The advantages that the SBK probes have over other fluorescent and MRI imaging agents make it an excellent candidate probe for diagnosing GBM and guiding surgical removal of tumor tissue. The mechanism of adhesion and the structural domains necessary for mediating cell-cell adhesion have been characterized for PTPμ and PTPρ [12, 14, 16, 17, 20, 72]. Combined with what is known about the crystal structure of the extracellular domain of PTPμ [23] and the surface polarity of all R2B RPTPs [18], the development of probes to PTPρ and PTPκ similar to the ones designed for PTPμ is feasible. Because PCP-2 mediates in vitro homophilic binding [12], although not cell-cell adhesion [18], it is theoretically possible to generate a homophilic binding PCP-2 probe to bind to its shed extracellular fragments. Studies evaluating the expression of shed R2B fragments in cancer will clarify whether these probes will be as diagnostically promising as those for PTPμ. We have also proposed additional models for generating diagnostic probes for other RPTPs [91].
5.2 Proteolysis of axon guidance cues
Axon guidance is a form of cell migration where the protruding tip of a growing neuron, the growth cone, extends long distances and passes many cells before reaching its target. The growth cone does this by responding to both cell surface and soluble guidance cues in its environment. The guidance cues generally belong to four families: netrins that bind to DCC receptors; Slits that bind Robo receptors; Semaphorins that bind Plexin and Neuropilin receptors; and Ephrins that bind Eph receptors [107]. Some Semaphorins and Ephrins are membrane bound, whereas the rest of the guidance cues tend to be secreted. Transcriptional regulation of the receptors and guidance cues was thought to be the main mechanism of regulating axon guidance [108]. More recently, post-translational modification of axon guidance molecules, including proteolysis, has been demonstrated [109]. This was initially demonstrated in the Drosophila mutant kuzbanian, which has midline guidance defects as a result of a mutation in a metalloprotease homologous to ADAM 10 [110]. The kuzbanian defects are caused by reduced cleavage and clearance of Robo from commissural axons [111]. Kuzbanian is also necessary for cleavage of Ephrin A2 on growth cones to allow for growth cone retraction following EphA3 binding on non-target cells [112]. Around the same time, the activity of an unidentified metalloprotease was shown to reduce spinal cord explant outgrowth in response to the soluble chemoattractant netrin, presumably by reducing the number of DCC receptors available to bind to netrin [113].
In more recent years, additional proteases have been demonstrated to cleave guidance cues: for example, ADAM17 cleaves Semaphorin5B in mouse dorsal root ganglion neurites [114]; β-site amyloid precursor protein (APP) cleaving enzymes 1 (BACE1) is required for mouse olfactory sensory neuron guidance [115], potentially via the cleavage of contactin-2 [116]; Proprotein Convertases Subtilisin Kexin Isozyme-1 (SKI-1) and furin cleavage of the repulsive guidance molecule RGMa is necessary to make it repellant [117]; calpain1 cleavage of Plexin-A1 is necessary for commissural axon crossing of the midline [118]; and γ-secretase activity is necessary for DCC processing in spinal motor neurons [119]. The growing consensus is that cleavage of membrane associated cues regulates axonal guidance, often by releasing growth cones following a repellant receptor-ligand interaction [111, 112], or for generating growth repellant cues [114, 117]. Cleavage can also, however, silence repulsive guidance cues [119].
5.2.1 Implications of R2B RPTP proteolysis on axon guidance
R2B RPTPs mediate axonal outgrowth from neurons when applied as substrates in vitro, and have been hypothesized to regulate neurite outgrowth and pathfinding during development. PTPμ protein is expressed in a high temporal low nasal pattern in the neural retina and a high anterior low posterior pattern in the tectum, and has been suggested to be part of the molecular gradient contributing to axon pathfinding of the retinotectal system [40]. PTPμ is a growth permissive substrate to retinal ganglion cell neurites that originate from the ventral nasal retina and is repulsive to neurites from the rest of the retina [42]. A soluble Fc chimera of PTPμ containing the entire extracellular domain of PTPμ can induce growth cone collapse from temporal retinal neurons [40, 43]. Tyrosine phosphatase activity of PTPμ in nasal and temporal retina is required for both the permissive and repulsive responses to a PTPμ substrate, respectively [42]. Finally, over-expression of wild-type PTPμ is sufficient to induce repulsion of nasal neurons [42].
A soluble chimera of the extracellular domain of PTPκ linked to an Fc protein promotes neurite outgrowth of cerebellar neurons growing on 3T3 fibroblast cells [49], demonstrating that a proteolyzed or shed form of PTPκ, that we now know exists in cells, could function during development [36]. Addition of a soluble version of the extracellular domain of PTPρ to primary culture of hippocampal neurons decreases synapse development [54]. The mechanism attributed to the PTPρ-Fc chimera is via the inhibition of cis interactions of PTPρ with the neuroligin-neurexin complex [54].
Given what we now know about R2B proteolysis, some of the earlier studies on their roles in axon guidance and neurite outgrowth could be interpreted in a new light. For instance, the expression studies of PTPμ in the retina were conducted using an intracellular antibody to PTPμ [40], and therefore, did not evaluate whether any cleaved extracellular fragments of PTPμ exist in the developing nervous system. Since soluble Fc chimeras of PTPμ induce growth cone collapse of temporal growth cones, shed PTPμ fragments could be repellant guidance cues [40, 43]. This would be analogous to the generation of a repulsive guidance cue by the cleavage and shedding of Semaphorin5B [114]. Evaluation of intracellular cleavage fragments of R2B RPTPs in development is also important. There is evidence that the intracellular stub of the guidance receptor DCC, generated following ADAM and γ-secretase cleavage, has functions that are both necessary for axon guidance and that differ from that of full-length unprocessed DCC [119]. Finally, cleavage of RGMa protein in the neural retina produces differential responses in nasal and temporal retina due to differences in expression of the RGMa fragment receptor, Neogenin [117]. Future studies of R2B RPTP function in neural development and axon guidance will need to consider the effects and repercussions of receptor proteolysis in the developmental milieu.
6. Perineural Invasion, Collective Cell Migration and Cell-Cell CAM cleavage
Perineural invasion (PNI) is a hallmark of many carcinomas in which cancer cells invade “in, around, and through” peripheral nerves in the vicinity of the tumor. Cancer cells use nerves as a channel to form clusters or glands of cancerous cells which can also be major routes of metastasis [120]. PNI was often seen as a minor pathway for cancer cell invasion and metastasis, behind vascular and lymphatic routes [120]. It turns out, though, that PNI is common in several carcinomas and is highly associated with poor prognosis. PNI is estimated to occur in 90–100% of pancreatic ductal adenocarcinoma, 80% of head and neck and prostate carcinoma, 50% of stomach cancer, and 33% of colorectal cancer [120, 121].
The study of the molecular mechanisms of PNI is only just beginning, yet a molecular framework is forming that has links to axon guidance. PNI likely occurs via chemotropism of cancer cells to nerves by GDNF [122], chemokines [123], and VEGF [124]. Additional guidance molecules that may regulate PNI include: plexin-B1 on cancer cells binding to Semaphorin4D on nerves [125]; mucin on cells binding to myelin associated glycoprotein (MAG) on nerves [126]; Slit2 in the tumor cells interacting with Robo receptor on nerves [127]; laminin rich nerve pathways that promote migration via the α6β1 integrin receptor [128]; and the L1-cell adhesion molecule [129]. Notably, proteolysis of Semaphorin4D has been observed in squamous cell carcinoma (a common cancer site of PNI) by MT1-MMP [130]. Whether Semaphorin4D proteolysis affects PNI needs to be addressed.
PNI has been described as a “continuous extension of tumor cells” [121], and brings to mind the collective migration of epithelial and mesenchymal cells often observed in development and cancer, known as collective cell migration [131, 132]. Collective cell migration of cancer cells occurs as strands of cells and as cell clusters [133]. We observed clusters and chains of migrating GBM cells in three-dimensional brain reconstructions of mouse orthotopic tumor models [134]. In these models, GBM cells migrate along blood vessels and white matter tracts that are composed of the axons of neurons [134].
To move together, the cells need to have collective adhesion, polarization, and guidance to cues to create a “supracellular” structure [133]. Homophilic cell-cell adhesion by classical cadherins, especially by N- and E-cadherin, is thought to be a major regulator of collective adhesion and migration [133, 135, 136]. R2B RPTPs are expressed in the nervous system, associate with cadherins, promote cell-cell adhesion, play a role in axon guidance and have known functions in cancer. We hypothesize that R2B RPTPs may function in PNI to promote the collective movement of tumor cells along nerve bundles. The SBK probe can label chains of migrating GBM cells in mouse brain tumors [103]. If PTPμ is also expressed in the microenvironment of carcinomas, the SBK probes may be useful tools for assessing the extent of PNI.
7. Conclusions and Future Directions
R2B RPTPs function in adhesion at adherens junctions, during development, and in cancer. It is becoming clear that post-translational regulation of R2B RPTPs is occurring in cancer. As many mechanisms that occur in development are often usurped in cancer, we hypothesize that proteolysis, glycosylation and phosphorylation will be relevant to developmental processes regulated by R2B RPTPs (Figure 2). Likewise, a greater consideration of the molecules and cells found in the tumor microenvironment and in the developmental milieu needs to be made. Cells in the tumor microenvironment may be influencing proteolysis or producing their own source of cell-cell CAM fragments, thereby creating a meshwork of migrational cues that can yield very different signaling events than when CAMs are preserved on the cell surface. This may also be happening during axon guidance and collective cell migration in development. The repercussions of proteolysis will likely be significant, as supported by the fact that the PTPκ PIC fragment produces the opposite effect on β-catenin signaling than that of full-length PTPκ [36].
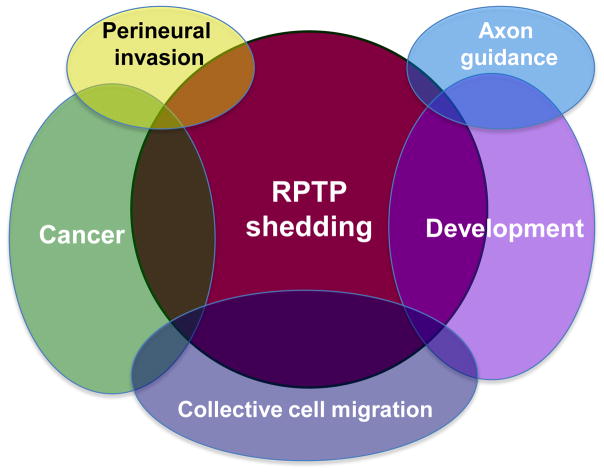
Figure 2.
RPTP proteolysis affects both development and cancer, occasionally by influencing the same cellular mechanism. For instance, proteolysis of proteins involved in collective cell migration is hypothesized to affect migration during development and in cancer. Proteolysis has also been shown to influence the neural developmental process of axon guidance. Finally, perineural invasion may be influenced by receptor proteolysis in cancer. Because the R2B RPTPs are implicated in many of these processes, their proteolysis is likely to have far reaching cellular implications in development and cancer.
Many questions remain: What are the consequences of proteolysis on the adherens junction R2B RPTP ‘spacer clamp’? What are the effects of shed extracellular RPTP fragments on adhesion, migration, and in cancer such as during PNI? Does the shed extracellular fragment function in developmental environments such as during axon guidance or collective cell migration [46, 49]? What are the effects of the cytoplasmic and nuclear localization of the cleaved intracellular R2B fragments? It is an exciting time to evaluate R2B RPTP function and regulation, and we look forward to future developments in understanding the effects of these complex proteolytic events.
Acknowledgments
We thank Dr. Polly Phillips-Mason for her comments on the manuscript. Funding was provided by the National Institutes of Health grant R01-NS063971, R01-CA179956 and by the Tabitha Yee-May Lou Endowment Fund for Brain Cancer Research.
Footnotes
Abbreviations: R2B RPTP, Type IIB Receptor protein tyrosine phosphatase; PTP, protein tyrosine phosphatase; MAM, meprin, A5, mu domain; Ig, immunoglobulin; FN III, fibronectin III; CAM, cell adhesion molecule; D1, membrane proximal phosphatase domain; D2, membrane distal phosphatase domain; LEC; Long-Evans Cinnamon;, head and neck squamous cell carcinoma; GBM, glioblastoma multiforme; PNI, perineural invasion; E-subunit, extracellular subunit; P-subunit, phosphatase subunit; PC5, proprotein convertase 5; PΔE, membrane-tethered PTPμ and PTPκ fragments; ICD, intracellular domain fragment of PTPμ; PIC, phosphatase intracellular domain fragment of PTPκ; 5-ALA, 5-Aminolevulinic acid.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tonks NK. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–78. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady-Kalnay SM, Tonks NK. Protein tyrosine phosphatases as adhesion receptors. Curr Opin Cell Biol. 1995;7:650–7. doi: 10.1016/0955-0674(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 3.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–36. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady-Kalnay S. Ig-superfamily phosphatases. In: Sonderegger P, editor. Ig Superfamily Molecules in the Nervous System. Zurich: Harwood Academic Publishers; 1998. [Google Scholar]
- 5.Tonks NK, Yang Q, Flint AJ, Gebbink MF, Franza BR, Jr, Hill DE, et al. Protein tyrosine phosphatases: the problems of a growing family. Cold Spring Harb Symp Quant Biol. 1992;57:87–94. doi: 10.1101/sqb.1992.057.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Brady-Kalnay S, Tonks NK. Receptor protein tyrosine phosphatases, cell adhesion and signal transduction. Adv Prot Phosphatases. 1994;8:241–71. [Google Scholar]
- 7.Gebbink MF, Verheijen MH, Zondag GC, van Etten I, Moolenaar WH. Purification and characterization of the cytoplasmic domain of human receptor-like protein tyrosine phosphatase RPTP mu. Biochemistry. 1993;32:13516–22. doi: 10.1021/bi00212a017. [DOI] [PubMed] [Google Scholar]
- 8.Ensslen-Craig SE, Brady-Kalnay SM. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev Biol. 2004;275:12–22. doi: 10.1016/j.ydbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Besco J, Popesco MC, Davuluri RV, Frostholm A, Rotter A. Genomic structure and alternative splicing of murine R2B receptor protein tyrosine phosphatases (PTPkappa, mu, rho and PCP-2) BMC Genomics. 2004;5:14. doi: 10.1186/1471-2164-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebbink MF, van Etten I, Hateboer G, Suijkerbuijk R, Beijersbergen RL, Geurts van Kessel A, et al. Cloning, expression and chromosomal localization of a new putative receptor-like protein tyrosine phosphatase. FEBS Lett. 1991;290:123–30. doi: 10.1016/0014-5793(91)81241-y. [DOI] [PubMed] [Google Scholar]
- 11.Jiang YP, Wang H, D’Eustachio P, Musacchio JM, Schlessinger J, Sap J. Cloning and characterization of R-PTP-kappa, a new member of the receptor protein tyrosine phosphatase family with a proteolytically cleaved cellular adhesion molecule-like extracellular region. Mol Cell Biol. 1993;13:2942–51. doi: 10.1128/mcb.13.5.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng J, Wu K, Armanini M, O’Rourke N, Dowbenko D, Lasky LA. A novel protein-tyrosine phosphatase related to the homotypically adhering kappa and mu receptors. J Biol Chem. 1997;272:7264–77. doi: 10.1074/jbc.272.11.7264. [DOI] [PubMed] [Google Scholar]
- 13.McAndrew PE, Frostholm A, White RA, Rotter A, Burghes AH. Identification and characterization of RPTP rho, a novel RPTP mu/kappa-like receptor protein tyrosine phosphatase whose expression is restricted to the central nervous system. Brain Res Mol Brain Res. 1998;56:9–21. doi: 10.1016/s0169-328x(98)00014-x. [DOI] [PubMed] [Google Scholar]
- 14.Brady-Kalnay SM, Flint AJ, Tonks NK. Homophilic binding of PTP mu, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J Cell Biol. 1993;122:961–72. doi: 10.1083/jcb.122.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebbink MF, Zondag GC, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993;268:16101–4. [PubMed] [Google Scholar]
- 16.Sap J, Jiang YP, Friedlander D, Grumet M, Schlessinger J. Receptor tyrosine phosphatase R-PTP-kappa mediates homophilic binding. Mol Cell Biol. 1994;14:1–9. doi: 10.1128/mcb.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Becka S, Zhang P, Zhang X, Brady-Kalnay SM, Wang Z. Tumor-derived extracellular mutations of PTPRT /PTPrho are defective in cell adhesion. Mol Cancer Res. 2008;6:1106–13. doi: 10.1158/1541-7786.MCR-07-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becka S, Zhang P, Craig SE, Lodowski DT, Wang Z, Brady-Kalnay SM. Characterization of the adhesive properties of the type IIb subfamily receptor protein tyrosine phosphatases. Cell Commun Adhes. 2010;17:34–47. doi: 10.3109/15419061.2010.487957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zondag GC, Koningstein GM, Jiang YP, Sap J, Moolenaar WH, Gebbink MF. Homophilic interactions mediated by receptor tyrosine phosphatases mu and kappa. A critical role for the novel extracellular MAM domain. J Biol Chem. 1995;270:14247–50. doi: 10.1074/jbc.270.24.14247. [DOI] [PubMed] [Google Scholar]
- 20.Brady-Kalnay SM, Tonks NK. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTP mu. J Biol Chem. 1994;269:28472–7. [PubMed] [Google Scholar]
- 21.Del Vecchio RL, Tonks NK. The conserved immunoglobulin domain controls the subcellular localization of the homophilic adhesion receptor protein-tyrosine phosphatase mu. J Biol Chem. 2005;280:1603–12. doi: 10.1074/jbc.M410181200. [DOI] [PubMed] [Google Scholar]
- 22.Aricescu AR, Hon WC, Siebold C, Lu W, van der Merwe PA, Jones EY. Molecular analysis of receptor protein tyrosine phosphatase mu-mediated cell adhesion. Embo J. 2006;25:701–12. doi: 10.1038/sj.emboj.7600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aricescu AR, Siebold C, Choudhuri K, Chang VT, Lu W, Davis SJ, et al. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science. 2007;317:1217–20. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- 24.Aricescu AR, Siebold C, Jones EY. Receptor protein tyrosine phosphatase mu: measuring where to stick. Biochem Soc Trans. 2008;36:167–72. doi: 10.1042/BST0360167. [DOI] [PubMed] [Google Scholar]
- 25.Cismasiu VB, Denes SA, Reiländer H, Michel H, Szedlacsek SE. The MAM (meprin/A5-protein/PTPmu) domain is a homophilic binding site promoting the lateral dimerization of receptor-like protein-tyrosine phosphatase mu. J Biol Chem. 2004;279:26922–31. doi: 10.1074/jbc.M313115200. [DOI] [PubMed] [Google Scholar]
- 26.Brady-Kalnay SM, Rimm DL, Tonks NK. Receptor protein tyrosine phosphatase PTPmu associates with cadherins and catenins in vivo. J Cell Biol. 1995;130:977–86. doi: 10.1083/jcb.130.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs M, Müller T, Lerch MM, Ulrich A. Association of human protein-tyrosine phosphatase κ with members of the armadillo family. J Biol Chem. 1996;271:16712–9. doi: 10.1074/jbc.271.28.16712. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Lian Z, Lerch MM, Chen Z, Xie W, Ullrich A. Characterization of PCP-2, a novel receptor protein tyrosine phosphatase of the MAM domain family. Oncogene. 1996;12:2555–62. [PubMed] [Google Scholar]
- 29.Hellberg CB, Burden-Gulley SM, Pietz GE, Brady-Kalnay SM. Expression of the receptor protein-tyrosine phosphatase, PTPmu, restores E-cadherin-dependent adhesion in human prostate carcinoma cells. J Biol Chem. 2002;277:11165–73. doi: 10.1074/jbc.M112157200. [DOI] [PubMed] [Google Scholar]
- 30.Yan HX, Yang W, Zhang R, Chen L, Tang L, Zhai B, et al. Protein-tyrosine phosphatase PCP-2 inhibits beta-catenin signaling and increases E-cadherin-dependent cell adhesion. J Biol Chem. 2006;281:15423–33. doi: 10.1074/jbc.M602607200. [DOI] [PubMed] [Google Scholar]
- 31.Novellino L, De Filippo A, Deho P, Perrone F, Pilotti S, Parmiani G, et al. PTPRK negatively regulates transcriptional activity of wild type and mutated oncogenic beta-catenin and affects membrane distribution of beta-catenin/E-cadherin complexes in cancer cells. Cell Signal. 2008;20:872–83. doi: 10.1016/j.cellsig.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 32.Burden-Gulley SM, Brady-Kalnay SM. PTPmu regulates N-cadherin-dependent neurite outgrowth. J Cell Biol. 1999;144:1323–36. doi: 10.1083/jcb.144.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady-Kalnay SM, Mourton T, Nixon JP, Pietz GE, Kinch M, Chen H, et al. Dynamic interaction of PTPmu with multiple cadherins in vivo. J Cell Biol. 1998;141:287–96. doi: 10.1083/jcb.141.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besco JA, Hooft van Huijsduijnen R, Frostholm A, Rotter A. Intracellular substrates of brain-enriched receptor protein tyrosine phosphatase rho (RPTPrho/PTPRT) Brain Res. 2006;1116:50–7. doi: 10.1016/j.brainres.2006.07.122. [DOI] [PubMed] [Google Scholar]
- 35.Sui XF, Kiser TD, Hyun SW, Angelini DJ, Del Vecchio RL, Young BA, et al. Receptor protein tyrosine phosphatase mu regulates the paracellular pathway in human lung microvascular endothelia. Am J Pathol. 2005;166:1247–58. doi: 10.1016/s0002-9440(10)62343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, et al. Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin’s transcriptional activity. Mol Cell Biol. 2006;26:3917–34. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan HX, He YQ, Dong H, Zhang P, Zeng JZ, Cao HF, et al. Physical and functional interaction between receptor-like protein tyrosine phosphatase PCP-2 and beta-catenin. Biochemistry. 2002;41:15854–60. doi: 10.1021/bi026095u. [DOI] [PubMed] [Google Scholar]
- 38.Zondag GC, Reynolds AB, Moolenaar WH. Receptor protein-tyrosine phosphatase RPTPmu binds to and dephosphorylates the catenin p120(ctn) J Biol Chem. 2000;275:11264–9. doi: 10.1074/jbc.275.15.11264. [DOI] [PubMed] [Google Scholar]
- 39.Sakamoto Y, Ogita H, Komura H, Takai Y. Involvement of nectin in inactivation of integrin alpha(v)beta(3) after the establishment of cell-cell adhesion. J Biol Chem. 2008;283:496–505. doi: 10.1074/jbc.M704195200. [DOI] [PubMed] [Google Scholar]
- 40.Burden-Gulley SM, Ensslen SE, Brady-Kalnay SM. Protein tyrosine phosphatase-mu differentially regulates neurite outgrowth of nasal and temporal neurons in the retina. J Neurosci. 2002;22:3615–27. doi: 10.1523/JNEUROSCI.22-09-03615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ensslen SE, Rosdahl JA, Brady-Kalnay SM. The receptor protein tyrosine phosphatase mu, PTPmu, regulates histogenesis of the chick retina. Dev Biol. 2003;264:106–18. doi: 10.1016/j.ydbio.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Ensslen-Craig SE, Brady-Kalnay SM. PTP mu expression and catalytic activity are required for PTP mu-mediated neurite outgrowth and repulsion. Mol Cell Neurosci. 2005;28:177–88. doi: 10.1016/j.mcn.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Ensslen SE, Brady-Kalnay SM. PTPmu signaling via PKCdelta is instructive for retinal ganglion cell guidance. Mol Cell Neurosci. 2004;25:558–71. doi: 10.1016/j.mcn.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Phillips-Mason PJ, Gates TJ, Major DL, Sacks DB, Brady-Kalnay SM. The receptor protein-tyrosine phosphatase PTPμ interacts with IQGAP1. J Biol Chem. 2006;281:4903–10. doi: 10.1074/jbc.M506414200. [DOI] [PubMed] [Google Scholar]
- 45.Phillips-Mason PJ, Kaur H, Burden-Gulley SM, Craig SE, Brady-Kalnay SM. Identification of phospholipase C gamma1 as a protein tyrosine phosphatase mu substrate that regulates cell migration. J Cell Biochem. 2011;112:39–48. doi: 10.1002/jcb.22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosdahl JA, Ensslen SE, Niedenthal JA, Brady-Kalnay SM. PTP mu-dependent growth cone rearrangement is regulated by Cdc42. J Neurobiol. 2003;56:199–208. doi: 10.1002/neu.10231. [DOI] [PubMed] [Google Scholar]
- 47.Rosdahl JA, Mourton TL, Brady-Kalnay SM. Protein kinase C delta (PKCdelta) is required for protein tyrosine phosphatase mu (PTPmu)-dependent neurite outgrowth. Mol Cell Neurosci. 2002;19:292–306. doi: 10.1006/mcne.2001.1071. [DOI] [PubMed] [Google Scholar]
- 48.Major DL, Brady-Kalnay SM. Rho GTPases regulate PTPmu-mediated nasal neurite outgrowth and temporal repulsion of retinal ganglion cell neurons. Mol Cell Neurosci. 2007;34:453–67. doi: 10.1016/j.mcn.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drosopoulos NE, Walsh FS, Doherty P. A soluble version of the receptor-like protein tyrosine phosphatase kappa stimulates neurite outgrowth via a Grb2/MEK1-dependent signaling cascade. Mol Cell Neurosci. 1999;13:441–9. doi: 10.1006/mcne.1999.0758. [DOI] [PubMed] [Google Scholar]
- 50.Shen P, Canoll PD, Sap J, Musacchio JM. Expression of a truncated receptor protein tyrosine phosphatase kappa in the brain of an adult transgenic mouse. Brain Res. 1999;826:157–71. doi: 10.1016/s0006-8993(99)01179-8. [DOI] [PubMed] [Google Scholar]
- 51.Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 52.Johnson KG, Holt CE. Expression of CRYP-alpha, LAR, PTP-delta, and PTP-rho in the developing Xenopus visual system. Mech Dev. 2000;92:291–4. doi: 10.1016/s0925-4773(99)00345-7. [DOI] [PubMed] [Google Scholar]
- 53.McAndrew PE, Frostholm A, Evans JE, Zdilar D, Goldowitz D, Chiu IM, et al. Novel receptor protein tyrosine phosphatase (RPTPrho) and acidic fibroblast growth factor (FGF-1) transcripts delineate a rostrocaudal boundary in the granule cell layer of the murine cerebellar cortex. J Comp Neurol. 1998;391:444–55. doi: 10.1002/(sici)1096-9861(19980222)391:4<444::aid-cne3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 54.Lim SH, Kwon SK, Lee MK, Moon J, Jeong DG, Park E, et al. Synapse formation regulated by protein tyrosine phosphatase receptor T through interaction with cell adhesion molecules and Fyn. EMBO J. 2009;28:3564–78. doi: 10.1038/emboj.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park AR, Oh D, Lim SH, Choi J, Moon J, Yu DY, et al. Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a substrate of PTPRT. J Cell Sci. 2012;125:4518–31. doi: 10.1242/jcs.105502. [DOI] [PubMed] [Google Scholar]
- 56.Jacobs FM, van der Linden AJ, Wang Y, von Oerthel L, Sul HS, Burbach JP, et al. Identification of Dlk1, Ptpru and Klhl1 as novel Nurr1 target genes in meso-diencephalic dopamine neurons. Development. 2009;136:2363–73. doi: 10.1242/dev.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kose H, Sakai T, Tsukumo S, Wei K, Yamada T, Yasutomo K, et al. Maturational arrest of thymocyte development is caused by a deletion in the receptor-like protein tyrosine phosphatase kappa gene in LEC rats. Genomics. 2007;89:673–7. doi: 10.1016/j.ygeno.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 58.Iwata R, Sasaki N, Agui T. Contiguous gene deletion of Ptprk and Themis causes T-helper immunodeficiency (thid) in the LEC rat. Biomed Res. 2010;31:83–7. doi: 10.2220/biomedres.31.83. [DOI] [PubMed] [Google Scholar]
- 59.Asano A, Tsubomatsu K, Jung CG, Sasaki N, Agui T. A deletion mutation of the protein tyrosine phosphatase kappa (Ptprk) gene is responsible for T-helper immunodeficiency (thid) in the LEC rat. Mamm Genome. 2007;18:779–86. doi: 10.1007/s00335-007-9062-0. [DOI] [PubMed] [Google Scholar]
- 60.Erdenebayar N, Maekawa Y, Nishida J, Kitamura A, Yasutomo K. Protein-tyrosine phosphatase-kappa regulates CD4+ T cell development through ERK1/2-mediated signaling. Biochem Biophys Res Commun. 2009;390:489–93. doi: 10.1016/j.bbrc.2009.09.117. [DOI] [PubMed] [Google Scholar]
- 61.Aerne B, Stoker A, Ish-Horowicz D. Chick receptor tyrosine phosphatase Psi is dynamically expressed during somitogenesis. Gene Expr Patterns. 2003;3:325–9. doi: 10.1016/s1567-133x(03)00038-3. [DOI] [PubMed] [Google Scholar]
- 62.Aerne B, Ish-Horowicz D. Receptor tyrosine phosphatase psi is required for Delta/Notch signalling and cyclic gene expression in the presomitic mesoderm. Development. 2004;131:3391–9. doi: 10.1242/dev.01222. [DOI] [PubMed] [Google Scholar]
- 63.Taniguchi Y, London R, Schinkmann K, Jiang S, Avraham H. The receptor protein tyrosine phosphatase, PTP-RO, is upregulated during megakaryocyte differentiation and Is associated with the c-Kit receptor. Blood. 1999;94:539–49. [PubMed] [Google Scholar]
- 64.Hardy S, Julien SG, Tremblay ML. Impact of oncogenic protein tyrosine phosphatases in cancer. Anticancer Agents Med Chem. 2012;12:4–18. doi: 10.2174/187152012798764741. [DOI] [PubMed] [Google Scholar]
- 65.Zhao S, Sedwick D, Wang Z. Genetic alteration of protein tyrosine phosphatases in human cancers. Oncogene. doi: 10.1038/onc.2014.326. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 67.Lui VW, Peyser ND, Ng PK, Hritz J, Zeng Y, Lu Y, et al. Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proc Natl Acad Sci U S A. 2014;111:1114–9. doi: 10.1073/pnas.1319551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laczmanska I, Karpinski P, Bebenek M, Sedziak T, Ramsey D, Szmida E, et al. Protein tyrosine phosphatase receptor-like genes are frequently hypermethylated in sporadic colorectal cancer. J Hum Genet. 2013;58:11–5. doi: 10.1038/jhg.2012.119. [DOI] [PubMed] [Google Scholar]
- 69.Liu F, You X, Chi X, Wang T, Ye L, Niu J, et al. Hepatitis B virus X protein mutant HBxDelta127 promotes proliferation of hepatoma cells through up-regulating miR-215 targeting PTPRT. Biochem Biophys Res Commun. 2014;444:128–34. doi: 10.1016/j.bbrc.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 70.Huang S, Cai M, Zheng Y, Zhou L, Wang Q, Chen L. miR-888 in MCF-7 side population sphere cells directly targets E-cadherin. J Genet Genomics. 2014;41:35–42. doi: 10.1016/j.jgg.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Pasquo A, Consalvi V, Knapp S, Alfano I, Ardini M, Stefanini S, et al. Structural stability of human protein tyrosine phosphatase rho catalytic domain: effect of point mutations. PLoS One. 2012;7:e32555. doi: 10.1371/journal.pone.0032555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang P, Becka S, Craig SE, Lodowski DT, Brady-Kalnay SM, Wang Z. Cancer-derived mutations in the fibronectin III repeats of PTPRT/PTPrho inhibit cell-cell aggregation. Cell Commun Adhes. 2009;16:146–53. doi: 10.3109/15419061003653771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A. 2010;107:2592–7. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun PH, Ye L, Mason MD, Jiang WG. Protein tyrosine phosphatase mu (PTP mu or PTPRM), a negative regulator of proliferation and invasion of breast cancer cells, is associated with disease prognosis. PLoS One. 2012;7:e50183. doi: 10.1371/journal.pone.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, et al. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009;11:767–78. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Assem M, Sibenaller Z, Agarwal S, Al-Keilani MS, Alqudah MA, Ryken TC. Enhancing diagnosis, prognosis, and therapeutic outcome prediction of gliomas using genomics. OMICS. 2012;16:113–22. doi: 10.1089/omi.2011.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakamura M, Kishi M, Sakaki T, Hashimoto H, Nakase H, Shimada K, et al. Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer Res. 2003;63:737–41. [PubMed] [Google Scholar]
- 78.Wang L, Sato-Otsubo A, Sugita S, Takase H, Mochizuki M, Usui Y, et al. High-resolution genomic copy number profiling of primary intraocular lymphoma by single nucleotide polymorphism microarrays. Cancer Sci. 2014;105:592–9. doi: 10.1111/cas.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barghorn A, Speel EJ, Farspour B, Saremaslani P, Schmid S, Perren A, et al. Putative tumor suppressor loci at 6q22 and 6q23-q24 are involved in the malignant progression of sporadic endocrine pancreatic tumors. Am J Pathol. 2001;158:1903–11. doi: 10.1016/S0002-9440(10)64658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Novellino L, Renkvist N, Rini F, Mazzocchi A, Rivoltini L, Greco A, et al. Identification of a mutated receptor-like protein tyrosine phosphatase kappa as a novel, class II HLA-restricted melanoma antigen. J Immunol. 2003;170:6363–70. doi: 10.4049/jimmunol.170.12.6363. [DOI] [PubMed] [Google Scholar]
- 81.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 488:660–4. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, Reynolds GM, et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111:292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- 84.Mo W, Zhang J, Li X, Meng D, Gao Y, Yang S, et al. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8:e56592. doi: 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun PH, Ye L, Mason MD, Jiang WG. Protein tyrosine phosphatase kappa (PTPRK) is a negative regulator of adhesion and invasion of breast cancer cells, and associates with poor prognosis of breast cancer. J Cancer Res Clin Oncol. 2013;139:1129–39. doi: 10.1007/s00432-013-1421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McArdle L, Rafferty M, Maelandsmo GM, Bergin O, Farr CJ, Dervan PA, et al. Protein tyrosine phosphatase genes downregulated in melanoma. J Invest Dermatol. 2001;117:1255–60. doi: 10.1046/j.0022-202x.2001.01534.x. [DOI] [PubMed] [Google Scholar]
- 87.Sun PH, Ye L, Mason MD, Jiang WG. Receptor-like protein tyrosine phosphatase kappa negatively regulates the apoptosis of prostate cancer cells via the JNK pathway. Int J Oncol. 2013;43:1560–8. doi: 10.3892/ijo.2013.2082. [DOI] [PubMed] [Google Scholar]
- 88.Agarwal S, Al-Keilani MS, Alqudah MA, Sibenaller ZA, Ryken TC, Assem M. Tumor derived mutations of protein tyrosine phosphatase receptor type k affect its function and alter sensitivity to chemotherapeutics in glioma. PLoS One. 2013;8:e62852. doi: 10.1371/journal.pone.0062852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang SE, Wu FY, Shin I, Qu S, Arteaga CL. Transforming growth factor {beta} (TGF-{beta})-Smad target gene protein tyrosine phosphatase receptor type kappa is required for TGF-{beta} function. Mol Cell Biol. 2005;25:4703–15. doi: 10.1128/MCB.25.11.4703-4715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McArdle L, Rafferty MM, Satyamoorthy K, Maelandsmo GM, Dervan PA, Herlyn M, et al. Microarray analysis of phosphatase gene expression in human melanoma. Br J Dermatol. 2005;152:925–30. doi: 10.1111/j.1365-2133.2005.06454.x. [DOI] [PubMed] [Google Scholar]
- 91.Craig SE, Brady-Kalnay SM. Tumor-Derived Extracellular Fragments of Receptor Protein Tyrosine Phosphatases (RPTPs) as Cancer Molecular Diagnostic Tools. Anticancer Agents Med Chem. 2011 doi: 10.2174/187152011794941244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Craig SE, Brady-Kalnay SM. Cancer cells cut homophilic cell adhesion molecules and run. Cancer Res. 2011;71:303–9. doi: 10.1158/0008-5472.CAN-10-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, et al. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009;69:6960–8. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Phillips-Mason PJ, Craig SE, Brady-Kalnay SM. A Protease Storm Cleaves a Cell-Cell Adhesion Molecule in Cancer: Multiple Proteases Converge to Regulate PTPmu in Glioma Cells. J Cell Biochem. 2014 doi: 10.1002/jcb.24824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, et al. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12:305–16. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campan M, Yoshizumi M, Seidah NG, Lee ME, Bianchi C, Haber E. Increased proteolytic processing of protein tyrosine phosphatase mu in confluent vascular endothelial cells: the role of PC5, a member of the subtilisin family. Biochemistry. 1996;35:3797–802. doi: 10.1021/bi952552d. [DOI] [PubMed] [Google Scholar]
- 97.Kaur H, Burden-Gulley SM, Phillips-Mason PJ, Basilion JP, Sloan AE, Brady-Kalnay SM. Protein tyrosine phosphatase mu regulates glioblastoma cell growth and survival in vivo. Neuro Oncol. 2012;14:561–73. doi: 10.1093/neuonc/nos066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim YS, Kang HY, Kim JY, Oh S, Kim CH, Ryu CJ, et al. Identification of target proteins of N-acetylglucosaminyl transferase V in human colon cancer and implications of protein tyrosine phosphatase kappa in enhanced cancer cell migration. Proteomics. 2006;6:1187–91. doi: 10.1002/pmic.200500400. [DOI] [PubMed] [Google Scholar]
- 99.Kim YS, Jung JA, Kim HJ, Ahn YH, Yoo JS, Oh S, et al. Galectin-3 binding protein promotes cell motility in colon cancer by stimulating the shedding of protein tyrosine phosphatase kappa by proprotein convertase 5. Biochem Biophys Res Commun. 2011;404:96–102. doi: 10.1016/j.bbrc.2010.11.071. [DOI] [PubMed] [Google Scholar]
- 100.Pichlmeier U, Bink A, Schackert G, Stummer W. Resection and survival in glioblastoma multiforme: an RTOG recursive partitioning analysis of ALA study patients. Neuro Oncol. 2008;10:1025–34. doi: 10.1215/15228517-2008-052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stummer W, Reulen HJ, Meinel T, Pichlmeier U, Schumacher W, Tonn JC, et al. Extent of resection and survival in glioblastoma multiforme: identification of and adjustment for bias. Neurosurgery. 2008;62:564–76. doi: 10.1227/01.neu.0000317304.31579.17. discussion-76. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen QT, Tsien RY. Fluorescence-guided surgery with live molecular navigation--a new cutting edge. Nat Rev Cancer. 2013;13:653–62. doi: 10.1038/nrc3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burden-Gulley SM, Qutaish MQ, Sullivant KE, Tan M, Craig SE, Basilion JP, et al. Single cell molecular recognition of migrating and invading tumor cells using a targeted fluorescent probe to receptor PTPmu. Int J Cancer. 2012 doi: 10.1002/ijc.27838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stummer W, Novotny A, Stepp H, Goetz C, Bise K, Reulen HJ. Fluorescence-guided resection of glioblastoma multiforme by using 5-aminolevulinic acid-induced porphyrins: a prospective study in 52 consecutive patients. J Neurosurg. 2000;93:1003–13. doi: 10.3171/jns.2000.93.6.1003. [DOI] [PubMed] [Google Scholar]
- 105.Zhao S, Wu J, Wang C, Liu H, Dong X, Shi C, et al. Intraoperative fluorescence-guided resection of high-grade malignant gliomas using 5-aminolevulinic acid-induced porphyrins: a systematic review and meta-analysis of prospective studies. PLoS One. 2013;8:e63682. doi: 10.1371/journal.pone.0063682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burden-Gulley SM, Zhou Z, Craig SE, Lu ZR, Brady-Kalnay SM. Molecular Magnetic Resonance Imaging of Tumors with a PTPmu Targeted Contrast Agent. Transl Oncol. 2013;6:329–37. doi: 10.1593/tlo.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–64. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 108.Dickson TC, Mintz CD, Benson DL, Salton SR. Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J Cell Biol. 2002;157:1105–12. doi: 10.1083/jcb.200111076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bai G, Pfaff SL. Protease regulation: the Yin and Yang of neural development and disease. Neuron. 2011;72:9–21. doi: 10.1016/j.neuron.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fambrough D, Pan D, Rubin GM, Goodman CS. The cell surface metalloprotease/disintegrin Kuzbanian is required for axonal extension in Drosophila. Proc Natl Acad Sci U S A. 1996;93:13233–8. doi: 10.1073/pnas.93.23.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schimmelpfeng K, Gogel S, Klambt C. The function of leak and kuzbanian during growth cone and cell migration. Mech Dev. 2001;106:25–36. doi: 10.1016/s0925-4773(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 112.Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact-mediated axon repellent. Science. 2000;289:1360–5. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- 113.Galko MJ, Tessier-Lavigne M. Function of an axonal chemoattractant modulated by metalloprotease activity. Science. 2000;289:1365–7. doi: 10.1126/science.289.5483.1365. [DOI] [PubMed] [Google Scholar]
- 114.Browne K, Wang W, Liu RQ, Piva M, O’Connor TP. Transmembrane semaphorin5B is proteolytically processed into a repulsive neural guidance cue. J Neurochem. 2012;123:135–46. doi: 10.1111/j.1471-4159.2012.07885.x. [DOI] [PubMed] [Google Scholar]
- 115.Cao L, Rickenbacher GT, Rodriguez S, Moulia TW, Albers MW. The precision of axon targeting of mouse olfactory sensory neurons requires the BACE1 protease. Sci Rep. 2012;2:231. doi: 10.1038/srep00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gautam V, D’Avanzo C, Hebisch M, Kovacs DM, Kim DY. BACE1 activity regulates cell surface contactin-2 levels. Mol Neurodegener. 2014;9:4. doi: 10.1186/1750-1326-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tassew NG, Charish J, Seidah NG, Monnier PP. SKI-1 and Furin generate multiple RGMa fragments that regulate axonal growth. Dev Cell. 2012;22:391–402. doi: 10.1016/j.devcel.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 118.Nawabi H, Briancon-Marjollet A, Clark C, Sanyas I, Takamatsu H, Okuno T, et al. A midline switch of receptor processing regulates commissural axon guidance in vertebrates. Genes Dev. 2010;24:396–410. doi: 10.1101/gad.542510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bai G, Chivatakarn O, Bonanomi D, Lettieri K, Franco L, Xia C, et al. Presenilin-dependent receptor processing is required for axon guidance. Cell. 2011;144:106–18. doi: 10.1016/j.cell.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer: a review of the literature. Cancer. 2009;115:3379–91. doi: 10.1002/cncr.24396. [DOI] [PubMed] [Google Scholar]
- 121.Marchesi F, Piemonti L, Mantovani A, Allavena P. Molecular mechanisms of perineural invasion, a forgotten pathway of dissemination and metastasis. Cytokine Growth Factor Rev. 2010;21:77–82. doi: 10.1016/j.cytogfr.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 122.Liu H, Li X, Xu Q, Lv S, Li J, Ma Q. Role of glial cell line-derived neurotrophic factor in perineural invasion of pancreatic cancer. Biochim Biophys Acta. 2012;1826:112–20. doi: 10.1016/j.bbcan.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 123.Marchesi F, Grizzi F, Laghi L, Mantovani A, Allavena P. Molecular mechanisms of pancreatic cancer dissemination: the role of the chemokine system. Curr Pharm Des. 2012;18:2432–8. doi: 10.2174/13816128112092432. [DOI] [PubMed] [Google Scholar]
- 124.Fromont G, Godet J, Pires C, Yacoub M, Dore B, Irani J. Biological significance of perineural invasion (PNI) in prostate cancer. Prostate. 2012;72:542–8. doi: 10.1002/pros.21456. [DOI] [PubMed] [Google Scholar]
- 125.Binmadi NO, Yang YH, Zhou H, Proia P, Lin YL, De Paula AM, et al. Plexin-B1 and semaphorin 4D cooperate to promote perineural invasion in a RhoA/ROK-dependent manner. Am J Pathol. 2012;180:1232–42. doi: 10.1016/j.ajpath.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 126.Swanson BJ, McDermott KM, Singh PK, Eggers JP, Crocker PR, Hollingsworth MA. MUC1 is a counter-receptor for myelin-associated glycoprotein (Siglec-4a) and their interaction contributes to adhesion in pancreatic cancer perineural invasion. Cancer Res. 2007;67:10222–9. doi: 10.1158/0008-5472.CAN-06-2483. [DOI] [PubMed] [Google Scholar]
- 127.Gohrig A, Detjen KM, Hilfenhaus G, Korner JL, Welzel M, Arsenic R, et al. Axon guidance factor SLIT2 inhibits neural invasion and metastasis in pancreatic cancer. Cancer Res. 2014;74:1529–40. doi: 10.1158/0008-5472.CAN-13-1012. [DOI] [PubMed] [Google Scholar]
- 128.Sroka IC, Anderson TA, McDaniel KM, Nagle RB, Gretzer MB, Cress AE. The laminin binding integrin alpha6beta1 in prostate cancer perineural invasion. J Cell Physiol. 2010;224:283–8. doi: 10.1002/jcp.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ben QW, Wang JC, Liu J, Zhu Y, Yuan F, Yao WY, et al. Positive expression of L1-CAM is associated with perineural invasion and poor outcome in pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2010;17:2213–21. doi: 10.1245/s10434-010-0955-x. [DOI] [PubMed] [Google Scholar]
- 130.Basile JR, Holmbeck K, Bugge TH, Gutkind JS. MT1-MMP controls tumor-induced angiogenesis through the release of semaphorin 4D. J Biol Chem. 2007;282:6899–905. doi: 10.1074/jbc.M609570200. [DOI] [PubMed] [Google Scholar]
- 131.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 132.Friedl P, Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 2008;68:7247–9. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- 133.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–57. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 134.Burden-Gulley SM, Qutaish MQ, Sullivant KE, Lu H, Wang J, Craig SE, et al. Novel cryo-imaging of the glioma tumor microenvironment reveals migration and dispersal pathways in vivid three-dimensional detail. Cancer Res. 2011;71:5932–40. doi: 10.1158/0008-5472.CAN-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Theveneau E, Mayor R. Cadherins in collective cell migration of mesenchymal cells. Curr Opin Cell Biol. 2012;24:677–84. doi: 10.1016/j.ceb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Canel M, Serrels A, Miller D, Timpson P, Serrels B, Frame MC, et al. Quantitative in vivo imaging of the effects of inhibiting integrin signaling via Src and FAK on cancer cell movement: effects on E-cadherin dynamics. Cancer Res. 2010;70:9413–22. doi: 10.1158/0008-5472.CAN-10-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]