Abstract
SoxC transcription factors play critical roles in many developmental processes, including neurogenesis, cardiac formation, and skeletal differentiation. In vitro and in vivo loss-of-function studies have suggested that SoxC genes are required for oculogenesis, however the mechanism was poorly understood. Here, we have explored the function of the SoxC factor Sox4 during zebrafish eye development. We show that sox4a and sox4b are expressed in the forebrain and periocular mesenchyme adjacent to the optic stalk during early eye development. Knockdown of sox4 in zebrafish resulted in coloboma, a structural malformation of the eye that is a significant cause of pediatric visual impairment in humans, in which the choroid fissure fails to close. Sox4 morphants displayed altered proximo-distal patterning of the optic vesicle, including expanded pax2 expression in the optic stalk, as well as ectopic cell proliferation in the retina. We show that the abnormal ocular morphogenesis observed in Sox4-deficient zebrafish is caused by elevated Hedgehog (Hh) signaling, and this is due to increased expression of the Hh pathway ligand Indian hedgehog b (ihhb). Consistent with these results, coloboma in sox4 morphants could be rescued by pharmacological treatment with the Hh inhibitor cyclopamine, or by co-knockdown of ihhb. Conversely, overexpression of sox4 reduced Hh signaling and ihhb expression, resulting in cyclopia. Finally, we demonstrate that sox4 and sox11 have overlapping, but not completely redundant, functions in regulating ocular morphogenesis. Taken together, our data demonstrate that Sox4 is required to limit the extent of Hh signaling during eye development, and suggest that mutations in SoxC factors could contribute to the development of coloboma.
Keywords: Sox4, Hedgehog signaling, Indian Hedgehog, zebrafish, coloboma, eye, choroid fissure
INTRODUCTION
Ocular coloboma is a developmental disorder that occurs when the choroid fissure, a transient opening that forms in the ventral portion of the optic cup, fails to properly close, causing a cleft in the inferonasal quadrant of the eye. Depending on the timing and location of the closure defect, coloboma can affect multiple regions of the developing eye, including the cornea, iris, ciliary body, retina, pigmented epithelium, and the optic nerve (Chang et al., 2006). Coloboma is often observed in conjunction with other ocular abnormalities, such as microphthalmia, anophthalmia, or anterior chamber defects, and may also be associated with more complex syndromes affecting multiple systems (Porges et al., 1992; Eccles and Schimmenti, 1999; Morrison et al., 2000; Guirgis and Lueder, 2003; Chang et al., 2006). Although coloboma represents an important cause of pediatric visual impairment, contributing to 3-11% of childhood blindness worldwide (Hornby et al., 2000), the molecular and cellular mechanisms underlying choroid fissure closure and the genetic etiology of coloboma remain poorly understood. This is partly because ocular coloboma is both phenotypically and genetically heterogeneous, and although numerous coloboma-causing genes have been identified, mutations in these genes account for only a minority of cases. Based on data from patient studies and from animal models, a coloboma gene network has been proposed, which highlights the complex relationships between several genes implicated in coloboma or associated ocular malformations (Gregory-Evans et al., 2004). The members of this network include eye field specific transcription factors (EFTFs), cell cycle regulators, and cell signaling molecules that collectively control cell proliferation, cell migration, cell fate specification, and cell death.
One of the central hubs of the coloboma gene network is the secreted molecule Sonic Hedgehog (Shh), one of the ligands for the evolutionarily conserved Hedgehog (Hh) signaling pathway. Hh signaling is critical for the correct morphogenesis, growth, and patterning of several embryonic tissues and organs, and also plays a key role in tissue regeneration, stem cell maintenance, and tumorigenesis (Ingham and McMahon, 2001; Briscoe and Therond, 2013). In the development of the visual system, Hedgehog (Hh) signaling is required at several stages. Early in development, Hh signals emanating from the ventral midline regulate the segregation of a single eye field into two bilateral optic primordia. As the optic vesicles evaginate from the forebrain, Hh signaling controls proximo-distal patterning of the optic vesicle into optic stalk and optic cup domains, respectively (Ekker et al., 1995). And after optic cup formation, Hh signaling within the developing retina regulates neurogenesis by controlling retinal progenitor cell proliferation and differentiation (Marti and Bovolenta, 2002). Reflecting its central role in oculogenesis, mutations in Shh are associated with several ocular defects, including cyclopia, microphthalmia, anophthalmia, and coloboma (Schimmenti et al., 2003; Amato et al., 2004; Gongal et al., 2011). Moreover, the downstream targets of Shh include several coloboma-causing genes (Sanyanusin et al., 1995; Slavotinek et al., 2012). However, less is known about the factors that lie upstream of Hh ligand expression – such genes may also contribute to the pathogenesis of coloboma.
We recently demonstrated that the SoxC transcription factor Sox11 is one of the upstream regulators of Hh signaling, and that in the zebrafish, Sox11 regulates choroid fissure closure as well as retinal neurogenesis by inhibiting the expression of Shh (Pillai-Kastoori et al., 2014). The Sox proteins are named for a shared motif called the SRY box, a high-mobility-group (HMG) DNA binding domain homologous to the DNA-binding domain of the mammalian sex-determining gene SRY. Based on amino acid identity within the HMG domain, Sox proteins are divided into 8 groups (A to H). The SoxC group includes Sox4, Sox11 and Sox12 (Bowles et al., 2000). SoxC family members display overlapping expression domains in several embryonic tissues, including neuronal and mesenchymal progenitor cells (Dy et al., 2008). The SoxC factors function redundantly in the development of some tissues, such as the nervous system, but also have distinct roles in the development of other tissues, such as the heart (Bergsland et al., 2006; Bhattaram et al., 2010; Penzo-Mendez, 2010; Paul et al., 2013). Moreover, Sox11 null mice survive several days longer than Sox4 null mice, suggesting non-redundant roles for these two proteins in early embryogenesis.
In this study, we examined the function of Sox4 in zebrafish ocular development. Like Sox11, Sox4 is expressed in multiple embryonic tissues, including the retina (Dy et al., 2008; Usui et al., 2013; Pillai-Kastoori et al., 2014). In the mouse, Sox4 expression initiates in the central retina near the optic nerve at E11.5. As retinal development proceeds, Sox4 is expressed in the ganglion cell layer (GCL) and the neuroblastic layer (NBL). Constitutive Sox4 null mice die at E14.5 due to severe cardiac malformation and defects in B lymphocyte differentiation (Schilham et al., 1996), precluding a more thorough assessment of Sox4's role in ocular development in this model. A conditional Sox4 knockout mouse in which Sox4 is deleted in the developing retina displayed a modest reduction in ganglion cell number, but no other ocular defects were reported, and the effect of Sox4 deletion on Hh signaling was not investigated (Jiang et al., 2013).
Zebrafish possess two co-orthologs of the mammalian Sox4 gene: sox4a and sox4b. Previously, microarray analysis of retinal mRNA from a zebrafish model of chronic rod photoreceptor degeneration and regeneration revealed that sox4a and sox4b mRNA levels are up-regulated in response to rod photoreceptor loss (Morris et al., 2011), suggesting they function in rod regeneration. However, the role of sox4a/b in embryonic zebrafish ocular development was not known. In this study, we show that Sox4 is required for choroid fissure closure and proper proximo-distal patterning of the optic vesicle in zebrafish. We also demonstrate that loss of Sox4 affects retinal progenitor cell proliferation. Furthermore, we show that similar to loss of Sox11, the ocular phenotypes of Sox4-deficient zebrafish are caused by elevated Hh signaling. However, in contrast to Sox11, we found that Sox4 primarily regulates expression of the Hh signaling ligand Indian Hedgehog (Ihh), rather than Shh. Therefore, our data demonstrate that Sox4 and Sox11 together control levels of Hh signaling during ocular development by regulating expression of distinct Hh ligands, and suggest that SoxC factors may be additional members of the coloboma gene network.
RESULTS
Sox4a and sox4b are expressed in periocular tissues and the developing retina
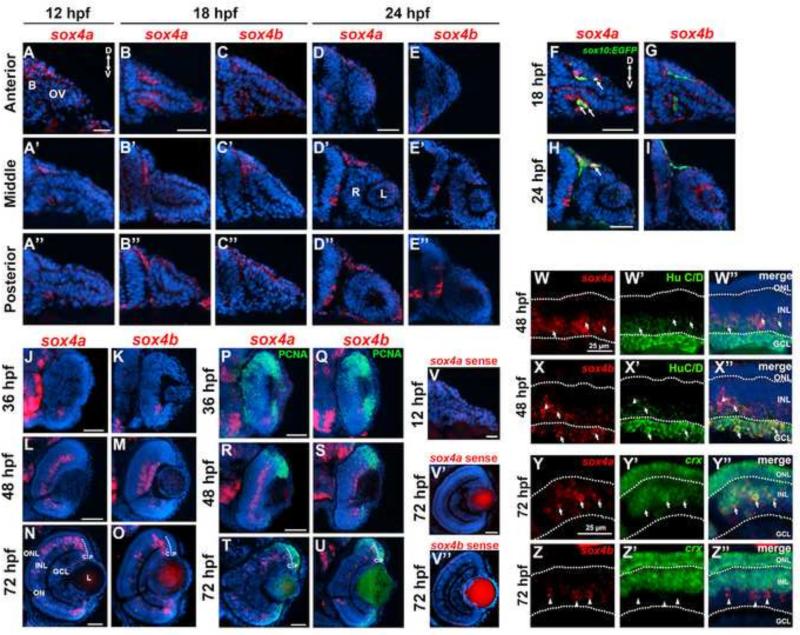
We performed fluorescent in situ hybridization (FISH) with antisense probes for sox4a and sox4b to determine their expression patterns in the forebrain and the eye during zebrafish embryonic development. Previous studies reported expression of sox4 as early as the 5 somite stage (approximately 12 hours post fertilization, hpf) in the lateral plate mesoderm and mid-trunk endoderm of zebrafish embryos, respectively (Mavropoulos et al., 2005). Using FISH, we detected sox4a expression in the dorsal forebrain and around the optic stalk at 12 hpf, (Figure 1A-A”). Sox4b was not detectable at this time point. At 18 hpf, during optic vesicle invagination to form the bi-layered optic cup, expression of sox4a was detected in cells on either side of and directly adjacent to the optic stalk (Figure 1B). More posteriorly, sox4a expression was observed in periocular cells in the dorsal diencephalon (Figure 1B’-B”). Expression of sox4b was also occasionally observed in this region, although not in all embryos analyzed (Figure 1C’-C”). Some of the sox4a-positive periocular cells were of neural crest origin, as they co-localized with GFP+ cells in the sox10:EGFP transgenic line (Hoffman et al., 2007) (Figure 1F, arrows). At 24 hpf, expression of both sox4 paralogs was observed in the ventral diencephalon adjacent to the retina (Figure 1D-E”), and sox4a expression persisted in the dorsal sox10:EGFP+ periocular mesenchyme (Figure 1H). Expression of sox4a and sox4b was first observed in the ventronasal retina at 36 hpf (Figure 1J-K), coinciding with the onset of retinal neurogenesis. These sox4+ cells did not co-localize with PCNA, a marker for proliferating cells, suggesting that they were postmitotic (Figure 1P-U). At 48 hpf, sox4a expression was detected in the inner half of the inner nuclear layer (INL), whereas sox4b was expressed in both the ganglion cell layer (GCL) and the inner portion of the INL (Figure 1L-M). Some of the sox4a/b-positive cells co-localized with the ganglion and amacrine cell marker HuC/D at 48 hpf (Figure 1W-X”). At 72 hpf, scattered expression of sox4a and sox4b was observed in the outer half of the INL, and expression of sox4a/b was also detected in the ciliary marginal zone (CMZ), the persistently neurogenic region at the periphery of retina (Figure 1N-O). The sox4a/b-positive cells in the CMZ were located in the central CMZ and mostly did not co-localize with PCNA (Figure 1T-U), suggesting that sox4a/b marks postmitotic neuronal precursors in this region. In the distal INL, rare sox4b-positive cells co-localized with the horizontal cell marker Prox1, however the sox4a-positive cells did not co-localize with horizontal, bipolar, or Müller cell markers (Figure S1). Instead, we found that some sox4a-positive cells in the outer INL also expressed crx (Figure 1Y-Y”), suggesting that sox4a may be expressed in some photoreceptor progenitors (Nelson et al., 2008). Taken together, the early expression of sox4 adjacent to the optic stalk and evaginating optic vesicle, and later expression within the developing retina, suggests that it functions during zebrafish ocular development.
Figure 1. Expression of sox4a and sox4b during ocular development.
(A-A”) Fluorescent in situ hybridization (FISH) for sox4a performed on transverse cryosections at 12 hpf. (B-D”) FISH for sox4a (B-B” and D-D”) or sox4b (C-C” and E-E”) performed on transverse cryosections taken at the level of the anterior, middle, and posterior optic vesicle at 18 or 24 hpf. (F-I) FISH with sox4a and sox4b probes performed on sections from sox10:EGFP transgenic embryos at 18 and 24 hpf . (J-O) FISH for sox4a or sox4b performed on transverse retinal cryosections at 36, 48, and 72 hpf. (P-U) FISH combined with immunohistochemistry for the proliferation marker PCNA at 24, 48, or 72 hpf. (V-V”) Control FISH with a sense probe for sox4a and sox4b at 12 and 72 hpf. (W-X”) FISH combined with immunohistochemistry for HuC/D, which labels ganglion and amacrine cells, at 48 hpf. (Y-Z”) Two-color FISH with sox4a/b and crx probes performed at 72 hpf. Arrows indicate co-localization, arrowheads indicate sox4a/b+ cells that did not co-localize with the indicated marker. All scale bars equal 50 μm. D, dorsal; V, ventral; B, brain; OV, optic vesicle; L, lens; R, retina; P, peripheral CMZ; C, central CMZ; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; ON, optic nerve.
Knockdown of sox4 causes ocular coloboma
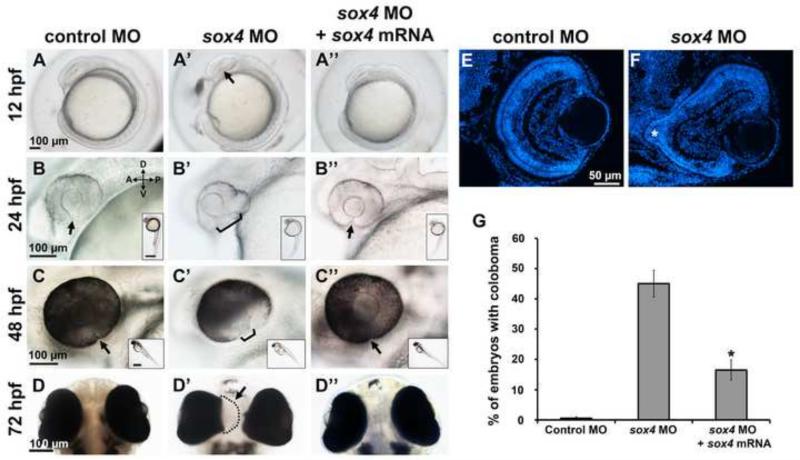
Translation blocking morpholinos (MOs) targeting the 5’-UTR upstream of the translation start site were used to knock down sox4a/b expression in zebrafish embryos. The efficiency of each sox4 MO was assessed by co-injecting an EF1α-EGFP plasmid containing the MO binding site. At 24 hpf, 91.94± 1.93% of the control MO injected embryos displayed GFP expression; in contrast, none of the sox4 MO injected embryos showed any GFP expression, indicating that the sox4 MOs are highly efficient in blocking target gene translation (Figure S2A-E). Morpholino-injected embryos were scored for ocular phenotypes at 24, 48, and 72 hpf by light microscopy. Since some toxicity was observed in sox4 morphants, a p53 MO was co-injected to inhibit non-specific cell death. Knockdown of sox4a resulted in ocular coloboma in 12.23± 2.89% of embryos (n= 17/139), whereas knockdown of sox4b caused coloboma in 9.24±2.79% of embryos (n= 11/119). Knockdown of both sox4a/b simultaneously caused coloboma in 47.84 ± 7.32% of embryos (n= 133/278), suggesting a synergistic effect on the phenotype (Figure S2F). In all subsequent experiments, sox4a and sox4b MOs were co-injected, and sox4a/b MO-injected embryos are referred to as sox4 morphants. Occasionally, pericardial edema and a curved tail were also observed in sox4 morphant embryos (data not shown). However, as these phenotypes can be produced by non-specific activity of MOs, we did not analyze them further. We observed a higher incidence of coloboma (76.92%, n= 130/169) in sox4 morphants without the p53 MO, suggesting that cell death may also contribute to the coloboma phenotype (data not shown).
We performed three separate experiments to confirm the specificity of the sox4 morphant coloboma phenotype. First, a second set of non-overlapping sox4 MOs was used, which produced a similar proportion of choroid fissure closure defects to the first set (Figure S2G-J). Second, sox4a and sox4b mRNAs (lacking the MO binding site) were co-injected with the sox4 MOs, and the embryos were scored for coloboma at 48 hpf. Co-injection of sox4a/b mRNA significantly reduced the incidence of coloboma in sox4 morphants to 16.49% ± 3.37% (Fisher's exact test, P<0.0001; Figure 2A”-D”, 2G), suggesting that this phenotype resulted from specific knockdown of sox4. Finally, we utilized the CRISPR/Cas9 system (Hruscha et al., 2013; Hwang et al., 2013; Jao et al., 2013; Talbot and Amacher, 2014) to target mutations to the sox4a and sox4b genomic loci. For each gene, single guide RNAs (sgRNAs) were designed to target two different sites within the coding region, neither of which overlapped with the MO target sequence. In founders injected with Cas9 mRNA and sox4a or sox4b sgRNAs, we observed coloboma in 31.19± 6.40% (n= 13/44) and 20.91± 8.14% (n= 9/48) of injected embryos, respectively (Figure S1M-O). In contrast, none (n= 0/201) of the uninjected controls (UIC) displayed coloboma, and only 1.33± 2.31% (n= 1/89) of embryos injected with a tyrosinase (tyr) sgRNA/Cas9 exhibited coloboma, although tyr sgRNA/Cas9 was very effective at inducing pigmentation defects, as previously described (Jao et al., 2013) (Figure S1K-L, S1O). High resolution melt curve analysis (HRMA) and sequencing of the sox4a and sox4b target regions in individual injected embryos confirmed that short indels were generated in the sox4 gRNA/Cas9 injected individuals with high efficiency (Figure S1P-Q and data not shown), with the mutation frequency ranging from 52.78% to 94.44%. These data suggest that the CRISPRs induced bi-allelic gene inactivation in mosaic injected embryos, as has been reported previously in zebrafish and mice (Jao et al., 2013; Yen et al., 2014; Zhang et al., 2014), resulting in ocular phenotypes similar to those seen in the sox4 morphants. Taken together, these results strongly support the specificity of the sox4 morphant coloboma phenotype.
Figure 2. Knockdown of sox4 causes coloboma.
Representative images of control MO-injected embryos (A-D), sox4 MO-injected embryos (A’-D’), and embryos injected with both sox4 MO and sox4 mRNA (A”-D”). A minimum of 5 embryos were imaged for each group. Images in A-C” are lateral views; images in D-D” were taken from the ventral side of the embryo. Insets show the whole body of the same embryo in the larger panel. Arrow in A’ indicates the horizontal crease; arrows and brackets in B-C” indicate the choroid fissure. (E-F) Transverse DAPI-stained sections of control and sox4 morphants at 72 hpf. The coloboma in the sox4 morphant retina is prominent (F, asterisk). (G) Quantification of the coloboma phenotype at 48 hpf. In control morphants, 0.61± 0.49% of embryos displayed coloboma (n= 2/329). In sox4 morphants, 45.01± 4.43% of embryos displayed coloboma (n= 257/571). Co-injection of sox4 mRNA significantly reduced the incidence of coloboma to 16.49± 3.37% (n= 31/188; Fisher's exact test, P< 0.0001).Scale bars for the insets in B-C” equal 500 μm.
The first sign of an ocular defect in sox4 morphants was detected at the 8-10 somite stage (12-14 hpf), when the horizontal crease (formed during the evagination of the optic vesicle), becomes visible. Compared to the control morphants, the sox4 morphants that developed ocular coloboma at 48 hpf always displayed a “crooked” horizontal crease at 12-14 hpf (Figure 2A’, arrow), suggesting that the coloboma resulted from an earlier ocular morphogenesis defect. At 24 hpf, the two lips on either side of the choroid fissure in the ventral retina were either wide open or bent backwards in sox4 morphants (Figure 2B’). The persistently open choroid fissure was more obvious at 48 and 72 hpf, because this region of the ventral retina appeared to lack pigmentation of the underlying retinal pigmented epithelium (RPE) (Figure 2C’). Ventral views of sox4 morphant embryos revealed a hole in the posterior RPE through which the retinal tissue extruded into the brain, which accounts for the apparent lack of pigmentation in this region when viewed laterally (Figure 2D’). Histological sections of 72 hpf control and sox4 morphant embryos clearly demonstrated the extrusion of colobomatous tissue through the open choroid fissure (Figure 2F, asterisk). Sox4-deficient retinas also displayed less mature lamination compared to control retinas, however all three retinal cell layers were present.
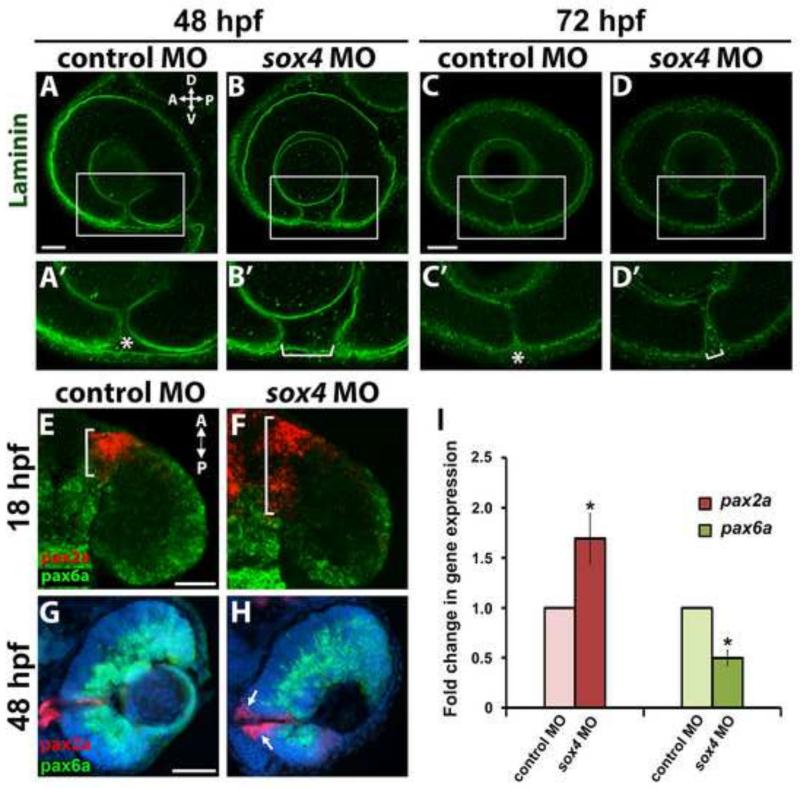
We immunolabeled control and sox4 morphant embryos with an antibody that recognizes laminin, which is present in the basement membrane surrounding the optic cup. As the nasal and temporal lips of the retina on either side of the choroid fissure fuse together, laminin is degraded and disappears. In control morphants at 48 hpf, laminin immunostaining revealed that the nasal and temporal lobes of the choroid fissure were closely apposed, but not yet fused (Figure 3A-A’). In contrast, the choroid fissure was much wider in sox4 morphants at this time (Figure 3B-B’). By 72 hpf in control morphants, only faint expression of a single layer of laminin was visible at the now closed choroid fissure; however, in sox4 morphants the choroid fissure remained open with two distinct layers of laminin outlining the unfused lips of the nasal and temporal retina (Figure 3C-D’).
Figure 3. Persistence of laminin expression at the choroid fissure and altered proximo-distal patterning of the optic vesicle in sox4 morphants.
(A-D’) Laminin immunostaining on control and sox4 morphant embryos at 48 and 72 hpf. Representative images from 15-20 individuals analyzed for each group are shown. (E-H) Double FISH for pax2a and pax6a at 18 and 48 hpf. (I) qPCR analysis revealed a 1.69-fold increase in pax2a expression and a 2.0-fold decrease in pax6a expression in sox4 morphant heads at 18 hpf (Student's t-test, P<0.05). All scale bars equal 50 μm. Asterisks in A’ and C’ indicate the closing or fused choroid fissure in control morphants; brackets in B’ and D’ indicate the open choroid fissure in sox4 morphants.
Proximo-distal patterning of the optic vesicle is altered in Sox4-deficient zebrafish
Coloboma can result from an abnormal or enlarged optic stalk, which may arise when there is a shift in the cell fate boundary between the optic stalk and the optic vesicle (Chang et al., 2006). We performed FISH on control and sox4 morphant sections to examine the expression of pax2a and pax6a, which mark the optic stalk and optic vesicle domains, respectively. In control morphants at 18 hpf, pax2a expression was restricted to a wedge of cells located between the optic vesicle and the brain, and pax6a expression was evident throughout the optic vesicle (Figure 3E). In sox4 morphants however, the region of pax2a expression was expanded into the optic vesicle, and there was a corresponding retraction of pax6a expression (Figure 3F). The expansion of pax2a expression in sox4 morphants persisted at 48 and 72 hpf, where it was particularly prominent in the ventral retina around the open choroid fissure (Figure 3G-H and data not shown). The changes in pax2a and pax6a expression in sox4 morphants were confirmed by quantitative RT-PCR (qPCR), which showed that in sox4 morphant heads at 18 hpf pax2a was significantly up-regulated (1.69±0.25 fold) while pax6a was significantly down-regulated (2.0±0.08 fold; Figure 3I). This result indicates that the proximo-distal patterning of the optic vesicle was disrupted in Sox4-deficient embryos.
Sox4-deficient retinas display ectopic cell proliferation
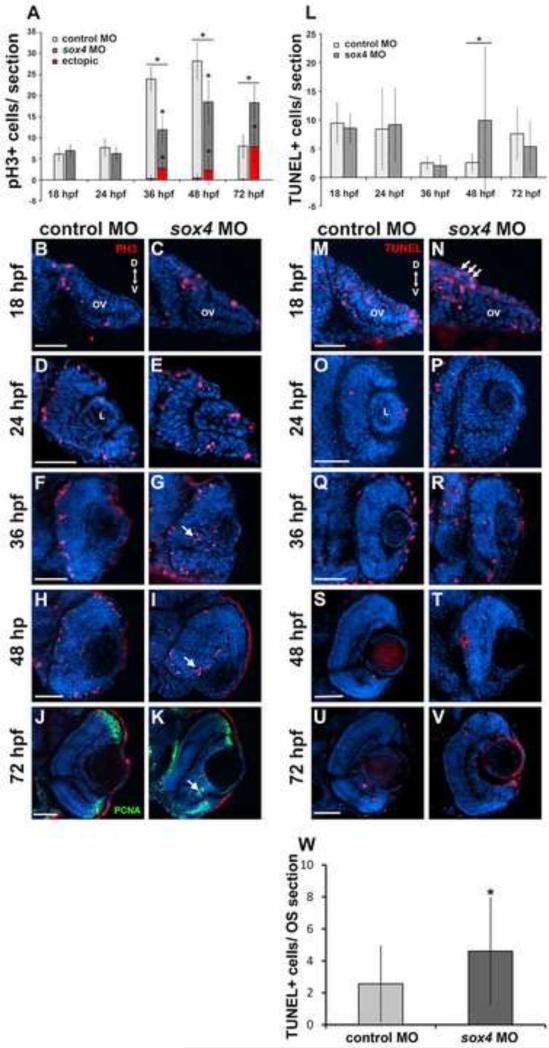
Altered retinal progenitor cell proliferation has been observed in coloboma models (Kim et al., 2007; Liu et al., 2012). We used an antibody to phosphohistone H3 (PH3) to label mitotic cells in control and sox4 morphant eyes. At 18 and 24 hpf, no apparent difference was observed between control and sox4 morphants (Figure 4A-E). At 36 and 48 hpf, the total number of PH3+ cells in the retina was significantly reduced in sox4 morphants compared to controls. Interestingly, some PH3+ cells were ectopically located in the inner retina of sox4 morphants, whereas in controls PH3+ cells were all aligned at the apical border of the retina (Figure 4A, 4F-I). At 72 hpf, when retinal differentiation is largely complete, PH3+ cells in control morphant retinas were confined to the peripheral CMZ (Figure 4J). Sox4 morphant retinas possessed PH3+ cells at the CMZ as well, but we also observed ectopic PH3+ cells in the GCL, which clustered with PCNA-positive cells (Figure 4K). These cells did not express the retinal ganglion cell marker Zn8, or the retinal progenitor cell gene pax6a (Figure S3). We also observed PCNA-positive cells within the colobomatous tissue protruding from the sox4 morphant retinas (Figure 4K). Taken together, these data suggest that overproliferation does not contribute to the coloboma phenotype of sox4 morphants, since no difference in cell proliferation was observed at 18 or 24 hpf, by which time abnormal ocular morphogenesis was evident. However, sox4 does appear to influence cell proliferation in the retina, as knockdown caused an overall reduction in the number of dividing cells as well as ectopic cell proliferation in the GCL.
Figure 4. Sox4 knockdown causes ectopic cell proliferation in the retina.
(A) Quantification of PH3-positive cells in control and sox4 morphant retinas from 18 to 72 hpf. 8-10 individuals were analyzed in each time point (Student's t-test, P< 0.01). (B-K) Representative transverse sections through the optic vesicle and retina of control and sox4 morphants immunolabeled for PH3 at the indicated time points. Ectopic PH3-positive cells are indicated by arrows. At 72 hpf, the ectopic PH3-positive cells in the GCL also clustered with PCNA-positive cells (K, arrow). (L) Quantification of TUNEL-positive cells in control and sox4 morphant optic vesicle and retina from 18 to 72 hpf. 10-20 individuals were analyzed for each time point (Student's t-test, P=0.017). (M-V) Representative transverse sections of control and sox4 morphant retinas labeled for TUNEL-positive cells from 18 to 72 hpf. (W) Quantification of TUNEL-positive cells in control and sox4 morphant optic stalk at 18 hpf. Arrows in N indicate increased TUNEL-positive cells in the optic stalk of an 18 hpf sox4 morphant. All scale bars equal 50 μm.
We performed a TUNEL assay to determine whether apoptosis was elevated in sox4 morphant retinas. No apparent difference was observed at 18, 24, and 72 hpf in the number of TUNEL-positive cells in the optic vesicle/retina between control and sox4 morphant eyes (Figure 4L). However, at 18 hpf the number of TUNEL-positive cells was significantly higher in the optic stalk region of sox4 morphants compared to controls (Figure 4N, arrows; Fig. 4W, Student's t-test, P< 0.05). Also, at 48 hpf sox4 morphant retinas possessed a highly variable, but significantly greater number of TUNEL-positive cells than control retinas (Figure 4L, 4S-T). Moreover, as mentioned previously we observed an increased proportion of embryos with coloboma when the p53 MO was omitted from the injection (data not shown). Taken together, these data suggest that apoptosis, particularly within the optic stalk region may contribute to the severity of the coloboma phenotype.
Hh signaling is elevated in Sox4-deficient zebrafish
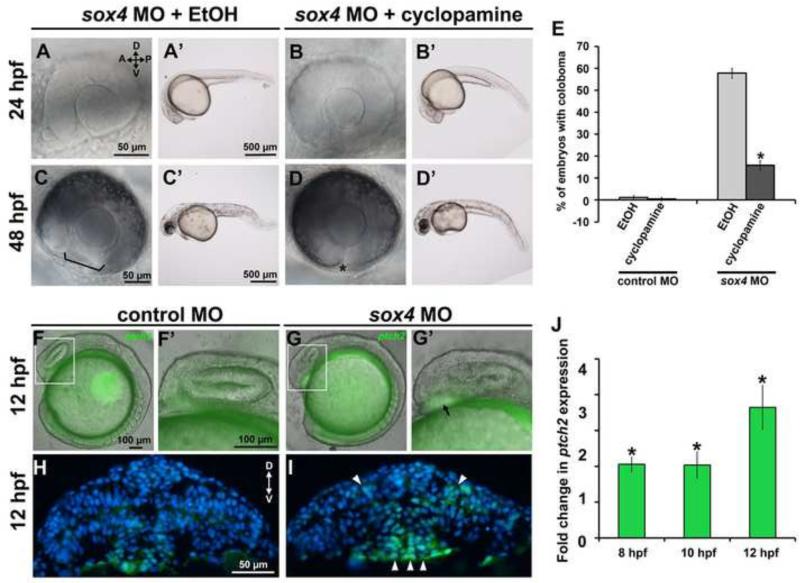
As mentioned previously, we have shown that knockdown of Sox11 causes coloboma due to an elevation of Hh signaling (Pillai-Kastoori et al., 2014). Moreover, the coloboma phenotype in both sox11 and sox4 morphants resembles other animal models of overactive Hh signaling (Lee et al., 2008; Bassett et al., 2010; Lee et al., 2012; Zhang et al., 2013). Therefore, we asked whether cyclopamine, a pharmacological Hh inhibitor, could reduce the incidence of coloboma in sox4 morphants. We treated sox4 morphants with 2 μM cyclopamine, a concentration which did not produce any ocular defects on its own (Figure 5E), and which was shown previously to rescue the coloboma phenotype in zebrafish blowout mutants (which carry a mutation in the ptch2 receptor) and sox11 morphants (Lee et al., 2008; Pillai-Kastoori et al., 2014). Sox4 morphants were exposed to either cyclopamine or an equivalent amount of ethanol vehicle from 5.5 to 13 hpf, and then analyzed at 48 hpf for the presence of coloboma. In vehicle-treated sox4 morphants, 57.78±2.28% of embryos (n= 104/180) displayed coloboma at 48 hpf, consistent with our previous results. In contrast, cyclopamine treatment suppressed the incidence of coloboma to 15.87±2.20% (n= 26/165; Fisher's exact test, P<0.0001; Figure 5A-E). Treatment of sox4 morphants with cyclopamine from 10 to 24 hpf also significantly decreased the proportion of embryos with coloboma, however the effect was not as strong as with the earlier treatment period (data not shown).
Figure 5. Hh signaling is elevated in Sox4-deficent zebrafish.
(A-D’) Representative eye and whole body images of vehicle-treated and cyclopamine-treated sox4 morphants at 24 and 48 hpf. (E) Quantification of the percentage of coloboma in vehicle- and cyclopamine-treated control and sox4 morphants (control morphants + vehicle: 1.14± 0.96% coloboma, n= 2/175; control morphants + cyclopamine: 0.49± 0.79% coloboma, n= 1/205; sox4 morphants + vehicle: 57.78± 2.28% coloboma, n= 104/180; sox4 morphants + cyclopamine: 15.87± 2.20% coloboma, n= 26/165; Fisher's exact test, P<0.0001). (F-I) Representative eye and body lateral images (F-G’) and transverse sections (H, I) of ptch2:EGFP(+/−) transgenic embryos injected with control (F-F’, H) or sox4 MO (G-G’, I) at 12 hpf. (J) qPCR analysis of ptch2 expression in embryos injected with control or sox4 MO at 8, 10, and 12 hpf. Relative transcript abundance was normalized to level of atp5h (three biological replicates; Student's t-test, P<0.01).
To confirm that Hh signaling was elevated in response to sox4 knockdown, we injected sox4 MOs into heterozygous ptch2:EGFP transgenic embryos, which report on Hh signaling by expressing GFP in cells that activate the Hh target gene ptch2 (Shen et al., 2013). Elevation of GFP in the ventral forebrain was observed in sox4 morphants as early as 12 hpf and through 72 hpf in sox4 morphants (Figure 5F-I and data not shown), confirming that Hh signaling was indeed upregulated in Sox4-deficient embryos. To quantify the elevation of endogenous ptch2 expression in sox4 morphants, we performed qPCR on mRNA from control and sox4 morphant embryos at 8, 10, and 12 hpf. We found that ptch2 was upregulated in sox4 morphants by more than 2-fold at all time points (Figure 5J). Taken together, these data indicate that knockdown of Sox4 results in an elevation in Hh signaling in the early embryo that leads to ocular morphogenesis defects and coloboma.
Sox4 negatively regulates expression of Indian hedgehog b (ihhb)
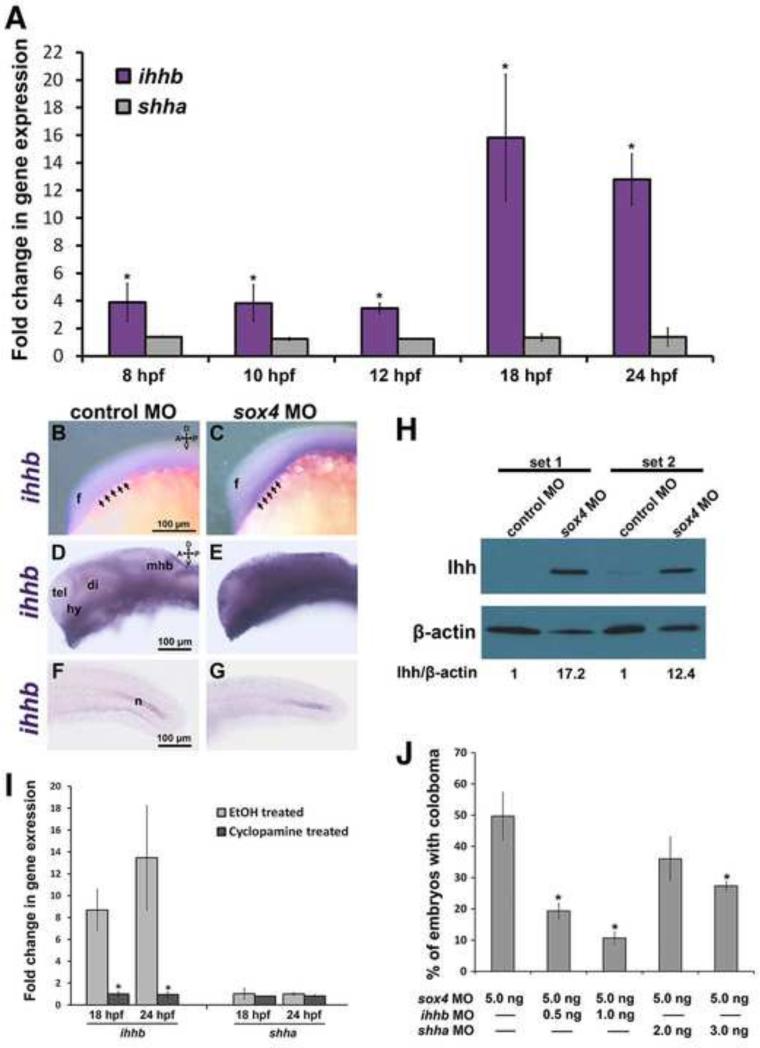
In Sox11-deficient zebrafish, elevation of Hh signaling is caused by an increase in expression of the Shh gene [shha; (Pillai-Kastoori et al., 2014)]. To determine whether Sox4 also regulates expression of shha or some other component of the Hh signaling pathway, we performed qPCR on mRNA prepared from the control and sox4 morphant embryos at 8, 10, and 12 hpf, and from the heads of control and sox4 morphant embryos at 18 and 24 hpf. Intriguingly, we found that expression of shha was not significantly different in sox4 morphants compared to controls at any time point. However, expression of the Hh ligand ihhb was significantly upregulated in sox4 morphants as early as 8 hpf and was strongly upregulated at 18 and 24 hpf (Figure 6A, Figure S4, Student's t-test, P<0.01). We also detected a more variable increase in expression of ptch1, ptch2, and gli1 in sox4 morphants at 18 and 24 hpf (Figure S4). We confirmed the upregulation of ihhb transcript in sox4 morphants at 12 and 24 hpf by whole mount in situ hybridization (WISH), which revealed that ihhb expression in sox4 morphants was upregulated in the ventral midline at 12 hpf (Figure 6C), and at 24 hpf ihhb expression was upregulated in the hypothalamus and ventral diencephalon, and had also expanded into regions of the telencephalon and dorsal diencephalon that did not express ihhb in controls (Figure 6E). However, the increase in ihhb expression in sox4 morphants was not ubiquitous, as ihhb expression in the posterior notochord was similar to controls at 24 hpf (Figure 6G). We also confirmed that Ihhb protein levels were elevated in 24 hpf sox4 morphant heads by Western blot (Figure 6H). Moreover, qPCR of mRNA prepared from cyclopamine-treated sox4 morphants revealed that expression of ihhb returned to control levels following cyclopamine treatment (Figure 6I). Taken together, these results demonstrate that knockdown of Sox4 causes an increase in Hh signaling primarily through upregulation of ihhb expression.
Figure 6. Sox4 negatively regulates ihhb expression.
(A) ihhb and shha qPCR of mRNA from control and sox4 morphant whole embryos at 8, 10 and 12 hpf, and heads at 18 and 24 hpf. Relative transcript abundance was normalized to level of atp5h or gapdh. Y axis represents the average ratio of normalized sox4 morphant to control expression (three biological replicates; Student's t-test, P<0.01). (B-G) Ihhb expression in control and sox4 morphants at 12 and 24 hpf detected by WISH. Representative images are presented from 15-20 individuals analyzed. Lateral images in B-E focus on the midline, therefore the eye is not visible. Arrows in B and C mark the ventral forebrain at the level of the optic vesicle. (H) Western blot analysis of protein lysates from control or sox4 morphant heads at 24 hpf. Two biological replicates were performed (sets 1 and 2) (I) qPCR of mRNA from vehicle- and cyclopamine-treated sox4 morphants (Student's t-test, P<0.01). (J) Quantification of the proportion of embryos with coloboma at 48 hpf in sox4 morphants co-injected with two different doses of either ihhb MO or shha MO (sox4 MO alone: 51.24± 2.7%, n= 209/404; sox4 MO + 0.5 ng/embryo ihhb MO: 19.33± 2.52%, n= 14/75; sox4 MO + 1.0 ng/embryo ihhb MO: 10.67± 2.08%, n= 9/91; sox4 MO + 2.0 ng/embryo of shha MO: 41.84± 5.62%, n= 53/125; sox4 MO + 3.0 ng/embryo of shha MO, 34.39± 5.03%, n= 79/229) Fisher's exact test, P<0.0001 for sox4 MO + ihhb MO; P= 0.1676 for sox4 MO + 2.0 ng/embryo of shha MO; P=0.0001 for sox4 MO + 3.0 ng/embryo of shha MO. f, forebrain; tel, telencephalon; di, diencephalon, hy, hypothalamus; mhb, midbrain-hindbrain boundary; n, notochord.
Next, we co-injected an ihhb MO together with the sox4 MOs and quantified the incidence of coloboma at 48 hpf. Co-injection of the ihhb MO significantly reduced the incidence of coloboma in sox4 morphants in a dosage dependent manner: 19.33± 2.52% (n= 14/75) of sox4 morphant embryos displayed coloboma when 0.5 ng of ihhb MO was injected (compared to 51.24± 2.70% with sox4 MO alone), whereas injecting 1.0 ng of ihhb MO suppressed coloboma to 10.67± 2.08% (n= 9/91; Figure 6J). We also co-injected a previously described shha MO (Nasevicius and Ekker, 2000) with the sox4 MOs to determine whether it could rescue the coloboma phenotype. When 2.0 ng of shha MO was injected the proportion of embryos with coloboma was not significantly different from that observed with sox4 MO alone (Figure 6J). However, 3.0 ng of shha MO did significantly reduce coloboma to 34.39 ± 5.03% (n= 79/229; Fisher's exact test, P=0.0001), although the extent of rescue was not as strong as with the ihhb MO. Together, these results confirm that Sox4 negatively regulates the expression of ihhb and that elevated ihhb expression in Sox4-deficient zebrafish causes coloboma.
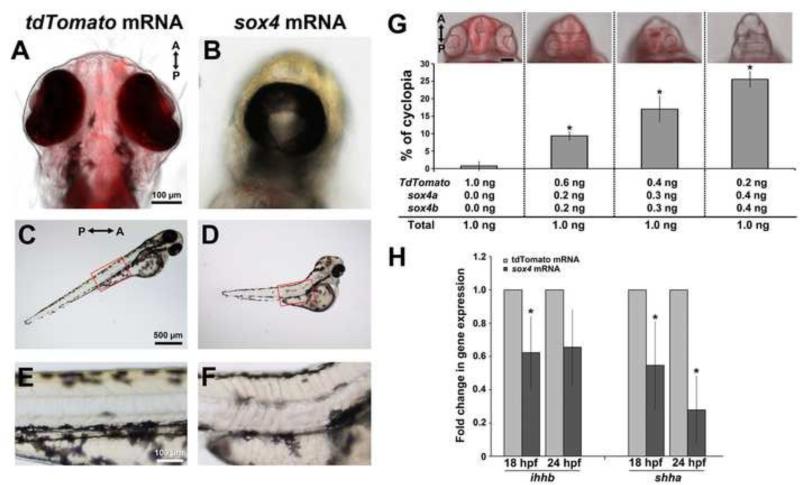
Since knockdown of Sox4 caused an elevation in ihhb expression and Hh signaling, we predicted that overexpression of Sox4 would result in a corresponding reduction in Hh activity. We injected in-vitro synthesized control or sox4a/b mRNA into wild type embryos, and scored the injected embryos at 24 hpf for the presence of cyclopia, a phenotype associated with reduced Hh activity from the midline (Ekker et al., 1995). Whereas only 0.78±1.34 % of embryos (n= 1/128) injected with a control Td-Tomato mRNA exhibited cyclopia, we detected cyclopia in 36.54 ± 5.10% (n= 38/104) of the sox4 mRNA-injected embryos (Figure 7A-B). In addition, we found that the cyclopic embryos often possessed a truncated body and U-shaped somites (Figure 7C-F), a phenotype that is also observed with reductions in Hh signaling (Currie and Ingham, 1996; Schauerte et al., 1998; Wolff et al., 2003). To confirm that the cyclopic phenotype was specific for overexpression of sox4, we injected the same total amount of mRNA containing different ratios of control Td-Tomato and sox4 mRNA. We observed that both the occurrence and severity of cyclopia increased with increasing concentration of sox4 mRNA (Figure 7G), strongly suggesting that the cyclopic phenotype was not an artifact of mRNA injection. qPCR performed on mRNA prepared from the heads of injected embryos confirmed that ihhb expression was reduced in the embryos overexpressing sox4 at 18 and 24 hpf, although the reduction was not statistically significant at 24 hpf (Figure 7H; Student's t-test, P= 0.0565). Surprisingly, although sox4 knockdown did not affect the expression level of shha, sox4 overexpression significantly reduced the expression of shha in the head. Taken together, these data suggest that sox4 can negatively regulate the expression of both ihhb and shha, and that overexpression of sox4 causes a reduction in midline Hh activity, leading to cyclopia.
Figure 7. Overexpression of sox4 inhibits Hh ligand expression and causes cyclopia.
(A-F) Representative images of embryos injected with control tdTomato or sox4 mRNA. (A-B) Representative images of the head were taken at 72 hpf from the ventral side. (C-F) Lateral views of body showing the somites. Insets in (C) and (D) were enlarged in (E) and (F). (G) Quantification of the percentage of cyclopia in embryos injected with different combinations of control and sox4 mRNA. 1.0 ng/embryo Td-tomato: 0.76± 1.31%, n= 1/108; 0.2 ng sox4a/b + 0.6 ng Td-tomato/embryo: 9.35± 1.17%, n= 13/139; 0.3 ng sox4a/b + 0.4 ng Td-tomato/embryo: 17.03± 3.69%, n= 11/67; 0.4 ng sox4a/b + 0.2 ng Td-tomato/embryo: 25.56± 2.20%, n= 25/97. (H) qPCR analysis of ihhb and shha expression in control and sox4 overexpressing heads at 18 and 24 hpf. In sox4 mRNA injected embryos, ihhb expression was significantly decreased by 1.61-fold at 18 hpf (Student's t-test, P<0.05) and 1.51-fold at 24 hpf (Student's t-test, P=0.057). Shha expression was also significantly reduced following sox4 mRNA injection at both 18 and 24 hpf (by 1.82-fold and 3.57-fold, respectively; Student's t-test, P<0.05).
Bmp7 is a potential intermediate between sox4 and ihhb
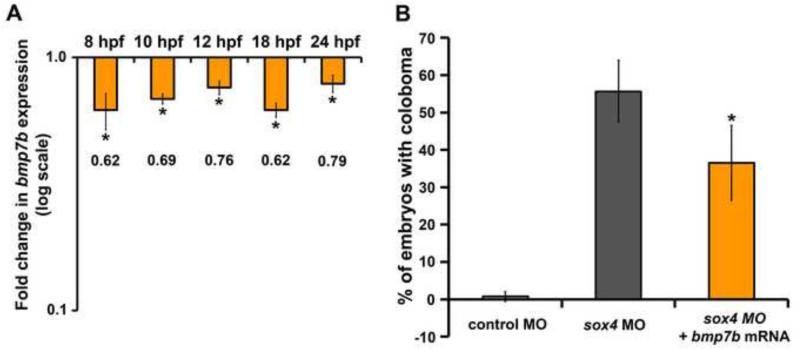
Our results support the hypothesis that Sox4 is a negative regulator of ihhb expression. However, SoxC transcription factors are known to act as transcriptional activators rather than repressors (van de Wetering et al., 1993; Bergsland et al., 2006). Therefore, we hypothesized that Sox4 indirectly inhibits ihhb expression by activating an intermediate repressor. One candidate Hh repressor is Bmp7. Bmp7 is known to modulate Hh signaling (both positively and negatively), was previously shown to be significantly down-regulated in Sox11-deficient zebrafish and Sox11 null mice, and bmp7 mRNA can partially rescue coloboma in sox11 morphants (Pathi et al., 1999; Seki and Hata, 2004; Manning et al., 2006; Bastida et al., 2009; Duench and Franz-Odendaal, 2012; Pillai-Kastoori et al., 2014). Moreover, Bmp7 mutant mice display ocular malformations such as microphthalmia and optic fissure defects (Morcillo et al., 2006). We performed qPCR for bmp7b transcript levels in controls and sox4 morphants at 8, 10, 12, 18 and 24 hpf. This analysis revealed that bmp7b expression was significantly downregulated at all time points in sox4 morphants (Figure 8A, Student's t-test, P< 0.01). We then injected bmp7b mRNA along with the sox4 MO and found that this partially rescued the coloboma phenotype of sox4 morphants (Figure 8B, Student's t-test, P< 0.01). These data suggest that Sox4 may negatively regulate Ihhb at least in part through the activation of Bmp7b. We also analyzed the expression of four additional regulators of Hh signaling: two Hh inhibitors (fgfr2 and kras), and two Hh activators (lhx8 and nkx6.1; (Cai et al., 2000; Flandin et al., 2011; Mukhopadhyay et al., 2013). Fgfr2 was downregulated in sox4 morphant heads at 18 hpf and lhx8 was upregulated in sox4 morphants at 12 hpf, suggesting that these genes may also function downstream of Sox4 and upstream of Hh signaling during eye development (Figure S5, Student's t-test, P<0.05).
Figure 8. Bmp7b expression is reduced in sox4 morphants.
(A) qPCR analysis revealed a significant reduction in bmp7b expression in sox4 morphant embryos at 8, 10, 12 hpf and heads at 18 and 24 hpf (Student's t-test, P<0.01). (B) bmp7b mRNA injection significantly reduced the proportion of embryos with coloboma in sox4 morphants. Control MO only: 0.74± 1.28%, n= 1/110; sox4 MO only: 55.66± 8.32%, n= 59/106; sox4 MO+ bmp7b mRNA: 35.56± 10.02%, n= 29/80 (Student's t-test, P< 0.05).
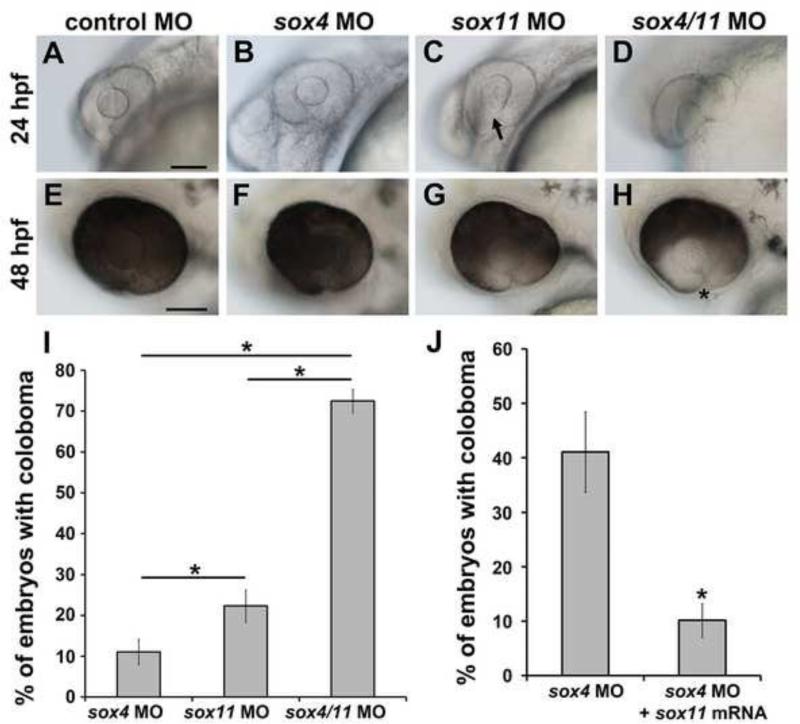
Sox4 and sox11 have overlapping functions in regulating choroid fissure closure
Our previous work demonstrated that Sox11-deficient zebrafish, similar to sox4 morphants, exhibit ocular coloboma and elevated Hh signaling. Moreover, sox4 mRNA injection can significantly reduce the incidence of coloboma in sox11 morphants (Pillai-Kastoori et al., 2014). To further investigate the functional overlap between Sox4 and Sox11, we injected sox4 and sox11 MOs, either alone or together, at half the usual dose. When a half-dose of sox4 MO was injected, only 11.05± 3.15% (n= 16/145) of morphants exhibited coloboma at 48 hpf. Similarly, when a half-dose of sox11 MO was injected, 22.31± 4.00% (n= 16/71) of the morphants exhibited coloboma. However, co-injection of half-doses of sox4 and sox11 MOs together resulted in 72.47± 2.95% (n= 51/70) of embryos with coloboma (Figure 9A-H, 9I), suggesting that knockdown of the two SoxC factors simultaneously produced a synergistic effect on the coloboma phenotype. Furthermore, injection of sox11a/b mRNA into sox4 morphants significantly suppressed the incidence of coloboma from 41.09±7.36% (n= 113/275) to 10.14± 3.09% (n= 28/276, Figure 9J). We did not detect abnormalities in lens development in sox4 morphants (Figure 9C, arrow). Moreover, the proportion of embryos with a malformed lens was not increased by addition of the sox4 MO (data not shown). Taken together, these results indicate that Sox4 and Sox11 have partially overlapping roles in regulating choroid fissure closure and can functionally compensate for one another during ocular morphogenesis, but are not functionally redundant with respect to lens development.
Figure 9. Synergistic effect of sox4 and sox11 knockdown on coloboma.
(A-H) Representative images of embryos injected with control MO (A, E), a half-dose of sox4 MO (B, F), a half-dose of sox11 MO (C, G), and half-doses of sox4 and sox11 MOs (D, H). (A, E) Half the normal dose of sox4 MO caused a low incidence of mild coloboma. (C, G) Half the normal dose of sox11 MO resulted in low incidence of coloboma and lens malformations at 24 hpf (C, arrow). (D, H) Injection of a half-dose of both sox4 and sox11 MO significantly increased the incidence of coloboma, and the coloboma phenotype was more severe compared to sox4 or sox11 morphants alone. (I) Quantification of the proportion of embryos with coloboma at 48 hpf (half-dose sox4 MO, 11.05± 3.15%, n= 16/145; half-dose sox11 MO, 22.31± 4.0%, n= 16/71; half-dose of both sox4 and sox11 MO, 72.47± 2.95%, n= 51/70; Fisher's exact test, P=0.0001). (J) Sox11 can compensate for the loss of Sox4. Injection of sox11 mRNA into sox4 morphants significantly reduced the incidence of coloboma from 41.09±7.36% (n= 113/275) to 10.14± 3.09% (n= 28/276; Fisher's exact test, P<0.0001). All scale bars equal 100 μm.
DISCUSSION
Although many genes have been identified that contribute to coloboma (Chang et al., 2006), the molecular mechanisms of choroid fissure closure have not been completely elucidated. It has been shown in animal models as well as in human patients that alterations in Hh signaling can cause coloboma (Schimmenti et al., 2003; Take-uchi, 2003; Lee et al., 2008; Lee et al., 2012). However, not much is known about how proper levels of Hh activity are maintained during eye development. In this study, we demonstrated that successful choroid fissure closure requires the activity of Sox4 to limit both the level and extent of expression of the Hh pathway ligand Ihh, permitting proper proximo-distal patterning of the optic stalk and vesicle. Together with our previous work demonstrating a role for Sox11 in controlling expression of Shh during ocular development (Pillai-Kastoori et al., 2014), our studies firmly place the SoxC transcription factors upstream of the Hh signaling pathway in the complex genetic network that regulates vertebrate choroid fissure closure.
The involvement of both Sox4 and Ihh in choroid fissure closure is a novel result, but is consistent with previous work demonstrating that related family members (Sox11 and Shh, respectively) also participate in this process. Although coloboma was not reported in the Sox4 null mutant mouse (Schilham et al., 1996; Cheung et al., 2000), the early lethality of that model likely precluded a thorough characterization of any ocular phenotypes. Coloboma was also not reported in a conditional mouse mutant lacking Sox4 expression within the developing eye (Jiang et al., 2013). However, our cyclopamine rescue data suggest that the coloboma phenotype of sox4 morphants is caused by disruption in early, midline-derived Hh signaling, likely due to loss of sox4 expression in the forebrain and/or periocular mesenchyme (POM). Therefore, alterations to Sox4 expression within the optic vesicle may not have been sufficient to cause coloboma. Finally, 65% of patients with CHARGE syndrome, a genetic disorder characterized by coloboma and cardiac abnormalities in addition to other birth defects, carry a mutation in CDH7 (Chang et al., 2006), which directly activates Sox4 and Sox11 (Feng et al., 2013). Therefore, it is possible that altered expression of Sox4 (and/or Sox11) underlies the coloboma phenotype observed in CHARGE syndrome patients.
Our data strongly suggest that knockdown of Sox4 results in an early elevation of midline-derived Hh signaling, which ultimately results in defects in ocular morphogenesis and coloboma. How might this occur? We observed that the expression of the optic stalk marker pax2a was significantly expanded into the optic vesicle of sox4 morphants. As Hh signaling has a well-established role in specifying the optic stalk region through regulation of Pax2 expression, our results suggest that the elevation in Hh signaling from the midline expands the region of the optic vesicle that is fated to become optic stalk tissue. The decrease in bmp7b expression in sox4 morphants may also contribute to the defect in proximo-distal patterning of the optic stalk and vesicle, as others have shown previously that Bmp7 is required prior to Hh signaling for proper expression of Pax2 in the optic disk (Morcillo et al., 2006). In conjunction with (or as a result of) reduced Bmp7 and expanded Pax2, we observed an increase in apoptosis in the optic stalk region at 18 hpf. Therefore, we propose that misspecification of the optic vesicle cells as optic stalk physically interferes with proper closure of the choroid fissure, resulting in coloboma, and that the abnormally specified optic stalk cells ultimately die by apoptosis.
Although many studies in animal models and humans have demonstrated the critical role of Shh in the morphogenesis and patterning of the developing eye (Amato et al., 2004), there are much less data on the role of Ihh, which is better known for its function in regulating skeletal and intestinal development (St-Jacques et al., 1999; Seki and Hata, 2004; Kosinski et al., 2010). However, some previous studies are consistent with our finding that Ihh levels influence ocular development. For example, overexpression of murine Ihh in zebrafish embryos caused expansion of pax2 expression in the optic stalk (Hammerschmidt et al., 1996), and in the mouse, Ihh was shown to be required for proper development of the RPE and peri-ocular tissues (Dakubo et al., 2008). Interestingly, although we observed a significant elevation (approximately 4-fold) of ihhb expression at 8-12 hpf in sox4 morphants, this increase was even more pronounced at 18 and 24 hpf (approximately 15-fold). This suggests that ihhb is itself a target of Hh signaling, resulting in a feedback loop of enhanced expression when Hh signaling is increased. A similar feedback loop with respect to Hh signaling and shha expression has been observed previously (Pillai-Kastoori et al., 2014), suggesting that this may be a general phenomenon of Hh signaling and Hh ligand expression.
Similar to other studies of SoxC family members, our results indicate that Sox4 and Sox11 have overlapping, but not completely redundant, functions in eye development. Thus, although sox4 and sox11 were able to compensate for each other when ectopically overexpressed, we observed distinct differences in their knockdown phenotypes. First, although knockdown of either sox11 or sox4 resulted in coloboma, abnormal lens development was only observed following knockdown of sox11. Second, the penetrance of the coloboma phenotype was higher in Sox11-deficient zebrafish than in Sox4-deficient zebrafish [~70% versus ~50%, respectively; this study and (Pillai-Kastoori et al., 2014)]. And third, whereas both sox11 and sox4 morphants displayed elevated Hh signaling, this occurred through increased expression of distinct Hh ligands (shha and ihhb, respectively).
These phenotypic differences may be explained by the differing expression patterns of sox11 and sox4. For example, whereas both sox4 and sox11 are expressed in the diencephalon adjacent to the optic vesicle at 18 and 24 hpf, sox4 (but not sox11) is also expressed in the periocular mesenchyme (POM). As the POM was previously shown to be a target of Ihh but not Shh signals in the mouse eye (Dakubo et al., 2008), sox4 expression in the POM may function in a negative feedback mechanism to limit the level or duration of Ihh signaling there. Similarly, sox11, (but not sox4) is expressed in the developing zebrafish lens, which may account for the unique lens phenotype in sox11 morphants. Finally, the differing penetrance of the coloboma phenotype between the two knockdown models may be due to differences in their transactivation properties, as previous work has shown that Sox11 is a stronger transcriptional activator than Sox4 (Dy et al., 2008).
Targeted disruption of Sox4 or Sox11 in the developing mouse retina results in a reduction in retinal ganglion cell (RGC) number and optic nerve thickness (Jiang et al., 2013). Although we detected expression of both sox4 and sox11 in the zebrafish GCL during retinal development, we did not observe a loss of ganglion cells in either sox4 or sox11 morphants (data not shown). It is possible that we failed to detect an early delay in RGC differentiation due to the much shorter window of retinal neurogenesis in the zebrafish compared with mouse (Cepko et al., 1996; Stenkamp, 2007). Alternatively, it is possible that knockdown of both sox4 and sox11 is required to alter RGC formation in the zebrafish.
Although we did not detect a loss of RGCs, we did observe ectopic cell proliferation in the GCL of the sox4 morphant retina. This phenotype has been observed in other models of overactive Hh signaling, such as zebrafish and mouse Ptch receptor mutants (Moshiri and Reh, 2004; Bibliowicz and Gross, 2011), and is consistent with the known role of the Hh pathway in promoting retinal progenitor cell proliferation. However, we also found that the total number of proliferating cells was reduced in the retinas of sox4 morphants at 36 and 48 hpf, whereas it was elevated at 72 hpf. These seemingly contradictory results could be explained by alterations in the cell cycle kinetics of RPCs, either via elevated Hh signaling [as described by (Locker et al., 2006)] or through a Hh-independent role of Sox4 in regulating cell cycle exit. Alternatively, the ectopically located proliferating cells in sox4 morphant retinas could indicate disruptions in the organization of the retinal neuroepithelial cells. Finally, it is not clear whether the altered cell proliferation in the retina is due to loss of sox4 expression (and the subsequent elevation in Hh signaling) from the midline, or from within the retina itself. Future studies may be able to separate the early and later functions of Sox4 in eye development using photo-morpholinos or inducible transgenes to control the timing of sox4 inactivation and rescue.
MATERIALS AND METHODS
Zebrafish strains and maintenance
All zebrafish (Danio rerio) strains were bred, raised, and maintained at 28.5° C on a 14 hour light:10 hour dark cycle according to established protocols (Westerfield, 2000). The Tg(gfap:GFP)mi2001 transgenic line (Bernardos et al., 2007), hereafter called gfap:GFP, was obtained from the Zebrafish International Resource Center (ZIRC, Eugene, OR). The Tg(GBS-ptch2:nlsEGFP) transgenic line (Shen et al., 2013), hereafter called ptch2:EGFP, was kindly provided by R. Karlstrom (University of Massachusetts, Amherst, MA). The Tg(−7.2sox10:EGFP)zf77 transgenic line (Hoffman et al., 2007), hereafter called sox10:EGFP, was generously provided by B.A. Link (Medical College of Wisconsin, Milwaukee, WI). Embryos were staged according to established developmental hallmarks (Kimmel et al., 1995). All animal procedures were carried out in accordance with guidelines established by the University of Kentucky Institutional Animal Care and Use Committee.
Whole mount in situ hybridization (WISH) and fluorescent in situ hybridization (FISH)
Antisense RNA probes were prepared by in vitro transcription of linearized plasmids containing a portion of the coding sequence of the gene of interest, using SP6, T7, or T3 polymerase and digoxigenin (DIG) or fluorescein (FITC) labeling mix (Roche Applied Science, Indianapolis, IN). The sox4a plasmid was prepared by cloning PCR products into the pGEM-T-easy vector (Promega, Madison, WI). The sequences of all PCR primers used in this study are given in Table S1. The sox4b plasmid (Mavropoulos et al., 2005) was generously provided by Bernard Peers (Université de Liège, Sart Tilman, Belgium). The ihhb plasmid (Chung et al., 2013) was generously provided by H.C. Park (Korea University, Ansan, Gyeonggido, Republic of Korea). The crx and pax6a plasmids (Shen and Raymond, 2004; Ochocinska and Hitchcock, 2007) were kindly provided by Y.F. Leung (Purdue University, West Lafayette, IN). The pax2a plasmid (Lee et al., 2008) was kindly provided by J.M. Gross (University of Texas, Austin, TX). WISH and FISH were performed as previously described (Forbes-Osborne et al., 2013; Pillai-Kastoori et al., 2014). Images were obtained on an inverted fluorescent microscope (Eclipse Ti-U; Nikon Instruments).
Morpholino and mRNA microinjection
All morpholinos (MOs) were obtained from Gene Tools, LLC (Philomath, OR) and injected into zebrafish embryos at the one- to two-cell stage. The following MOs were used in this study: standard control MO: 5’-CCTCTTACCTCAGTTACAATTTATA-3’; sox4a MO1: 5’-GCGCTAAGAGTCTTTCTTCTTCACT-3’; sox4b MO1: 5’-ACGCGCCTTCAGTCGTGCTTAGTGC-3’; ihhb MO: 5’-CGCCGCCGCCGTGGAGAGTCTCAT-3’ (Lewis et al., 2005); shha MO: 5’-CAGCACTCTCGTCAAAAGCCGCATT-3’ (Nasevicius and Ekker, 2000). The specificity of the sox4 morphant phenotypes was confirmed with a second, non-overlapping set of sox4 morpholinos (sox4a MO2: 5’-CAACAGTCTCAACTTTTAATTGCGC-3’; sox4b MO2: 5’-GAGACTCAGTCTGATTGCAGCACAC-3’). Embryos were injected with 5.3 ng each of sox4a MO1 and sox4b MO1, 10.5 ng each of sox4a MO2 and sox4b MO2, or 10.5 ng of standard control MO. Both sox4 MO1 and MO2 generated similar phenotypes. Unless stated otherwise, all data presented in this study were from embryos injected with sox4a MO1 and sox4b MO1. A p53 morpholino (Robu et al., 2007) was co-injected (at 1.5-fold the amount of sox4 MOs) to suppress cell death (p53 MO 5’- GCGCCATTGCTTTGCAAGAATTG-3’).
To determine the efficiency of the sox4 MOs, PCR fragments corresponding to the 5’ UTRs of sox4a and sox4b mRNA and containing the sox4a MO and sox4b MO target sequence, respectively, were amplified and cloned upstream and in frame with the GFP gene in the pEF1α:GFP plasmid (Addgene, Cambridge, MA). One-cell stage zebrafish embryos were injected with 50 pg/embryo of pEF1α-sox4a MO:GFP and 50 pg/embryo of pEF1α-sox4b MO:GFP plasmids in the presence or absence of sox4a MO1 and/or sox4b MO1. GFP expression in injected embryos was analyzed by fluorescence microscopy at 24 hpf.
For mRNA rescue and overexpression experiments, the zebrafish sox4a/b and sox11a/b coding sequences (lacking the MO target sites) were PCR amplified from 48 hpf complimentary DNA (cDNA) and cloned into the pGEM-T-easy vector (Promega, Madison, WI). The capped mRNAs were synthesized with the mMessage mMACHINE transcription kit (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Control Td-tomato mRNA was synthesized from pRSET-B-td-Tomato (kindly provided by D.A. Harrison, University of Kentucky, Lexington, KY). For mRNA rescue experiments, 0.5 ng/embryo of sox4a and sox4b mRNA or 1.0 ng/embryo of sox11a and sox11b mRNA was co-injected with the sox4a/b MOs. For mRNA overexpression experiments, 0.5 ng/embryo of sox4a and sox4b mRNA or an equimolar amount of Td-tomato mRNA was injected into wild type embryos. Alternatively, a total of 1 ng of mRNA containing different ratios of control Td-Tomato and sox4 mRNA was injected. Bmp7b mRNA was synthesized from a pCRII-bmp7b plasmid (a kind gift from Dr. S. Fabrizio, The Novartis Institutes for Biomedical Research, Cambridge, MA). 1.0 ng/embryo of bmp7b mRNA was injected into sox4 morphants.
CRISPR sgRNA and Cas9 mRNA synthesis and injection
Sox4a and sox4b CRISPR target sites were identified and the corresponding sgRNA oligos were designed using the ZiFiT online software (www.zifit.partners.org/ZiFiT/; Table S1). Oligo pairs (100μM) for each sgRNA were mixed with NEBuffer4 (New England Biolabs, lpswich, MA), incubated in boiling water for 5 minutes, followed by 2 hours annealing at room temperature, and then ligated with linearized pDR274 vector (Addgene, Cambridge, MA) at 16°C overnight. Recombinant plasmid was digested with DraI to drop out the sgRNA template, followed by PCR amplification (Table S1) using the KOD Hot Start Master Mix (Millipore, Billerica, MA) and purification using QIAquick PCR Purification Kit (Qiagen, Valencia, CA). sgRNA was generated using the MEGAscript T7 Transcription Kit (Life Technologies, Carlsbad, CA). To prepare the tyr sgRNA, pT7tyrgRNA (Addgene, Cambridge, MA) was linearized with BamHI and sgRNA was synthesized using MEGAscript T7 Transcription Kit (Life Technologies). To generate the Cas9 mRNA, pCS2-nCas9n plasmid (Addgene, Cambridge, MA) was linearized with NotI and capped Cas9 mRNA was synthesized using the mMESSAGE mMACHINE SP6 Transcription Kit (Life Technologies). To confirm the quality of sgRNA and Cas9 mRNA, RNA was mixed with formamide, heated at 72°C for 5 minutes and run on a 1% (wt/vol) agarose gel. The following sgRNA and Cas9 mRNA doses were microinjected into embryos at the one-cell stage: 100 pg/embryo of sox4a or sox4b sgRNA + 200 pg/embryo of Cas9 mRNA; 50 pg/embryo of tyr sgRNA + 150 pg/embryo of Cas9 mRNA.
HRMA analysis
To isolate genomic DNA from uninjected or sgRNA/Cas9 injected individual embryos, 24 hpf dechorionated embryos were placed into individual wells of a 96-well plate containing 20 μl of 1X ThermoPol Buffer (New England Biolabs, lpswich, MA). The plate was placed in a PCR cycler at 95°C for 10 minutes, after which 5 μl of 10 mg/ml Proteinase K (Sigma, St. Louis, MO) was added to each well and the plate was incubated at 55°C for 1 hour and 95°C for 10 minutes. HRMA analysis was performed on a LightCycler 96 Real-Time PCR System (Roche, Indianapolis, IN) using LightCycler 480 High Resolution Melting Master (Roche), following the manufacturer's instructions. Primer sequences used for HRMA are listed in Table S1.
Immunohistochemistry and TUNEL assay
Immunohistochemistry was performed on cryosections or whole zebrafish embryos as previously described (Fadool, 2003; Forbes-Osborne et al., 2013). Images were obtained on an inverted fluorescent microscope (Eclipse Ti-U; Nikon Instruments) using the 20X objective. The following primary antibodies and dilutions were used: anti-PCNA (mouse, 1:100, Santa Cruz Biotechnology, Dallas, Texas), which labels cells in G1/S phase; anti-PH3 (rabbit, 1:500, Millipore, Billerica, MA), which labels cells in G2/M phase; anti-Zn-8 (mouse, 1:10, ZIRC, Eugene, OR), which labels ganglion cells; anti-Prox1 (rabbit, 1:1000, Acris, San Diego, CA), which recognizes horizontal cells; anti-PKCα (rabbit, 1:100, Santa Cruz Biotechnology, Dallas, Texas), which labels bipolar cells. Alexa fluor-conjugated secondary antibodies (Invitrogen, Grand Island, NY) and Cy-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were all used at a dilution of 1:200. Sections were counterstained with DAPI (1:10,000, Sigma, St. Louis, MO) to visualize cell nuclei. For laminin immunostaining, 48 and 72 hpf zebrafish embryos were hybridized with anti-laminin (mouse, 1:60, Sigma, St. Louis, MO), and imaged on a laser scanning confocal microscope (Leica TCS SP5). TUNEL assay was performed on retinal cryosections using the ApopTag Fluorescein Direct In Situ Apoptosis Detection Kit (Millipore, Billerica, MA) according to the manufacturer's instructions.
Real-Time quantitative RT-PCR
Total RNA was extracted from the whole body of control and sox4 morphant embryos at 8, 10, and 12 hpf or the heads of control and sox4 morphant embryos, or TdTomato and sox4 mRNA injected embryos, at 18 and 24 hpf using TRIzol reagent (Invitrogen, Grand Island, NY). RNA was reverse-transcribed using the GoScript Reverse Transcriptase System (Promega, Madison, WI). Real time PCR was performed using Maxima SYBR Green qPCR master mix (Thermo Scientific, Waltham, MA) on an iCycler iQ Real Time PCR Detection system (Bio-Rad, Hercules, CA), or using FastStart Essential DNA Green Master (Roche, Indianapolis, IN) on a LightCycler 96 Real-Time PCR System (Roche, Indianapolis, IN). For all experiments, three biological replicates were analyzed, and relative transcript abundance was normalized to expression of the housekeeping genes gapdh or atp5h.
Cyclopamine treatments
Cyclopamine (Sigma, St. Louis, MO) was resuspended at 1mM in 100% ethanol and diluted with fish water to a final concentration of 0.2 μM. For vehicle controls, 0.2% ethanol in fish water was used. Embryos injected with standard control MO or sox4 MOs were exposed to cyclopamine or ethanol from 5.5 to 13 hpf and then placed into fresh fish water until 24 hpf.
SDS PAGE and Western blots
Protein was extracted from pools of 80-100 24 hpf control and sox4 morphant heads. Protein concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA). Ten μg of total protein per sample was diluted 1:1 with Laemmli buffer and 2.5% β-mercaptoethanol, boiled for 10 minutes, and then separated by SDS-PAGE on 10% polyacrylamide gels. Resolved proteins were transferred to nitrocellulose membranes and blocked in 1X TBS/0.05% TWEEN/5% BSA for 1 hour at room temperature prior to incubation with either anti-IHH antibody (rabbit polyclonal, 1:800, Sigma, St. Louis, MO), or anti-β-Actin (rabbit polyclonal, 1:2000, Abcam, Cambridge, MA) as a loading control. The IHH antibody immunogen sequence partially overlaps with zebrafish Ihha, Ihhb and Shhb protein sequences. Membranes were washed and incubated in goat anti-rabbit-peroxidase secondary antibody (1:3000, Sigma, St. Louis, MO). Blots were developed using the Pierce ECL Western Blotting Substrate (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions.
Statistical Analyses
Significance was calculated using a two-tailed Student's t-test or Fisher's exact test, with P<0.05 being considered significant. For all graphs, data are represented as the mean ± the standard deviation (s.d.).
Supplementary Material
HIGHLIGHTS.
Sox4 is required for choroid fissure closure, and Sox4 deficiency causes ocular coloboma.
Knockdown of Sox4 alters proximo-distal patterning of the optic stalk and vesicle.
Knockdown of Sox4 results in elevated Hh signaling.
Sox4 negatively regulates expression of the Hh ligand gene indian hedgehog (ihhb).
ACKNOWLEDGEMENTS
The authors would like to thank Sara Perkins for zebrafish care, and Marie Forbes-Osborne, Joanna Ng, Abirami Krishna, and the laboratory of Dr. Vincent Cassone and Dr. Ashley Seifert for technical assistance. The authors would also like to give special thanks to Dr. Jared C. Talbot (S. Amacher lab, Ohio State University, Columbus, OH) and Dr. Jeffrey J. Essner (Iowa State University, Ames, IA) for sharing their CRISPR protocols.
FUNDING
This work was supported by a grant from the National Institutes of Health (R01EY021769, A.C.M.), the Pew Biomedical Scholar Program (A.C.M.), and the Lyman T. Johnson and Academic Year graduate fellowships from the University of Kentucky (W.W. and L.P.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS:
A.C.M. and W.W. conceived and designed the experiments, W.W., L.P.K., and S.G.W. performed the experiments, A.C.M., W.W., L.P.K., and S.G.W. analyzed the data, and A.C.M. and W.W. wrote the paper.
REFERENCES
- Amato MA, Boy S, Perron M. Hedgehog signaling in vertebrate eye development: a growing puzzle. Cellular and molecular life sciences : CMLS. 2004;61:899–910. doi: 10.1007/s00018-003-3370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Williams T, Zacharias AL, Gage PJ, Fuhrmann S, West-Mays JA. AP-2alpha knockout mice exhibit optic cup patterning defects and failure of optic stalk morphogenesis. Human molecular genetics. 2010;19:1791–1804. doi: 10.1093/hmg/ddq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastida MF, Sheth R, Ros MA. A BMP-Shh negative-feedback loop restricts Shh expression during limb development. Development (Cambridge, England) 2009;136:3779–3789. doi: 10.1242/dev.036418. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, Depamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nature communications. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibliowicz J, Gross JM. Ectopic proliferation contributes to retinal dysplasia in the juvenile zebrafish patched2 mutant eye. Investigative ophthalmology & visual science. 2011;52:8868–8877. doi: 10.1167/iovs.11-8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Developmental biology. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews. Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Cai J, Xu X, Yin H, Wu R, Modderman G, Chen Y, Jensen J, Hui CC, Qiu M. Evidence for the differential regulation of Nkx-6.1 expression in the ventral spinal cord and foregut by Shh-dependent and -independent mechanisms. Genesis. 2000;27:6–11. doi: 10.1002/1526-968x(200005)27:1<6::aid-gene20>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Blain D, Bertuzzi S, Brooks BP. Uveal coloboma: clinical and basic science update. Current opinion in ophthalmology. 2006;17:447–470. doi: 10.1097/01.icu.0000243020.82380.f6. [DOI] [PubMed] [Google Scholar]
- Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain research. Molecular brain research. 2000;79:180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- Chung AY, Kim S, Kim E, Kim D, Jeong I, Cha YR, Bae YK, Park SW, Lee J, Park HC. Indian hedgehog B function is required for the specification of oligodendrocyte progenitor cells in the zebrafish CNS. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:1728–1733. doi: 10.1523/JNEUROSCI.3369-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie PD, Ingham PW. Induction of a specific muscle cell type by a hedgehog-like protein in zebrafish. Nature. 1996;382:452–455. doi: 10.1038/382452a0. [DOI] [PubMed] [Google Scholar]
- Dakubo GD, Mazerolle C, Furimsky M, Yu C, St-Jacques B, McMahon AP, Wallace VA. Indian hedgehog signaling from endothelial cells is required for sclera and retinal pigment epithelium development in the mouse eye. Developmental biology. 2008;320:242–255. doi: 10.1016/j.ydbio.2008.05.528. [DOI] [PubMed] [Google Scholar]
- Duench K, Franz-Odendaal TA. BMP and Hedgehog signaling during the development of scleral ossicles. Developmental biology. 2012;365:251–258. doi: 10.1016/j.ydbio.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic acids research. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles MR, Schimmenti LA. Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clinical genetics. 1999;56:1–9. doi: 10.1034/j.1399-0004.1999.560101.x. [DOI] [PubMed] [Google Scholar]
- Ekker SC, Ungar AR, Greenstein P, von Kessler DP, Porter JA, Moon RT, Beachy PA. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Current biology : CB. 1995;5:944–955. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- Fadool JM. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Developmental biology. 2003;258:277–290. doi: 10.1016/s0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]
- Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C, Liu HK. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell stem cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes-Osborne MA, Wilson SG, Morris AC. Insulinoma-associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Developmental. 2013;380:157–171. doi: 10.1016/j.ydbio.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongal PA, French CR, Waskiewicz AJ. Aberrant forebrain signaling during early development underlies the generation of holoprosencephaly and coloboma. Biochimica et biophysica acta. 2011;1812:390–401. doi: 10.1016/j.bbadis.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Gregory-Evans CY, Williams MJ, Halford S, Gregory-Evans K. Ocular coloboma: a reassessment in the age of molecular neuroscience. Journal of medical genetics. 2004;41:881–891. doi: 10.1136/jmg.2004.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirgis MF, Lueder GT. Choroidal neovascular membrane associated with optic nerve coloboma in a patient with CHARGE association. American journal of ophthalmology. 2003;135:919–920. doi: 10.1016/s0002-9394(02)02293-6. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Bitgood MJ, McMahon AP. Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes & Development. 1996;10:647–658. doi: 10.1101/gad.10.6.647. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. Journal of experimental zoology. Part B, Molecular and developmental evolution. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Hornby SJ, Gilbert CE, Rahi JK, Sil AK, Xiao Y, Dandona L, Foster A. Regional variation in blindness in children due to microphthalmos, anophthalmos and coloboma. Ophthalmic epidemiology. 2000;7:127–138. [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development (Cambridge, England. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Ding Q, Xie X, Libby RT, Lefebvre V, Gan L. Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. The Journal of biological chemistry. 2013;288:18429–18438. doi: 10.1074/jbc.M113.478503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Goodman J, Anderson KV, Niswander L. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Developmental cell. 2007;13:87–102. doi: 10.1016/j.devcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kosinski C, Stange DE, Xu C, Chan AS, Ho C, Yuen ST, Mifflin RC, Powell DW, Clevers H, Leung SY, et al. Indian hedgehog regulates intestinal stem cell fate through epithelial-mesenchymal interactions during development. Gastroenterology. 2010;139:893–903. doi: 10.1053/j.gastro.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Willer JR, Willer GB, Smith K, Gregg RG, Gross JM. Zebrafish blowout provides genetic evidence for Patched1-mediated negative regulation of Hedgehog signaling within the proximal optic vesicle of the vertebrate eye. Developmental biology. 2008;319:10–22. doi: 10.1016/j.ydbio.2008.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cox BD, Daly CM, Lee C, Nuckels RJ, Tittle RK, Uribe RA, Gross JM. An ENU mutagenesis screen in zebrafish for visual system mutants identifies a novel splice-acceptor site mutation in patched2 that results in Colobomas. Investigative ophthalmology & visual science. 2012;53:8214–8221. doi: 10.1167/iovs.12-11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Bates J, Eisen JS. Regulation of iro3 expression in the zebrafish spinal cord. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;232:140–148. doi: 10.1002/dvdy.20215. [DOI] [PubMed] [Google Scholar]
- Liu C, Bakeri H, Li T, Swaroop A. Regulation of retinal progenitor expansion by Frizzled receptors: implications for microphthalmia and retinal coloboma. Human molecular genetics. 2012;21:1848–1860. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker M, Agathocleous M, Amato MA, Parain K, Harris WA, Perron M. Hedgehog signaling and the retina: insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006;20:3036–3048. doi: 10.1101/gad.391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning L, Ohyama K, Saeger B, Hatano O, Wilson SA, Logan M, Placzek M. Regional morphogenesis in the hypothalamus: a BMP-Tbx2 pathway coordinates fate and proliferation through Shh downregulation. Developmental cell. 2006;11:873–885. doi: 10.1016/j.devcel.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Marti E, Bovolenta P. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends in neurosciences. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, Motte P, Martial JA, Peers B. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Developmental biology. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Morcillo J, Martinez-Morales JR, Trousse F, Fermin Y, Sowden JC, Bovolenta P. Proper patterning of the optic fissure requires the sequential activity of BMP7 and SHH. Development (Cambridge, England) 2006;133:3179–3190. doi: 10.1242/dev.02493. [DOI] [PubMed] [Google Scholar]
- Morris AC, Forbes-Osborne MA, Pillai LS, Fadool JM. Microarray analysis of XOPS-mCFP zebrafish retina identifies genes associated with rod photoreceptor degeneration and regeneration. Investigative ophthalmology & visual science. 2011;52:2255–2266. doi: 10.1167/iovs.10-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DA, FitzPatrick DR, Fleck BW. Iris coloboma with iris heterochromia: a common association. Archives of ophthalmology. 2000;118:1590–1591. doi: 10.1001/archopht.118.11.1590. [DOI] [PubMed] [Google Scholar]
- Moshiri A, Reh TA. Persistent progenitors at the retinal margin of ptc+/−mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:229–237. doi: 10.1523/JNEUROSCI.2980-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Krishnaswami SR, Cowing-Zitron C, Hung NJ, Reilly-Rhoten H, Burns J, Yu BD. Negative regulation of Shh levels by Kras and Fgfr2 during hair follicle development. Developmental biology. 2013;373:373–382. doi: 10.1016/j.ydbio.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nature genetics. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nelson SM, Frey RA, Wardwell SL, Stenkamp DL. The developmental sequence of gene expression within the rod photoreceptor lineage in embryonic zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:2903–2917. doi: 10.1002/dvdy.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochocinska MJ, Hitchcock PF. Dynamic expression of the basic helix-loop-helix transcription factor neuroD in the rod and cone photoreceptor lineages in the retina of the embryonic and larval zebrafish. The Journal of comparative neurology. 2007;501:1–12. doi: 10.1002/cne.21150. [DOI] [PubMed] [Google Scholar]
- Pathi S, Rutenberg JB, Johnson RL, Vortkamp A. Interaction of Ihh and BMP/Noggin signaling during cartilage differentiation. Developmental biology. 1999;209:239–253. doi: 10.1006/dbio.1998.9181. [DOI] [PubMed] [Google Scholar]
- Paul MH, Harvey RP, Wegner M, Sock E. Cardiac outflow tract development relies on the complex function of Sox4 and Sox11 in multiple cell types. Cellular and molecular life sciences : CMLS. 2013 doi: 10.1007/s00018-013-1523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. The international journal of biochemistry & cell biology. 2010;42:425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai-Kastoori L, Wen W, Wilson SG, Strachan E, Lo-Castro A, Fichera M, Musumeci SA, Lehmann OJ, Morris AC. Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis. PLoS genetics. 2014;10:e1004491. doi: 10.1371/journal.pgen.1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porges Y, Gershoni-Baruch R, Leibu R, Goldscher D, Zonis S, Shapira I, Miller B. Hereditary microphthalmia with colobomatous cyst. American journal of ophthalmology. 1992;114:30–34. doi: 10.1016/s0002-9394(14)77409-4. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS genetics. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont ME, Sullivan MJ, Dobyns WB, Eccles MR. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nature genetics. 1995;9:358–364. doi: 10.1038/ng0495-358. [DOI] [PubMed] [Google Scholar]
- Schauerte HE, van Eeden FJ, Fricke C, Odenthal J, Strahle U, Haffter P. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development (Cambridge, England) 1998;125:2983–2993. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- Schimmenti LA, de la Cruz J, Lewis RA, Karkera JD, Manligas GS, Roessler E, Muenke M. Novel mutation in sonic hedgehog in non-syndromic colobomatous microphthalmia. American journal of medical genetics. Part A. 2003;116A:215–221. doi: 10.1002/ajmg.a.10884. [DOI] [PubMed] [Google Scholar]
- Seki K, Hata A. Indian hedgehog gene is a target of the bone morphogenetic protein signaling pathway. The Journal of biological chemistry. 2004;279:18544–18549. doi: 10.1074/jbc.M311592200. [DOI] [PubMed] [Google Scholar]
- Shen MC, Ozacar AT, Osgood M, Boeras C, Pink J, Thomas J, Kohtz JD, Karlstrom R. Heat-shock-mediated conditional regulation of hedgehog/gli signaling in zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:539–549. doi: 10.1002/dvdy.23955. [DOI] [PubMed] [Google Scholar]
- Shen YC, Raymond PA. Zebrafish cone-rod (crx) homeobox gene promotes retinogenesis. Developmental biology. 2004;269:237–251. doi: 10.1016/j.ydbio.2004.01.037. [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Chao R, Vacik T, Yahyavi M, Abouzeid H, Bardakjian T, Schneider A, Shaw G, Sherr EH, Lemke G, et al. VAX1 mutation associated with microphthalmia, corpus callosum agenesis, and orofacial clefting: the first description of a VAX1 phenotype in humans. Human mutation. 2012;33:364–368. doi: 10.1002/humu.21658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL. Neurogenesis in the Fish Retina. 2007;259:173–224. doi: 10.1016/S0074-7696(06)59005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Take-uchi M. Hedgehog signalling maintains the optic stalk-retinal interface through the regulation of Vax gene activity. Development (Cambridge, England) 2003;130:955–968. doi: 10.1242/dev.00305. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Amacher SL. A Streamlined CRISPR Pipeline to Reliably Generate Zebrafish Frameshifting Alleles. Zebrafish. 2014;11:583–585. doi: 10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui A, Iwagawa T, Mochizuki Y, Iida A, Wegner M, Murakami A, Watanabe S. Expression of Sox4 and Sox11 is regulated by multiple mechanisms during retinal development. FEBS letters. 2013;587:358–363. doi: 10.1016/j.febslet.2012.12.017. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. The EMBO journal. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 4th ed. Univ. of Oregon Press; Eugene: 2000. [Google Scholar]
- Wolff C, Roy S, Ingham PW. Multiple Muscle Cell Identities Induced by Distinct Levels and Timing of Hedgehog Activity in the Zebrafish Embryo. Current Biology. 2003;13:1169–1181. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Swinton PG, Martin JF, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Developmental biology. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Golubkov VS, Han W, Correa RG, Zhou Y, Lee S, Strongin AY, Dong PD. Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Developmental biology. 2014;395:96–110. doi: 10.1016/j.ydbio.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Huang H, Cao P, Wang Z, Chen Y, Pan Y. Sma-and Mad-related protein 7 (Smad7) is required for embryonic eye development in the mouse. The Journal of biological chemistry. 2013;288:10275–10285. doi: 10.1074/jbc.M112.416719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.