Abstract
Intrinsically disordered proteins (IDPs) are prevalent in macromolecular assemblies and are thought to mediate protein recognition in complex regulatory processes and signaling pathways. The formation of a polybivalent scaffold is a key process by which IDPs drive early steps in macromolecular assemblies. Three intrinsically disordered proteins, IC, Swallow and Nup159, are core components, respectively, of cytoplasmic dynein, bicoid mRNA localization apparatus, and nuclear pore complexes. In all three systems, the hub protein LC8 recognizes on the IDP, short linear motifs that are fully disordered in the apo form, but adopt a β-strand when bound to LC8. The IDP/LC8 complex forms a bivalent scaffold primed to bind additional bivalent ligands. Scaffold formation also promotes self-association and/or higher order organization of the IDP components at a site distant from LC8 binding. Rigorous thermodynamic analyses imply that association of additional bivalent ligands is driven by entropic effects where the first binding event is weak but subsequent binding of additional ligands occurs with higher affinity. Here, we review specific examples of macromolecular assemblies in which polybivalency of aligned IDP duplexes not only enhances binding affinity and results in formation of a stable complex but also compensates unfavorable steric and enthalpic interactions. We propose that polybivalent scaffold assembly involving IDPs and LC8-like proteins is a general process in the cell biology of a class of multi-protein structures that are stable yet fine-tuned for diverse cellular requirements.
Keywords: Intrinsically disordered proteins, LC8, macromolecular assembly, bivalency, poly-bivalent scaffold, enthalpy-entropy compensation
1. Introduction
Multivalent interactions involving the linked associations of ligands binding to multiple sites on a receptor confer increased affinity compared to binding to monovalent receptors. Multivalent interactions function in diverse biological systems, including antibody antigen associations, interactions of multisubunit toxins such as cholera toxin and botulinum neurotoxin with their target cells, lectin-carbohydrate and protein-glycan interactions. Bivalent interactions are a subset of multivalent interactions involving associations of bivalent ligands with a receptor having two binding sites for that ligand.
While multivalent interactions are common and well described for ordered receptors and small ligands [1–3] their role in intrinsically disordered proteins (IDPs) is only recently appreciated in supramolecular protein assembly [4, 5], and in polyphosphorylation [6]. A key feature of bivalent interactions of disordered proteins in macromolecular assembly is that, in the process, two monovalent IDP chains are incorporated into a bivalent macromolecular receptor or scaffold with each chain carrying one binding site. The two IDP chains may be linked by binding a dimeric protein with two symmetrical binding sites, by self-association, or by a synthetic linkage (Figure 1). For such a disordered protein system with multisite recognition motifs for different bivalent ligands, alignment of two chains, either by binding a bivalent ligand, or by self-association, results in a duplex with multiple additional bivalent sites. We refer to this duplex as a polybivalent scaffold. For all cases (bivalent or polybivalent), the outcome is the same: the first binding event pays the entropic cost so that a subsequent event occurs with lower entropic penalty when compared to a monovalent ligand. In summary, bivalent events organize two disordered chains into a bivalent, partially disordered system that has higher binding affinity for other protein(s) at other recognition motif(s). The partial disorder resides in the incorporated IDP chains, which retain disordered regions.
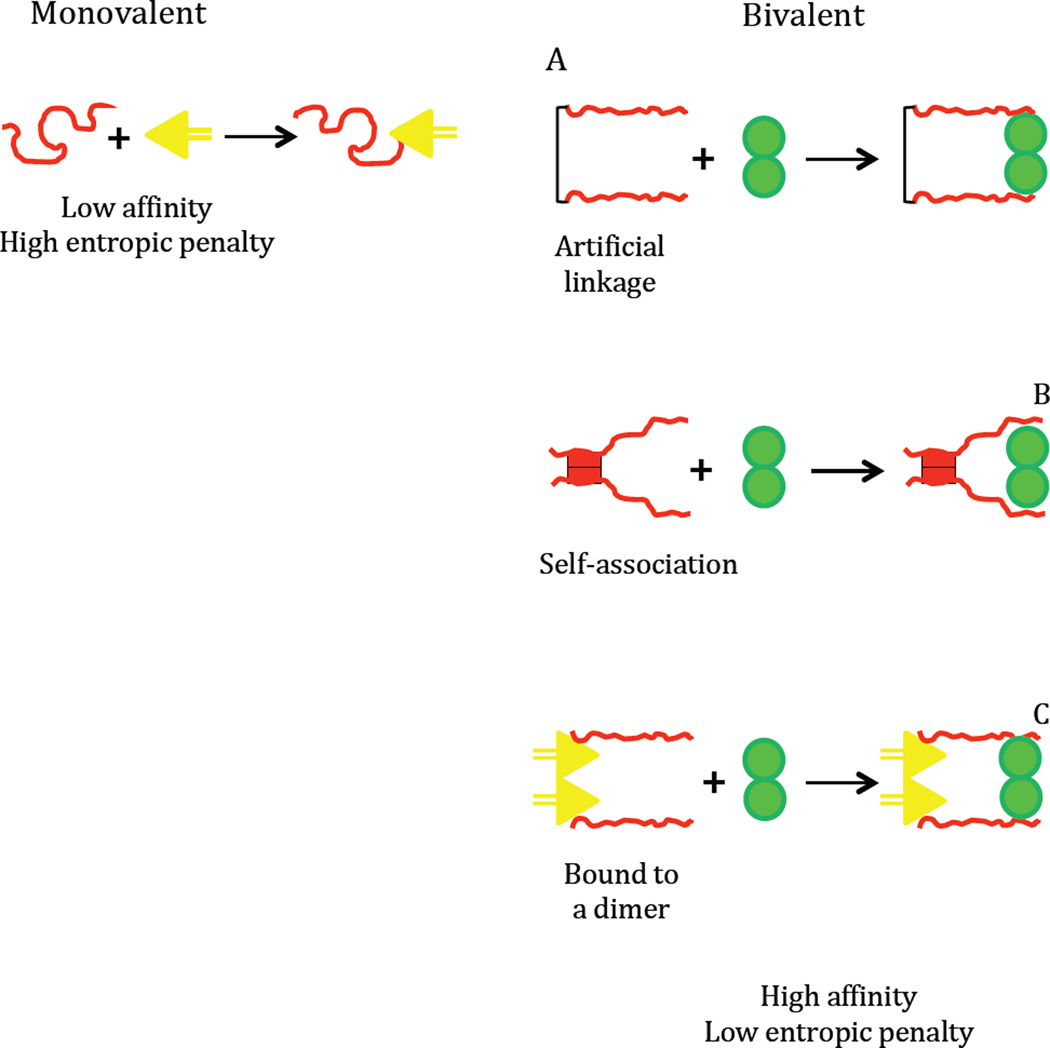
Figure 1.
Scheme illustrating bivalent interactions in disordered proteins. A high entropic penalty is associated with monovalent interactions. Aligning two chains of a disordered protein by an artificial linker, black line (A), through inter-chain (self-association) interactions, red bars (B), or by their interactions with a dimeric protein, yellow triangles (C) reduces the entropic penalty of a second binding event, green spheres.
Binding enhancement arising from bivalency/polybivalency has been demonstrated in assembly of Swallow/LC8 complex [7, 8], the nuclear pore protein Nup159/LC8 complex [9] and the dynein intermediate chain/light chains complex [4, 5, 10, 11]. A common feature among the three systems is that a segment of their disordered regions has one or more recognition motifs for the hub protein LC8 [12]. On Swallow, one LC8 dimer binds a recognition sequence C-terminal to a weak coiled-coil region and promotes stable coiled-coil self-association. On nucleoporin Nup159, five LC8 dimers bind at positions N-terminal to a predicted coiled-coil and form a rigid beads-on-a-string rod, possibly with a kink. On dynein intermediate chain, IC, binding of one LC8 dimer creates a bivalent IC duplex with enhanced affinity to another LC8-like dimer, Tctex1, and promotes self-association of IC at a distant domain. In these examples, bivalency involves not only interactions between intrinsically disordered chains and bivalent protein dimers but also self-association interactions.
Nup159 and IC are canonical examples of polybivalent scaffolds because Nup159 binds multiple LC8 dimers, and IC binds several bivalent ligands. The multiple bivalent sites provide the potential for mutual enhancement of affinity for an additional ligand(s), as well as for coiled-coil interchain interactions. Initial binding to these multiple bivalent sites is associated with a large entropic penalty. By pre-paying this entropic cost, bivalency either significantly enhances binding affinity of subsequent bivalent ligands, or energetically compensates unfavorable interactions associated with complex formation. Each of these scenarios is described below.
2. Bivalency enhances binding affinity and results in formation of a stable complex
To evaluate the thermodynamic parameters of binding enhancement of the IC component of the dynein cargo complex, we used a simplified IC segment, ICLL, that has two recognition motifs for LC8 separated by a 3–5 residues linker. ICLL is a variant of wild type IC with the Tctex1 recognition sequence replaced by the LC8 recognition sequence; the resulting disordered ICLL has two identical LC8 recognition motifs. ICLL is a suitable model because Tctex1 and LC8 are structural homologs with similar tertiary and quaternary structures, and similar binding affinity to IC.
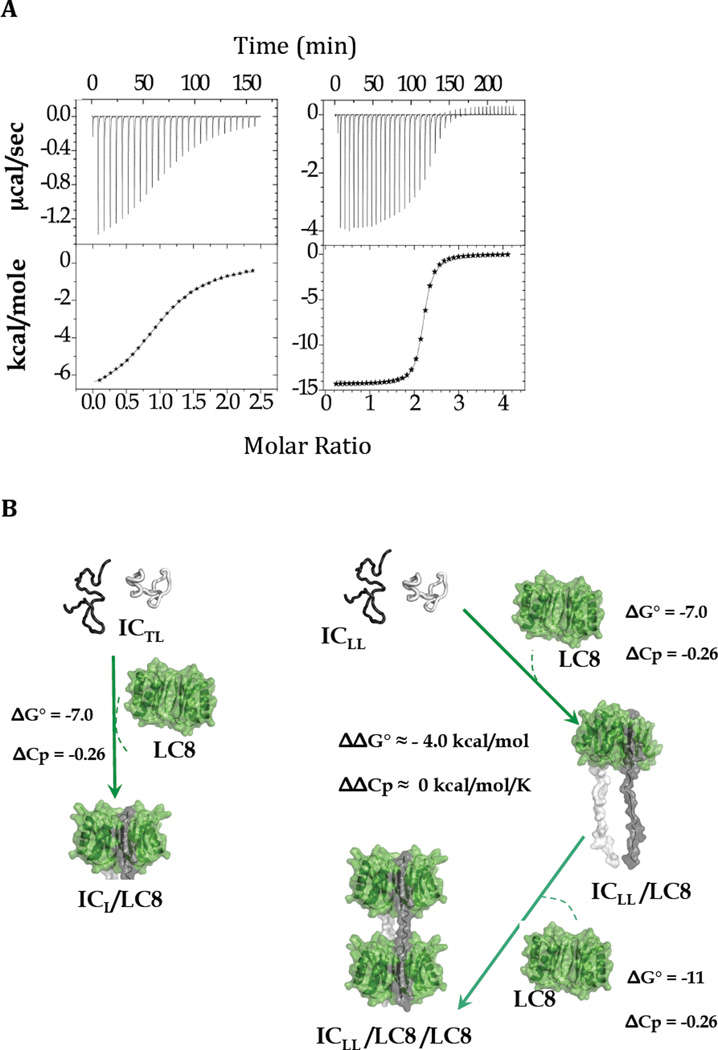
Isothermal titration calorimetry, ITC, does not resolve individual binding events for ICLL binding to two LC8 dimers. Instead, the output is a macroscopic Kd value of 0.5 µM and a stoichiometry of two ICLL chains per two LC8 dimers (Figure 2) with an average value for ΔG° of −9.0 kcal/mol, and a ΔCp of −0.26 kcal/mol/K. Distinct thermodynamic parameters for each binding are inferred using the assumption that the thermodynamic parameters of the first binding are similar to LC8 binding a monovalent IC variant with a single LC8 site (Figure 2A, left). By this reasoning, the inferred thermodynamic parameters for the second binding are ΔG° of −11.0, and ΔCp of −0.26 kcal/mol/K, corresponding to an affinity enhancement for the second LC8 binding of 1000-fold (ΔΔG° of −4.0 kcal/mol), arising solely from a favorable change in entropy Δ(-TΔS°) of −4.0 kcal/mol in the 20–35 °C temperature range (Figure 3, left), as expected from a full bivalency effect [13].
Figure 2.
Thermodynamics of LC8 binding to monovalent and bivalent IC. (A) Representative ITC thermograms (top panels) and binding isotherms (bottom panels) for titration of ICTL (left) and ICLL (right) with LC8. (B) Binding free energies ΔG° (kcal/mol), and ΔCp (kcal/mol/K) are given for each step. The difference in free energy and heat capacity changes between the second and first binding events of LC8 to ICLL (right) are expressed as ΔΔG° and ΔΔCp, respectively, and are shown in the center. Binding parameters for the first ICLL/LC8 binding event are assumed to be similar to those to ICL (left). Binding parameters for the second ICLL/LC8 binding event are inferred from average values determined from direct measurements. Figures adapted from [5].
Figure 3.
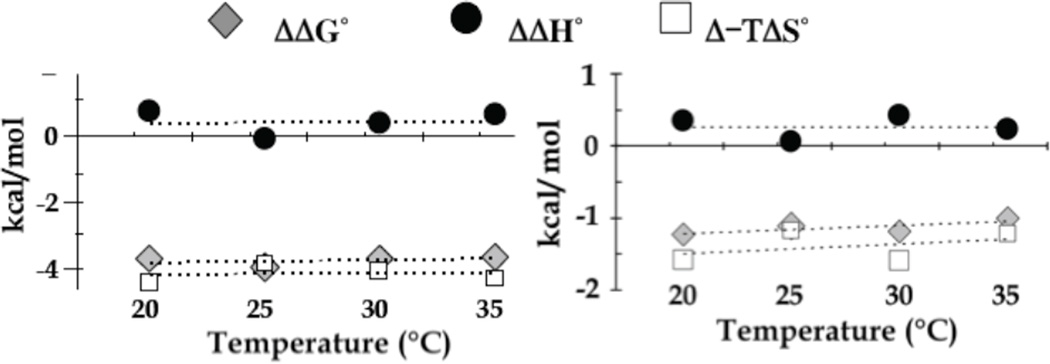
Changes in thermodynamic parameters between first and second LC8 binding events (ΔΔG°, grey diamonds; ΔΔH°, black circles; and Δ-TΔS°, white squares). Differences in association parameters at different temperatures between LC8 binding to monovalent apo-ICLL and LC8 binding to bivalent ICLL/LC8 (left), and between LC8 binding to monovalent SwaMONOMER and LC8 binding to bivalent SwaDIMER (right). The absence of temperature dependence (ΔΔCp of zero) shows that binding enhancement (ΔΔG°) is not due to enthalpic change (ΔΔH° of zero) but due to entropic change (Δ-TΔS° of same value as ΔΔG°). Figures adapted from [5], and [8].
Another possibility for creating a bivalent IDP scaffold is linkage through self-association (Figure 1). For an LC8 binding partner called Swallow, also primarily disordered, the two Swallow chains are linked by a weakly predicted coiled-coil N-terminal to the LC8 recognition motif. To determine the origin of LC8 binding enhancement due to Swallow self-association, binding thermodynamics were measured for two Swallow constructs designed to decouple self-association from LC8 binding [8]. SwaDIMER, an engineered variant with coiled-coil stabilizing mutations, is a tightly associated bivalent binding partner with two aligned recognition sequences for LC8, and SwaMONOMER, an engineered variant with coiled-coil destabilizing mutations is a disordered monovalent chain with one LC8 recognition motif. As with ICLL, the binding enhancement of 7-fold in SwaDIMER relative to SwaMONOMER is primarily of entropic origin (ΔΔG° of −1.1 kcal/mol, TΔΔS° of −1.2 kcal/mol, and ΔΔH° and ΔΔCp of 0) (Figure 3, right).
A third possibility for creating a bivalent IDP scaffold is by covalent cross-linking (Figure 1). A bivalent IC, referred to as 219Cdimer, was produced by introducing a disulfide cross-link at residue 219, 81 residues distant from the LC8 recognition sequence, and immediately preceding a weak self-association domain [11]. Binding affinity of the 219Cdimer to LC8 was enhanced 6–7-fold relative to monovalent IC. Binding enhancement is again primarily of entropic origin (ΔΔG° of −1.1, TΔΔS° of −0.8, and ΔΔH° of −0.3 kcal/mol).
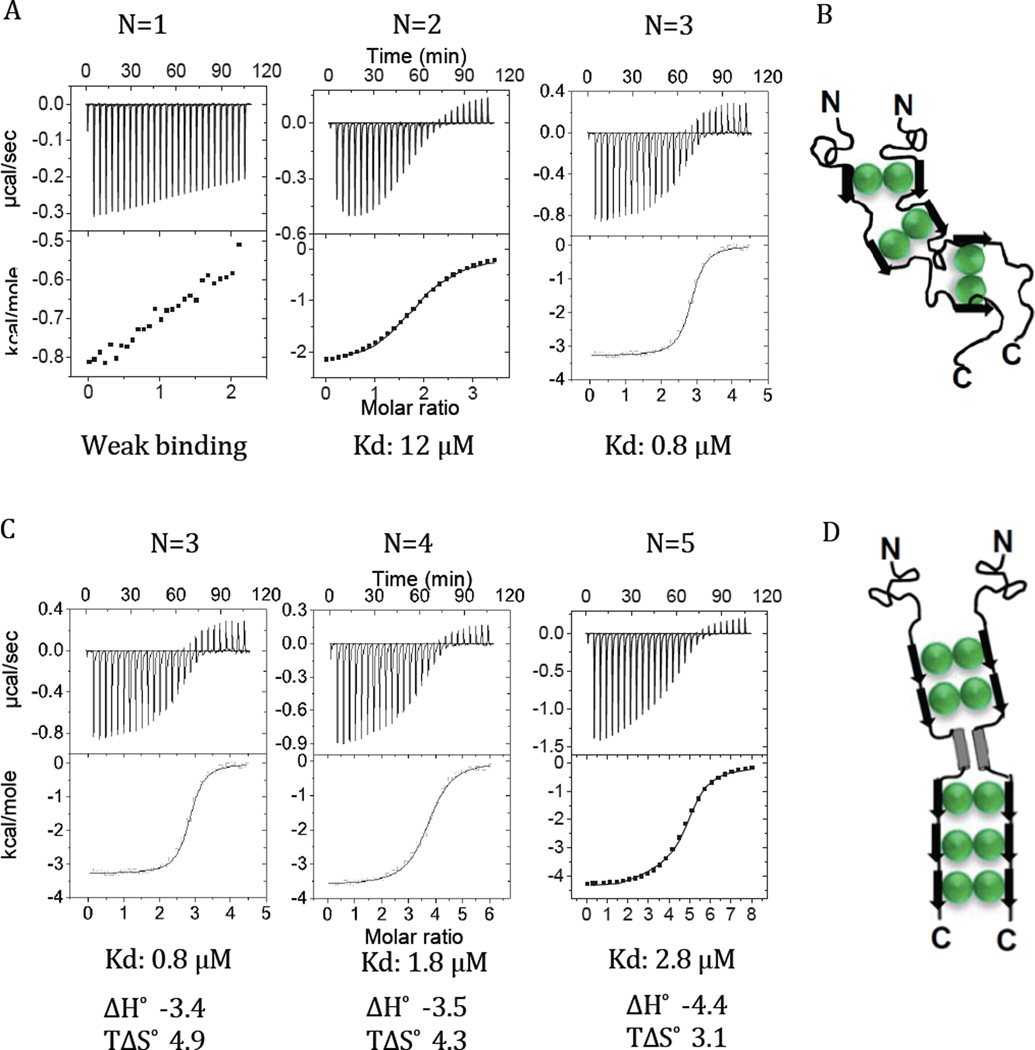
Nucleoporin Nup159, a subunit of the yeast nuclear pore complex (NPC), functions both as a structural component of the NPC, and as a docking site for transiently associated nuclear transport factors [14]. Nup159 – part of the Nup159-Nup82-Nsp1 nucleoporins subcomplex – also binds Dyn2, the yeast ortholog of LC8 [15]. The carboxy-terminal half of Nup159 includes five Dyn2/LC8 recognition motifs (called QT sites) adjacent to a predicted coiled-coil domain. Thermodynamic measurements of Dyn2 binding to a designed series of Nup159 variants, having increasing numbers of Dyn2/LC8 recognition motifs (Figure 4), elegantly demonstrate that the third motif (QT3) does not, as expected [15], bind Dyn2; our studies suggest that instead, residues in QT3 are involved in self-association [9]. The interaction of Dyn2 with monovalent Nup159, where only one Dyn2/LC8 recognition motif is present, is very weak and not fit by a binding model (Figure 4). When two recognition motifs are present, the average binding affinity is enhanced, presumably because the first Dyn2 binding forms a bivalent Nup159 that binds the second Dyn2 with higher affinity. The average binding affinity is significantly enhanced with three binding motifs (compare Kd of 12 µM with 0.8 µM, Figure 4), again clearly underscoring the role of entropic bivalency effects in enhancing binding affinity.
Figure 4.
Dyn2-Nup159 binding parameters and ITC data. Binding thermograms, isotherms, and computed thermodynamic parameters show both the effect of bivalency on binding enhancement and compensation of unfavorable interactions. (A) Representative isothermal titration plots of Dyn2 with one, two and three recognition motifs, respectively, with the corresponding binding constant shown at the bottom of each plot. The interaction with one recognition motif is too weak for data fitting. (B) A hypothetical model of the bound complex composed of two disordered chains of Nup159 (black) and three Dyn2 dimers (green) with Nup159 contact residues adopting β-strand conformations (black arrows). (C) Representative isothermal titration plots of Dyn2 with three, four and five recognition motifs, respectively, with the corresponding binding constant shown at the bottom of each plot, along with the enthalpic and entropic contributions for each binding events. (D) A model of the wild type Nup 159 complex with 5 Dyn2 dimers. The ITC-measured differences in the contribution of the entropy term suggest more flexibility with three Dyn2 (B), but more order and rigidity with five Dyn2 (D). Figure is adapted from [9].
3. Bivalency may compensate energetically unfavorable interactions
Nup159 is a canonical example of a disordered protein with multiple recognition motifs for LC8-like dimers, in this case Dyn2, that are separated by short linkers, and the only example that stacks five LC8-like dimers like beads on a string in the fully bound complex. With five interaction sites for Dyn2, the expectation is an increase in affinity for subsequent binding events with every additional recognition site, as observed from differences between the first, second and third binding events (Figure 4A). Interestingly, and contrary to expectations, this enhancement trend stops and even reverses as the fourth and fifth interaction sites are included (N=4 and 5, Figure 4C), suggesting that the fourth and fifth binding events result in a somewhat less stable assembly with a final affinity of 2.8 µM.
An intriguing observation from these studies is that even though naturally occurring Nup159 contains five sites for Dyn2, only three sites are optimal for forming a stable Nup159/Dyn2 complex. One feasible explanation for the presence of five sites is that binding of Dyn2 at the last two motifs imparts rigidity to the assembled complex (Figure 4D). A considerable loss of degrees of freedom when the last two motifs are bound is inferred from the increased order (decreased entropy) of the system that accompanies binding (TΔS of 4.9 to 4.3 to 3.1, Figure 4C). An increase in unfavorable entropy results in decreased stability even though the enthalpic contribution is more favorable.
We propose that the requirement for binding 5 rather than 3 Dyn2 dimers is to provide entropy/enthalpy balance to a stable yet entropically unfavorable complex. The first three binding sites are required for forming a complex stable enough to balance the entropically unfavorable increase in rigidity with the fourth and fifth sites [9]. In this respect Nup–Dyn2 assembly highlights the importance of entropic considerations in disordered proteins in fine-tuning assembly of multipart complexes.
Similar fine-tuning is observed with the assembly of dynein intermediate chain IC with dimeric LC8 and dimeric Tctex1. Binding of either dimer enhances binding of the second dimer; the enhancement is significantly lower than observed for ICLL. A 50-fold enhancement in WT IC (ICTL) compared to a 1000-fold enhancement observed with ICLL which has two identical LC8 sites suggests additional unfavorable interactions occur during the second binding event that offset the full gain from bivalency, further illustrating that in the WT protein, modulation of affinity optimizes the balance of stability and reversibility.
The lower enhancement in ICTL is accompanied by a significant change in difference in heat capacity between the first and second binding events (ΔΔCp). Since a change in heat capacity is commonly associated with burial of nonpolar surface event [16], the implication of the ΔΔCp value is that surface area buried in the ternary complex of IC/Tctex1/LC8 is larger than in the binary complex of IC/Tctex1 or IC/LC8, suggesting that structural changes accompany formation of the ternary complex. We attribute the lower enhancement of LC8 binding to ICTL/Tctex1 versus ICLL/LC8 to additional disorder-to-order transition at the ICTL/Tctex1 interface. The unfavorable packing against Tctex1 in the ternary complex acts as an attenuator of the favorable bivalency effects of the system. In contrast, in the IC/LC8/LC8 variant system, and in Swallow, ΔΔCp is zero (Figures 2 and 3), indicating a full entropic gain and no accompanying structural/enthalpic changes.
4. Bivalent IDP duplex formation
IC/Tctex1/LC8 complex formation was the first model system used to probe residue-level changes during the assembly of a bivalent IDP, in this case IC. A domain of IC that includes recognition motifs for light chains Tctex1 and LC8 is, by NMR chemical shift criteria, fully disordered with limited propensity for secondary structure [4, 17]. A high level of disorder is also manifested by negative heteronuclear NOEs [4]. Further, measurments of NMR peak intensity upon titration with LC8 monitor selective peak attenuation at the residue level. Peaks corresponding to residues in the LC8 recognition sequence fully disappear while the rest of the backbone peaks, specifically those corresponding to a region where the second light chain binds, show no detectable change in chemical shifts, resonance broadening or 15N relaxation [4]. Thus the two IC chains in a bivalent complex, except for the 10–12 residues at the LC8 site that in the bound form adopt β-strand structure [18], appear by NMR criteria to remain fully disordered and similar to monovalent IC. Therefore, neither structural nor dynamics changes at the second binding site explain the observed bivalency, consistent with its being an entropic effect.
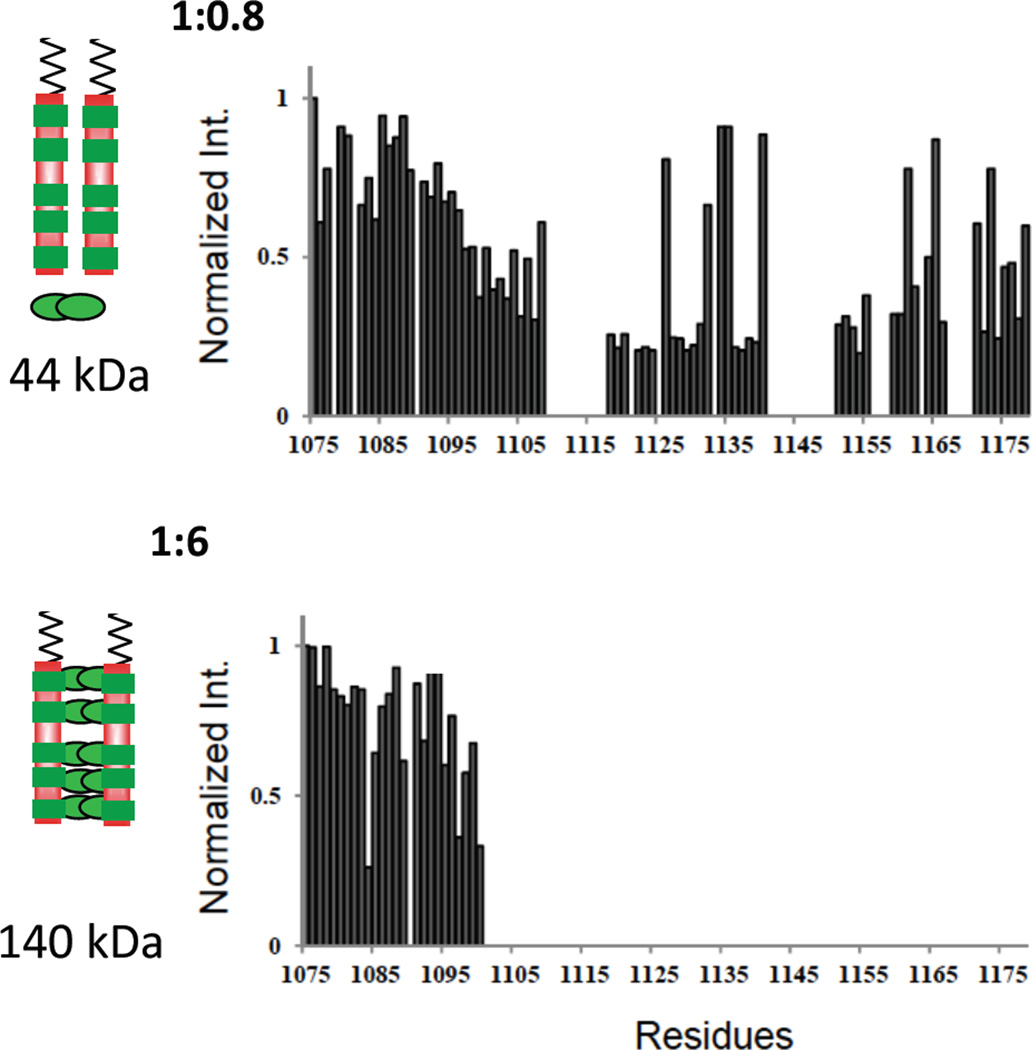
Nup159/Dyn2 complex formation, where multiple Dyn2 dimers bind two Nup159 chains, presents a more complex system to probe residue-level changes during formation of a bivalent IDP. NMR titration experiments monitoring NH peak intensity of Nup159 residues at substoichiometric concentrations of Dyn2 reveal new information about the processes of bivalent duplex formation, namely, a pattern of selective reduction in NH peak intensity along the chain containing the five recognition motifs (Figure 5). The loss of peak intensity arises from conformational changes affecting peak broadening (see the legend to Figure 5). We take the underlying conformational change(s) as due to specific binding interactions of a Nup residue with Dyn2 and/or to the increased tumbling time of that NH in the resultant complex. Taken together with ITC data (Figure 4), these results suggest that initial binding of Dyn2 aligns two Nup chains in a flexible, collapsed state. We envision a nascent scaffold formed from the first bound Dyn2 as a dynamic ensemble of interconverting conformations in which one or two Nup molecules transiently interact with a Dyn2 molecule, but are not stably bound. In the ensemble present in samples having substoichiometric ratios (e.g. Figure 5 top), Nup/Dyn2 interactions selectively attenuate NH peaks to give three clearly distinguishable NH groups. The first group completely disappears from the spectrum as indicated by zero intensity; the second group is partially attenuated, with relative intensity range of 0.25–0.6; the third group shows little, if any, attenuation at a relative intensity around 0.8. Loosely bound Dyn2 dimer, reversibly interacts with a number of sites all along the Nup chain, but preferentially interacts with NH sites in group 1. Group 2 peaks are partially attenuated because at least part of the time those NHs transiently interact and/or tumble with a larger complex. Group 3 NHs do not appreciably interact with Dyn2 and remain locally quite flexible. The possibility of a significant population of misaligned Nup duplexes (e.g. a Dyn2 dimer binding one Nup chain at motif QT2 and the second Nup chain at QT4) – conceivably associated with similarly non-specific, higher order oligomerization as observed in other multivalent systems [19] – is unlikely. First, misaligned Nup/Dyn2 duplexes are not consistent with the residue-specific pattern of attenuation/disappearance in Figure 5 (top). Misaligned duplexes, non-specifically linking any two QT motifs would give an ensemble average in which the relative peak intensity of NHs along the chain is rather evenly attenuated. Higher order oligomerization of misaligned duplexes would, similarly, not show selective peak broadening. Further, and significantly, ITC experiments (Figure 4) show no indication of non-cooperative interaction or higher order aggregation.
Figure 5.
Residue-specific NMR titration of Nup159 NH peak intensity with added Dyn2. Relative NH peak intensity versus residue numbers of non-proline Nup159 NH peaks in NMR spectra of Dyn2-bound Nup159 in the sequence 1075–1178. Shown are Nup:Dyn2 molar ratios of 1:0.8 (top) and 1:6 (bottom). Peak intensities are relative to the intensity of the same peak in apo Nup159, which is taken as one. This figure is adapted from [9] which also reports results at a 1:2 ratio. The models on the left show two chains of Nup159 (red) with five binding sites for Dyn2 (green). In the top model, a single Dyn2 dimer, indicated at the bottom, is added to two molar equivalents of Nup monomer, and its effect is manifested throughout the Nup159 chain, as indicated regions of attenuated intensity all along the Nup chain. In the bottom model, Dyn2 dimers saturate the five Dyn2 binding sites. The observed loss of peak intensity arises from peak broadening due to exchange, on the NMR time scale, of an NH between different local environments (e.g., in bound and unbound conformations), and/or from an increased population within the ensemble of Nup conformations having significantly greater rotational correlation time [20]. In the latter case, the NH being measured might, for example, fractionally exist in several-to-many conformations such that the same NH when in disordered and locally flexible conformations gives a sharp ‘random coil’ signal, and when in more collapsed and/or aggregated conformations with a significantly longer tumbling time and faster relaxation time gives a broadened signal.
5. Polybivalent scaffold assembly: a general process in the cell biology of multi-protein structures
The phenomenon of polybivalent assembly became apparent in experiments flowing from the ‘LC8 hub’ hypothesis which posits that LC8 functions as a regulatory hub protein in a number of essential systems by facilitating the assembly and stabilization of its primarily disordered partners [12]. Underpinning the initial hypothesis were sequence analyses of over 20 LC8-partner proteins showing that LC8 recognition motif(s) lie within a highly disordered segment and, more importantly, that the protein as a whole has a high degree of intrinsic disorder. Experimental tests of the hypothesis reveal that LC8 partners have in common several other striking features, evident in three systems: dynein IC, Swallow and Nup159 [4, 5, 7–11]. Although their functions are unrelated, all are large and complex proteins that appear to assemble other proteins lengthwise along two identical disordered chains, aligned in parallel by binding of LC8 to create an elongated, bivalent duplex with enhanced affinity for the other ligands of the assembly. IC includes binding sites for three light chains and several dynein regulator protein dimers; Swallow includes a putative RNA-binding domain at the N-terminus and a γ-tubulin recognition site at the C-terminal; and Nup159 includes binding sites for Nup82, Nsp1 and Dbp5. In the LC8 hub model, the essential function of LC8 in higher order assembly of these complexes is to form scaffold systems which are stable, yet responsive, assembly even when binding of any single ligand is moderate to weak. The responsiveness of the scaffold stems from its polybivalency which renders the association of bound ligands easily reversible and readily adaptable to fluctuations in cellular environment. Retained disorder in the scaffold contributes to favorable entropy of the system.
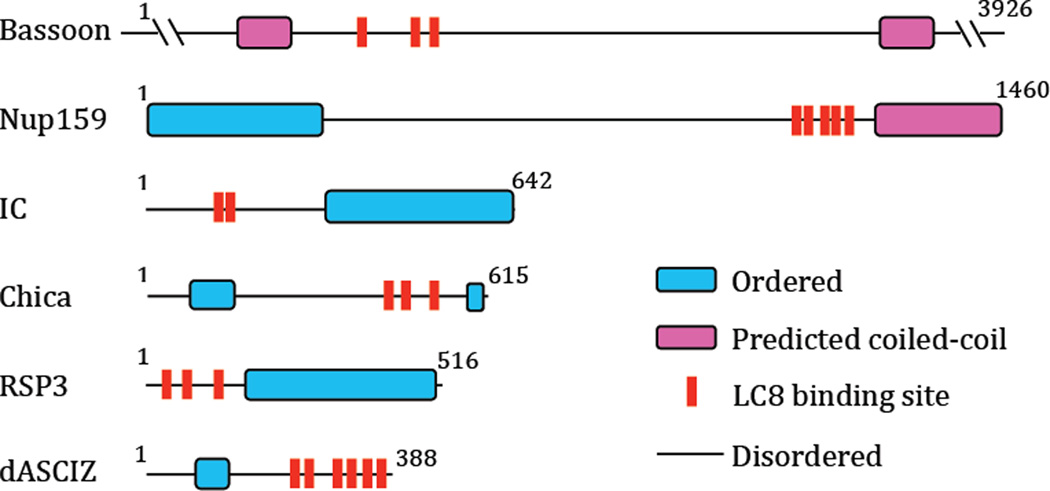
For other LC8 binding partners that have both multi-recognition motifs and weak self-association domains, such as the examples in Figure 6, we hypothesize that bivalency effects analogous to those in LC8 binding to IC and Nup159 also occur. In these examples, multiple LC8 recognition motifs are all localized in intrinsically disordered domains of the respective IDP. Unique aspects of these polybivalent assemblages depend on the binding affinity of the IDP motif for the LC8-like protein, the length of the linkers separating recognition motifs, the degree of disorder retained in the linkers, and the extent of unfavorable interactions in the functional complex.
Figure 6.
Examples of LC8 binding partners having multiple recognition motifs in intrinsically disordered regions. Sequence-based predictions of order, disorder, coiled-coil and LC8 binding motifs are shown for residues 1000 – 2500 of Bassoon, a protein involved in the organization of the cytomatrix at the nerve terminals active zone; the intermediate chain (IC) subunit of the cytoplasmic dynein molecular motor; yeast nucleoporin, Nup159; Chica, a spindle-associated adaptor protein; the flagellar radial spoke protein 3, RSP3, and the Drosophila homologue of ASCIZ (dASCIZ), a DNA damage response and transcription factor. In all these examples, the putative LC8 sites are located within segments with high sequence disorder. Putative motifs are based on the following references: bassoon [21], Nup159 [9], IC[5], Chica[22], RSP3[23] and dASCIZ[24].
In summary, the novel ideas proposed here lead to our conclusion that disordered proteins which bind LC8-like proteins function in a new aspect of the cell biology of macromolecular assemblies. IDP/LC8 complexes form scaffolds that provide mutual enhancement of affinity for additional ligands, as well as for coiled-coil or self-association interactions. The polybivalent nature of the scaffold produces an assemblage that is suitably stable even when the association constant of any single ligand, or self-associating coiled-coil, is moderate to weak; provides ready reversibility of individual ligands; allows fine tuning for changing local cellular environments; and permits formation of a rigid structure. Collectively these properties lend the assemblage a high degree of regulatory flexibility and functional adaptability.
Highlights.
IDPs form polybivalent scaffolds that provide mutual enhancement of affinity for additional ligands, as well as for coiled-coil or self-association interactions.
Bivalency compensates for unfavorable steric and enthalpic interactions.
Bivalency is not accompanied by a detectable change in structure or NMR measured dynamics.
The polybivalent nature of the scaffold produces an assemblage that is suitably stable even when the association constant of any single ligand, or coiled-coil, is moderate to weak.
The polybivalent assemblage function in providing a high degree of regulatory flexibility and functional adaptability.
Acknowledgments
Funding
This work was supported, in whole or in part, by National Institutes of Health Grant GM 084276.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krishnamurthy VM, Semetey V, Bracher PJ, Shen N, Whitesides GM. Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J Am Chem Soc. 2007;129:1312–1320. doi: 10.1021/ja066780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mack ET, Snyder PW, Perez-Castillejos R, Bilgicer B, Moustakas DT, Butte MJ, et al. Dependence of avidity on linker length for a bivalent ligand-bivalent receptor model system. J Am Chem Soc. 2012;134:333–345. doi: 10.1021/ja2073033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands as probes of signal transduction. Angew Chem Int Ed Engl. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benison G, Nyarko A, Barbar E. Heteronuclear NMR identifies a nascent helix in intrinsically disordered dynein intermediate chain: implications for folding and dimerization. J Mol Biol. 2006;362:1082–1093. doi: 10.1016/j.jmb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Hall J, Karplus PA, Barbar E. Multivalency in the assembly of intrinsically disordered dynein intermediate chain. J Biol Chem. 2009;284:33115–33121. doi: 10.1074/jbc.M109.048587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Hare M, Hays TS, Barbar E. Dynein light chain LC8 promotes assembly of the coiled-coil domain of swallow protein. Biochemistry. 2004;43:4611–4620. doi: 10.1021/bi036328x. [DOI] [PubMed] [Google Scholar]
- 8.Kidane AI, Song Y, Nyarko A, Hall J, Hare M, Lohr F, et al. Structural features of LC8-induced self-association of swallow. Biochemistry. 2013;52:6011–6020. doi: 10.1021/bi400642u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyarko A, Song Y, Novacek J, Zidek L, Barbar E. Multiple recognition motifs in nucleoporin Nup159 provide a stable and rigid Nup159-Dyn2 assembly. J Biol Chem. 2013;288:2614–2622. doi: 10.1074/jbc.M112.432831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyarko A, Hare M, Hays TS, Barbar E. The intermediate chain of cytoplasmic dynein is partially disordered and gains structure upon binding to light-chain LC8. Biochemistry. 2004;43:15595–15603. doi: 10.1021/bi048451+. [DOI] [PubMed] [Google Scholar]
- 11.Nyarko A, Barbar E. Light chain-dependent self-association of dynein intermediate chain. J Biol Chem. 2011;286:1556–1566. doi: 10.1074/jbc.M110.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbar E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry. 2008;47:503–508. doi: 10.1021/bi701995m. [DOI] [PubMed] [Google Scholar]
- 13.Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci U S A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 15.Stelter P, Kunze R, Flemming D, Hopfner D, Diepholz M, Philippsen P, et al. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol. 2007;9:788–796. doi: 10.1038/ncb1604. [DOI] [PubMed] [Google Scholar]
- 16.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 17.Morgan JL, Song Y, Barbar E. Structural dynamics and multiregion interactions in Dyneindynactin recognition. J Biol Chem. 2011;286:39349–39359. doi: 10.1074/jbc.M111.296277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benison G, Karplus PA, Barbar E. Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J Mol Biol. 2007;371:457–468. doi: 10.1016/j.jmb.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavanagh J, Fairbrother WJ, III, AGP, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd ed. academic Press; 2006. [Google Scholar]
- 21.Fejtova A, Davydova D, Bischof F, Lazarevic V, Altrock WD, Romorini S, et al. Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J Cell Biol. 2009;185:341–355. doi: 10.1083/jcb.200807155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol. 2012;198:1039–1054. doi: 10.1083/jcb.201202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, Diener DR, Sivadas P, Rosenbaum JL, Yang P. The versatile molecular complex component LC8 promotes several distinct steps of flagellar assembly. J Cell Biol. 2012;198:115–126. doi: 10.1083/jcb.201111041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaytseva O, Tenis N, Mitchell N, Kanno SI, Yasui A, Heierhorst J, et al. The Novel Zinc Finger Protein dASCIZ Regulates Mitosis in Drosophila via an Essential Role in Dynein Light Chain Expression. Genetics. 2013 doi: 10.1534/genetics.113.159541. 10.1534/genetics.113.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]