Abstract
Receptor-based pharmacophore modeling is an efficient computer-aided drug design technique that uses the structure of the target protein to identify novel leads. However, most methods consider protein flexibility and desolvation effects in a very approximate way, which may limit their use in practice. The Site-Identification by Ligand Competitive Saturation (SILCS) assisted pharmacophore modeling protocol (SILCS-Pharm) was introduced recently to address these issues as SILCS naturally takes both protein flexibility and desolvation effects into account by using full MD simulations to determine 3D maps of the functional group-affinity patterns on a target receptor. In the present work, the SILCS-Pharm protocol is extended to use a wider range of probe molecules including benzene, propane, methanol, formamide, acetaldehyde, methylammonium, acetate and water. This approach removes the previous ambiguity brought by using water as both the hydrogen-bond donor and acceptor probe molecule. The new SILCS-Pharm protocol is shown to yield improved screening results as compared to the previous approach based on three target proteins. Further validation of the new protocol using five additional protein targets showed improved screening compared to those using common docking methods, further indicating improvements brought by the explicit inclusion of additional feature types associated with the wider collection of probe molecules in the SILCS simulations. The advantage of using complementary features and volume constraints, based on exclusion maps of the protein defined from the SILCS simulations, is presented. In addition, re-ranking using SILCS-based ligand grid free energies is shown to enhance the diversity of identified ligands for the majority of targets. These results suggest that the SILCS-Pharm protocol will be of utility in rational drug design.
Introduction
Pharmacophore modeling is a widely used computer-aided drug design (CADD) approach that, in addition to docking methods, is used in virtual screening (VS) studies1, 2. Compared to the energy function driven docking methods, it is based on the pattern of functional groups that are crucial for interactions of ligands with the protein target. These so-called pharmacophore features, and the resulting pharmacophore models, may be used to screen against a compound database to identify ligands with functional groups that match the pharmacophore features, an approach that is often superior to ligand docking VS3, 4. While pharmacophores may be developed based on the structure of known ligands, if the target protein structure is known, receptor-based pharmacophores can be constructed without knowledge of any known ligands of the target. Methods to develop receptor-based pharmacophores include the multi-copy simultaneous search (MCSS) derived pharmacophore method,5 the GRID molecular interaction fields (MIFs) based method6 and the recent hydration-site-restricted pharmacophore (HSRP) method7.
While there have been a number of successes using receptor-based pharmacophore modeling8–10, the effectiveness of those methods may be limited due to neglect of protein flexibility and desolvation effects. This is due to available methods being based on only a single or limited number of receptor conformations and being performed in vacuum or with a limited representation of the aqueous solvent environment, as discussed previously11, 12. More recent works using receptor-based pharmacophore modeling methods have begun to take these concerns into account, usually by utilizing molecular dynamics (MD) simulations13–15. But effective use of information present in MD simulations to further refine pharmacophore models is still an active area of research.
The site identification by ligand competitive saturation (SILCS) approach is a method that maps the functional group requirements of proteins, including contributions from protein flexibility and desolvation. Recently, a SILCS assisted pharmacophore modeling protocol (SILCS-Pharm)16 was introduced by us. The SILCS technique17 naturally takes both protein flexibility and desolvation effects into account by using MD simulations in an aqueous solution that contains a collection of probe molecules. During the simulation the probe molecules compete with water and with each other for binding sites on the protein. The binding information is then converted into probability maps of the functional group-binding patterns on the target (FragMaps) by binning the residences of probe molecule atoms into a 3D grid that encompasses the target receptor. The FragMaps may then be Boltzmann transformed into a free energy representation, termed grid free energy (GFE) FragMaps which enable its quantitative use18. Thus, the upfront calculated SILCS GFE FragMaps are an informative way to consider both protein flexibility and aqueous solvation contributions to chemical group binding that can be used for various aspects of receptor-based CADD16–21, including in the context of the SILCS-Pharm method.
SILCS-Pharm converts the GFE FragMaps into pharmacophore features to enable the use of SILCS in terms of pharmacorphore models. The spatial distribution of the GFE FragMaps is used to calculate feature GFEs (FGFE), which is the sum of the voxel GFEs that comprise a FragMap feature, where the voxels are based on a 3D grid that is used to define the spatial distribution of the grid free energy information in the FragMaps. The FragMap features act as the basis of the pharmacophore features and serve as a score to prioritize the identified features in an automatic fashion. Validation of the original SILCS-Pharm protocol used three representative protein targets along with ligands and decoys from the Dictionary of Useful Decoys (DUD)22 with the method showing improved performance versus docking based VS using the common docking programs DOCK23 and AutoDock24 and one recently developed receptor-based pharmacophore modeling technique developed by Lill and coworkers7. While the method was shown to offer improvements over those methods, the original SILCS-Pharm protocol has limitations associated with the SILCS simulations being performed using benzene and propane as molecular probes for aromatic and aliphatic functionalities with explicit water as the probe for both hydrogen bond donors and acceptors. Thus, only four basic pharmacophore feature types are possible. In addition, there is ambiguity in differentiating between donor and acceptor FragMap features caused by the dual role played by water. Finally, by using water to define the hydrogen bond donor and acceptor features disallows accounting for the energetic cost of displacing water from the associated binding sites as well as the energetic penalty associated with desolvation of the donors and acceptors upon interacting with the protein.
To overcome the above limitations, in the present work the SILCS-Pharm protocol is extended to use the FragMaps generated using the recently extended SILCS setup25. In the extended SILCS method, besides benzene and propane, a wider range of probe molecules, included methanol, formamide, acetaldehyde, methylammonium and acetate are used which enables the SILCS-Pharm to cover more types of pharmacophore features. The use of explicit probe molecules for hydrogen bond donors and acceptors allows the new SILCS-Pharm protocol to generate more clearly defined hydrogen bond donor and acceptor features and avoids the ambiguity brought by the use of water as the probe for hydrogen bond interactions. Moreover, the competition between probe molecules and waters in the new SILCS protocol and the use of explicit probe molecules instead of water for donors and acceptors allows the generated pharmacophore features to take into account desolvation of both the probe molecules and the protein. The new protocol is validated using eight protein targets and associated ligands and decoys from the DUD database. Of the three protein targets that were tested using the original protocol, improvements are seen using the new SILCS-Pharm protocol. Moreover, the SILCS-based pharmacophore approach as compared to docking based VS using DOCK 4.0, AutoDock 4 and AutoDock Vina26 shows improved or comparable results indicating its potential for use in CADD. Single point and SILCS driven Monte Carlo (SILCS-MC) sampling based ligand grid free energy (LGFE) re-ranking was also performed and was shown to be able to further enhance the pharmacophore results, indicating the general utility of SILCS in VS.
Methods
Extended SILCS-Pharm protocol
GFE FragMaps from SILCS simulations (see below) are used as inputs for SILCS-Pharm to build the pharmacophore features. Similar to the previous protocol16, the new SILCS-Pharm protocol contains four steps to generate a pharmacophore model: (1) voxel selection; (2) voxel clustering and FragMap feature generation; (3) FragMap feature to pharmacophore feature conversion; and (4) generation of pharmacophore hypotheses (i.e. models) for VS. The first step is designed to identify crucial binding patterns from the GFE FragMaps within a specified binding region. As the GFE represents the binding strength of a functional group at a specific location on the protein surface, it allows for voxels with the most favorable interactions to be identified based on a user assigned GFE cutoff. Clustering is then performed on the selected voxels in the second step to identify interaction patterns yielding FragMap features. In the third step, the FragMap features are classified, combined and converted into SILCS pharmacophore features. Finally, all the pharmacophore features are prioritized using FGFE score, from which pharmacophore hypotheses are generated and evaluated.
Presented in Table 1 are the FragMaps types and the corresponding FragMap features. The FragMap features are then converted to commonly used pharmacophore features3, 4, which are also listed in Table 1. Conversion from FragMap features to pharmacophore features involves analysis to determine, for example, whether a FragMap associated with the hydrogen or oxygen of the alcohol in methanol should be assigned a hydrogen bond donor (HBDON) or acceptor (HBACC) pharmacophore feature based on the geometric criteria described below.
Table 1.
Correspondence between FragMap features and pharmacophore features used in SILCS-Pharm.
| FragMaps and FragMap features | Pharmacophore featuresa |
|---|---|
| APOLAR (AROM+ALIP) | AROM|ALIP |
| HBDON | - |
| HBACC | HBACC |
| POS | - |
| NEG | NEG |
| AROM | AROM |
| ALIP | ALIP |
| HBDONp | HBDON |
| POSp | POS |
| MEOO | - |
| MEOH | - |
| FORN | - |
Only basic pharmacophore features are shown here for hydrogen bond donor and acceptors, donor and acceptor joint pharmacophore features are also available as described in the text.
The considered FragMaps and corresponding FragMap features are as follows. Five generic FragMap types are considered: (1) generic nonpolar (APOLAR, benzene and propane carbons); (2) generic neutral donor (HBDON, methanol and formamide polar hydrogens; (3) generic neutral acceptor (HBACC, methanol, formamide, and acetaldehyde oxygens); (4) positive donor (POS, polar methylammonium hydrogens); and (5) negative acceptor (NEG, acetate oxygens). In addition, seven specific FragMaps are constructed: (6) aromatic (AROM, benzene carbons); (7) aliphatic (ALIP, propane carbons); (8) generic positive donor nitrogen parent atom (POSp, methylammonium nitrogen); (10) methanol oxygen atom (MEOO); (11) methanol hydrogen atom (MEOH); and (12) formamide nitrogen (FORN).
Conversion of FragMaps to FragMap features in step 2 is based on a clustering algorithm. A hierarchical clustering algorithm was used previously with a user defined cluster member distance parameter to determine voxels that belong to the same cluster. A default value of 1 Å was used for hydrogen bond donor and acceptor voxel clustering since the voxel size is 1 Å × 1 Å × 1 Å, such that only neighboring voxels are included in a cluster. However, a larger distance parameter, in the range of 2.8 Å, was used for aromatic and aliphatic voxel such that the clusters included both neighboring as well as near but discrete voxels. However, as part of this study it was found that GFE cutoffs alone can serve as the sole parameter to select neighboring voxels to define a cluster by simply setting the cluster member distance parameters to 1 Å such that only neighboring voxels define a given cluster. In cases where discrete voxels are adjacent to a cluster they are treated as separate FragMap features.
Step 3 involves conversion of the FragMap features to pharmacophore features. First, the hydrophobic features (APOLAR, AROM and ALIP) are considered. If an APOLAR FragMap feature overlaps with both AROM and ALIP FragMap features, then an AROM|ALIP joint pharmacophore feature is defined. If an APOLAR FragMap feature only overlaps with AROM FragMap feature, then an AROM pharmacophore feature will be defined. Otherwise, an ALIP pharmacophore feature is defined. HBACC and NEG FragMap features are directly converted into respective HBACC and NEG pharmacophore features.
Conversion of hydrogen bond donor pharmacophore features is more complex. First, overlaps between HBDON and HBDONp FragMap features are identified. Only those HBDONp FragMap features that have overlap with HBDON features are retained as that overlap indicates that the HBDONp is a true representative of a hydrogen bond donor. Next, MEOO and MEOH FragMap features are identified and only those MEOO FragMap features that overlap with MEOH features are retained, again as they represent true hydrogen bond donors. Finally, overlap of the remaining HBDONp FragMap features with the remaining MEOO FragMap features is identified. If the HBDONp FragMap feature overlaps with both MEOO and FORN FragMap features, then the HBDONp FragMap feature is assigned as a HBDON pharmacophore feature. Or if the HBDONp FragMap features overlap only with an FORN feature, then the FORN feature is used to define a HBDON pharmacophore feature.
Finally, it is possible that both HBACC and HBDON pharmacophore features or both charged and neutral features are found in the same location. These locations are then defined as joint hydrogen bond features. The possible joint polar pharmacophore features include HBDON|POS, HBACC|NEG, HBDON|HBACC, (HBDON|POS)|HBACC, HBDON|(HBACC|NEG), and (HBDON|POS)|(HBACC|NEG).
SILCS simulations and FragMap preparation
SILCS simulations were performed using the new SILCS setup25 for the 8 target proteins. FragMaps for HIV protease (HIVPR), Factor Xa (FXa) and P38MAP kinase (P38 MAP) were obtained from our previous study25, while FragMaps for remaining targets were calculated as part of the present study. Crystal structures were obtained from the Protein Data Bank (PDB)27 for the targets Dihydrofolate reductase (DHFR, PDB ID:3DFR), Fibroblast Growth Factor Receptor 1 kinase (FGFr1, PDB ID:3KY2, Adenosine deaminase (ADA, PDB ID:1NDW), Estrogen Receptor Alpha Ligand-Binding Domain (ER, PDB ID:3ERT) and AmpC beta-lactamase (AmpC, PDB ID:1XGJ) and used to initialize the SILCS simulations. For holo structures, the ligands were removed while coordinated metal ions and crystal waters were retained. The Reduce software28 was used to determine the optimal protonation states of histidine and side-chain orientations of asparagine and glutamine residues. GROMACS29 tools were used to prepare the simulation systems involving protein, water and small probe molecules. Ten simulation systems with randomly positioned solutes at approximately 0.25 M each were simulated for 40 ns using GROMACS29 with the systems being described using CHARMM22 protein force field30 with CMAP backbone correction31, CHARMM General force field (CGenFF)32, 33 and the TIP3P water model34 modified for the CHARMM force field35. For ADA, distance restraints with force constants of 1000 kJ/mol/nm2 were applied between the zinc ion and the four coordinating residues during the MD simulation; analysis shows that the structure of the zinc ion and the coordinating ligands were well maintained in the simulations.
FragMaps were generated by binning selected solute atoms into voxels of a 1 Å spaced grid spanning the simulation systems. 3D normalized probability distributions were obtained by dividing the voxel occupancies computed in the presence of the protein by the respective values in bulk. The normalized distributions were Boltzmann-transformed to free energies for each FragMap type to yield GFE FragMaps. The convergence of the FragMaps was monitored by calculating overlap coefficients (OC) as previously described25. The ten trajectories for each SILCS simulation were divided into two groups as trajectories 1–5 and trajectories 6–10, and FragMaps from each group were separately computed and the OC was calculated between the two groups. As shown in the Supporting Information Table S1, all FragMaps for all the targets have OC values greater than 0.7, indicating good convergence as previously discussed25.
Pharmacophore VS and performance evaluation
VS was performed using the MOE software package36. All ligands or decoys for a target were extracted from the DUD database in mol2 format and were then converted into MOE database files. Ligand and decoy conformations were searched using the “Conformation Import” application in MOE36 and up to 100 low-energy conformations defined by the MMFF94x force field37 were retained for each molecule using the default MOE settings. SILCS pharmacophore models given by SILCS-Pharm were prepared in MOE pharmacophore query file format and used for VS with the default settings. The root-mean-square deviation (RMSD) between the matched features in a query molecule and the SILCS pharmacophore model was used as an activity score and the best matched conformation for each molecule with the smallest RMSD among all 100 conformations was then used as the pharmacophore predicted binding pose.
The performance of the new SILCS-Pharm protocol was compared with three other docking based VS methods using DOCK 4.023, AutoDock 424 and AutoDock Vina26. For the DOCK 4.0 VS, an in-house protocol, as applied in a number of CADD projects38, 39, was used and the sum of electrostatic and van der Waals (vdW) energies as defined in DOCK 4.0 used for final compound ranking. For AutoDock 4 and AutoDock Vina VS, mol2-formatted files for ligands and decoys as well as pdb-formatted files for the crystal protein structures from DUD were converted into pdbqt-formatted files to generate AutoDock atom types24. For AutoDock 4, energy grid map files with probe atoms covering all possible atom types within the database were generated and used to guide the docking search. The Lamarckian genetic algorithm (LGA)40 was adopted for the docking run and 20 independent runs with a maximum of 1,750,000 energy evaluations and 27,000 GA generations were conducted. AutoDock 4 energy scores, including electrostatic, vdW and desolvation terms,40 of the top 20 conformations for each molecule were averaged and the mean value was used for the final score ranking. For AutoDock Vina, the energy grid maps were calculated on-the-fly within a docking run and 20 binding modes were generated for each database molecule. The final score for each molecule used for AutoDock Vina was given by the mean score value averaged from the empirical energy scores26 of the top 20 predicted binding conformations. As used in our previous study16 and in the DUD paper22, enrichment plots, showing the percentage of ranked ligands at any given percentage of ranked database were employed to evaluate the VS performance. Enrichment factor (EF) reflecting the ability of a method to find more true positives while maintaining a low level of false positive rate is calculated following ranking of all the ligands and decoys, as follows:
| (1) |
where subset is defined by the percentage of the ranked decoys, Nligands_in_subset, Nligands, Ndecoys_in_subset and Ndecoys are the number of active ligands in a subset, total number of active ligands, number of decoys in a subset and total number of decoys. EFs at 1 % (EF1), 10 % (EF10) and 20 % (EF20) of the ranked decoys, which represent early and late stage enrichment performance, were calculated. The overall enrichment performance considering the whole database was also assessed by calculating the area-under-the-curve (AUC), which was evaluated from the Receiver Operating Characteristic (ROC) curve41. Similar to the previous study16, since not all molecules in the database are assigned an RMSD score in a pharmacophore VS runs due to the fact that they do not have the correct number and type of features, failed ligands and decoys are ranked at the end of the ranking list with decoys ranked above ligands to allow for a direct and unbiased comparison with the docking results, where all molecules have a score and can be ranked.
LGFE calculation and SILCS-MC sampling
To test the utility of including energetic information to supplement the SILCS pharmacophores, LGFE scores,25 were used to re-rank the pharmacophore modeling results. The LGFE was defined using:
| (2) |
where the summation is over all GFE FragMap types, T, and applicable atoms assigned to specific FragMap types, iT, NC is the number of GFE FragMap classified atoms and NH is the total number of non-hydrogen heavy atoms. The FragMap assignment is based on an atom classification rule file that translates CGenFF atom types into the FragMap classes (Table S4 of the supporting information).
Two types of LGFE scores were used depending on whether only a single conformation or an ensemble of conformations from MC sampling were used. The single point (SP) LGFE was calculated by using the conformations directly obtained from the pharmacophore search. To allow for local conformational relaxation and generation of an ensemble of conformations, the SILCS-MC25 approach was used. This uses the GFE FragMaps to drive the MC sampling and CGenFF parameters32, 33 to describe the intramolecular energy terms. For each compound, ten SILCS-MC runs with different random initialization seeds starting from the pharmacophore conformation were conducted for 5000 steps with small MC step sizes (molecular translation, rotation and dihedral rotation step sizes are 0.05 Å, 1 and 1 degrees) to minimize the extent that ligand would shift from the starting conformation. The LGFEs of all MC snapshots across the ten runs were Boltzmann averaged to generate the final SILCS-MC based LGFE score. A GFE upper energy cutoff of 3 kcal/mol was used in both types of LGFE calculations, such that both favorable and unfavorable GFE FragMap contributions contribute to the LGFEs.
Results and Discussion
SILCS Pharmacophore Models
An in-house FORTRAN program was used for the extended SILCS-Pharm protocol to generate SILCS pharmacophore models for the eight protein targets. The choice of the GFE cutoffs, which controls the selection of voxels to be used in FragMap features for subsequent generation of the pharmacophore features, was made on a case-by-case basis. This involved visual inspection of the FragMaps at different GFE cutoffs. The GFE cutoffs were then adjusted to achieve well-separated clusters of voxels of a specific type so that the total number of resulting features within the binding region was less than 8. GFE cutoffs used for the eight targets are listed in Table S2 in the supporting information along with the number of features. Once the GFE cutoffs were selected, the pharmcophore features were generated as described in the methods. We note that in our previous study, the use of only water to define hydrogen bond donor and acceptor pharmacophore features required the use of detailed geometric criteria. The use of a wider range of FragMaps types greatly simplifies the assignment of pharmacophore features, with the only complications involving the identification of true hydrogen bond donor pharmacophore features.
In addition to the pharmacophore features, volume constraints associated with “forbidden regions” are considered in the final pharmacophore models. Forbidden regions are defined on the 3D grid as voxels where the SILCS solutes as well as water do not sample during the SILCS simulations (i.e. voxels with zero solute or water occupancies considering all atoms including hydrogens). Thus, instead of, for example, using the protein surface to define regions were ligands cannot sample during VS, an exclusion map representing the forbidden region from the SILCS simulations is used to define a volume constraint in the pharmacophore model. This exclusion map may be considered a better alternative to more traditional representations of the protein surface since it takes the protein flexibility into account in an explicit way. Essentially, the excluded volume associated with the protein is being defined based on solute and water inaccessibility in the context of protein flexibility rather that by the space occupied by the protein in any given or all conformations.
Selection of the final SILCS pharmacophore model for screening was based on the use of the partial matching mode in VS, where the ligands only have to match a subset of “key” pharmacophore features. The key pharmacophore features are defined as those features with the most favorable FGFE scores, where the user can define the number of key features. The remaining pharmacophore features in the binding region are then defined as complementary features, which may or may not be matched in VS. Thus, during VS, only those compounds that have the correct number and types of functional groups that match the key pharmacophore features are selected. Alignment of the compounds with the key pharmacophore features as well as with complementary features if possible is then performed. Matching of complementary features in addition to the key features allows for more possible binding modes to be identified. Final RMSD scores are based on all the key and complementary features matched for a given ligand.
In summary, five advantages over the original SILCS-Pharm protocol are present in the new approach. First, charged features are introduced yielding 6 instead of 4 basic pharmacophore features. Second, with the use of explicit probes for hydrogen bond donors and acceptors instead of water, hydrogen bond donor features are more accurately described. Third, simple overlap of FragMap features are used to define the shape of joint features instead of the more complex geometric description used before. Fourth, exclusion maps are used to construct volume constraints that define the shape of the binding region, thereby including protein flexibility in the definition of the binding site shape. As shown below, the inclusion of the excluded region decreases the number false positive hits making the SILCS pharmacophore models more specific. Last, the inclusion of all SILCS pharmacophore features in the model and the use of partial matching during pharmacophore VS is anticipated to further improve the hit rate.
Test Set
To test the extended SILCS-Pharm protocol, eight protein targets were selected (Table 2). All the targets have the corresponding ligands and decoys in the DUD database22. The proteins include HIV protease (HIVPR), Factor Xa (FXa), dihydrofolate reductase (DHFR), fibroblast growth factor receptor kinase 1 (FGFR1), P38 mitogen activated protein kinase (P38 MAP), adenosine deaminase (ADA), estrogen receptor (ER) and AmpC β-lactamase (AmpC), and were chosen based on several considerations. HIVPR, FXa and DHFR were selected because they were used to test the previous SILCS-Pharm method, allowing for comparison with the present, extended method. The eight proteins are from different families according to the DUD classification allowing the new protocol to be tested on more target types. The numbers of active ligands and decoys vary from 21 to 256 and 732 to 8387 for the tested targets, respectively, which may help to reduce biases due to dataset size effects. In addition, the selected proteins have different difficulties as reflected by the enrichment performances of docking using DOCK22 with the enrichment factor varying from 0 (no enrichment) to 20 (good enrichment).
Table 2.
DUD data sets for the protein targets used in virtual screening.
| Targets | Classification | DOCK 3.5 EF1a | Number of Ligands | Number of Decoys | ||
|---|---|---|---|---|---|---|
|
| ||||||
| All molecules | Unique moleculesb | All molecules | Unique moleculesb | |||
| HIVPR | Other Enzymes | 3.7 | 62 | 53 | 2038 | 1885 |
| FXa | Serine Proteases | 14.6 | 146 | 142 | 5745 | 5095 |
| DHFR | Folate Enzymes | 21.7 | 410 | 201 | 8367 | 7145 |
| FGFr1 | Kinases | 0 | 120 | 118 | 4550 | 4205 |
| P38 MAP | Kinases | 2.1 | 454 | 256 | 9141 | 8387 |
| ADA | Metalloenzymes | 12.9 | 39 | 23 | 927 | 821 |
| ERantagonist | Nuclear Hormone Receptors | 12.7 | 39 | 39 | 1448 | 1395 |
| AmpC | Other Enzymes | 17.1 | 21 | 21 | 786 | 732 |
Published 1% Enrichment factors for the DOCK 3.5 results from the DUD paper22 which can be treated as an indicator for the test case difficulty.
In the DUD data sets, some molecules are represented in multiple forms considering different tautomer and protonation states. For the final VS result, only the most favorable scored state based on the most favorable energy score for docking and lowest RMSD for pharmacophore matching for each molecule was used.
SILCS Pharmacophore Features for the Tested Targets
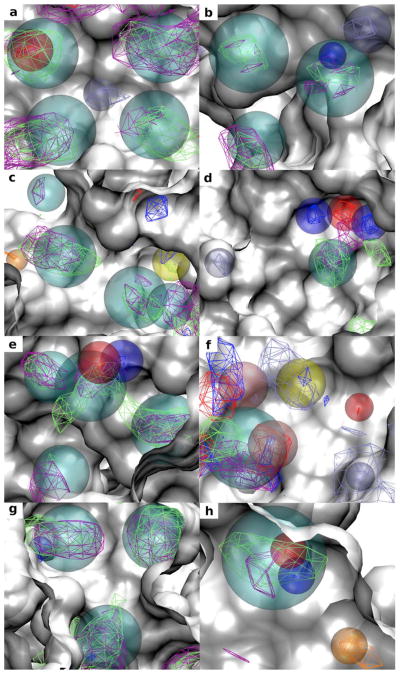
Figure 1 shows the SILCS-Pharm derived pharmacophore features together with the corresponding FragMaps contoured at the GFE cutoff levels used to generate the features. Consistent with steps 1 and 2 all FragMaps within the binding pockets have been identified as pharmacophore features and are fully covered by the pharmacophore feature spheres. Besides the basic features, various joint pharmacophore features are seen in all the targets and have been generated correctly. For example, both aromatic (in purple) and aliphatic (in green) FragMaps can be found at the same locations as the AROM|ALIP joint features colored in cyan. For DHFR (Figure 1c) both neutral (blue) and charged (iceblue) donor FragMaps can be found in a deeply buried subsite and this is represented as a HBDON|POS joint pharmacophore feature colored in yellow. For ADA (Figure 1f) both neutral hydrogen bond donor (blue) and acceptor (red) as well as charged donor (iceblue) FragMaps can be seen at the top left region, and this forms a (HBDON|POS)|HBACC feature colored in pink. In the lower left region both neutral hydrogen bond donor and acceptor features are present and result in a HBDON|HBACC feature (pink). Figure 1 also shows that the generated feature spheres can penetrate into the protein surface of the crystal structure used to initialize the SILCS simulation, indicating that protein flexibility is considered in the SILCS pharmacophore features.
Figure 1.
FragMaps and identified SILCS pharmacophore features within the binding pockets of the eight protein targets: (a) HIVPR (PDB 1G2K); (b) FXa (PDB 1FJS); (c) DHFR (PDB 3DFR); (d) FGFr1 (PDB 3KY2); (e) P38 MAP (PDB 1OUY); (f) ADA (PDB 1NDW); (g) ER (PDB 3ERT); (h) AmpC (PDB 1XGJ). FragMap contours are displayed at the GFE cutoffs used to generate the pharmacophore features. The color of the AROM, ALIP, HBDONp, HBACC, POSp and NEG FragMaps are purple, green, blue, red, iceblue and orange, respectively. Pharmacophore features are shown by transparent spheres. The color of the AROM|ALIP, HBDON, HBACC, POS, NEG and HBDON|POS pharmacophore features are cyan, blue, red, iceblue, orange and yellow. The joint pharmacophore feature (HBDON|POS)|HBACC (top left) and HBDON|HBACC (lower left) are colored in pink. The protein surfaces are based on the crystal structures used to initialize the SILCS simulation (white). Protein atoms occluding the view of the pocket are removed to facilitate visualization.
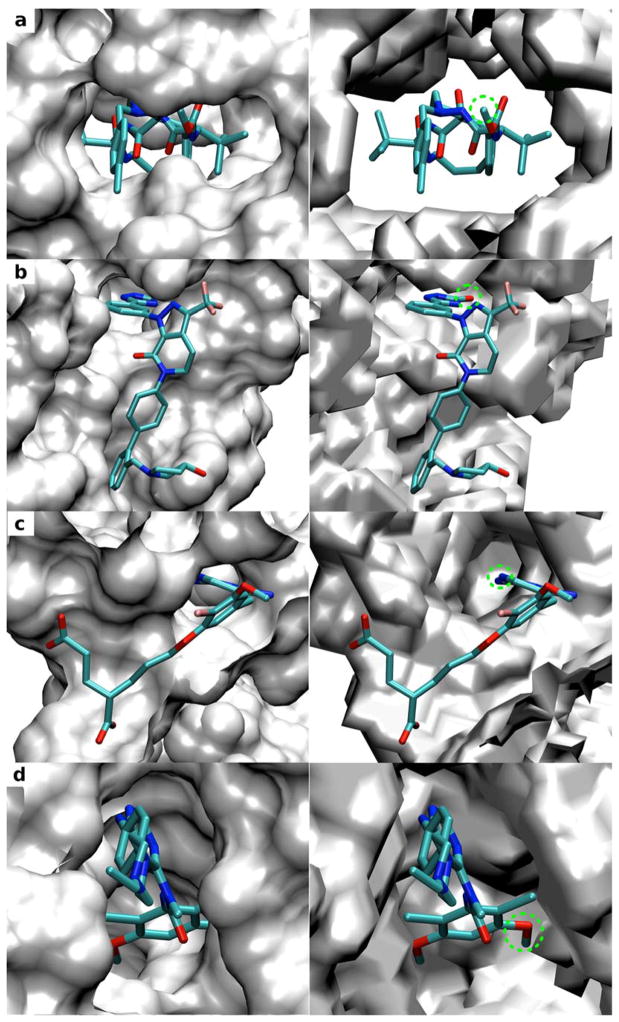
To more clearly analyze the importance of the inclusion of protein flexibility we compare the SILCS exclusion maps with the surfaces of the crystal protein structures used to initialize the SILCS simulations (Four targets are shown in Figure 2, with results for all targets in Figure S1 in the supporting information). Crystal orientations of selected ligands from protein complexes other than those used for SILCS simulation are also shown for each target in the figure. Clashes are seen between the ligand atoms with the surface of the rigid protein structure indicating that such crystal binding modes of the ligands cannot be predicted when using rigid protein structure. In contrast, the exclusion maps define a much broader binding region and the presented binding modes of the ligands are allowed indicating the importance of including protein flexibility during pharmacophore model development as well as binding mode prediction. Table 3 shows the calculated surface area (SA) for the exclusion map and crystal protein surface. The much lower SA values for the exclusion maps further indicates how additional accessible area is available when using the exclusion maps versus that using the rigid protein structure.
Figure 2.
Comparison of the solvent accessible surfaces of crystal protein structures used to initialize the SILCS simulation (left panel) and SILCS exclusion maps (right panel) for the four targets: (a) HIVPR (PDB 1G2K); (b) FXa (PDB 1FJS); (c) DHFR (PDB 3DFR); (d) FGFr1 (PDB 3KY2). Results for other targets can be found in Figure S1 in the supporting information. The crystal binding orientation of a selected ligand for each target presents in protein-ligand complex other than the one used for SILCS simulation is also shown: (a) HIVPR (PDB 3ZPS); (b) FXa (PDB 3FFG); (c) DHFR (PDB 1DIU); (d) FGFr1 (PDB 3TT0). The green dashed circle indicates ligand atoms that have clashes with the protein surface but not the exclusion map.
Table 3.
Surface area (SA in Å2) calculated for the crystal protein structures and exclusion maps of the eight targets.
| Target | Protein surface | Exclusion map |
|---|---|---|
| HIVPR | 20170 | 11440 |
| FXa | 28909 | 17465 |
| DHFR | 16975 | 8426 |
| FGFr1 | 31598 | 15206 |
| P38 MAP | 36332 | 17100 |
| ADA | 35941 | 20066 |
| ER | 25280 | 10549 |
| AmpC | 35790 | 19826 |
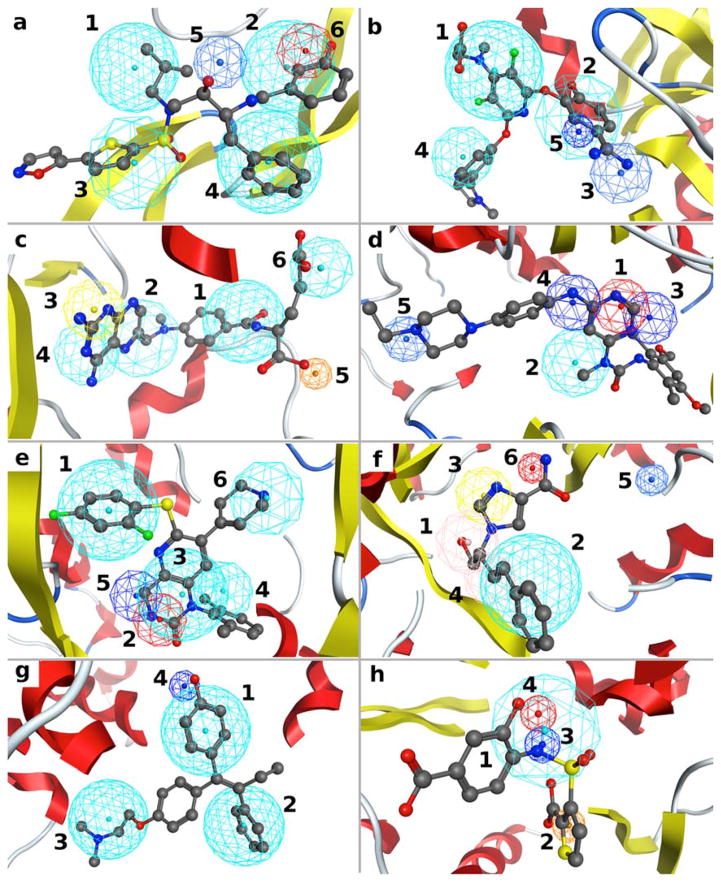
SILCS FragMaps have been shown to recapitulate crucial ligand-protein interactions and reproduce the crystal binding modes of ligands for a number of proteins.17, 18, 25 For the protein targets studied here, FragMaps are also found to reproduce crucial binding modes as indicated by the overlaps between the SILCS FragMap-derived pharmacophore features and functional groups in selected crystallographically identified ligands (Figure 3). Ranking of the pharamcophore features for each target based on the FGFE scores is also shown in Figure 3. Such ranking may serve as an indicator for the importance of a feature and may potentially be used as weighting factors when doing scoring during VS.
Figure 3.
SILCS pharmacophore models for the eight protein targets with the crystal orientations of a representative ligand for each target: (a) HIVPR (PDB 3SAC); (b) FXa (PDB 1FJS); (c) DHFR (PDB 3DFR); (d) FGFr1 (PDB 3TT0); (e) P38 MAP (PDB 1OUY); (f) ADA (PDB 1NDW); (g) ER (PDB 3ERT); (h) AmpC (PDB 1XGJ). Protein atoms occluding the view of the pocket are removed to faacilitate visualization. The colors of the features are the same as used in Figure 2. Numbering indicates the rank ordering of the pharmacophore features based on the FGFE scores.
In general, the pharmacophore features recapitulate interactions that have been shown experimentally to be important for ligand binding and FGFE can qualitatively rank the relative feature importance. For HIVPR (Fig. 3a), the four hydrophobic features were the highest ranked, consistent with the fact that these four nonpolar binding pockets were known to be important for binding42, 43. The fifth feature is a POS feature occupying the catalytic site where the ligand hydrogen bond donor groups, usually neutral hydroxyl groups, interact with the two Asp residues. As discussed previously25, this inconsistency is due to the Asp residues being deprotonated in the SILCS simulation, while only one Asp residue may be deprotonated44. For FXa (Fig. 3b), two of the three top ranked hydrophobic features are located in the well-studied S1 and S4 pockets which were shown to be crucial for binding of the known inhibitors45, 46 and the third ranked POS feature also reproduces the commonly observed binding pattern among the known inhibitors45. For DHFR (Fig. 3c), features 1, 2 and 4 reproduce the hydrophobicity of the core region in the buried binding pocket and feature 3 is a HBDON|POS joint feature corresponding to the surrounding charged Asp and neutral Thr residues that have both been shown to be important for ligand binding47.
For FGFr1 (Fig. 3d), the three top ranked hydrogen bond donor and acceptor features 1, 3 and 4 reproduce the known binding patterns that involve critical hydrogen bond interactions with the Ala and Glu backbone carbonyl and amide groups of the hinge region at the ATP-binding pocket48. For P38 MAP the three top ranked features 1, 2 and 3 reproduce the three functionalities that are conserved among the majority of P38 MAP inhibitors (Fig. 3e)49–51. The last ranked hydrophobic features 4 and 6 and neutral hydrogen bond donor feature 5 represent functional groups that are present in some, but not all inhibitors49, 50. For ADA (Fig. 3f), top ranked hydrophobic feature 2, hydrogen bond donor and acceptor joint features 1 and 4 all reproduce known binding interactions,52, 53 which involve hydrophobic residues, His17 and Asp19, respectively. For target ER (Fig. 3g), the three top ranked hydrophobic features and the donor feature ranked 4 are consistent with the known binding modes of ER antagonists54, 55. As seen in Fig. 3h the top ranked hydrophobic feature 1 mimics the stacking interaction with Tyr221 residue56, 57 and the second ranked feature NEG recapitulates the oxyanion hole caused by the catalytic residue Ser6456, 57. Neutral hydrogen bond donor and acceptor features 3 and 4 are related to the amide recognition region composed of Ala318 and Asn152 in the binding pocket and are common functional patterns found in AmpC ligands56, 57. The above analysis indicates that the SILCS-Pharm models are able to qualitatively reproduce important features known to be required for the binding of ligands to the eight targets. In addition, the FGFE ranking is an indicator of the importance of a feature for binding.
Feature Prioritization using FGFE
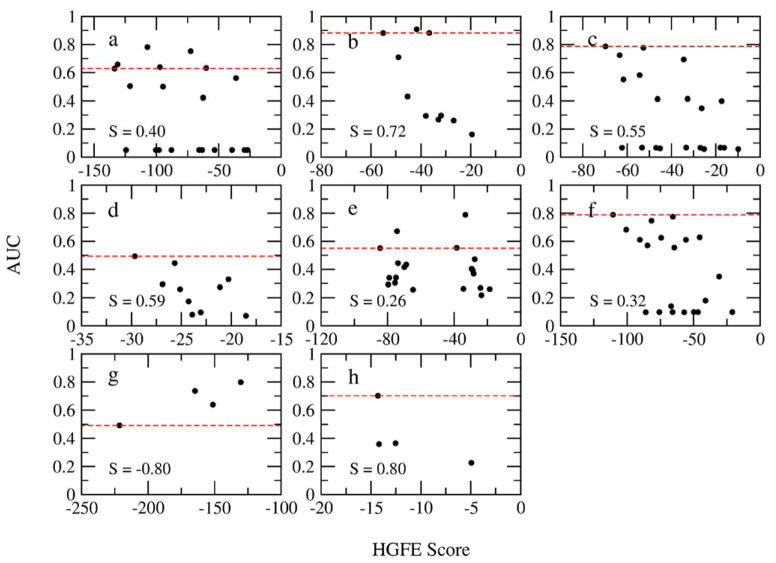
Receptor-based pharmacophore modeing methods usually first generate all possible pharmacophore elements on the protein target surface and then, using various approaches (e.g. hydration site analysis7), select a subset for use in VS. In SILCS-Pharm, the FGFE may be used to prioritize identified features. Here, we quantitatively test the use of FGFE. Assuming three key pharmacophore feature models, all possible combinations out of the total number of SILCS-Pharm identified features (Table S2 in the supporting information) were considered for each target and VS was performed using all constructed models under full matching mode. The AUC for each VS was then evaluated and used to rank the model quality. Relationships between the hypothesis GFE (HGFE, which is the sum of FGFEs of all features in a model) and AUC were then plotted to investigate if the FGFE can serve as an indicator of feature importance and thereby be used to select a phamacophore model for VS. Figure 4 presents the AUC values as a function of the HGFE scores for three key feature containing pharmacophore models. Analysis of models with larger numbers of features was not done as ER and AmpC only have four available features. In general, the most favorable pharmacophore model based on the HGFE scores is among the top models as ranked by AUC. The most significant exceptions occur with HIVPR, P38 MAP and ER. The results with HIVPR (Fig. 4a) may have contributions from the protonation state of the Asp residues in the catalytic site (see above) while the known conformational flexibility of P38 MAP may play a role in that protein (Fig. 4e). However, in both cases the top scoring model based on HGFE is among the top three or four AUC-ranked models. The discrepancy with ER may be associated with the small number of models (Fig. 4g). We note that the correlations are generally quite poor as reflected by the low correlation coefficient values, which may be indicative of individual pharmacophore features being poor predictors of active compounds. For example, in the case of HIVPR, many of the models with favorable HGFEs have AUC values that are under 0.1. All of these models contain the POS feature which was discussed above as an inappropriate feature for the target. Thus, while the correlations are generally weak, the overall ability of HGFE scores to yield high AUC values is satisfactory, indicating its utility in selecting pharmacophore models for VS. Accordingly, this approach was used for the final analysis.
Figure 4.
Relationship between HGFEs of all possible pharmcophore models containing three features and AUCs from VS for each target. (a) HIVPR; (b) FXa; (c) DHFR; (d) FGFr1; (e) P38 MAP; (f) ADA; (g) ER; (h) AmpC. The Spearman correlation coefficients S are shown. The red dashed line represents the AUC value for the model with the most favorable HGFE for each target.
VS using SILCS Pharmacophore Models
SILCS pharmacophore models were tested in VS using DUD data sets for the eight proteins listed in Table 2. The models for each target contain all identified SILCS pharmacophore features and volume constraints with the number of key features varying. The tested models were chosen to have at least three key features and only those features with the most favorable FGFE are labeled as key features. For example, if four features are identified for a target, then a 3-key features model is a model where the three top FGFE ranked features are assigned as key features. And it is possible to have all the features in a model assigned as key features. Accordingly, a total of N-2 models for a target that has N identified features can be constructed and tested in VS under partial matching mode.
As the pharmacophore generation scheme presented in the preceding paragraph allows for the creation of multiple models, enrichment performance for all possible models was determined for each target. As shown in Table S3 of the supporting information, that range of variability is significant. In all cases the choice of 3 or 4 key features yields the highest enrichment while the use of 5 or more key features leads to a significant decrease in enrichment. Thus, in situations where no priori information on active ligands is available, there is the possibility of selecting a less than ideal model for VS, though the selection of 3 or 4 key features is recommended. In addition, as presented below, the use of LGFE re-ranking leads to the selection of alternate active ligands versus the use of RMSD selection alone, suggesting that both approaches should be used and the results combined when selecting compounds for experimental assays. The remainder of the manuscript focuses on the best performing model for each target.
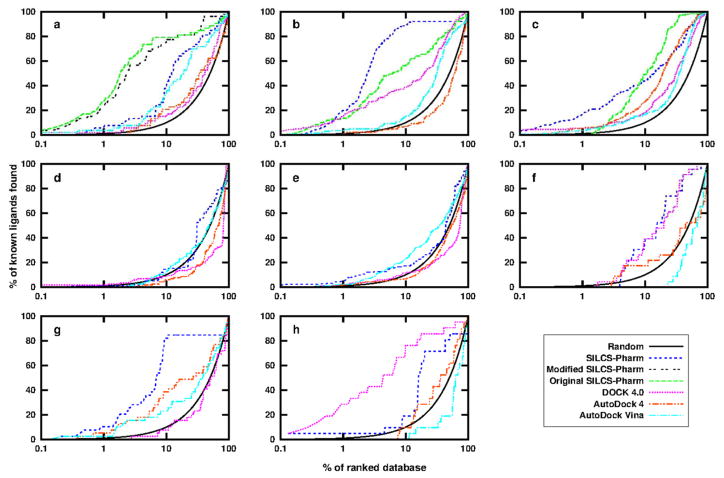
Enrichment results for the best performing pharmacophore model are shown in Figure 5 and Table 4 and compared with docking results using available docking programs. For the first three targets, the results using the extended SILCS-Pharm protocol were also compared with previously reported VS results using the original SILCS-Pharm protocol16. In addition, comparisons are made with available results from Lill and coworkers’ work7.
Figure 5.
Enrichment plots of the SILCS pharmacophore modeling using DUD data sets for the eight protein targets: (a) HIVPR; (b) FXa; (c) DHFR; (d) FGFr1; (e) P38 MAP; (f) ADA; (g) ER; (h) AmpC. Results using DOCK 4.0, AutoDock 4 and AutoDock Vina are also shown for comparison. For the first three targets, the results from our former SILCS-Pharm study are shown. The black line indicates random selection of compounds from the database. The X axis is in logarithmic scale to show the early stage performance more clearly.
Table 4.
Comparison of enrichments for different docking and pharmacophore modeling methods.
| Targets | Methods | EF1 | EF10 | EF20 | AUC |
|---|---|---|---|---|---|
| HIVPR | 4F SILCS-Pharma | 6.7 | 4.1 | 3.6 | 0.78 |
| 5F Modified SILCS-Pharmb | 77.1 | 9.2 | 4.4 | 0.89 | |
| Original SILCS-Pharmc | 99.6 | 9.8 | 4.3 | 0.88 | |
| DOCK 4.0 | 2.0 | 1.5 | 1.2 | 0.58 | |
| AutoDock 4 | 0 | 2.1 | 1.5 | 0.56 | |
| AutoDock Vina | 4.2 | 3.6 | 2.8 | 0.73 | |
| FPPd | 9.3 | 4.7 | 3.5 | 0.81 | |
| HSRPd | - | - | - | <0.80e | |
|
| |||||
| FXa | 4F SILCS-Pharma | 41.9 | 11.4 | 5.1 | 0.91 |
| Original SILCS-Pharmc | 20.7 | 6.5 | 3.5 | 0.81 | |
| DOCK 4.0 | 26.3 | 4.2 | 2.6 | 0.76 | |
| AutoDock 4 | 0.7 | 0.7 | 0.7 | 0.42 | |
| AutoDock Vina | 3.8 | 1.5 | 1.6 | 0.64 | |
| FPPd | 12.2–29.7 | 5.6–6.7 | 3.5–3.9 | 0.81–0.87f | |
| HSRPd | - | - | - | <0.83e | |
|
| |||||
| DHFR | 3F SILCS-Pharma | 29.3 | 4.8 | 3.0 | 0.79 |
| Original SILCS-Pharmc | 0.0 | 5.5 | 4.0 | 0.87 | |
| DOCK 4.0 | 6.3 | 1.8 | 1.8 | 0.66 | |
| AutoDock 4 | 6.3 | 2.8 | 2.8 | 0.76 | |
| AutoDock Vina | 3.8 | 1.7 | 1.2 | 0.65 | |
| FFPd | 2.0 | 1.5 | 1.3 | 0.51 | |
| HSRPd | - | - | - | <0.62e | |
|
| |||||
| FGFr1 | 3F SILCS-Pharma | 0.8 | 1.5 | 0.8 | 0.55 |
| DOCK 4.0 | 1.7 | 0.8 | 0.7 | 0.33 | |
| AutoDock 4 | 0.0 | 0.6 | 0.4 | 0.38 | |
| AutoDock Vina | 0.0 | 1.2 | 1.2 | 0.50 | |
|
| |||||
| P38 MAP | 3F SILCS-Pharma | 6.4 | 1.8 | 1.3 | 0.57 |
| DOCK 4.0 | 2.0 | 1.1 | 0.7 | 0.39 | |
| AutoDock 4 | 2.0 | 0.7 | 0.8 | 0.45 | |
| AutoDock Vina | 2.5 | 2.4 | 1.8 | 0.59 | |
|
| |||||
| ADA | 3F SILCS-Pharma | 0.0 | 3.8 | 3.0 | 0.80 |
| DOCK 4.0 | 0.0 | 4.3 | 2.7 | 0.79 | |
| AutoDock 4 | 0.0 | 1.8 | 1.3 | 0.46 | |
| AutoDock Vina | 0.0 | 0.0 | 0.0 | 0.36 | |
|
| |||||
| ERantagonist | 4F SILCS-Pharma | 14.3 | 10.3 | 4.6 | 0.81 |
| DOCK 4.0 | 0.0 | 0.8 | 0.8 | 0.46 | |
| AutoDock 4 | 6.0 | 4.2 | 2.5 | 0.64 | |
| AutoDock Vina | 2.8 | 2.4 | 1.7 | 0.59 | |
|
| |||||
| AmpC | 3F SILCS-Pharma | 5.0 | 2.0 | 3.6 | 0.70 |
| DOCK 4.0 | 104.6 | 9.5 | 4.7 | 0.87 | |
| AutoDock 4 | 0.0 | 1.0 | 1.4 | 0.58 | |
| AutoDock Vina | 0.0 | 0.0 | 0.5 | 0.35 | |
The results of the best performing SILCS pharmacophore model based on AUC with a specific number of key features (e.g. 4F means 4 top FGFE ranked features are considered as key features in the model) are shown.
The best modified model for HIVPR with the POS feature changed into a HBDON|POS feature.
The best results from the original SILCS-Pharm work16.
Full Protein Pharmacophore (FPP) and Hydration Site Restricted Pharmacophore (HSRP) results from Lill and coworkers work7.
The upper AUC values for HSRP models using different parameters are estimated from figure 7 in Ref.7 as no exact values were reported.
Three protein structures were used for FXa in Ref.7, so the range of AUC values is shown here.
For HIVPR, the best pharmacophore model has four key features and its performance is slightly worse than the best previous results using the original SILCS-Pharm protocol. However, the best previous model contained 6 features, while the original 4-feature model, which has the four hydrophobic features, had significantly worse performance (EF1=2.0, EF10=2.1, EF20=1.4, AUC=0.56) than the new 4-feature model. This is due to the features from the new SILCS-Pharm being more accurately defined than in the original method. As discussed above, the fifth feature, POS, in the current model may not be suitable due to both the catalytic Asp residues being charged in the SILCS simulations, whereas one of the residues is likely neutral which would favor a neutral HBDON feature in that region44. Accordingly, we tested a modified model with feature POS being changed to HBDON|POS to allow the matching of both neutral and charged donors at this location. The resulting 5-feature model has a similar performance as the best original model, emphasizing the importance of the protonation state selected for the SILCS simulation. For FXa, the new model has better performance than the original SILCS-Pharm model as indicated by the increased EFs and AUC. For DHFR, concerns about the zero value for EF1 existed in the original SILCS-Pharm study, but this is no longer an issue in the new model where EF1 = 29.3 though the AUC is slightly worse than the original result. These results indicate that the extended SILCS-Pharm model shows more robust performance versus the original SILCS-Pharm results, indicating the advantage of using multiple probe molecules during pharmacophore model development.
The quality of the extended SILCS-Pharm approach was further validated by comparing results with those from Dock 4.0, AutoDock 4 and AutoDock Vina. For all the targets tested, except AmpC, SILCS-Pharm outperformed the three docking programs as reflected by the larger EF and AUC values. For AmpC, though SILCS-Pharm yielded better results than the two AutoDock programs, it was outperformed by DOCK 4.0. It is interesting to note that the DOCK 3.5 result for AmpC presented in the DUD paper was worse than the current SILCS-Pharm result. The improved result for AmpC using DOCK 4.0 may due to the use of the in-house developed docking protocol,38, 39 where the CHARMM parameters30 instead of the original DOCK parameters are used to determine the binding energy and guide the docking.
Complementary Features and Volume Constraints
While the success of the extended SILCS-Pharm may be largely attributed to the use of multiple solute types in the SILCS simulation, the use of complementary features and volume constraints may also contribute to the improved VS results. The contributions by these two factors were tested by using models in their absence for VS. For each target, the best performing model was considered and two new models were created with removal of either the complementary features or the volume constraints and tested in VS. Table 5 lists the percentage loss on the AUC value using the two modified models compared with the full model for each target. Though generally not large, some decreases in the AUCs occurred for some targets when the complementary features or the volume constraints were absent suggesting that the two factors indeed helped with enrichment during VS. The observed small change may be due to the DUD data sets being designed to have decoys that have physiochemical properties similar to the ligands, such that the sizes and functional features of the ligands and decoys for each target are similar. Thus, the complementary features used to select ligands with more matches and the volume constraints used to avoid oversized compounds that do not fit into the binding site have limited roles in compound selection in this case. However, it is anticipated that the two terms will make additional contributions in practical use, where compounds with very dissimilar properties and shapes are being screened. Moreover, given that the new terms do not add much additional computational cost to VS, even a relatively small improvement warrants their inclusion. For example, VS against the 5745 decoys of FXa took 190 and 170 seconds, respectively, using pharmacophore models with and without volume constraints.
Table 5.
Percentage loss (%) of AUC using the best performing model as listed in table 4 without complementary features or without volume constraints for each target.
| Targets | HIVPR | FXa | DHFR | FGFr1 | P38 MAP | ADA | ERantagonist | AmpC |
|---|---|---|---|---|---|---|---|---|
| without complementary features | 0 | 0 | 0 | 0 | 2 | 0 | -a | 0 |
| without volume constraints | 1 | 0 | 0 | 11 | 2 | 1 | 0 | 0 |
The best model for ER contains all four identified features and thus all features are key features, such that models without complementary features cannot be created.
LGFE Re-ranking
Though for most test cases, an AUC of more than 0.7 was observed when using SILCS-Pharm, low AUCs were found for two targets. These two targets, FGFr1 and P38 MAP, show an AUC lower than 0.6 implying the two targets are challenging cases for VS, which is also consistent with the previous DUD docking results (EF1 is 0 and 2.1)22. As shown in Figure 3 and discussed above, the SILCS-Pharm features for these two targets are consistent with the binding modes of the known ligands. This suggests the features in the pharmacophore model are appropriate for VS and should be able to identify the known ligands and differentiate them from decoys. The poor performance may thus imply that the RMSD score used to rank compounds is inadequate. Previously, LGFE, which is the sum of all atom GFE scores for a ligand, was proposed for scoring and shown to reproduce various experimental binding affinity data sets satisfactorily18, 20, 25. Here, normalized LGFE was used to re-rank the obtained pharmacophore RMSD-selected conformations from SILCS-Pharm VS for all the targets and to determine if the use of LGFE ranking leads to improvements in the enrichment. Calculation of the LGFE scores is defined in equation 2. The current scoring represents a refinement of that previously reported25 with no weighting factors being used, the scores being normalized for the number of classified versus total non-hydrogen atoms in the ligand and new definitions of the atom types based only on non-hydogen atoms as presented in Table S4 of the supportining information. In addition to SP LGFE scores based on the RMSD-selected conformation, local relaxation of the ligands was performed using GFE based SILCS-MC sampling25 from which Boltzmann-averaged LGFE scores were obtained.
Table 6 shows the AUCs for the results with ligand ranking based on SP LGFE scores, SILCS-MC sampled LGFE score and the RMSD criteria. For the two worst RMSD based ranking cases, FGFr1 and P38 MAP, the SP LGFE re-ranking improves the enrichment with AUCs increased from 0.55 to 0.75 for FGFr1 and from 0.57 to 0.75 for P38 MAP. For most of the other cases, similar or slightly worse results are seen when using SP LGFE re-ranking. Similarly, use of the relaxed SILCS-MC LGFE scores yielded ambiguous results with respect to the RMSD and SP LGFE scores. For HIVPR, the heavily decreased performance using LGFE for scoring is likely again due to the wrong protonation state used for the SILCS simulations as described before.
Table 6.
AUCs using RMSD, SP LGFE and SILCS-MC sampled LGFE for ranking for the eight targets. D%a is the percentage of different active ligands in the top 10% ranked compounds identified by LGFE score that were not identified by RMSD score are also shown.
| Ranking score | HIVPR | FXa | DHFR | FGFr1 | P38 MAP | ADA | ER | AmpC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | D%a | AUC | D%a | AUC | D%a | AUC | D%a | AUC | D%a | AUC | D%a | AUC | D%a | AUC | D%a | |
| RMSD | 0.78 | - | 0.91 | - | 0.79 | - | 0.55 | - | 0.57 | - | 0.80 | - | 0.81 | - | 0.70 | - |
| SP LGFE | 0.54 | 4 | 0.78 | 8 | 0.67 | 9 | 0.75 | 14 | 0.75 | 43 | 0.58 | 4 | 0.86 | 46 | 0.55 | 0 |
| MC LGFE | 0.49 | 2 | 0.90 | 17 | 0.70 | 6 | 0.66 | 13 | 0.73 | 30 | 0.64 | 0 | 0.83 | 41 | 0.62 | 0 |
D%=Ndiff/Nligands*100%, where Ndiff is the number of active ligands identified by LGFE score among the top 10% LGFE ranked compounds that are not found by RMSD score among the top 10% RMSD ranked compounds, and Nligands are the total number of active ligands in the database for the target.
Given ambiguous results associated with LGFE ranking, analysis was undertaken to test if LGFE ranking may identify alternate ligands with a high probability of binding to the target protein versus those from RMSD ranking alone. Included in Table 6 is the percentage of different ligands identified using by the use of LGFE scores. This result suggests that the most optimal approach for the selection of ligands in a practical VS would be a combination of ligands from RMSD and LGFE rankings. To test this, the top 10% of the ranked compounds from RMSD or LGFE scoring were obtained and the percentage of active ligands amongst those compounds determined (i.e. if all the active compounds were found in the top 10%, the result would be 100 in Table 7). RMSD and LGFE results were combined by simply taking the top 10% list from both rankings and determining the number of active compounds among the combined set. As shown in Table 7, the combination of RMSD and LGFE based ranking always yielded an equivalent or larger percentage of active ligands over the individual methods. This result is consistent with previous findings about consensus scoring58 that the identification of true positives is enhanced and more diverse sets of ligands are identified using consensus ranking. Concerning computational costs, taking P38 MAP, which includes 454 ligands, the SP LGFE calculations required 2,700 CPU seconds using a single core on an AMD Opteron 2350 processor equipped node while the 5000 steps SILCS-MC sampled LGFEs required almost the same amount of calculation time. Thus, it is recommended that the final selection of compounds be based on the combined final top ranked lists from RMSD and LGFE ranking, with the SP LGFE scores yielding the greatest improvement with the exception of FXa.
Table 7.
Percentage of active ligands identifieda in the top 10% ranked compounds using RMSD, single point LGFE and SILCS-MC sampled LGFE ranking alone as well as the consensus ranking using both RMSD and LGFE for the eight targets.
| Ranking score | HIVPR | FXa | DHFR | FGFr1 | P38 MAP | ADA | ER | AmpC |
|---|---|---|---|---|---|---|---|---|
| RMSD | 38 | 25 | 44 | 14 | 17 | 39 | 38 | 10 |
| SP LGFE | 19 | 11 | 20 | 16 | 48 | 4 | 85 | 0 |
| MC LGFE | 11 | 22 | 20 | 13 | 33 | 4 | 77 | 0 |
| RMSD + SP LGFE | 42 | 33 | 53 | 29 | 60 | 43 | 85 | 10 |
| RMSD + MC LGFE | 40 | 42 | 50 | 27 | 47 | 39 | 79 | 10 |
Percentage of active ligands=Nligands_in_10%/Nligands*100%, where Nligands_in_10% is the number of active ligands identified among the top 10% ranked compounds and Nligands is the total number of active ligands in the database for the target.
Conclusion
Using additional solutes in the SILCS simulations enables the SILCS-Pharm protocol to be extended and reformulated. Advantages over the original approach were validated using eight protein targets with their DUD test sets. The new protocol not only defines the pharmacophore features more accurately by including explicit hydrogen bond donor and acceptor solutes, but also includes more specific features allowing for the definition of more feature types. With the use of complementary features and volume constraints, the VS results show that the extended SILCS-Pharm improves upon the original protocol and outperforms three commonly used docking programs in most cases suggesting its potential utility for CADD. Single point and SILCS-MC sampling based LGFE re-rankings, which introduce energetic criteria into compound ranking, were tested and shown to enhance the SILCS-Pharm results with respect to the identification of alternate ligands for experimental assay. Thus, as pharmacophore-based VS is very efficient and far less time consuming than other VS methods, SILCS-Pharm can be considered for use in CADD projects when screening a large databases of compounds as well as to facilitate ligand docking in general. Furthermore, given the capability of the SILCS approach to facilitate ligand optimization18, 20, 25, which would be performed using the same GFE FragMaps as used for the SILCS-Pharm, the overall economy and utility of the SILCS approach in ligand design may be significant.
Supplementary Material
Acknowledgments
This work was supported by NIH grant CA107331, Maryland Industrial Partnerships Award 5212, University of Maryland Center for Biomolecular Therapeutics and the Samuel Waxman Cancer Research Foundation. The authors acknowledge computer time and resources from the Computer-Aided Drug Design (CADD) Center at the University of Maryland, Baltimore.
Footnotes
Conflict of Interest. A.D.M., Jr. is co-founder and chief scientific officer of SilcsBio LLC.
Supporting Information. Overlap coefficients used to check convergences of the SILCS simulations, GFE cutoffs used to develop SILCS pharmacophore features, enrichment performances for all pharmacophore models of the eight tested targets, set of rules for conversion between CGenFF atom types and SILCS FragMap types for LGFE calculation, and figure showing SILCS exclusion maps for all eight tested targets. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yu W, Guvench O, MacKerell AD. Computational Approaches for the Design of Protein–Protein Interaction Inhibitors. In: Zinzalla G, editor. Understanding and Exploiting Protein–Protein Interactions As Drug Targets. Future Science Ltd; London, UK: 2013. pp. 99–102. [Google Scholar]
- 2.Zhong S, Oashi T, Yu W, Shapiro P, MacKerell AD. Prospects of Modulating Protein–Protein Interactions. In: Gohlke H, editor. Protein-Ligand Interactions. Wiley KGaA; Weinheim, Germany: 2012. pp. 295–329. [Google Scholar]
- 3.Leach AR, Gillet VJ, Lewis RA, Taylor R. Three-Dimensional Pharmacophore Methods in Drug Discovery. J Med Chem. 2009;53:539–558. doi: 10.1021/jm900817u. [DOI] [PubMed] [Google Scholar]
- 4.Yang SY. Pharmacophore Modeling and Applications in Drug Discovery: Challenges and Recent Advances. Drug Discovery Today. 2010;15:444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Joseph-McCarthy D, Alvarez JC. Automated Generation of MCSS-Derived Pharmacophoric DOCK Site Points for Searching Multiconformation Databases. Proteins: Struct, Funct Bioinf. 2003;51:189–202. doi: 10.1002/prot.10296. [DOI] [PubMed] [Google Scholar]
- 6.Cross S, Baroni M, Goracci L, Cruciani G. GRID-Based Three-Dimensional Pharmacophores I: FLAPpharm, A Novel Approach for Pharmacophore Elucidation. J Chem Inf Model. 2012;52:2587–2598. doi: 10.1021/ci300153d. [DOI] [PubMed] [Google Scholar]
- 7.Hu B, Lill MA. Protein Pharmacophore Selection Using Hydration-Site Analysis. J Chem Inf Model. 2012;52:1046–1060. doi: 10.1021/ci200620h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurczab R, Bojarski AJ. New Strategy for Receptor-Based Pharmacophore Query Construction: A Case Study for 5-HT7 Receptor Ligands. J Chem Inf Model. 2013;53:3233–3243. doi: 10.1021/ci4005207. [DOI] [PubMed] [Google Scholar]
- 9.Indarte Mn, Liu Y, Madura JD, Surratt CK. Receptor-Based Discovery of a Plasmalemmal Monoamine Transporter Inhibitor via High-Throughput Docking and Pharmacophore Modeling. ACS Chemical Neuroscience. 2010;1:223–233. doi: 10.1021/cn900032u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng J, Lee KW, Sanchez T, Cui M, Neamati N, Briggs JM. Dynamic Receptor-Based Pharmacophore Model Development and Its Application in Designing Novel HIV-1 Integrase Inhibitors. J Med Chem. 2005;48:1496–1505. doi: 10.1021/jm049410e. [DOI] [PubMed] [Google Scholar]
- 11.Lerner MG, Bowman AL, Carlson HA. Incorporating Dynamics in E. coli Dihydrofolate Reductase Enhances Structure-Based Drug Discovery. J Chem Inf Model. 2007;47:2358–2365. doi: 10.1021/ci700167n. [DOI] [PubMed] [Google Scholar]
- 12.Guha S, Majumbar D. Effect of Hydration on the Conformational Properties and Pharmacophoric Pattern of Several GABA Mediators. J Mol Struct THEOCHEM. 1992;257:451–473. [Google Scholar]
- 13.Bowman AL, Makriyannis A. Approximating Protein Flexibility through Dynamic Pharmacophore Models: Application to Fatty Acid Amide Hydrolase (FAAH) J Chem Inf Model. 2011;51:3247–3253. doi: 10.1021/ci200371z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Zhou S, Yu K, Gao B, Jiang H, Zhen X, Fu W. Molecular Modeling of the 3D Structure of 5-HT1AR: Discovery of Novel 5-HT1AR Agonists via Dynamic Pharmacophore-Based Virtual Screening. J Chem Inf Model. 2013;53:3202–3211. doi: 10.1021/ci400481p. [DOI] [PubMed] [Google Scholar]
- 15.Thangapandian S, John S, Lee Y, Kim S, Lee KW. Dynamic Structure-Based Pharmacophore Model Development: A New and Effective Addition in the Histone Deacetylase 8 (HDAC8) Inhibitor Discovery. Int J Mol Sci. 2011;12:9440–9462. doi: 10.3390/ijms12129440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, Lakkaraju S, Raman EP, MacKerell A., Jr Site-Identification by Ligand Competitive Saturation (SILCS) Assisted Pharmacophore Modeling. J Comput-Aided Mol Des. 2014;28:491–507. doi: 10.1007/s10822-014-9728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guvench O, MacKerell AD., Jr Computational Fragment-Based Binding Site Identification by Ligand Competitive Saturation. PLoS Comput Biol. 2009;5:e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raman EP, Yu W, Guvench O, MacKerell AD. Reproducing Crystal Binding Modes of Ligand Functional Groups Using Site-Identification by Ligand Competitive Saturation (SILCS) Simulations. J Chem Inf Model. 2011;51:877–896. doi: 10.1021/ci100462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raman EP, Vanommeslaeghe K, MacKerell AD. Site-Specific Fragment Identification Guided by Single-Step Free Energy Perturbation Calculations. J Chem Theory Comput. 2012;8:3513–3525. doi: 10.1021/ct300088r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao X, Yap J, Newell-Rogers M, Peddaboina C, Jiang W, Papaconstantinou H, Jupitor D, Rai A, Jung KY, Tubin R, Yu W, Vanommeslaeghe K, Wilder P, MacKerell A, Fletcher S, Smythe R. The Novel BH3 Alpha-Helix Mimetic JY-1-106 Induces Apoptosis in a Subset of Cancer Cells (Lung Cancer, Colon Cancer and Mesothelioma) by Disrupting Bcl-xL and Mcl-1 Protein-Protein Interactions with Bak. Mol Cancer. 2013;12:42. doi: 10.1186/1476-4598-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster TJ, MacKerell AD, Guvench O. Balancing Target Flexibility and Target Denaturation in Computational Fragment-Based Inhibitor Discovery. J Comput Chem. 2012;33:1880–1891. doi: 10.1002/jcc.23026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang N, Shoichet BK, Irwin JJ. Benchmarking Sets for Molecular Docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing TA, Makino S, Skillman AG, Kuntz I. DOCK 4.0: Search Strategies for Automated Molecular Docking of Flexible Molecule Databases. J Comput-Aided Mol Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 24.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman EP, Yu W, Lakkaraju SK, MacKerell AD. Inclusion of Multiple Fragment Types in the Site Identification by Ligand Competitive Saturation (SILCS) Approach. J Chem Inf Model. 2013;53:3384–3398. doi: 10.1021/ci4005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking With A New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: A Computer-Based Archival File for Macromolecular Structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 28.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and Glutamine: Using Hydrogen Atom Contacts in the Choice of Side-Chain Amide Orientation. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 29.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, Flexible, and Free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 30.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 31.Mackerell AD, Feig M, Brooks CL. Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 32.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu W, He X, Vanommeslaeghe K, MacKerell AD. Extension of the CHARMM General Force Field to Sulfonyl-Containing Compounds and Its Utility in Biomolecular Simulations. J Comput Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 35.Durell SR, Brooks BR, Ben-Naim A. Solvent-Induced Forces between Two Hydrophilic Groups. J Phys Chem. 1994;98:2198–2202. [Google Scholar]
- 36.Molecular operating environment (MOE), 2012.10. Chemical Computing Group Inc; Montreal, Canada: 2012. [Google Scholar]
- 37.Halgren TA. Merck Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 38.Zhong S, Chen X, Zhu X, Dziegielewska B, Bachman KE, Ellenberger T, Ballin JD, Wilson GM, Tomkinson AE, MacKerell AD. Identification and Validation of Human DNA Ligase Inhibitors Using Computer-Aided Drug Design. J Med Chem. 2008;51:4553–4562. doi: 10.1021/jm8001668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Farès C, Arrowsmith CH, Yang SN, Garcia M, Coop A, MacKerell AD, Jr, Privé GG, Melnick A. A Small-Molecule Inhibitor of BCL6 Kills DLBCL Cells In Vitro and In Vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated Docking Using A Lamarckian Genetic Algorithm and An Empirical Binding Free Energy Function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 41.Zweig MH, Campbell G. Receiver-Operating Characteristic (ROC) Plots: A Fundamental Evaluation Tool in Clinical Medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]
- 42.Lam PY, Jadhav PK, Eyermann CJ, Hodge CN, Ru Y, Bacheler LT, Meek JL, Otto MJ, Rayner MM, Wong YN, et al. Rational Design of Potent, Bioavailable, Nonpeptide Cyclic Ureas as HIV Protease Inhibitors. Science. 1994;263:380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]
- 43.Schaal W, Karlsson A, Ahlsén G, Lindberg J, Andersson HO, Danielson UH, Classon B, Unge T, Samuelsson B, Hultén J, Hallberg A, Karlén A. Synthesis and Comparative Molecular Field Analysis (CoMFA) of Symmetric and Nonsymmetric Cyclic Sulfamide HIV-1 Protease Inhibitors. J Med Chem. 2000;44:155–169. doi: 10.1021/jm001024j. [DOI] [PubMed] [Google Scholar]
- 44.Smith R, Brereton IM, Chai RY, Kent SBH. Ionization States of the Catalytic Residues in HIV-1 Protease. Nat Struct Mol Biol. 1996;3:946–950. doi: 10.1038/nsb1196-946. [DOI] [PubMed] [Google Scholar]
- 45.Adler M, Davey DD, Phillips GB, Kim SH, Jancarik J, Rumennik G, Light DR, Whitlow M. Preparation, Characterization, and the Crystal Structure of the Inhibitor ZK-807834 (CI-1031) Complexed with Factor Xa. Biochemistry. 2000;39:12534–12542. doi: 10.1021/bi001477q. [DOI] [PubMed] [Google Scholar]
- 46.Maignan S, Guilloteau JP, Pouzieux S, Choi-Sledeski YM, Becker MR, Klein SI, Ewing WR, Pauls HW, Spada AP, Mikol V. Crystal Structures of Human Factor Xa Complexed with Potent Inhibitors. J Med Chem. 2000;43:3226–3232. doi: 10.1021/jm000940u. [DOI] [PubMed] [Google Scholar]
- 47.Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J. Crystal Structures of Escherichia Coli and Lactobacillus Casei Dihydrofolate Reductase Refined at 1.7 A Resolution. I. General Features and Binding of Methotrexate. J Biol Chem. 1982;257:13650–13662. [PubMed] [Google Scholar]
- 48.Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, Brueggen J, Jensen MR, Schnell C, Schmid H, Wartmann M, Berghausen J, Drueckes P, Zimmerlin A, Bussiere D, Murray J, Graus Porta D. Discovery of 3-(2,6-Dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), A Potent and Selective Inhibitor of the Fibroblast Growth Factor Receptor Family of Receptor Tyrosine Kinase. J Med Chem. 2011;54:7066–7083. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 49.Fitzgerald CE, Patel SB, Becker JW, Cameron PM, Zaller D, Pikounis VB, O’Keefe SJ, Scapin G. Structural basis for p38[alpha] MAP Kinase Quinazolinone and Pyridol-Pyrimidine Inhibitor Specificity. Nat Struct Mol Biol. 2003;10:764–769. doi: 10.1038/nsb949. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Canagarajah BJ, Boehm JC, Kassisà S, Cobb MH, Young PR, Abdel-Meguid S, Adams JL, Goldsmith EJ. Structural Basis of Inhibitor Selectivity in MAP Kinases. Structure. 1998;6:1117–1128. doi: 10.1016/s0969-2126(98)00113-0. [DOI] [PubMed] [Google Scholar]
- 51.Shewchuk L, Hassell A, Wisely B, Rocque W, Holmes W, Veal J, Kuyper LF. Binding Mode of the 4-Anilinoquinazoline Class of Protein Kinase Inhibitor: X-ray Crystallographic Studies of 4-Anilinoquinazolines Bound to Cyclin-Dependent Kinase 2 and p38 Kinase. J Med Chem. 1999;43:133–138. doi: 10.1021/jm990401t. [DOI] [PubMed] [Google Scholar]
- 52.Terasaka T, Kinoshita T, Kuno M, Nakanishi I. A Highly Potent Non-Nucleoside Adenosine Deaminase Inhibitor: Efficient Drug Discovery by Intentional Lead Hybridization. J Am Chem Soc. 2003;126:34–35. doi: 10.1021/ja038606l. [DOI] [PubMed] [Google Scholar]
- 53.Terasaka T, Kinoshita T, Kuno M, Seki N, Tanaka K, Nakanishi I. Structure-Based Design, Synthesis, and Structure–Activity Relationship Studies of Novel Non-nucleoside Adenosine Deaminase Inhibitors. J Med Chem. 2004;47:3730–3743. doi: 10.1021/jm0306374. [DOI] [PubMed] [Google Scholar]
- 54.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The Structural Basis of Estrogen Receptor/Coactivator Recognition and the Antagonism of This Interaction by Tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 55.Blizzard TA, DiNinno F, Morgan JD, II, Chen HY, Wu JY, Kim S, Chan W, Birzin ET, Yang YT, Pai LY, Fitzgerald PMD, Sharma N, Li Y, Zhang Z, Hayes EC, DaSilva CA, Tang W, Rohrer SP, Schaeffer JM, Hammond ML. Estrogen Receptor Ligands. Part 9: Dihydrobenzoxathiin SERAMs with Alkyl Substituted Pyrrolidine Side Chains and Linkers. Bioorg Med Chem Lett. 2005;15:107–113. doi: 10.1016/j.bmcl.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 56.Powers RA, Morandi F, Shoichet BK. Structure-Based Discovery of a Novel, Noncovalent Inhibitor of AmpC β-Lactamase. Structure. 2002;10:1013–1023. doi: 10.1016/s0969-2126(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 57.Tondi D, Morandi F, Bonnet R, Costi MP, Shoichet BK. Structure-Based Optimization of a Non-β-lactam Lead Results in Inhibitors That Do Not Up-Regulate β-Lactamase Expression in Cell Culture. J Am Chem Soc. 2005;127:4632–4639. doi: 10.1021/ja042984o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R, Wang S. How Does Consensus Scoring Work for Virtual Library Screening? An Idealized Computer Experiment. J Chem Inf Comput Sci. 2001;41:1422–1426. doi: 10.1021/ci010025x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.