Abstract
Background & Aims
Phosphoinositides (PIs) bind and regulate localization of proteins via a variety of structural motifs. PI 4,5-bisphosphate (PI[4,5]P2) interacts with and modulates the function of several proteins involved in intracellular vesicular membrane trafficking. We investigated interactions between PI(4,5)P2 and hepatitis C virus (HCV) nonstructural protein 5A (NS5A) and effects on the viral life cycle.
Methods
We used a combination of quartz crystal microbalance (QCM), circular dichroism, molecular genetics, and immunofluorescence to study specific binding of PI(4,5)P2 by the HCV NS5A protein. We evaluated the effects of PI(4,5)P2 on NS5A’s function by expressing wild-type or mutant forms of Bart79I or FL-J6/JFH-5’C19Rluc2AUbi21 RNA in Huh7 cells. We also studied the effects of strategies designed to inhibit PI(4,5)P2 on HCV replication in these cells.
Results
The N-terminal amphipathic helix of NS5A bound specifically to PI(4,5)P2, inducing a conformational change that stabilized the interaction between NS5A andTBC1D20, which is required for HCV replication. A pair of positively charged residues within the amphipathic helix (the BAAPP domain) was required for PI(4,5)P2 binding and replication of the HCV RNA genome. A similar motif was found to be conserved across all HCV isolates, as well as amphipathic helices of many pathogens and apolipoproteins.
Conclusions
PI(4,5)P2 binds to HCV NS5A to promote replication of the viral RNA genome in hepatocytes. Strategies to disrupt this interaction might be developed to inhibit replication of HCV and other viruses.
Keywords: QCM, antiviral strategies, signaling molecule, phospholipid
Introduction
Hepatitis C virus (HCV) is an important cause of chronic liver disease. Many key details of the viral life cycle remain unknown. The HCV non-structural protein 5A (NS5A) harbors an N-terminal amphipathic helix (AH) that is necessary and sufficient for mediating NS5A’s association with cellular-derived and model membranes 1-3. This membrane association appears to involve a host cell membrane protein partner 2 and is essential for membrane-associated viral RNA replication 1, 3. NS5A has also been found to interact with regulators of host cell vesicular membrane trafficking machinery 4, 5. Phosphoinositides (PIs) have long been known to mediate key intracellular signaling pathways 6-8, and more recently have also been recognized as playing important roles in the subcellular localization of PI-interacting proteins that bind PIs via a variety of structural motifs 9, 10. In particular, PIs such as phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) are recognized by, and can modulate the function of, several proteins involved in intracellular vesicular membrane trafficking 11, 12. We therefore hypothesized that the NS5A AH might also bind PIs and that this interaction is essential for viral replication. Indeed we found that while the AH’s amphipathic nature is the primary determinant of NS5A’s association with lipid membranes, the NS5A AH also harbors a motif that specifically binds PI(4,5)P2, that such binding induces a key conformational change within the AH, and that PI(4,5)P2 binding is essential for viral genome replication. Moreover, this novel PI(4,5)P2-binding motif is found in a variety of pathogens. These results provide a molecular explanation for recent reports 13-16 of PI-kinases involvement in the HCV life cyle, and suggest novel potential antiviral therapeutic strategies.
Materials and Methods
Vesicles Containing Polymerized PIPs
The molar ratios of PIPs -diyne, PC-diyne, and PE-diyne (5:65:30 molar ratios) were combined in an 8 mL glass vial, to which one ml chloroform was added, followed by drying on a SpeedVac overnight. (Multiple vials of dried lipid mixture were prepared in advance and stored at −20°C until needed). For each □mol of total lipid, PBS buffer (150 mM NaCl, pH 7.5) made with Milli-Q water was added to make a 1 mM final solution. Probe sonication (~2-3 watts) at 60°C for ~60 minutes was followed by extrusion through 400 nm polycarbonate track-etched membrane using the conventional vesicle extrusion method. The solution was transferred to a beaker and placed in a slightly larger beaker in an ice bath, followed by irradiation for 60-90 minutes with a hand-held UV lamp (254 nm) placed directly on top of the larger beaker, with the solution turning faintly red. Resulting PIPs-containing vesicles were characterized by their average size distribution and zeta potential with dynamic light scattering and zeta potential measurements, respectively.
Peptides
The peptides used in this study were synthesized by Anaspec Corporation (San Jose, CA, USA). The sequences of the non-structural 5A (NS5A) protein derived amphipathic α-helical peptide (AH) and non-amphipathic non-helical control peptide (NH) are H-Ser-Gly-Ser-Trp-Leu-Arg-Asp-Val-Trp-Asp-Trp-Ile-Cys-Thr-Val-Leu-Thr-Asp-Phe-Lys-Thr-Trp-Leu-Gln-Ser-Lys-Leu-Asp-Tyr-Lys-Asp-NH2 and H-Ser-Gly-Ser-Trp-Leu-Arg-Asp-Asp-Trp-Asp-Trp-Glu-Cys-Thr-Val-Leu-Thr-Asp-Asp-Lys-Thr-Trp-Leu-Gln-Ser-Lys-Leu-Asp-Tyr-Lys-Asp-NH2, respectively (the introduced charged amino acids differentiating NH from AH are indicated in bold underline). Synthesized BAAPP domain mutant AH peptides have the same sequence as AH except for substitution of alanine for lysine at position 20 (AH-K20A), 26, (AH-K26A), or both (AH-K20AK26A).
Dynamic Light Scattering
Dynamic light scattering (DLS) was performed on a 90Plus Particle Size Analyzer and the results were analyzed by digital autocorrelation software (Brookhaven Instruments Corporation, New York, USA). All measurements were taken at a scattering angle of 90°so that the reflection effe ct is minimized. All autocorrelation functions obtained were also analyzed by CONTIN and Non-Negatively Constrained Least Squares (NNLS) algorithms to check for multimodal distributions.
E4 Quartz Crystal Microbalance-Dissipation (E4 QCM-D)
QCM-D measurements were performed essentially as previously described 2, and as detailed in the Supplementary Information. We employed the Sauerbrey equation 38 to convert frequency to a real mass of bound peptide. All experiments were repeated at least three times, with a standard deviation of less than 5 %. For QCM-D experiments involving ER membranes, the latter were purchased from Celsis In Vitro Technologies and enriched by incubation with solutions of either PI, PI(3,5)P2, or PI(4,5)P2 prior to loading into the QCM chambers.
Circular Dichroism (CD) Measurements
Circular dichroism (CD) measurements were carried out as described in the Supplementary Information. The scans obtained with ellipticity (Θ) were converted to mean molar residue ellipticity ([Θ]) as previously described 17. Spectra were processed with CD6 software, baseline corrected, and smoothed using a third-order least square polynomial fit. The fraction helicity (fH) of peptide in solution was determined as previously described 18.
Plasmids
Bart79I, a high-efficiency subgenomic replicon of HCV 19, harbors the neomycin resistance gene (neo) and the HCV nonstructural proteins. The Bart-Luciferase plasmid, Bart79I-luc, was cloned from the Bart79I parent 3 and the pGL3-Basic parent (Promega), as described in the Supplementary Information. FL-J6/JFH-5'C19Rluc2AUbi--a monocistronic, full-length HCV genome that expresses Renilla luciferase (Rluc) and was derived from the previously described infectious genotype 2a HCV genome J6/JFH1 20--was a gift from Dr. Charles Rice at Rockefeller University 21. The nucleotide sequence AAG that encodes for lysine at amino acid positions 20 and 26 of NS5A was changed to GCG (encoding for alanines) through the use of Quick-Change™XL site-directed mutagenesis kit (Stratagene, La Jolla,CA) as described by the manufacturer and confirmed by sequencing.
Immunofluorescence Microscopy
Was performed as described in the Supplementary Information.
Purification of Recombinant HCV NS5A and TBC1D20
Full length NS5A and TBC1D20 proteins were purified from bacteria as described elsewhere 22, 23.
Colony Formation Assays
These were performed using 5 micrograms of in vitro-transcribed wild type and mutant Bart79I RNAs as previously described 3 and detailed in the Supplementary Information.
Viral Sequencing Analysis
TRIzol reagent (Gibco BRL) was used to extract total RNA from the pool containing the Huh7 cells that survived the colony formation assays following electroporation with in vitro-transcribed wild type or mutant Bart79I RNAs, and the HCV RNA sequences were determined as described in the Supplementary Information.
Transient Replication Assays
Wild type or BAAPP domain mutant versions of Bart79I-luciferase or FL-J6/JFH-5'C19Rluc2AUbi 21 RNAs were electroporated into Huh7 cells followed by determination of luciferase activity at 8, 48, 96, and 144 hr post electroporation as described in the Supplementary Information.
Cell Viability Assays
Cells were incubated with cell culture media containing 10 % alamarBlueTM (Biosource International, Inc., Camarillo, CA) for 2 hours. Relative cell viabilities were compared by measuring the absorbance of cell culture media at 544 nm.
Identification of Putative BAAPP Domain-containing Proteins
Putative BAAPP domains were identified in the following manner: protein sequences were analyzed using Jpred3 24, to predict their secondary structure. Regions found to form alpha helices were then plotted in a helix wheel format (http://www.tcdb.org/progs/pepwheel.php), to determine if the helix contained a hydrophobic face and positive charges (basic amino acids lysine (K), arginine (R), or histidine (H)) that flanked the hydrophobic face.
Results
The NS5A amphipathic helix specifically binds PI(4,5)P2.
To test the hypothesis that the NS5A AH binds PIs, we determined the ability of a synthetic peptide corresponding to the NS5A AH to bind to polymerized lipid vesicles containing different PIs, using the quartz crystal microbalance with dissipation monitoring (QCM-D) technique 2 (see also supplementary Fig. 1). QCM-D provides for very sensitive and reproducible real-time measurements of mass binding to a target oscillating quartz crystal. The greater the mass bound, the greater the recorded decrease in resonating frequency of the quart crystal nanosensor 25, 26. For our QCM-D assay, target polymerized lipid vesicles are first deposited on an oscillating SiOx quartz crystal nanosensor. The binding mass of peptide to this vesicle platform is then determined as a function of the change in resonance frequency of the piezoelectric AT-cut crystal 25, 26. As shown in Fig. 1a, significant binding of the NS5A AH to PI(4,5)P2-containing vesicles was observed. To determine the specificity of the observed binding, lipid vesicles containing phosphatidylinositol (PI), phosphatidylinositol 3-phosphate (PI(3)P), phosphatidylinositol 4-phosphate (PI(4)P), phosphatidylinositol 5-phosphate (PI(5)P), phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2), phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), or phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3),were used in parallel assays. As shown in Fig. 1a, the NS5A AH exhibits a high degree of binding specificity for PI(4,5)P2. Related phosphatidylinositol bisphosphates such as PI(3,4)P2 and PI(3,5)P2, which have a similar number of negative charges to PI(4,5)P2, exhibited only minimal binding to the NS5A AH. This demonstrates that the interaction of the NS5A AH with PI(4,5)P2 is not derived simply from a positive electrostatic attraction for multivalent, negatively charged phosphatidylinositol bisphosphates. Further, no specific binding to the NS5A AH was observed with other phosphoinositides containing 1 or 3 phosphate-modified inositol headgroups, such as PI(3)P, PI(4)P, PI(5)P, and PI(3,4,5)P (see Fig. 1a). Readily-available control proteins reported to have binding specificity for some of these PI substrates did, however, perform as expected (see Fig 1b). QCM-D monitoring allows for determination of binding kinetics, and the Kd for the NS5A AH association with PI(4,5)P2 vesicles was found to be 4.5 μM (see Supplementary Fig. 2). By comparison, the Kd for the ~120 amino acid pleckstrin homology (PH) domain--one of the best characterized to date PI(4,5)P2, binding domains--has been reported to be quite similar (2 μM ~ 30 μM) 27, 28.
Fig. 1. The HCV NS5A AH specifically binds lipid vesicles containing PI(4,5)P2.
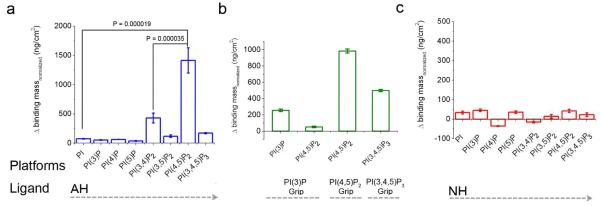
QCM-D measurements, quantifying mass changes due to the binding of the NS5A AH peptide to vesicles containing various PIs. The system consists of polymerized vesicles containing the PIs on a SiOx solid substrate. We employed the Sauerbrey equation 41 to convert frequency to a real mass of bound peptide. (a) The NS5A AH peptide (~ 8 μM) binds significantly more to vesicles containing PI(4,5)P2 than those containing PI, PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2 or PI(3,4,5)P3, where the lower amount of binding is consistent with prior work using model lipid vesicles alone 2. (b) As positive controls, we used three different GripTM (Echelon Biosciences Inc.) proteins that are known to bind PI(3)P, PI(4,5)P2, and PI(3,4,5)P3, respectively. PI(3)P Grip contains recombinant p40PX domain GST-tagged protein, PI(4,5)P2 Grip contains recombinant PLC-δ1 PH domain GST-tagged protein, and PI(3,4,5)P3 Grip contains recombinant GRP1-PH domain GST-tagged protein 42. (c) As a negative control, we employed the NH peptide, in which three point mutations were introduced into the NS5A AH to disrupt amino acids on the hydrophobic face 3. Notably, the NH peptide shows no significant binding to any of the target lipid vesicles. Error bars represent standard deviations, p values from Student’s T test. Please note the scale differences between the figures.
In contrast, a negative control peptide (termed NH), derived from the NS5A AH in which three charged point mutations were introduced into the hydrophobic face, exhibited minimal binding to any of the PI-containing substrates (Fig. 1c).
To provide additional evidence supporting the specific binding of the NS5A AH to PI(4,5)P2, and to confirm that the results obtained with the isolated NS5A AH peptide appropriately reflect the behavior of this AH segment when present in the context of the full length NS5A protein, we assessed the binding of full length NS5A to PI(4,5)P2 and related phosphoinositides (PI(3,4)P2 and PI(3,5)P2,). As shown in Supplementary Figure 3, the full length NS5A protein exhibits substantial binding to PI(4,5)P2 . Again, as in the experiments with peptides, preferential binding to PI(4,5)P2 over the other PI-bisphosphate isoforms was observed.
NS5A binds PI(4,5)P2 via a novel structural motif, termed a “BAAPP domain.”
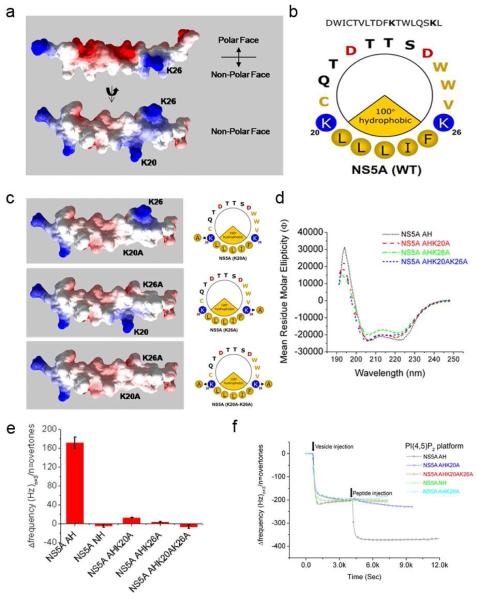
Although a variety of structural motifs are known to bind PI(4,5)P2,9, the NS5A AH does not appear to conform to any of these. As shown in Fig. 2a and 2b, inspection of the NS5A AH revealed a pair of basic amino acids (Lys 20 and Lys 26) that flank the hydrophobic face of the AH. As such, they are oriented towards the lipid bilayer with which the AH likely interacts in a monotypic fashion29. We hypothesized that these positively charged lysines might be ideally suited to interact with the negatively charged phosphates of the PI(4,5)P2 lipid headgroups. To test this hypothesis, we synthesized mutant versions of the NS5A AH peptide in which one or both of these lysines were mutated to alanines (Fig. 2c). CD measurements confirmed that these mutations did not alter the helical nature of the AH (Fig. 2d). These mutations did, however, dramatically impair the ability of the corresponding peptides to bind PI(4,5)P2-containing vesicles (Fig. 2e and f). Taken together, these results suggest that the PI(4,5)P2 binding domain in the NS5A AH represents a novel structural motif for PI(4,5)P2 binding that we term the Basic Amino Acid PI(4,5)P2 Pincer or “BAAPP” domain.
Fig. 2. PI(4,5)P2 binding to the NS5A AH is mediated by a pair of conserved positively charged amino acids, or Basic Amino Acid PI(4,5)P2 Pincer (BAAPP) domain.
(a) A molecular surface model of the BAAPP domain of the NS5A AH (genotype 1b) was created using the program DeepView / Swiss-PdbViewer (V3.7) 43, based on the NMR structure 1R7C. The surface is colored by electrostatic potential, which was calculated using default parameters and including only charged residues. Blue denotes positive electrostatic potentials, white denotes neutral potentials, and red denotes negative electrostatic potentials. (b) Helix wheel plot of the BAAPP domain in the NS5A AH, with hydrophobic face denoted by the yellow pie slice and filled yellow circles. Positively charged residues that flank the hydrophobic face are indicated by the filled-in blue circles. (c) Left, molecular surface models of BAAPP mutants in the NS5A AH. Mutations (as indicated, K20A, K26A, and K20AK26A) in the molecular structure were modeled using DeepView. Right, helix wheel plots of BAAPP mutants in the NS5A AH. (d) Far-UV circular dichroism (CD) analyses of the NS5A AH and the single mutant (K20A and K26A) and double-mutant (K20AK26A) peptides. The CD spectra were recorded in 10 mM PBS buffer, pH 7.5. (e) The binding kinetics of the NS5A AH peptide and mutant variants (~ 19 μM) to PI(4,5)P2 containing vesicles, using the same technique as in Fig. 1. (f) A comparison of the total frequency changes (Δf) due to binding of NS5A AH peptide and mutant variants, as presented in (e).
HCV replication sites are localized to sites enriched in PI(4,5)P2.
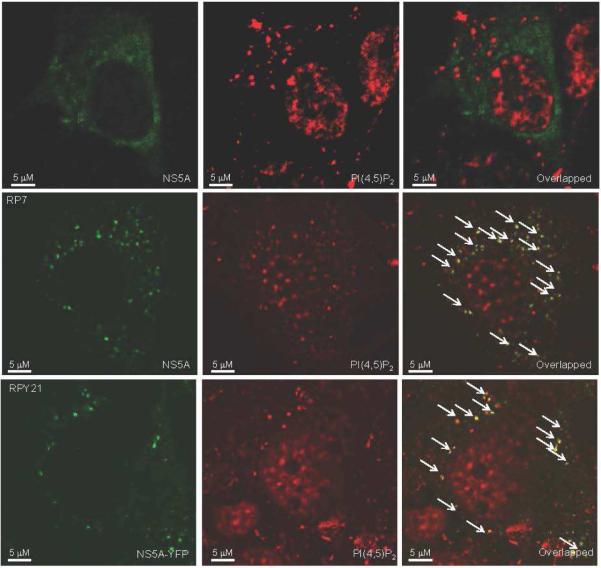
Because lysines 20 and 26 are highly conserved across HCV isolates, in spite of a fair amount of sequence variation within the AH, this suggested that PI(4,5)P2 binding by the NS5A AH is important for the HCV life cycle. We hypothesized that NS5A and PI(4,5)P2 might therefore be colocalized at the site of viral replication. As shown in Fig. 3, similar to previous work 30, a monoclonal antibody against PI(4,5)P2 demonstrates both a nuclear staining pattern and distinct small speckles distributed in the cytoplasm. Because HCV replication is known to be localized to similar appearing cytoplasmic speckle-like structures, we hypothesized the latter might now be found to be enriched in PI(4,5)P2. Indeed, in cells harboring HCV replicons, significant colocalization of NS5A with PI(4,5)P2 was observed (Fig. 3a). To eliminate the possibility of antibody-specific effects, we performed similar experiments with a YFP-tagged version of NS5A (Fig. 3b), which showed identical results, with over 90% of the NS5A-staining speckles found to co-stain with the anti-PI(4,5)P2 antibody. Expression of just an isolated NS5A failed to co-localize with the PI(4,5)P2 speckles (Fig. 3c). Moreover, the intracellular distribution of NS5A was unaffected by mutation of the BAAPP domain (Supplementary Figure 4A). Therefore, NS5A with an intact BAAPP domain appears to co-localize with PI(4,5)P2 only in the context of a replicating genome where NS5A is localized to HCV replication complex sites. This suggests that HCV replication complexes are established at PI(4,5)P2 sites or the HCV replication complex promotes formation of PI(4,5)P2 sites. Either way, PI(4,5)P2 represents a new marker for HCV replication complexes. These results also suggest that PI(4,5)P2 binding by NS5A might mediate an interaction important for viral replication. We hypothesized that such an interaction might involve a PI(4,5)P2-induced conformational change in the NS5A AH.
Fig. 3. NS5A colocalizes with PI(4,5)P2 at sites of HCV replication.
Subcellular localization of PI(4,5)P2 was examined by immunofluorescence using a monoclonal antibody specific for PI(4,5)P2 and Alexa 594-conjugated secondary antibody in (a) RP7 cells harboring Bart79I replicating HCV subgenomic replicons, where NS5A was detected with a mouse monoclonal antibody specific for NS5A and Alexa 488-conjugated secondary antibody, and in (b) RPY21 cells harboring replicating HCV replicons with an YFP-tagged version of NS5A . White arrows indicate sites of colocalization of NS5A and PI(4,5)P2 , shown in yellow. Quantitation of confocal images revealed that 91% of NS5A-staining speckles co-stain with the anti-PI(4,5)P2 antibody (c) When expressed alone off of a plasmid vector, NS5A fails to colocalize with the PI(4,5)P2 speckles. Scale bar = 5μM. Magnification 100X.
PI(4,5)P2 binding induces an important conformational change in NS5A
To test the above hypothesis, CD measurements of the NS5A AH peptide were performed in the presence or absence of PI(4,5)P2-containing lipid vesicles. As shown in Fig. 4a, a dramatic alteration of the helical structure of the NS5A AH was observed upon interaction with PI(4,5)P2. No such changes were noted with vesicles only or vesicles containing PI(3,4)P2. Therefore, PI(4,5)P2 binding appears to mediate a conformational change in the NS5A AH.
Fig. 4. PI(4,5)P2 induces a conformational change in the NS5A AH and stabilizes the NS5A AH-mediated interaction with TBC1D20.
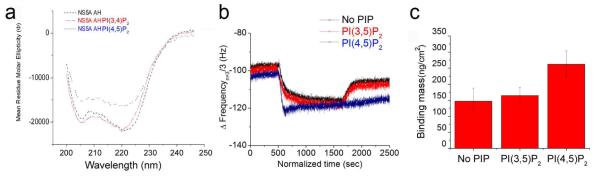
(a) Far-UV circular dichroism (CD) analyses of the NS5A AH peptide in the presence or absence of PI(4,5)P2 containing polymerized vesicles, demonstrating that a conformational change occurs upon addition of PI(4,5)P2 containing vesicles, but not PI(3,4)P2 containing vesicles. (b) The normalized frequency change (Δf) upon addition of TBC1D20 to three different platforms containing human liver derived microsomal membranes--as is, or after enrichment with either PI(3,5)P2 or PI(4,5)P2—followed by preloading of NS5A prior to addition of TBC1D20 (each flowed into the QCM-D chambers at ~1μg/ml). The kinetics of the frequency change (Δf) depict that human liver derived membranes alone, or following PI(3,5)P2 enrichment, fail to promote stable association of TBC1D20 with NS5A full length protein. The PI(4,5)P2 enriched membrane, however, promoted a strong interaction of NS5A with TBC1D20, so that there was no such dissociation. (c) Calculated bound mass density from binding of TBC1D20 to NS5A on the three different platforms after equilibration (2500 sec).
NS5A has been reported to interact with a variety of host proteins. Among these is TBC1D20, a GTPase activating protein for Rab1 5, 23. TBC1D20 is required for efficient HCV replication and the interaction between TBC1D20 and NS5A is mediated by the latter’s N-terminal AH 5. To test the hypothesis that the PI(4,5)P2-induced AH conformational change might regulate the NS5A’s interaction with TBC1D20, we performed QCM-D binding experiments of TBC1D20 to NS5A bound to human liver-derived microsomal membranes, with or without prior PI(4,5)P2 enrichment. As shown in Fig. 4b, the kinetics of the TBC1D20:NS5A interaction exhibited a striking dependence on PI(4,5)P2. In particular, the rate of TBC1D20 absorption to membrane bound NS5A was specifically increased when the membranes were enriched in PI(4,5)P2 . Moreover, the rate of TBC1D20 desorption from NS5A was even more dramatically affected when the membranes were enriched in PI(4,5)P2 , as opposed to the control membranes enriched with PI(3,5)P2. This is reflected in the corresponding calculated values for the respective kon and koff rate constants, which together indicate a prolonged and stable interaction between bound TBC1D20 and the membrane-bound NS5A in the presence of PI(4,5)P2 (Fig. 4b, Supplementary Fig. 5). That no such effect was observed upon substitution of PI(3,5)P2 for PI(4,5)P2, indicates the selectivity of PI(4,5)P2’s effect on the TBC1D20:NS5A interaction. The binding mass result demonstrated that PI(4,5)P2 promotes the binding of TBC1D20 to NS5A up to 57 % more on PI(4,5)P2 enriched membranes than on PI(3,5)P2 enriched membranes (Fig. 4c). There were no differences in TBC1D20 binding to PI(4,5)P2 enriched vs. unenriched membranes in the absence of prior NS5A addition (Supplementary Fig. 6).
The NS5A BAAPP domain mediates a critical role in HCV genome replication
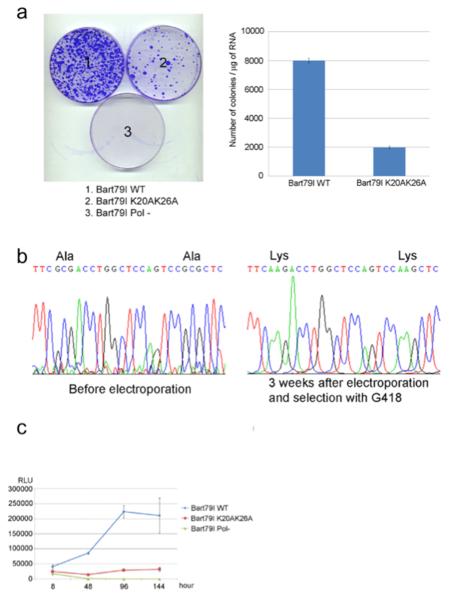
To test the hypothesis that the ability of NS5A to engage in the interaction with PI(4,5)P2 is essential for genome replication, we first performed standard HCV colony formation assays using high efficiency second generation replicons encoding wild-type or mutant (K20AK26A) NS5A 19. As shown in Fig. 5a, while the wild-type replicon yielded numerous colonies and the negative control replicon containing a lethal mutation in the polymerase gene yielded none, ~75% fewer colonies were obtained with the K20AK26A mutant replicon compared to the wild-type one. When the replicons in the colonies surviving on the mutant plate were sequenced, they were found to have reverted to wild-type during the ~3 week selection process (Fig. 5b). This suggests that (a) such reversion was essential for viral fitness, and (b) the ability of NS5A to bind PI(4,5)P2 is important for efficient genome replication. To directly test this hypothesis, we performed transient HCV replication assays with luciferase reporter-linked wild-type or K20AK26A mutant replicons. As shown in Fig. 5c, mutation of the PI(4,5)P2 interaction domain of the AH NS5A indeed severely impaired HCV genome replication. Similar results were observed upon introduction of these BAAPP domain mutations into an infectious clone of HCV (Supplementary Figure 7). Although PI(4,5)P2 interaction with the HIV Gag matrix protein has been implicated in virus particle assembly 31 to our knowledge, this is the first example of PI(4,5)P2 mediating viral genome replication.
Fig. 5. PI(4,5)P2 mediates HCV RNA genome replication.
(a) Colony formation assay. Huh7 cells were electroporated with 5 μg of in vitro-transcribed wild type Bart79I, mutant Bart79I encoding NS5A (K20AK26A), or Bart79I (Pol-) RNAs followed by selection with 750 Dg/ml of G418 for three weeks. Surviving colonies were stained with crystal violet and the number of colonies was counted from three different plates to calculate average number of colonies and standard deviation. (b) Reversion to wild-type sequence. Left panel shows the sequence of input mutant (K20AK26A) HCV replicon RNA. Sequence analysis of replicon RNA isolated from colonies growing on mutant plate from colony formation assay is shown in the right panel. Note that the mutant GCG alanine-encoding codons at positions 20 and 26 of NS5A have reverted back to the wild-type AAG sequences encoding lysine residues. (c) Luciferase reporter-linked transient HCV replication assay. Huh7 cells were electroporated with 10 μg of in vitro-transcribed wild type Bart79I-luc, mutant Bart79I-luc (K20AK26A), or Bart79I-luc (Pol-) RNAs. Firefly luciferase activities were measured at 8, 48, 96, and 144 hours post electroporation. RLU stands for relative light unit.
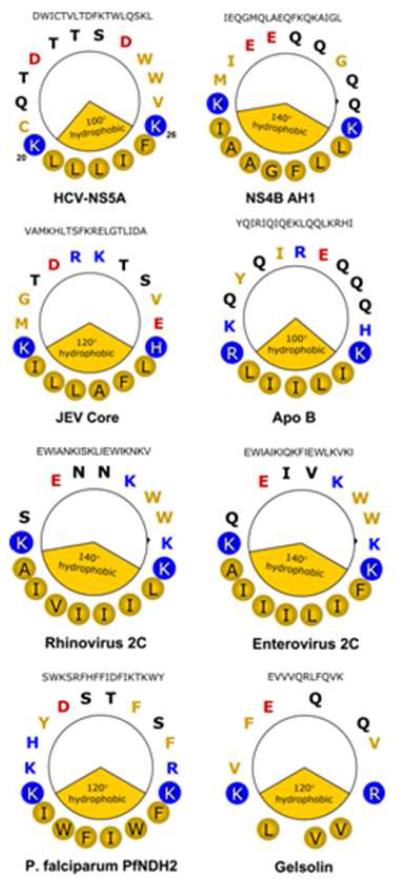
Finally, an analysis of sequences from a variety of organisms revealed that putative BAAPP domains are present in other viral proteins (see Fig. 6). BAAPP domains are also found in a variety of host proteins, including apolipoproteins (Fig. 6). Analysis of several of these BAAPP domains using methods analogous to those described here (e.g., Figs. 1 and 2) reveals that they also mediate binding to PI(4,5)P2 (see Supplementary Fig. 8)
Fig. 6. A Basic Amino Acid PI(4,5)P2 Pincer (BAAPP) domain is found in the AHs of other pathogens and host cell proteins.
Putative Basic Amino Acid PI(4,5)P2 Pincer (BAAPP) domain in the AHs of other proteins. Hydrophobic face denoted by the yellow pie slice and filled yellow circles. Positively charged residues that flank the hydrophobic face are indicated by the filled-in blue circles, with blue denoting that they are positively charged residues. Abbreviations: HCV: Hepatitis C Virus; NS5A: Non-Structural Protein 5A; NS4B AH1: Non-Structural Protein 4B, Amphipathic Helix 1; JEV: Japanese Encephalitis Virus; Apo: Apolipoprotein.
Discussion
The above experiments reveal an exciting role for phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) in the HCV life cycle and provide intriguing insights into the relevant molecular mechanism of action. In particular, we discovered that the HCV NS5A amphipathic helix specifically binds PI(4,5)P2 (Fig. 1) and that this binding is mediated by a novel structural motif, the BAAPP domain (Fig. 2a and 2b). PI(4,5)P2 binding induces an important conformational change in NS5A ( Fig. 4a), affecting the latter’s interaction with a key regulator of HCV replication (Fig. 4b). Importantly, disrupting the ability of the BAAPP domain to bind PI(4,5)P2 profoundly impairs HCV genome replication ( Fig.5). This is the first example of PI(4,5)P2 mediating the replication of a viral genome, yet the apparent widespread presence of BAAPP domains in pathogen proteins (Fig. 6) suggests that BAAPP domain-mediated PI(4,5)P2 binding plays equally important roles in other pathogen life cycles, and that our findings might be translated into novel anti-infective therapies.
Previous work implicated the involvement of a host cell membrane protein(s) (the identity of which remains unknown) in NS5A’s binding to intracellular membranes, as well as an inherent membrane associating activity of the NS5A AH with model membranes 2. Consistent with this previous work 2, we observed significant binding of the NS5A AH to model lipid vesicles alone (Fig. 1), which reflects the strong membrane associating activity provided by this amphipathic segment of NS5A. The current findings, however, also extend our understanding of NS5A’s interaction with membranes. In particular, we discovered that the NS5A AH can specifically bind PI(4,5)P2--both in the context of an isolated AH peptide (Fig. 1) and when contained within full length NS5A (Supplementary Fig. 3). The role of this PI(4,5)P2 binding does not appear to be a primary determinant for either NS5A membrane association or intracellular localization (Fig. 3, Supplementary Fig. 4). Rather, PI(4,5)P2 binding induces a conformational change within the AH (Fig. 4a) that alters the interaction with a key NS5A binding partner (Fig. 4b and c).
Our results also provide new insights into, and a molecular mechanism to account for, the relevance of recent data showing the sensitivity of HCV replication to siRNA-mediated knockdown of PI4K-IIIalpha and PI4K-IIIbeta, the reported interaction with, and activation of, these PI(4)P-generating kinases by viral proteins 13, 14, 16, 32, 33, as well as the heretofore unexplained role for the reported presence of PI(4)P at sites of HCV replication 34. In particular, in addition to PI4 kinases involvement in recruiting PI4P-binding proteins such as OSBP 35, and modulation of NS5A phosphorylation 36, our results suggest that the source of the PI(4,5)P2 relied upon by HCV is likely derived via the PI(4)P produced by one or both of these members of the PI4-kinases family. Together, the above suggests a model whereby the HCV polyproteins initially become membrane associated via PI(4,5)P2-independent mechanisms, and subsequently HCV replication complexes are established at sites that become locally enriched in PI(4,5)P2 (likely via hijacking of the requisite PI-kinases to the future replication sites by NS5A (e.g. 13, 16) and possibly other proteins). This latter PI(4,5)P2 is sensed by NS5A, triggering a key regulatory switch. This model is supported by the data demonstrating that NS5A is seen associated with PI(4,5)P2 only in the context of active replication (see Fig. 3), PI(4,5)P2 binding by NS5A induces a conformational change in the NS5A AH (see Fig. 4a), and that interaction of PI(4,5)P2 with NS5A regulates NS5A’s association with TBC1D20 (see Fig. 4b and c)—a host cell partner required for viral genome replication 5.
Importantly, NS5A is not the only protein containing a BAAPP domain. Residues 150-169 of the host protein gelsolin have previously been identified as comprising a structurally unclassified type of PI(4,5)P2 binding domain 37. NMR studies have demonstrated that this region forms an AH that is capable of binding PI(4,5)P2,38. This information, along with a helix wheel representation of this region, reveals that this region can now be classified as a BAAPP domain (Fig. 6).
Moreover, we speculate that the BAAPP domains we have identified in apolipoproteins (see Figure 6) may mediate interaction with PI(4,5)P2 and that this interaction plays an important role in the genesis of certain lipoprotein particles, with localized PI(4,5)P2 domains representing a common platform for the initial stages of VLDL lipoprotein and HCV particle assembly. In particular, HCV may either compete for or hijack limiting components of host cell PI(4,5)P2-associated machinery to help effectuate viral assembly. This could account for the reciprocal relationship observed between serum levels of VLDL and HCV titer before and after successful treatment of HCV (39, personal observations).
Finally, these results suggest a variety of new potential antiviral strategies. For example, neomycin is known to be a ligand of PI(4,5)P2 40. As such, either neomycin or analogues thereof could be considered as inhibitors of PI(4,5)P2-BAAPP interactions. A complementary approach would be to pharmacologically deplete PI(4,5)P2 at HCV replication sites, such as by stimulating a phosphatase involved in PI(4,5)P2 degradation, or inhibiting a kinase upon which HCV depends for its source of PI(4,5)P2. Moreover, because there appear to be back-up mechanisms to such contemplated disruption of PI(4,5)P2 metabolism that are available to the host but not the virus, relevant inhibitors with sufficient therapeutic indices can be readily contemplated. Such inhibitors might represent a valuable new class of antiviral agents to be included in future therapeutic cocktails designed to maximize pan-genotypic efficiency of, and minimize resistance to, therapies for treating hepatitis C or other BAAPP domain harboring pathogens.
Supplementary Material
Acknowledgments
see Supplementary Information.
Grant support: Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (to JSG); NIH RO1 DK064223, RO1 AI087917, RO1 AI099245 and U19 AI109662; American Liver Foundation Postdoctoral Fellowship Award; NIH T32 AI070502, T32 GM007365 and T32 AI007328; Stanford Dean’s Fellowship; Israel Science Foundation Bikura Postdoctoral Fellowship; and a Stanford Digestive Disease Center Pilot Study Award.
Abbreviations
- PIs
phosphoinositides
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- AH
amphipathic helix
- QCM
quartz crystal microbalance
- BAAPP
Basic Amino Acid PI(4,5)P2 Pincer
- PI4K-III
Type-III Phosphatidylinositol 4-Kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: Patent applications covering this technology and antiviral approaches have been filed by Stanford University. The authors have no other conflicts to declare.
References
Author names in bold designate shared co-first authorship.
- 1.Brass V, Bieck E, Montserret R, et al. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem. 2002;277:8130–9. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 2.Cho N-J, Cheong KH, Lee C, et al. Binding dynamics of hepatitis C virus’ NS5A amphipathic peptide to cell and model membranes. J. Virol. 2007;81:6682–9. doi: 10.1128/JVI.02783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elazar M, Cheong KH, Liu P, et al. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 2003;77:6055–6061. doi: 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A. 2004;101:13038–43. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sklan EH, Staschke K, Oakes TM, et al. A Rab-GAP TBC domain protein binds hepatitis C virus NS5A and mediates viral replication. J Virol. 2007;81:11096–105. doi: 10.1128/JVI.01249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 7.Nishizuka Y. Turnover of inositol phospholipids and signal transduction. Science. 1984;225:1365–70. doi: 10.1126/science.6147898. [DOI] [PubMed] [Google Scholar]
- 8.Toker A. Phosphoinositides and signal transduction. Cell Mol Life Sci. 2002;59:761–79. doi: 10.1007/s00018-002-8465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci. 2005;118:2093–104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 10.De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–92. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 11.Itoh T, Koshiba S, Kigawa T, et al. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–51. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 12.Lee E, Marcucci M, Daniell L, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–6. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 13.Ahn J, Chung KS, Kim DU, et al. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J Biochem Mol Biol. 2004;37:741–8. doi: 10.5483/bmbrep.2004.37.6.741. [DOI] [PubMed] [Google Scholar]
- 14.Berger KL, Cooper JD, Heaton NS, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–82. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu NY, Ilnytska O, Belov G, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiss S, Rebhan I, Backes P, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Marco V, De Marco A, Goldie KN, et al. Hoenger A. Dimerization properties of a Xenopus laevis kinesin-II carboxy-terminal stalk fragment. EMBO Rep. 2003;4:717–22. doi: 10.1038/sj.embor.embor884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JS, Scholtz JM. Energetics of polar side-chain interactions in helical peptides: salt effects on ion pairs and hydrogen bonds. Biochemistry. 1998;37:33–40. doi: 10.1021/bi972026h. [DOI] [PubMed] [Google Scholar]
- 19.Blight KJ, Kolykhalov A, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2001;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005 doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 21.Tscherne DM, Jones CT, Evans MJ, et al. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–41. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L, Sineva EV, Hargittai MR, et al. Purification and characterization of hepatitis C virus non-structural protein 5A expressed in Escherichia coli. Protein Expr Purif. 2004;37:144–53. doi: 10.1016/j.pep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Sklan EH, Serrano RL, Einav S, et al. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. The Journal of biological chemistry. 2007;282:36354–61. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- 24.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Josse F, Shana ZA, Radtke DE, et al. Analysis of piezoelectric bulk-acoustic-wave resonators as detectors in viscous conductive liquids. IEEE Trans Ultrason Ferroelectr Freq Control. 1990;37:359–68. doi: 10.1109/58.105242. [DOI] [PubMed] [Google Scholar]
- 26.Kosslinger C, Drost S, Aberl F, et al. A quartz crystal biosensor for measurement in liquids. Biosens Bioelectron. 1992;7:397–404. doi: 10.1016/0956-5663(92)85038-c. [DOI] [PubMed] [Google Scholar]
- 27.Czech MP. PIP2 and PIP3: complex roles at the cell surface. Cell. 2000;100:603–6. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- 28.McLaughlin S, Wang J, Gambhir A, et al. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–75. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 29.Penin F, Brass V, Appel N, et al. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835–43. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 30.Osborne SL, Thomas CL, Gschmeissner S, et al. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J Cell Sci. 2001;114:2501–11. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 31.Ono A, Ablan SD, Lockett SJ, et al. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–94. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger KL, Kelly SM, Jordan TX, et al. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–83. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tai AW, Salloum S. The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS One. 2011;6:e26300. doi: 10.1371/journal.pone.0026300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Hong Z, Lin W, et al. ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication. PLoS One. 2012;7:e32135. doi: 10.1371/journal.pone.0032135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amako Y, Sarkeshik A, Hotta H, et al. Role of oxysterol binding protein in hepatitis C virus infection. J Virol. 2009;83:9237–46. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiss S, Harak C, Romero-Brey I, et al. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 2013;9:e1003359. doi: 10.1371/journal.ppat.1003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu FX, Sun HQ, Janmey PA, et al. Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J Biol Chem. 1992;267:14616–21. [PubMed] [Google Scholar]
- 38.Xian W, Vegners R, Janmey PA, et al. Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles. Biophys J. 1995;69:2695–702. doi: 10.1016/S0006-3495(95)80140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siagris D, Christofidou M, Theocharis GJ, et al. Serum lipid pattern in chronic hepatitis C: histological and virological correlations. J Viral Hepat. 2006;13:56–61. doi: 10.1111/j.1365-2893.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang BM, Weiner ND, Takada A, et al. Characterization of aminoglycoside-lipid interactions and development of a refined model for ototoxicity testing. Biochem Pharmacol. 1984;33:3257–62. doi: 10.1016/0006-2952(84)90087-x. [DOI] [PubMed] [Google Scholar]
- 41.Sauerbrey G. Verwendung von Schwingquarzen zur Waegun duenner Schichten und zur Mikrowaegung. Z. Physik. 1959;155:206. [Google Scholar]
- 42.Ferguson CG, James RD, Bigman CS, et al. Phosphoinositide-containing polymerized liposomes: stable membrane-mimetic vesicles for protein-lipid binding analysis. Bioconjug Chem. 2005;16:1475–83. doi: 10.1021/bc050197q. [DOI] [PubMed] [Google Scholar]
- 43.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.