Abstract
Dual-specificity phosphatase 2 (Dusp2; also called phosphatase of activated cells 1, PAC1) is highly expressed in activated immune cells. We examined whether a potential inhibitor of Dusp2, salubrinal, prevents inflammatory cytokine expression in immune cells and arthritic responses in a mouse model of anti-collagen antibody-induced arthritis (CAIA). Salubrinal is a synthetic chemical that inhibits de-phosphorylation of eukaryotic translation initiation factor 2 alpha (eIF2α). In this study, we examined the effects of salubrinal on expression of inflammation linked genes as well as a family of DUSP genes using genome-wide microarrays, qPCR, and RNA interference. We also evaluated the effects of salubrinal on arthritic responses in CAIA mice using clinical and histological scores. The results revealed that salubrinal decreased inflammatory gene expression in macrophages, T lymphocytes, and mast cells. Dusp2 was suppressed by salubrinal in LPS-activated macrophages as well as PMA/ionomycin-activated T lymphocytes and mast cells. Furthermore, a partial silencing of Dusp2 downregulated IL1β and Cox2, and the inflammatory signs of CAIA mice were significantly suppressed by salubrinal. Collectively, this study presents a novel therapeutic possibility of salubrinal for inflammatory arthritis such as RA through inhibition of Dusp2.
Keywords: Salubrinal, CAIA, microarray, Inflammation, Dusp2 (PAC1)
1. Introduction
Dual-specificity phosphatases (DUSPs) are a family of phosphatases that de-phosphorylate both phosphotyrosine and phosphoserine/phosphothreonine residues [1]. Among 30 putative DUSP genes, many of them regulate mitogen-activated protein kinases (MAPKs) and thus referred to as MAPK phosphatases [2]. Dusp2 (PAC1) is one of those MAPK phosphatases, and it is predominantly expressed in immune cells [2, 3]. Dusp2 is highly expressed in immune cells, in particular, of patients with inflammatory arthritis, such as in rheumatoid arthritis (RA), and Dusp2 knockout mice present a significant reduction in inflammatory responses [4].
Because of its elevated expression in RA, we investigated herein a possibility of regulating RA symptoms through the inhibition of Dusp2. RA is the most common type of autoimmune arthritis, in which the immune system mistakenly attacks joint tissues [5]. There is no cure for RA, but synthetic agents such as methotrexate and hydroxychloroquine are commonly prescribed as disease-modifying antirheumatic drugs [6], as well as biologic response modifiers such as Etanercept and Adalimumab [7, 8]. To our knowledge, however, little is known about agents that modulate expression of Dusp2.
In the current study, we focused on a synthetic chemical agent, salubrinal, as a candidate drug for the modulation of Dusp2. Salubrinal is a 480-Da agent that inhibits a serine phosphatase, protein phosphatase 1 (PP1), and elevates the phosphorylation of eukaryotic translation initiation factor 2 alpha (eIF2α) [9]. Through eIF2α-mediated transcriptional and translational regulation, salubrinal has been shown to exhibit multiple beneficial effects on skeletal tissue. First, salubrinal downregulates expression and activity of MMP13 in chondrocytes by inactivating p38 and NFκB signaling pathways [10]. Second, the elevated phosphorylation of eIF2α by salubrinal activates translation of activating transcription factor 4 (ATF4), which stimulates osteoblastogenesis and bone formation [11]. Third, salubrinal suppresses receptor activator of nuclear factor kappa-B ligand (RANKL)-induced osteoclastogenesis followed by bone resorption by inactivating nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1) in bone marrow-derived cells [12, 13]. We performed herein genome-wide expression analysis in order to evaluate un-examined roles of salubrinal in the transcriptional regulation of varying phosphatases including Dusp2 and other Dusp family genes.
In this study, we conducted in vitro analysis using 4 sources of immune cells (RAW264.7 macrophages, primary macrophages, Jurkat T lymphocytes, and HMC-1.1 mast cells). Salubrinal’s effects on inflammatory responses were examined through genome-wide microarray experiments followed by a principal component analysis (PCA). PCA highlighted a set of genes which are most significantly affected by administration of salubrinal, including DUSP family genes. To examine physiological effects of salubrinal in inflammatory arthritis, we conducted in vivo analysis using a mouse model of anti-collagen antibody-induced arthritis (CAIA) [14]. The CAIA model offers several key advantages over the classic collagen-induced arthritis (CIA) model, including rapid disease onset and synchronicity [15]. To evaluate salubrinal’s role in the suppression of inflammatory responses in CAIA mice, we quantified inflammatory symptoms using a clinical scoring system and a histological scoring system.
2. Materials and methods
2.1 Cell Culture
Mouse bone marrow cells and RAW264.7 macrophages were cultured in αMEM with 10% FBS and antibiotics. Bone marrow cells were cultured with 10 ng/ml M-CSF (macrophage colony-stimulating factor; PeproTech, Rocky Hills, NC, USA) for 3 days, and the surface-attached cells were used as primary macrophages. Jurkat T lymphocytes and HMC-1.1 mast cells were cultured in RPMI 1640 and IMDM with 1-thioglycerol, respectively. RAW264.7 cells were activated by 0.1 or 1 µg/ml lipopolysaccharide (LPS), while Jurkat cells by 100 nM phorbol myristate acetate (PMA) and 1 µM ionomycin.
2.2 Induction of Anti-collagen Antibody Induced Arthritis (CAIA) and Clinical Score
Using Balb/c female mice (~nine weeks old), CAIA was induced by intravenous injection of a 2 mg cocktail of ArthritoMAb™ antibodies (MD Bioproducts, St Paul, MN, USA) on day 0 followed by intraperitoneal injection of 100 µg LPS on day 3 [14, 15]. Mice were randomly divided into a placebo group and a salubrinal-treated group. Salubrinal (2.0 mg/kg) was intravenously administered daily from day 0, while a solvent (49.5% PEG 400 and 0.5% Tween 80 in PBS) was administered to the placebo group.
The progression of CAIA was evaluated using a clinical score [16]: “0.25” = swelling in a single digit; “0.5” = swelling in more than one digit; “1” = swelling and erythema of the paw; “2” = swelling of the paw and ankle; and “3” = complete inflammation of the paw. The maximum possible score for each mouse was 12. We also measured thickness of fore and hind paws.
2.3 Histological Evaluation
Hind paws were harvested and decalcified in 10% EDTA for 2 weeks. They were embedded in paraffin, sectioned at 4 µm thickness, and stained with hematoxylin and eosin (H&E). The progression of CAIA was histologically evaluated using the scoring system [17]: “0” = normal; “1” = weak leukocyte infiltration but no erosion; “2” = modest infiltration and weak erosion; “3” = severe infiltration and invasion of bones; and “4” = loss of bone integrity.
2.4 Microarray
Genome-wide expression analysis was conducted using RAW264.7 cells (Mouse Gene 2.0 ST arrays, Affymetrix) as well as Jurkat cells (Human Gene 2.0 ST arrays, Affymetrix). Three groups for RAW264.7 cells were CN (control), LPS, and Sal (LPS + Sal), while for Jurkat cells they were CN (control), PMA (PMA/ionomycin), and Sal (PMA/ionomycin + Sal). The concentration of LPS, PMA, ionomycin, and salubrinal were 0.1 µg/ml, 100 nM, 1 µM, and 10 µM, respectively, and these agents were administered 0 h. Each group consisted of triplicate samples, which were harvested 6 h.
We selected a group of “activated” genes, whose mRNA levels were lowered by LPS or PMA/ionomycin (p < 0.05) and the decrease was reversed by salubrinal (p < 0.05). We also identified a group of “suppressed” genes, whose mRNA levels were elevated by the inducing agents (p < 0.05) and the elevation was suppressed by salubrinal (p < 0.05). Principal component analysis (PCA) was performed on those activated and suppressed genes [18]. The nine samples in total were positioned in the plane of the first and second principal axes.
Using WebGestalt, salubrinal-driven signaling pathways were identified [19]. Based on the hypergeometric statistical test with a Bonferroni correction (p<0.05), we selected pathways that significantly enriched the appearance of the activated or suppressed genes. We also plotted the location of each gene on the first and second principal component axes and highlighted the genes involved in three representative pathways.
2.5 Quantitative Real-time PCR
Total RNA was extracted and reverse transcription was conducted with high-capacity cDNA reverse transcription kits (Applied Biosystems, Carlsbad, CA, USA). Quantitative real-time PCR was performed using Power SYBR green PCR master mix kits (Applied Biosystems). We evaluated mRNA levels of Cox2, Dusp2, IL1β, IL2, IL13, and TNF using the PCR primers (Table 1). GAPDH was used for internal control [20].
Table 1.
Real-time PCR primers and siRNA used in this study.
| mouse | ||
| target | forward primer | backward primer |
| Cox2 | 5’-CCCCCACAGTCAAAGACACT-3’ | 5’-CTCATCACCCCACTCAGGAT-3’ |
| Dusp2 | 5’-TGGAAATCTTGCCCTACCTG-3 | 5’-CTCCTGGAACCAGGCACTTA-3’ |
| IL1β | 5’-GCCCATCCTCTGTGACTCAT-3’ | 5’-AGGCCACAGGTATTTTGTCG-3’ |
| GAPDH | 5’-TGCACCACCAACTGCTTAG-3’ | 5’-GGATGCAGGGATGATGTTC-3’ |
| human | ||
| target | forward primer | backward primer |
| Dusp2 | 5’-TCTTGCCCTACCTGTTCCTG-3’ | 5’-GGCACTGATCTCCACCATCT-3’ |
| IL2 | 5’-GCAACTCCTGTCTTGCATTG-3’ | 5’-GCCTTCTTGGGCATGTAAAA-3’ |
| IL13 | 5’-GTACTGTGCAGCCCTGGAAT-3’ | 5’-TTTACAAACTGGGCCACCTC-3’ |
| TNF | 5’-CAGAGGGCCTGTACCTCATC-3’ | 5’-GGAAGACCCCTCCCAGATAG-3’ |
| GAPDH | 5’-GCACCGTCAAGGCTGAGAAC-3’ | 5’-ATGGTGGTGAAGACGCCAGT-3’ |
| siRNA | ||
| Dusp2 | 5’-GCAUCACAGCAGUUCUCAA-3’ | |
| non-specific control | 5’-UGUACUGCUUACGAUUCGG-3’ | |
2.6 Knockdown of Dusp2 by siRNA
RAW264.7 cells were treated with siRNA specific to Dusp2 or nonspecific control siRNA (Life Technologies) (Table 1). Cells were transiently transfected in Opti-MEM I medium using Lipofectamine RNAiMAX (Life Technologies). The efficiency of silencing was assessed with PCR 48 h after transfection.
2.7 Statistical Analysis
For in vitro experiments, 3 to 4 independent experiments were conducted. Data were expressed as mean ± S.D. Statistical significance was evaluated using Student’s t-test at p<0.05. Single and double asterisks indicate p<0.05 and p<0.01, respectively.
3. Results
3.1 Suppression of Inflammatory Cytokines and Cox2 by Salubrinal
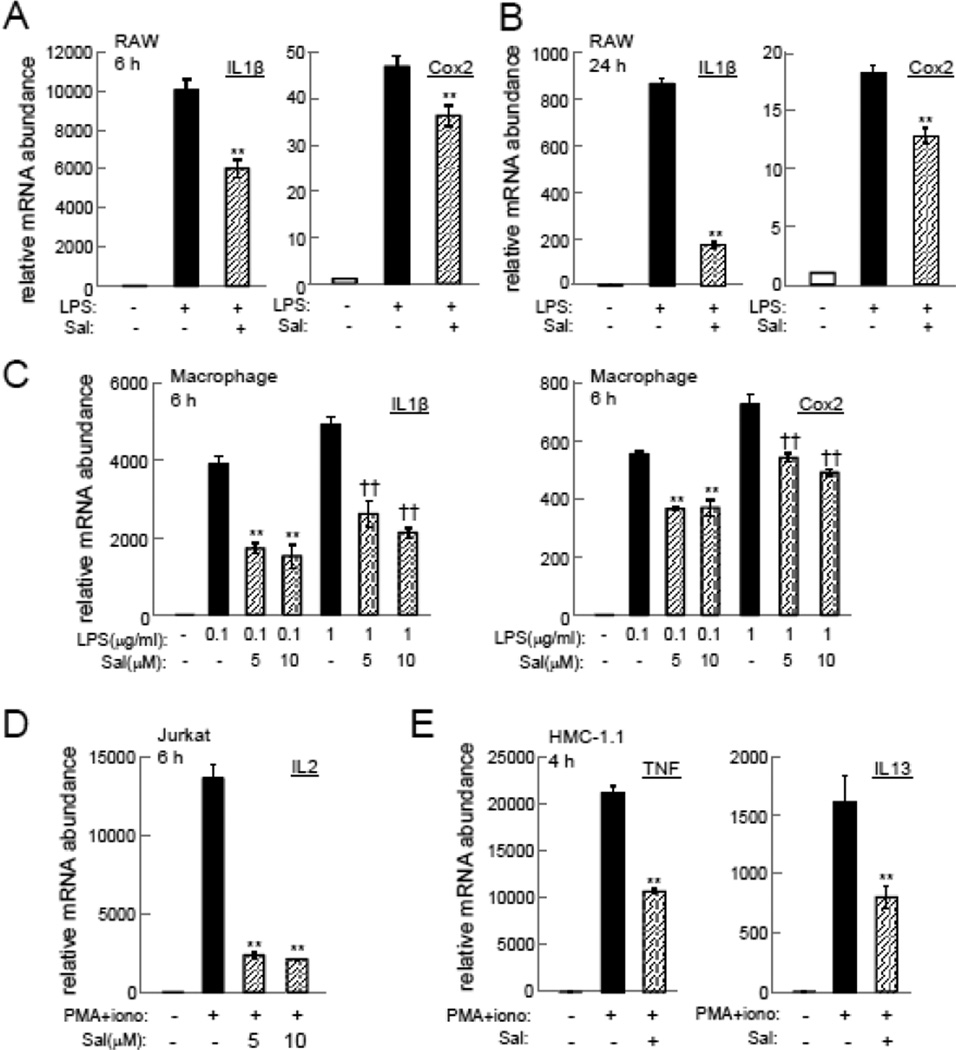
Incubation of RAW264.7 macrophages with 10 µM salubrinal for 6 h and 24 h significantly suppressed IL1β and Cox2 mRNAs (Figure 1A and B). The suppression of these mRNAs was also observed in primary macrophages by 5 or 10 µM salubrinal (Fig. 1C). Furthermore, the elevation of IL2 mRNA in Jurkat cells as well as the increase in TNF and IL13 mRNAs in HMC-1.1 cells by PMA/ionomycin was attenuated by 5 or 10 µM salubrinal (Fig. 1D & E).
Figure 1.
Suppression of inflammatory genes by salubrinal (Sal). (A&B) Effects of 10 µM salubrinal on IL1β and Cox2 mRNAs in RAW264.7 cells at 6 h and 24 h, respectively. (C) Effects of 5 or 10 µM salubrinal on IL1β and Cox2 mRNAs in primary macrophages at 6 h. (D) Effects of 5 or 10 µM salubrinal on IL2 mRNA in Jurkat cells at 6 h. (E) Effects of 10 µM salubrinal on TNF and IL13 mRNAs in HMC-1.1 cells at 4 h.
3.2 Genome-wide Modulation of Inflammatory Responses by Salubrinal
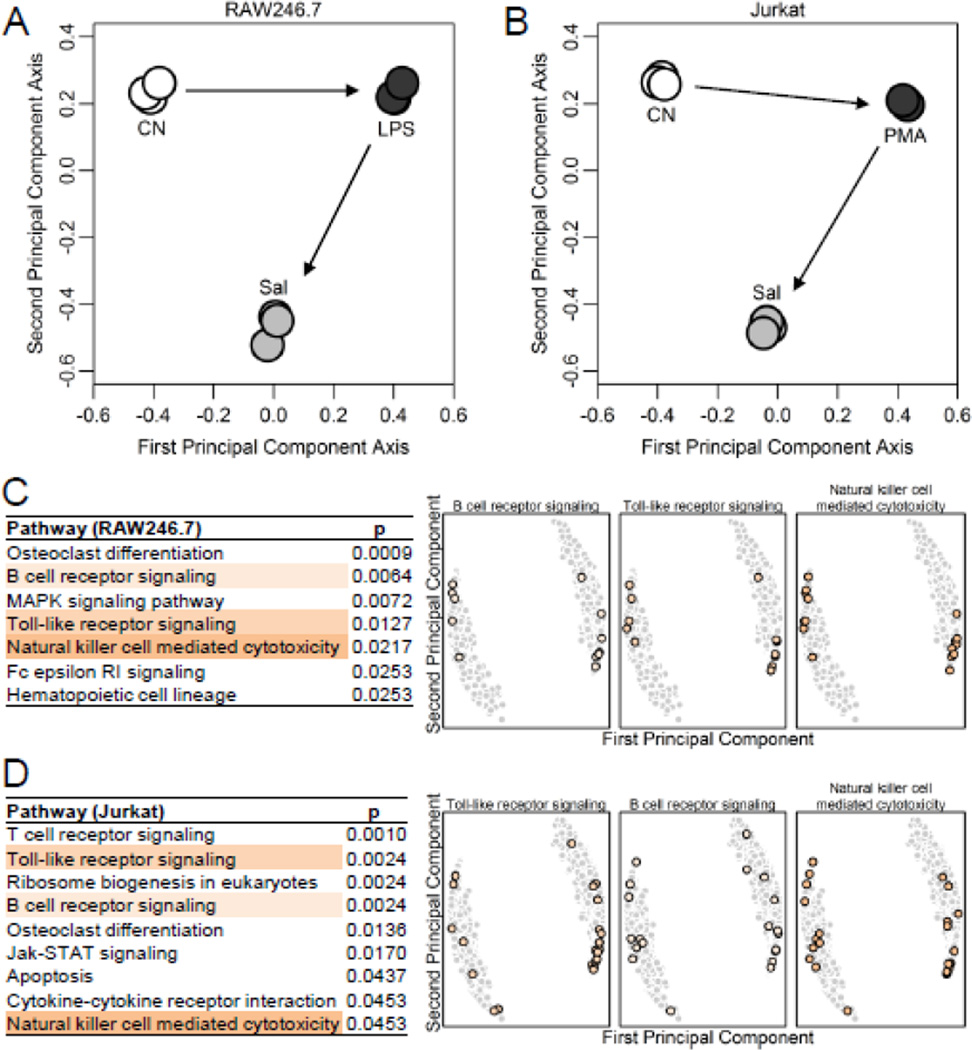
PCA revealed that along the first primary component axis, LPS or PMA/ionomycin driven alterations was partially restored by salubrinal (Fig. 2A & B). A pathway analysis showed the enrichment of several inflammation-related pathways in both the RAW246.7 and Jurkat cells (Fig. 2C & D & Fig. S1). Three pathways (B cell receptor signaling, Toll-like receptor signaling, and Natural killer cell mediated cytotoxicity) were commonly enriched between the two cell types. The genes involved in each pathway were identified on the first and second principal component axes.
Figure 2.
PCA and pathway prediction. (A) PCA for RAW264.7 cells. (B) PCA for Jurkat cells. (C&D) Enriched KEGG pathways by genes activated and suppressed by salubrinal in RAW264.7 and Jurkat cells, respectively. Three pathways, common to RAW264.7 and Jurkat cells, are highlighted. Genes in each pathway are identified on the first and second principal component axes.
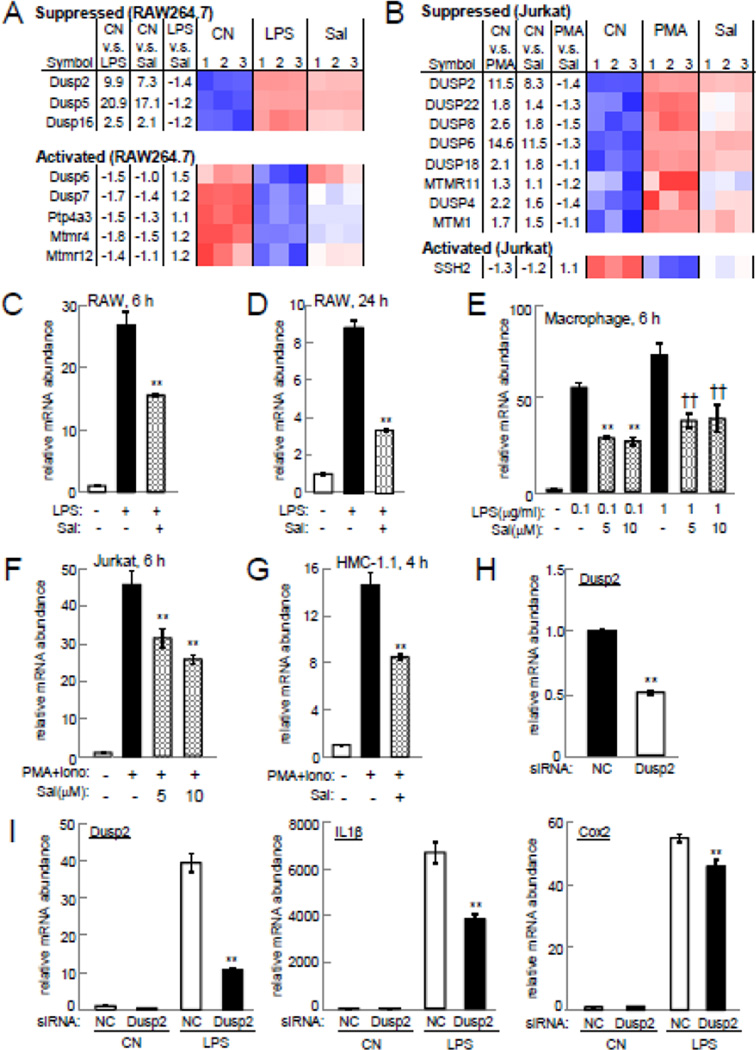
When we focused on the genes commonly altered in both types of cells and selected either “activated” or “suppressed” by salubrinal, a consensus expression pattern emerged, and Dusp2 was found to be suppressed in both cell lines (Fig. 3A). The activated and suppressed genes were identified on the first and second principal component axes in RAW246.7 cells (Fig. 3B) and Jurkat cells (Fig. 3C). Of note, Dusp2 was located near the lower end of the suppressed gene cluster in both cell types.
Figure 3.
(A) Heatmaps showing the top 10 genes highly activated or suppressed by 10 µM salubrinal commonly in RAW264.7 and Jurkat cells. Note that CN=control, and Sal=salubrinal in the presence of LPS (RAW264.7) or PMA/ionomycin (Jurkat). (B) Genes activated (red) or suppressed (blue) in RAW264.7 cells on the first and second principal component axes. (C) Genes activated (red) or suppressed (blue) in Jurkat cells on the first and second principal component axes.
3.3 Salubrinal-driven Suppression of Dusp2
Among the Dusp genes, the genes suppressed in RAW264.7 cells included Dusp2, Dusp5, and Dusp16, while in Jurkat cells they were Dusp2, Dusp4, Dusp6, Dusp8, Dusp18, and Dusp22 (Fig. 4A & B). Dusp2 was the only gene whose upregulation by LPS or PMA/ionomycin was suppressed by salubrinal in two cell lines. No Dusp genes were activated in both RAW264.7 and Jurkat cells. In RAW264.7, primary macrophages, Jurkat T lymphocytes, and HMC-1.1 mast cells, qPCR confirmed that the mRNA level of Dusp2 was elevated by LPS or PMA/ionomycin and its upregulation was decreased by 5 or 10 µM salubrinal (Fig. 4C – G).
Figure 4.
Salubrinal-driven suppression of Dusp2 upregulation. Note that CN=control, and Sal=salubrinal in the presence of LPS (RAW264.7) or PMA/ionomycin (Jurkat). (A) Heatmap of DUSP genes in RAW264.7 cells. (B) Heatmap of DUSP genes in Jurkat cells. (C&D) Levels of Dusp2 mRNA in RAW264.7 cells at 6 h and 24 h, respectively. (E) Dusp2 mRNA level in primary macrophages at 6 h. (F) Dusp2 mRNA level in Jurkat cells at 6 h. (G) Dusp2 mRNA level in HMC-1.1 cells at 4 h. (H) Dusp2 mRNA level in RAW264.7 cells treated with Dusp2 siRNA. (I) Levels of Dusp2, IL1β, and Cox2 mRNAs in RAW264.7 cells treated with Dusp2 siRNA in the presence and absence of LPS at 6 h.
3.4 Downregulation of IL1β and Cox2 by Partial Silencing of Dusp2
RNA interference using siRNA specific to Dusp2 revealed that partial silencing of Dusp2 significantly suppressed LPS-driven upregulation of IL1β and Cox2 in RAW264.7 cells (Fig. 4H & I). In the presence of LPS, treatment with Dusp2 siRNA reduced the level of IL1β mRNA by 43%, and Cox2 by 17%.
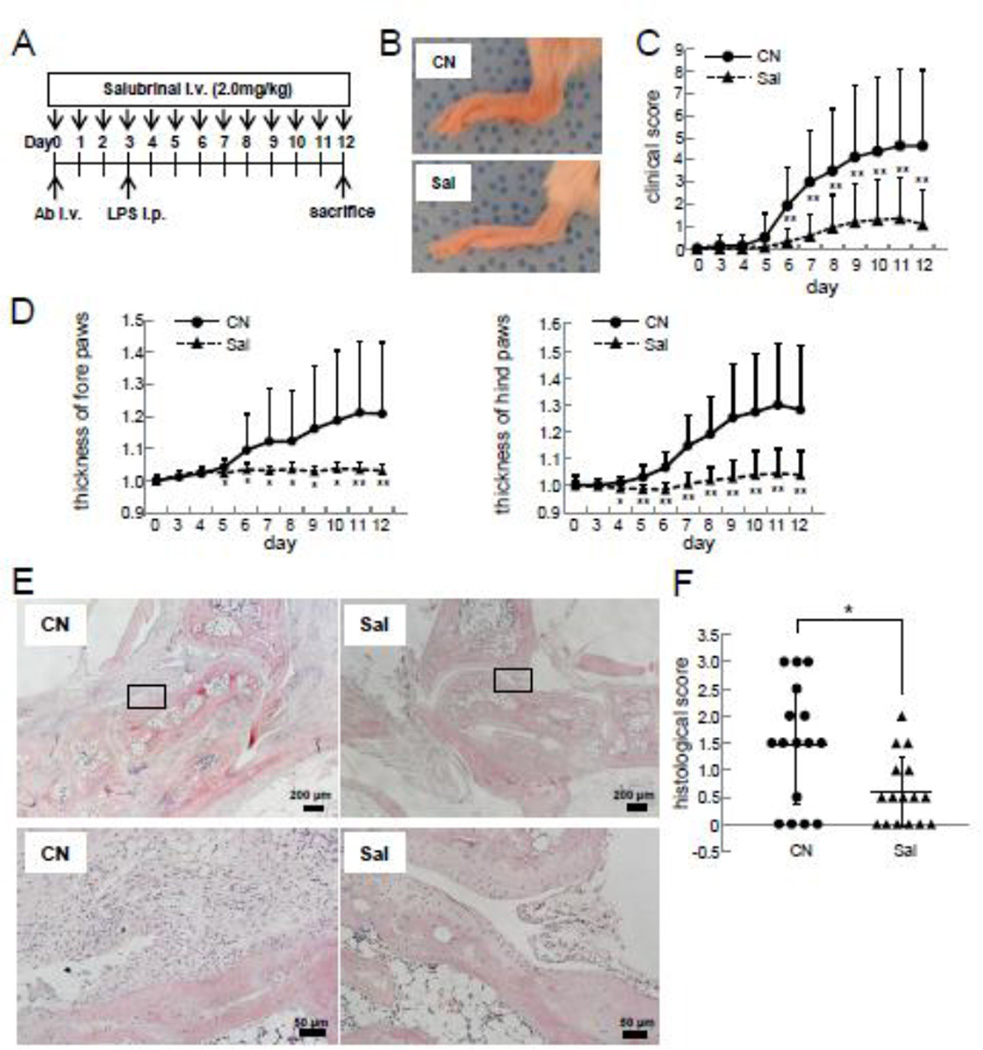
3.5 Salubrinal-driven Suppression of the Progression of CAIA
Salubrinal significantly suppressed inflammation of the paws of CAIA mice (Fig. 5A & B). For instance, the clinical scores were 1.94±1.7 (placebo) and 0.31±0.6 (salubrinal) on day 6; and 4.63±3.4 (placebo) and 1.09±1.6 (salubrinal) on day 12 (Fig. 5C). Consistent with the clinical scores, the thickening of the paws was also reduced in the salubrinal-treated group (Fig. 5D & E). Furthermore, salubrinal reduced the histological scores from 1.47±1.10 (N=16; placebo) to 0.59±0.64 (N =16; salubrinal) (p=0.01) (Fig. 5E & F).
Figure 5.
Suppression of the progression of CAIA by salubrinal. Note that CN=placebo, and Sal=salubrinal. (A) CAIA induction and salubrinal administration. (B) Hind paws 12 days after induction of CAIA. (C) Clinical scores. (D) Temporal changes in the thickness of fore and hind paws. (E) H&E staining of the hind paws. The scale bars are 200 µm (upper) and 50 µm (lower). (F) Histological scores. The single and double asterisks indicate p<0.05 and p<0.01, respectively.
4. Discussion
We showed that salubrinal downregulated the expression of Dusp2 and inflammatory genes in immune cells in vitro, and its administration to CAIA mice significantly reduced inflammatory responses in vivo. In the in vitro assays, genome-wide microarray analysis revealed that Dusp2 may potentially mediate salubrinal’s partial attenuation of the inflammatory effects of LPS and PMA/ionomycin in RAW264.7 and Jurkat cell cultures. In the in vivo analysis with CAIA mice, inflammation was induced by the administration of a cocktail of antibodies that recognize auto-antigenic epitopes in type II collagen, which offers a rapid disease onset and synchronicity over the classic collagen-induced arthritis (CIA) model. The clinical and histological scores as well as thickness measurements of the paws in CAIA mice support the efficacy of salubrinal in suppression of inflammation. On day 12, for instance, the administration of salubrinal improved the clinical scores from 4.63±3.4 to 1.09±1.6, and the histological scores from 1.47±1.10 to 0.59±0.64.
Genome-wide expression analysis using PCA and the classification of suppressed and activated genes revealed that salubrinal significantly suppressed Dusp2 expression. In DUSP genes highlighted in Figure 4, 4 DUSP genes (DUSP1, DUSP2, DUSP4, and DUSP5) are considered to be MAPK phosphatases that dephosphorylate Erk, Jnk, and p38 MAPK [2, 3]. Among those 4 DUSP genes, Dusp2 is known to be an activator of inflammatory responses in T and B cells [4]. Expression analysis revealed that Dusp2 was the only DUSP gene that was downregulated by salubrinal in both RAW264.7 and Jurkat cells. For instance, Dusp5 was reduced by salubrinal in RAW264.7 cells and Dusp4 in Jurkat cells, but not vice versa. Salubrinal activated Dusp6 in RAW264.7 cells but inhibited it in Jurkat cells.
We have previously shown using C28/I2 chondrocytes that salubrinal downregulates cytokine-induced inflammatory responses through the inactivation of NFκB and p38 MAPK [10]. Using RAW264.7 cells, it is also reported that salubrinal suppresses expression of pro-inflammatory cytokines by inactivating NFκB signaling [21]. In cystic fibrosis airway cells that induce an inflammatory lung disease, salubrinal is reported to reduce inflammatory responses through eIF2α-mediated downregulation of p38 MAPK [22]. We also observed in C28/I2 chondrocytes that salubrinal reduces activity of Src tyrosine kinases through the elevation of eIF2α phosphorylation [23]. Of note, Src kinases play important roles in production and activation of various cytokines [24]. Collectively, a salubrinal-Dusp2 regulatory axis that links Dusp2, p38 MAPK, NFκB, and Src kinases through eIF2α signaling is yet to be identified.
Genes involved in inflammatory responses differ depending on the types of immune cells. Among three cell lines (RAW264.7, Jurkat T lymphocytes, and HMC-1.1 mast cells) together with primary macrophages, salubrinal suppressed the elevation of IL1β and Cox2 in each cell type. Salubrinal reduced PMA/ionomycin-driven IL2 expression in Jurkat lymphocytes and decreased the level of TNF and IL13 mRNAs in HMC-1.1 mast cells. Of note, these cytokines (IL1β, IL2, IL13, and TNF) are selectively activated in different types of immune cells [25, 26]. Biologic drugs against TNFα, CD20, and IL1/IL6 receptors have been developed for treatment of RA [27, 28]. They can lessen RA symptoms and delay radiographic progression, but they may also present increased risk of infections.
Besides the suppression of inflammatory responses, the protection of bone erosions at the joint margins and in subchondral bone is critically important for patients with RA [29]. Salubrinal’s advantage is that it attenuates bone destruction by eIF2α-mediated downregulation of NFATc1, a master transcription factor for osteoclastogenesis [11]. Furthermore, salubrinal can downregulate MMP13, a collagenase that digests not only aggrecan in cartilage but also collagen in bone [10].
In order to further evaluate salubrinal as a drug candidate for treatment of inflammatory arthritis such as RA, there are several further studies to be done. First, it is necessary to understand salubrinal’s effect not only on immune cells but also on synovial fibroblasts, since synovial fibroblasts in the rheumatoid synovium are aggressive drivers of cartilage destruction [30]. Second, although the CAIA mouse model can induce inflammatory responses in a short period of time, it is necessary to monitor the efficacy of salubrinal over an extended period of time and determine appropriate dosage and treatment duration. Third, it is important to discover whether salubrinal can be administered with other agents, such as disease-modifying antirheumatic drugs or biological response modifiers, to induce synergistic therapeutic effects [31].
In summary, this study demonstrates that administration of salubrinal reduces expression of Dusp2 as well as inflammatory cytokines such as IL1β, IL2, IL13, and TNF in immune cells. Furthermore, salubrinal suppresses inflammatory responses in CAIA mice. Although it is premature to conclude with the current pre-clinical study, further studies on toxicology and dose dependence could warrant the development of preventive and therapeutic uses of salubrinal for inflammatory arthritis such as RA.
Supplementary Material
Highlights.
Salubrinal prevents inflammatory responses in a mouse model of arthritis.
Genome-wide analysis reveals Dusp2 as a primary target of salubrinal.
IL1β was downregulated by salubrinal and a partial silencing of Dusp2.
Acknowledgments
The authors appreciated J.H. Butterfield for kindly providing HMC-1.1 cells and M. Hamamura for technical assistance. The microarray analysis was supported by CTSA U54-TR001012.
Fundings
The microarray analysis was supported by CTSA U54-TR001012.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conception and experimental design: Hamamura K, Sudo A, Yokota H. data collection and interpretation: Hamamura K, Nishimura A, Chen A, Takigawa S. drafted manuscript: Hamamura K, Chen A, Yokota H.
Competing interests
None
References
- 1.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Biochem. J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 2.Jeffrey KL, Camps M, Rommel C, Mackay CR. Nat. Rev. Drug Discovery. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- 3.Lang R R, Hammer M, Mages J. J. Immonol. 2006;177:7497–7504. doi: 10.4049/jimmunol.177.11.7497. [DOI] [PubMed] [Google Scholar]
- 4.Jeffrey KL, Brummer T, Rolph MS, Liu SM, Callejas NA, Grumont RJ, Gillieron C, Mackay F, Grey S, Camps M, Rommel C, Gerondakis SD, Mackay CR. Nature Immunol. 2006;7:274–283. doi: 10.1038/ni1310. [DOI] [PubMed] [Google Scholar]
- 5.Arend WP. Arthritis Rheum. 2001;45:101–106. doi: 10.1002/1529-0131(200102)45:1<101::AID-ANR90>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 6.Joshi P, Dhaneshwar SS. Inflamm. Allergy Drug Targets. 2014;13:249–261. doi: 10.2174/187152811304140915152102. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Reino J. Rheumatology (Oxford) 2012;51(Suppl 5):v31–v37. doi: 10.1093/rheumatology/kes116. [DOI] [PubMed] [Google Scholar]
- 8.Selmi C, Generali E, Massarotti M, Bianchi G, Scire CA. Immunol. Res. 2014;60:277–288. doi: 10.1007/s12026-014-8565-5. [DOI] [PubMed] [Google Scholar]
- 9.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 10.Hamamura K, Lin CC, Yokota H. Osteoarthritis Cartilage. 2013;21:764–772. doi: 10.1016/j.joca.2013.02.657. [DOI] [PubMed] [Google Scholar]
- 11.Hamamura K, Tanjung N, Yokota H. J. Bone Miner. Metab. 2013;31:618–628. doi: 10.1007/s00774-013-0450-0. [DOI] [PubMed] [Google Scholar]
- 12.Hamamura K, Chen A, Tanjung N, Takigawa S, Sudo A, Yokota H. Cell. Signal. 2015;27:353–362. doi: 10.1016/j.cellsig.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Yokota H, Hamamura K, Chen A, Dodge TR, Tanjung N, Abedinpoor A, Zhang P. BMC Musculoskeletal Disorders. 2013;14:197. doi: 10.1186/1471-2474-14-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandakumar KS, Holmdahl R. J. Immunol. Methods. 2005;304:126–136. doi: 10.1016/j.jim.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Khachigian LM. Nature Protocols. 2006;1:2512–2516. doi: 10.1038/nprot.2006.393. [DOI] [PubMed] [Google Scholar]
- 16.Gould DJ, Bright C, Chernajovsky Y. Arthritis Res. Ther. 2004;6:R103–R113. doi: 10.1186/ar1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang LH, Huang HS, Wu PT, Jou IM, Pan MH, Chang WC, Wang DD, Wang JM. PLoS ONE. 2012;7:e45378. doi: 10.1371/journal.pone.0045378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamamura K, Chen A, Nishimura A, Tanjung N, Sudo A, Yokota H. Cell. Signal. 2014;26:2358–2369. doi: 10.1016/j.cellsig.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Duncan D, Shi Z, Zhang B. Nucl. Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamamura K, Yokota H. FEBS Lett. 2007;581:1769–1774. doi: 10.1016/j.febslet.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrestha N, Bahnan W, Wiley DJ, Barber G, Fields KA, Schesser K. J. Biol. Chem. 2012;287:28738–28744. doi: 10.1074/jbc.M112.375915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blohmke CJ, Maver ML, Tang AC, Hirschfeld AF, Fjell CD, Sze MA, Falsafi R, Wang S, Hsu K, Chilvers MA, Hogg JC, Hancock RE, Turvey SE. J. Immunol. 2012;189:5467–5475. doi: 10.4049/jimmunol.1103661. [DOI] [PubMed] [Google Scholar]
- 23.Wan Q, Xu W, Yan J, Yokota H, Na S. PLOS One. 2014;9:e105699. doi: 10.1371/journal.pone.0105699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page TH, Smolinska M, Gillespie J, Uraniak AM, Foxwell BM. Curr. Mol. Med. 2009;9:69–85. doi: 10.2174/156652409787314507. [DOI] [PubMed] [Google Scholar]
- 25.Bencsath M, Blaskovits A, Borvendeg J. Pathol. Oncol. Res. 2003;9:24–29. doi: 10.1007/BF03033710. [DOI] [PubMed] [Google Scholar]
- 26.Morris JC, Waldmann TA. Ann. Rheum. Dis. 2000;59:i109–i114. doi: 10.1136/ard.59.suppl_1.i109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bingham CO., 3rd Bull. NYU Hosp. Jt. Dis. 2008;66:210–215. [PubMed] [Google Scholar]
- 28.Agarwal SK. J. Manag. Care Pharm. 2011;17(9 Suppl B):S14–S18. doi: 10.18553/jmcp.2011.17.s9-b.S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung SM, Kim KW, Yang CW, Park SH, Ju JH. J. Immunol. Res. 2014:263625. doi: 10.1155/2014/263625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefevre S, Meier FM, Neumann E, Muller-Ladner U. Curr. Pharm. Des. 2014;21:130–141. doi: 10.2174/1381612820666140825122036. [DOI] [PubMed] [Google Scholar]
- 31.Bauerova K, Paulovicova E, Mihalova D, Svik K, Ponist S. Toxicol. Ind. Health. 2009;25:329–335. doi: 10.1177/0748233709102945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.