Summary
A transposon insertion screen implicated the yejH gene in the repair of ionizing radiation-induced damage. The yejH gene, which exhibits significant homology to the human transcription-coupled DNA repair gene XPB, is involved in the repair of double strand DNA breaks. Deletion of yejH significantly sensitized cells to agents that cause double strand breaks (ionizing radiation, UV radiation, ciprofloxacin). In addition, deletion of both yejH and radA hypersensitized the cells to ionizing radiation, UV, and ciprofloxacin damage, indicating that these two genes have complementary repair functions. The ΔyejH ΔradA double deletion also showed a substantial decline in viability following an induced double-strand DNA break, of a magnitude comparable to the defect measured when the recA, recB, recG, or priA genes are deleted. The ATPase activity and C-terminal zinc finger motif of yejH play an important role in its repair function, as targeted mutant alleles of yejH did not rescue sensitivity. We propose that yejH be re-named radD, reflecting its role in the DNA repair of radiation damage.
Keywords: yejH, radD, radA, ionizing radiation, DSBR
Introduction
Cellular DNA is routinely subjected to environmental, chemical, and metabolic damage. DNA backbone breakage can lead to double-strand breaks, which must be repaired in order for the genome to be replicated. There are several common sources of strand breaks. Ionizing radiation (IR) can generate breaks primarily via the generation of reactive oxygen species such as hydroxyl radicals (Bresler et al., 1979, Swarts et al., 2007, Ward, 1988). The reactive oxygen byproducts of aerobic metabolism can similarly give rise to strand breaks (Collins et al., 2005, Mikkelsen & Wardman, 2003). UV irradiation causes base pair lesions that can lead to transient strand breakage during nucleotide excision repair (Sinha & Hader, 2002). Protein-DNA adducts, caused by chemicals such as the gyrase-inhibiting quinolones, also lead to strand breaks following transcription, replication, or proteolysis (Drlica et al., 2008).
In bacteria, double-strand break repair (DSBR) is mediated through the recombinational DNA repair pathway catalyzed by the RecBCD helicase/exonuclease (Anderson & Kowalczykowski, 1997, Dillingham & Kowalczykowski, 2008, Spies & Kowalczykowski, 2006, Taylor & Smith, 2003), RecA recombinase (Cox, 2000, Cox et al., 2000, Cox, 2007, Kowalczykowski & Eggleston, 1994, Lusetti & Cox, 2002), and RuvABC resolvase (Kuzminov, 1999). While this process is relatively well understood, it is possible that additional in vivo components have not yet been identified. In addition, the function of some proteins already implicated in DSBR is poorly understood. For example, loss of the radA gene function clearly sensitizes cells to ionizing radiation (Diver et al., 1982, Byrne et al., 2014a). The radA gene product appears to play a role in processing branched DNA recombination intermediates, similar to recG, although this role has not been clearly defined (Beam et al., 2002).
The current recognized repertoire of E. coli DNA repair genes has been compiled in screens carried out over a period of nearly four decades (Konrad, 1977, Mahdi & Lloyd, 1989, Volkert & Nguyen, 1984, Kolodner et al., 1985, Ohta et al., 1999, Clark & Margulies, 1965, Howard-Flanders, 1968, Modrich, 1987). Screens to identify genes involved in radiation resistance were part of these efforts. The recN and recG genes have a demonstrated role in radiation resistance as well as DNA double strand break repair, and they were originally assigned a “rad” nomenclature (radB and radC, respectively) until their functions were further explored (Lombardo & Rosenberg, 2000, Sargentini & Smith, 1986).
Modern screening technologies provide ever more robust pathways to identify previously overlooked genes playing a role in almost any pathway or process of interest. We can more readily carry out saturating screens using disrupting, traceable inserts in every non-essential gene (van Opijnen et al., 2009). Using a transposon insertion library, we were able to identify all non-essential genes in Escherichia coli that are involved in responding to ionizing radiation damage (Byrne et al., 2014a). While the identified genes covered a range of DNA repair and protein metabolism factors, one that caught our attention was the previously uncharacterized gene yejH. Although yejH and the uvr proteins probably address different types of radiation-induced DNA damage, deleting yejH from the founder strain yielded a radiation sensitivity phenotype similar to that seen for uvrA/B deletions (Byrne et al., 2014a).
In this report, we investigate the function of yejH. We establish a role for the yejH gene product in the repair of double strand breaks that largely overlaps that of the genes radA and recG. The results justify a replacement of the generic and functionally uninformative yejH gene name with the more appropriately descriptive designation radD.
Results
Identification of yejH as a potential radiation repair gene
The yejH gene was identified during a genome-wide transposon-insertion screen for all non-essential genes with a role in recovery from ionizing radiation (IR) (Byrne et al., 2014a). BLAST searching revealed that the closest homolog to YejH/RadD is the archaeal or human XPB, a superfamily 2 helicase important for transcription initiation and transcription-coupled nucleotide excision repair (Fuss & Tainer, 2011). YejH/RadD contains all seven of the superfamily 2 helicase motifs (I, Ia, and II-VI), including the Walker A motif associated with ATP hydrolysis (I), indicating a possible helicase function (Fig. 1). Although YejH does not contain the N-terminal DNA recognition domain (DRD) found in XPB, it does contain a cluster of cysteines in the C-terminus. Utilizing the motif prediction program SVMProt (http://jing.cz3.nus.edu.sg/cgibin/svmprot.cgi; (Cai et al., 2003)), the structure of this cluster correlates most closely with a zinc binding motif (99% correlation with Zn, relative to 68% with Fe). This structural feature may assist with DNA binding.
Figure 1. Alignment of RadD and XPB.
The closest homologue to RadD is the human or archaeal XPB protein, which is involved in transcription initiation and transcription-coupled nucleotide excision repair. RadD contains all seven helicase motifs typical of the superfamily 2 helicases. Although RadD lacks the N-terminal “DNA recognition domain” found in XPB, it contains a C-terminal putative zinc finger motif.
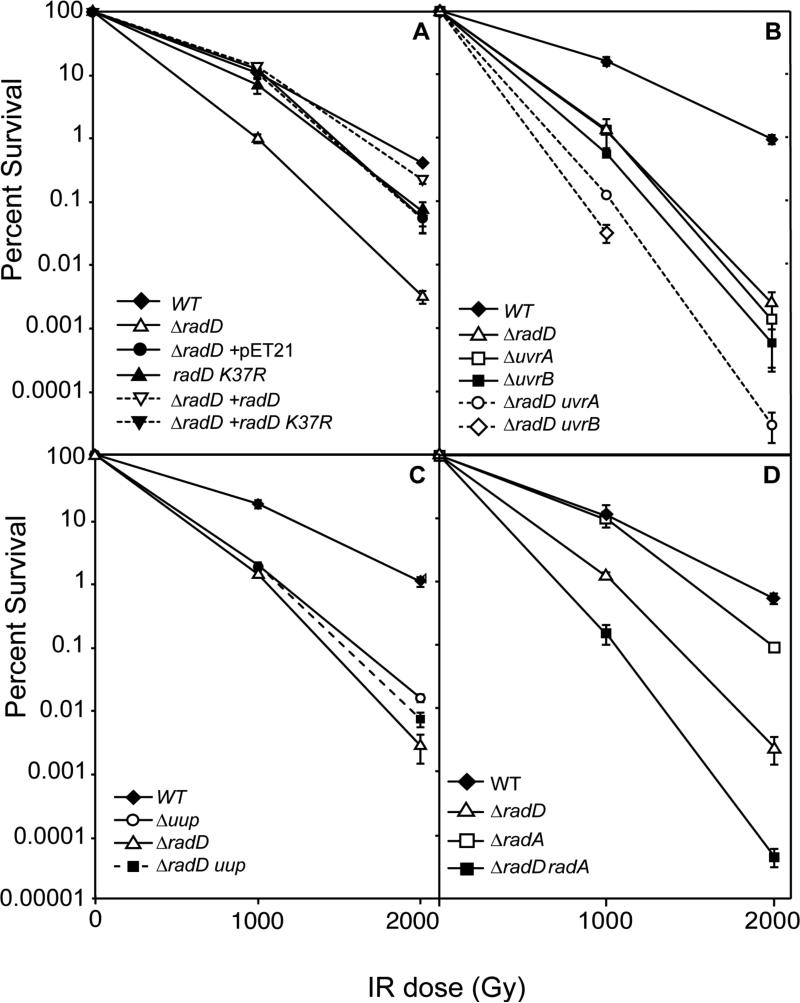
The effect of yejH/radD gene inactivation on IR survival was confirmed by deleting the yejH gene and observing increased radiation sensitivity ((Byrne et al., 2014a), and Fig. 2). The D37 for the ΔyejH/radD strain was 602 Gy (Fig. S1). This may be compared to a D37 of 1015 Gy for the founder strain used as the control strain in this study (Fig. S1), which includes a deletion of the cryptic e14 prophage that is lost rapidly in trials to generate radiation resistance by directed evolution (Harris et al., 2009, Byrne et al., 2014b). Deletion of e14 has a small but significant positive effect on IR sensitivity (Harris et al., 2009, Byrne et al., 2014b). It is deleted in all strains used in the present study to eliminate any effects its spontaneous (and unobserved) loss might cause in experiments involving irradiation. For the remainder of this report, we simply refer to the yejH gene as radD and the founder Δe14 strain as wildtype.
Figure 2. The function of radD is needed after exposure to ionizing radiation (IR).
Wildtype (founder Δe14) and deletion strains (Table 1) were exposed to high doses of ionizing radiation. Cell viability was determined after each dose and used to calculate percent survival. A. The effects of IR on strains lacking the function of the radD gene or with a putative ATPase mutation (K37R) are shown. The “+” indicates complementation with the indicated radD gene variant expressed at background levels on the plasmid pET21 without induction, or empty vector control. B. The effects of IR on strains lacking the function of the genes radD uvrA or B, or both radD and one of the uvr genes. The effects of the loss of radD and the uvrA/B genes together are further assessed in Fig. S2. C. The effects of IR on strains lacking the function of the genes radD uup, or both are shown. D. The effects of IR on strains lacking the function of the genes radD radA, or both radD and radA are shown. The effects of the loss of radD and the radA genes together are further assessed in Fig. S2.
The putative ATP hydrolytic function of radD contributes to radiation damage repair
A mutation was made in the conserved lysine of the Walker A motif (K37R), a change classically associated with the elimination of ATPase function (Moarefi et al., 2000). This mutant was inserted onto the genome in its normal chromosomal location, as well as on a plasmid for protein expression (Tables 1&2). Irradiated E. coli containing the radD K37R mutant in place of wildtype radD on the chromosome showed an intermediate level of survival between that of wildtype and the ΔradD strain (Fig. 2A), suggesting that the putative ATPase deficient mutant can perform some but not all of the functions of radD in responding to radiation damage.
Table 1.
Table of strains used
| Strain name | Genotype | Reference |
|---|---|---|
| EAW9 | MG1655 ΔrecA recG– | This study |
| EAW 7704 | Founder Δe14 | (Byrne et al., 2014a) |
| EAW 232 | Founder Δe14 ΔradD | (Byrne et al., 2014a) |
| EAW 252 | Founder Δe14 ΔradA | (Byrne et al., 2014a) |
| EAW 278 | Founder Δe14 radA K37R | This study |
| EAW 368 | FounderΔe14 ΔradD recG–. | This study |
| EAW 370 | Founder Δe14 ΔradD ΔradA | This study |
| EAW 404 | DL2006 ΔradD | This study |
| EAW 406 | DL2006 ΔradA | This study |
| EAW 416 | DL2573 ΔradD | This study |
| EAW 418 | DL2573 ΔradA | This study |
| EAW 424 | DL2006 ΔradD ΔradA | This study |
| EAW 425 | DL2573 ΔradD ΔradA | This study |
| EAW 522 | Founder Δe14 ΔradD ΔrecG | This study |
| DL2006 | BW27784 ΔPsbcDC PBAD-sbcDC lacZ::pal246 cynX::GmR | Eykelenboom et al, 2008 |
| DL2573 | BW2+7784 ΔPsbcDC PBAD-sbcDC lacZ+ cynX::GmR | Eykelenboom et al, 2008 |
| EAW 175 | CAG5052 ΔmetA ΔilvO | This study |
| EAW 174 | SS3388 ΔaroB | This study |
| EAW 477 | EAW 174 recG– | This study |
| EAW 478 | EAW 174 ΔradD | This study |
| EAW 479 | EAW 174 ΔradA | This study |
| EAW 480 | EAW 174 ΔruvB | This study |
| EAW 482 | EAW 174 ΔradD ΔradA | This study |
| CAG5052 | KL227. btuB3191::Tn10 metB1 relA1 9′ ->6′ | (Singer et al., 1989) |
| SS3388 | JC13509 ΔattB::psulA-GFP ΔmetE 100::Kan | gift from Steven Sandler |
Table 2.
Table of plasmids used
| Strain name | Description | Reference |
|---|---|---|
| pEAW 724 | pET21a RadD | This study |
| pEAW 752 | pET21a RadD 355aa truncation | This study |
| pEAW 755 | pET21a RadD K37R | This study |
| pEAW 977 | pET21a RadD C437A | This study |
| pEAW 915 | pACYC184 with a recN promoter in front of Super-Glo GFP | This study |
To confirm that the IR sensitivity phenotype was indeed due to lack of the radD gene, an expression plasmid containing wildtype radD was transformed into the ΔradD strain. This plasmid was able to rescue the phenotype of irradiated ΔradD cells to nearly wildtype levels (Fig. 2A). Expression of the gene was not induced with IPTG, indicating that a low background level of protein expression is sufficient to rescue the phenotype. Survival of irradiated ΔradD cells with a plasmid containing the radD K37R mutant was less, albeit very similar to the genomic radD K37R mutant. An empty vector control produced similar levels of resistance to that of the plasmid expressing radD K37R (but less than one expressing the wild type radD), suggesting that vector-mediated expression of the radD K37R mutant protein did not confer any significant increase in IR resistance. Overall, the effects of the presence or absence of the wild type radD gene indicates that elimination of radD function is responsible for the observed IR sensitivity phenotype. A possible effect of the RadD K37R mutant protein on IR survival is not confirmed by these results.
radD and radA have complementary functions in radiation damage repair
To further explore radD functions, the radD gene was deleted in combination with several other genes. A ΔradD mutation increased the effects of ΔuvrA or ΔuvrB (Fig. 2B). The increase in sensitivity is substantial, approximately additive (Fig. S2). The uvrA and uvrB gene products are involved in nucleotide excision repair and crosslink repair (Sancar & Rupp, 1983, Sladek et al., 1989). In contrast, as shown below, the radD gene product is involved in some aspect of DNA double strand break repair, and the relationship with uvrAB was not further explored. The radD deletion was also combined with a deletion of the uup gene, as the loss of uup confers sensitivity to ionizing radiation at levels similar to that seen when the radD function is lost (Byrne et al., 2014a). A ΔradD Δuup strain was no more sensitive to ionizing radiation than ΔradD alone (Fig. 2C). This may indicate that uup and radD participate in a joint pathway, but this has not been further explored. A ΔradD ΔrecG strain grew very slowly and accumulated suppressor mutations rapidly. A ΔradD recG– strain behaved similarly. We isolated multiple examples of the suppressors from both double mutant strains. Similar to suppressors of recG deficiency that were previously isolated by the Lloyd (Al Deib et al., 1996) and Kogoma (Kogoma et al., 1996) laboratories, all but one of the sequenced suppressors appeared in the gene priA and are listed in Table 3. One of these, priA A520P, appeared twice (once in the set obtained from each of the double mutant strains) and is identical to a priA suppressor of recG deficiency isolated previously (Al Deib et al., 1996). We presume that the priA changes eliminate the PriA helicase activity without eliminating primosome assembly as observed in the earlier studies. One of our priA alleles (priA IN W689 (RW); insertion of codons encoding RW after codon 689) suppressed the UV sensitivity of a ΔrecG strain (Fig. S3). However, the same priA allele increased the UV sensitivity of a ΔradD strain (Fig. S3). We conclude that the suppressors work primarily by suppressing the effects of the recG deficiency rather than mitigating the effects of the radD deletion. The one suppressor not found in priA has not yet been identified.
Table 3.
Suppressor mutations in the priA gene, arising in ΔradD ΔrecG (EAW 522) or ΔradD recG– (EAW 368) strains.
| Mutation | PriA substitution |
|---|---|
| Suppressors of ΔradD recG | |
| 832 T→G | S278A |
| 1136 G→C | R379P |
| 2182 G→T | D728Y |
| 1558 G→C | A520P** |
| 1470 A→G | T491A |
| CGCTGG | IN W689(RW)* |
| Suppressors of ΔradD ΔrecG | |
| 1286 G→C | G429A |
| 1861 G→C | A621P |
| 904 G→A | G302F |
| 1480 C→T | L494F |
| 1558 G→C | A520P** |
Insertion of two new codons encoding RW, after codon 689. Note that this is a new repetition of codons 686-687 and 688-689, which encode a tandem repeat of the sequence RW (i.e., RWRW→RWRWRW).
Mutation also found previously in response to recG deficiency (Al Deib et al., 1996).
The combination of ΔradD and ΔradA produced a significant and nearly additive decrease in survival post-irradiation (Fig. 2C and Fig. S2). The function of radA is poorly understood, but a link with RecA protein and DNA double strand break repair has been evident (Beam et al., 2002). This suggests the radD and radA genes have complementary functions in the cellular response to radiation damage. The results of combining ΔradD with ΔradA appeared the most immediately informative and formed much of the basis of the continued work described below.
radD and radA also respond to UV irradiation damage
We continued to investigate the effects of the ΔradD and ΔradDΔradA genotypes by exploring UV irradiation. In contrast to a previous report (Beam et al., 2002), we were able to consistently demonstrate UV sensitivity (albeit quite modest) in the ΔradA strain (Fig. 3). This is likely due to the higher doses of UV irradiation used in the current study.
Figure 3. The function of radD is needed to respond to UV radiation.
The radD and radA mutant strains (Table 1) were exposed to UV radiation. UV dosage is validated in Fig S3. A. Strains were plated, exposed to UV radiation, and colonies counted to obtain viability data, which was normalized against the zero dose point to obtain percent survival. B. The effects of UV irradiation on strains with elevated levels of the RadD or RadD K37R protein are shown. The RadD proteins were expressed at background levels from pET21. C and D. Strains were spot plated on LB prior to UV exposure to show a qualitative viability defect. Spots (left to right in each series of five) represent a serial dilution of 1:10, ending in 10−6. The “+” indicates complementation with the indicated radD gene variant expressed at background levels on the plasmid pET21 without induction, or empty vector control.
As with IR, the ΔradD and ΔradA strains both exhibited only small defects in viability as single mutants when exposed to high doses of UV. However, the ΔradDΔradA strain displayed a greatly enhanced, and in this case slightly synergistic, sensitivity (Fig. 3A and S2). The effects of the two deleted genes together are somewhat greater here than observed in the accompanying article (Deani Cooper, Daniel C. Boyle and Susan T. Lovett, accompanying paper), most likely due to the higher doses of UV irradiation used in our study. In contrast to the IR results, the ΔradD strain showed a somewhat less severe effect than the ΔradA strain, indicating that the two enzymes may target different types of damage. The UV dose levels utilized in Fig. 3 were directly validated (Fig. S4).
To complement the UV sensitivity phenotype, we provided the wildtype radD gene on an expression plasmid (Table 2). Due to the modest difference in UV sensitivity observed between founder and ΔradD strains in response to UV, we chose to complement the ΔradDΔradA strain to produce an effect that was potentially more readily measurable. Indeed, the ΔradDΔradA strain containing radD on a plasmid restores UV viability to a level that is within error of that observed with the ΔradA strain (Fig. 3A). Strikingly, adding back the Walker A mutant radD K37R to the ΔradDΔradA strain resulted in an increased sensitivity to UV irradiation. This suggests that RadD K37R may be binding to but not processing a DNA intermediate or protein-DNA complex, blocking its processing by alternative pathways. These results were confirmed by adding the radD K37R plasmid into the wild type strain and observing a dominant negative effect following UV irradiation (Fig 3B). The addition of wildtype radD into the wild type strain also had a somewhat negative effect at the highest dose of UV, suggesting that increased levels of RadD may interfere with some DNA repair events.
Based on the sequence of RadD (Fig. 1), the helicase domain is likely conserved in the core of the protein, while the C-terminus may be involved in protein-DNA or protein-protein interactions. To determine the importance of these regions, two additional radD alleles were generated and inserted into the expression plasmid to be used in complementation tests (Table 2). A RadD core enzyme was generated by truncation shortly after helicase motif VI, removing all C-terminal residues after amino acid 355. A RadD C437A mutant changed one of the four cysteines of the putative zinc finger to alanine. These two mutants, along with the Walker A mutant RadD K37R, were tested for their ability to complement the UV sensitivity of the ΔradDΔradA strain, using spot plating to observe qualitative viability defects. Although the constructs were designed to eliminate a distinct domain or motif, we cannot rule out that these mutations could have affected proper protein folding, Unlike the wildtype radD gene, none of the three mutant alleles were able to complement the viability defect (Fig. 3C), suggesting that ATP hydrolysis, the C-terminus, and the zinc finger motif are all important for responding to UV irradiation damage. An empty vector control also exhibited no complementation (Fig. 3D). As seen in Figure 3A&C, complementation with the K37R mutation again made the cells somewhat more sensitive to UV than the ΔradDΔradA strain. The other two variants did not produce this effect (Fig. 3C). The latter result may reflect a general loss of structural integrity due to the mutations, or a targeted loss of a DNA binding activity.
radD and radA are synergistic in their response to ciprofloxacin treatment
To further confirm the type of damage to which ΔradD mutants are susceptible, we implemented a radiation-free method that is known to induce double-strand breaks. We chose ciprofloxacin, an inhibitor of gyrase that traps covalent protein-DNA adducts, leading to double-strand breaks during replication, transcription, or proteolysis.
Wildtype and mutant strains were grown and spot plated on LB plates containing increasing concentrations of ciprofloxacin. Unlike the irradiation experiments, colonies grown on ciprofloxacin plates were of widely varying sizes, making colony counting impractical. Therefore, only qualitative results are shown. At the lower dose of ciprofloxacin (0.005 μg/mL), the founder, ΔradD, and ΔradA strains exhibit no defect. In contrast, the ΔradDΔradA strain exhibits a dramatic decrease in viability (Fig. 4A). At the higher dose (0.01 μg/mL ciprofloxacin), the wildtype strain begins to show a growth defect, indicated by the smaller size of the colonies. This is expected, as this dose exceeds the reported minimal inhibitory concentration (MIC) of wildtype E. coli (0.004 μg/mL) (Andrews, 2001). However, the ΔradD and ΔradA strains clearly exhibit a viability defect compared to wild type, similar to that seen for the double mutant at the lower dose (Fig. 4A). At the higher dose, the ΔradDΔradA strain is completely inviable. These results indicate that radD and radA are also important for repairing enzymatically-induced, as opposed to radiation-induced, DNA strand breaks. These results have been corroborated (Deani Cooper, Daniel C. Boyle and Susan T. Lovett, accompanying paper).
Figure 4. Deletion of radD renders cells sensitive to ciprofloxacin.
Cells were grown to log phase, serially diluted 1:10, and spot plated on LB plates containing varying amounts of ciprofloxacin. A. The effects of ciprofloxacin on cells lacking the function of radD, radA, or both are shown. B and C. Complementation of a strain lacking both radD and radA function by either RadD or RadD variants expressed at background levels on pET21. An empty vector control is provided in panel C. The “+” indicates complementation with the indicated radD gene variant expressed at background levels on the plasmid pET21 without induction, or empty vector control.
As with radiation-induced damage, plasmids containing wildtype and mutant radD were transformed into the ΔradDΔradA strain to test for complementation. Only the wildtype radD could rescue ciprofloxacin sensitivity (Fig. 4B), indicating that full-length, wildtype radD gene is needed to repair ciprofloxacin-induced damage. As seen in the UV sensitivity tests in Figure 3, an attempt at complementation with the K37R variant appeared to slightly increase sensitivity to ciprofloxacin. The empty vector control provided no measurable complementation (Fig. 4C)
radD and radA are important for responding to an induced double-strand break
Because the radD and radA deletion strains appear to be susceptible to types of damage that are known to cause double-strand DNA breaks, we utilized a system that induces a single and site-specific DNA double-strand break in the genome. Due to a palindrome artificially inserted into the lacZ gene and an arabinose-induced promoter in front of the sbcCD genes, cells plated on arabinose will incur a double-strand break during replication (Eykelenboom et al., 2008). Viability will be compromised if the break is not efficiently repaired. The single deletions of radD or radA do not produce a substantial decline in viability, although both exhibit a slight growth defect manifested by smaller colony size. Following the pattern established in earlier experiments, the ΔradDΔradA strain exhibits an obvious viability defect (Fig. 5), comparable to that previously seen for deletions of recA, recG, ruvAB, or priA (Eykelenboom et al., 2008). As in previous cases, wildtype radD introduced on a plasmid rescues this phenotype, while complementation with any of the mutant variants (or the empty vector) does not (Fig. 5).
Figure 5. The functions of radD and radA are needed for repairing an induced double-strand break.
The presence of a palindrome sequence (pal+) and arabinose will induce a targeted double-strand break in the lacZ gene (Eykelenboom et al., 2008). If not efficiently repaired, this break will lead to a low viability phenotype. The effects of this targeted double strand break on strains lacking the function of radD, radA, or both are shown. Complementation of a strain lacking both radD and radA function by either RadD or RadD variants expressed at background levels on pET21 is also shown. An empty vector control is provided in panel B. The “+” preceding a gene or plasmid name indicates complementation with the indicated radD gene variant expressed at background levels on the plasmid pET21 without induction, or empty vector control.
The ΔradDΔradA strain exhibits elevated levels of SOS response
In bacteria, DNA strand breaks lead to the induction of a number of repair genes in a process known as the SOS response (Walker et al., 2000, Michel, 2005). To test for an increased induction of the SOS response due to persistent strand breaks, a reporter plasmid in which the GFP protein is expressed from the recN promoter (pEAW 915, Table 2) was transformed into each of the single and double mutant strains. The recN gene is strongly induced at an early stage of the bacterial SOS response (Finch et al., 1985). In the absence of exogenous damage, a small but reproducible induction of GFP was detectable in the mutant strains (Fig. 6B). These strains exhibited very little difference in overall growth rate (Fig. 6A), but showed increased SOS beginning around mid-log phase (at 200 min, OD600 ~0.5). This may correspond to the increase in cells entering stationary phase. However, if this is the case, the cause is not yet evident. The effect is exacerbated in the double mutant ΔradDΔradA strain. The levels of SOS induction in the double mutant ΔradDΔradA strain are significant but very modest. Induction of the SOS response by antagonists such as ciprofloxacin produces an SOS response that is more than an order of magnitude greater on this scale and with the same assay (Fig. S5).
Figure 6. A ΔradD ΔradA double mutant shows low levels of constitutive SOS response.
Single and double mutant strains containing a plasmid expressing GFP under SOS control were grown in LB. A. Growth was monitored at 600 nm. B. SOS gene induction was measured with excitation at 474 nm and emission at 509 nm. C. Specific fluorescence, defined as measured fluorescence from panel B divided by the OD600 taken from panel A is shown. Due to the error inherent in dividing very small numbers, specific fluorescence is not shown for times prior to 100 min.
Unlike radA, deletion of radD does not significantly change levels of conjugational recombination
It was previously reported that deletion of radA decreases levels of RecA-mediated conjugational recombination (Beam et al., 2002). To determine the effect of a radD deletion on conjugational recombination, HFR recipient strains were constructed with deletions for radA, radD, both radA and radD, recG, and ruvB (Table 1), with the recG and ruvB mutants serving as negative controls. Following conjugation and plating, the resulting colonies were counted and normalized to the level of our HFR recipient control strain (EAW 174, Table 1). As previously reported, the radA single deletion displayed slightly less recombination than the control strain. However, the ΔradD and ΔradDΔradA strains had recombination levels that were within error of the control strain (Fig. 7). This represents the first phenotype for which radD mutants differ from the radA deletion strain, and indicates that radD does not participate in pathways that directly contribute to conjugational recombination.
Figure 7. The radD deletion does not significantly inhibit conjugational recombination.
The HFR recipient strains were constructed with deletions of known or predicted recombination genes (radD, radA, radD/radA, recG, ruvB see Table 1). See Methods for details.
Discussion
The work in this report establishes a role for radD (formerly known as yejH) in the cellular systems that repair DNA double strand breaks. The elimination of RadD function sensitizes the cells to a series of agents or conditions that cause DNA double strand breaks and modestly promotes induction of the SOS response. Deletions of radD exhibit synergistic effects with deletions of radA, and we hypothesize that the RadD protein plays a direct role in DNA double strand break repair. This would entail a specialized function for the protein that does not contribute to conjugational recombination, where radD function has no observable effect. Given their approximately additive sensitivity to DNA damaging agents (even synergistic with respect to UV), it appears that radD and radA serve a previously overlooked, overlapping or complementary function in the repair of double strand breaks in DNA. That function likely also complements that of several other enzymes as described below.
Although little is known about the role of radA, it has been proposed that the enzyme is involved in processing recombination intermediates (Beam et al., 2002). It appears that radA works in the recA repair pathway, as the phenotypes of radA mutants are recA-dependent (Diver et al., 1982). As recombinational repair is the main pathway of DSB repair in bacteria, it would follow that enzymes involved in DSB repair are part of the recA pathway. The radA and radD genes are not homologous. However, they share several features that are likely important in DNA repair, including ATP hydrolysis motifs (Walker A and B) and putative Zn fingers (Fig. 1 and (Beam et al., 2002)). The original identifying mutation in radA was a cysteine-to-tyrosine mutation in the putative zinc finger that caused an increased sensitivity to radiation (Diver et al., 1982). We have shown that the Zn finger and Walker A motifs of radD are important for surviving radiation exposure.
Interestingly, strains lacking radA or radD gene function respond differently to IR and UV irradiation, especially with respect to complementation with radD K37R. After exposure to IR, the ΔradD strain is more sensitive than ΔradA. The radD K37R provides a partial rescue of this sensitivity when present on the chromosome. However, this effect could not be confirmed, since the same mutant protein expressed from a plasmid exhibits the same degree of apparent rescue as does an empty vector control. This suggests the role of RadD in DNA double strand break repair is largely (if not entirely) dependent on its ATPase activity. In contrast, after exposure to UV, the ΔradD strain is somewhat less sensitive than ΔradA. Unlike its effects in the IR experiments, complementation attempts with radD K37R actually increase UV sensitivity, exhibiting a dominant negative effect. Thus, the ATPase-deficient RadD may interfere with some repair function after UV irradiation. Whereas some studies have suggested that the effects of UV and IR are interchangeable (Arrage et al., 1993), the two sources do cause different direct effects. IR causes DNA strand breaks via ionization and the creation of reactive oxygen species. UV largely promotes the formation of pyrimidine dimers that may indirectly lead to some strand breaks when replication forks encounter the template breaks or other barriers created transiently during repair (Khan & Kuzminov, 2012). These differences may help explain the failure to detect the effects of radD function in earlier genetic screens for DNA repair genes.
Evidence increasingly suggests the presence of a complex system that supports the work of RecA protein in DNA double strand break repair, processing the branched DNA intermediates that are generated by RecA. Significant components include the RecG, RuvABC, RecQ/Topoisomerase III, UvrD, and RadA proteins. Additional proteins and enzymes, notably RecBCD and RecFOR, prepare DNA substrates prior to RecA action and promote RecA loading. With this study and an accompanying report (Cooper et al, accompanying paper), RadD now joins this system. The RecG, RuvABC, and RecQ/Topoisomerase III proteins appear to have specific DNA structures as their reaction targets (Adams et al., 1994, Asai & Kogoma, 1994, Baharoglu et al., 2006, Bennett & West, 1995, Fonville et al., 2010, Lloyd et al., 1988, Mahdi et al., 2003, Harmon & Kowalczykowski, 1998). In DNA double strand break repair, UvrD helicase functions in removing RecA filaments from the DNA (Centore & Sandler, 2007, Centore et al., 2009, Veaute et al., 2005). The RadA and RadD proteins could function on DNA substrates, or alternatively might be involved in the remodeling of protein complexes bound to DNA as DNA double strand break repair progresses. Genetic evidence for involvement of these proteins in an interconnected network continues to accumulate, although enough distinctions are present to indicate specialized roles for each. A ΔradAΔrecGΔruvA (or ΔradAΔrecGΔruvC) triple mutant displays a deficiency in conjugational recombination comparable to a ΔrecA mutant (Beam et al., 2002). We have found that ΔradD, when combined with a recG insertion mutant, is slow growing and quickly accumulates suppressors even though ΔradD ΔruvB mutants grow normally (S.H. Chen & E.A. Wood, unpublished results). In addition, several of the phenotypes seen for the ΔradDΔradA mutants are similar to those seen for ΔrecG and ΔruvAB mutants, including radiation sensitivity and viability defects following an induced double-strand break (Eykelenboom et al., 2008).
The major phenotype of mutants lacking radD function is a deficiency in recovery from the effects of ionizing radiation. However, this sheds only limited light on the precise molecular function of RadD. Additional observations may provide a clue. The homology of RadD to archaeal and human XPB proteins, with their demonstrated involvement in transcription and transcription-coupled DNA repair, among other functions (Fuss & Tainer, 2011, Schaeffer et al., 1993) may suggest a role at the interface of DNA double strand break repair and transcription. Recent results from Vibrio cholerae potentially provide a more direct link between RadD and transcription complexes (Baharoglu et al., 2014). In V. cholerae, ΔyejH/radD, ΔrnhA, and Δmfd mutant strains grow slowly in the presence of tobramycin (which stalls RNA polymerases among other effects). More intriguing, overexpression of V. cholerae yejH/radD in E. coli suppresses the UV sensitivity of an E. coli Δmfd mutation (Baharoglu et al., 2014), suggesting that yejH/radD can replace the RNA polymerase displacement activity of the Mfd protein (Haines et al., 2014, Mahdi et al., 2003, Park et al., 2002, Sancar & Reardon, 2004, Savery, 2007, Smith et al., 2012, Trautinger et al., 2005). The V. cholerae and E. coli YejH/RadD proteins are 58% identical, including the helicase motifs and the C-terminal cysteine residues, and 65% similar. What need would arise for RNA polymerase displacement in DNA double strand break repair? In a bacterial genome, double strand breaks introduced stochastically during irradiation or by any mechanism have a significant chance of occurring within a gene that is being actively transcribed. The fate of RNA polymerase complexes when they encounter a double strand break has not been carefully explored. Limited stability of RNA polymerases stalled at DNA strand breaks was observed in one study (Nudler et al., 1996). However, the chloride containing buffers used in the study are known to destabilize RNA polymerase and are not a good mimic of in vivo conditions (Leirmo et al., 1987, Record et al., 1985, Shaner et al., 1983). If RNA polymerases remain stably bound to DNA at the sites of double strand breaks, their removal may be a prerequisite to repair. The degree to which RNA polymerases stalled at DNA ends represent a barrier to DNA double strand break repair is currently unknown and requires further exploration.
Experimental procedures
Strain construction
A modification of the procedure devised by Datsenko and Wanner (Datsenko & Wanner, 2000) was used to make chromosomal gene knockouts. The plasmid pEAW507 was used as a template in a PCR. pEAW507 consists of a pJFS42 mutant FRT-KanR-wt FRT cassette in an Ampicillin resistant backbone (Senecoff et al., 1988). pEAW507 was used because, after eliminating the KanR, the mutant FRT remaining on the chromosome cannot react with any other FRTs used in subsequent gene modifications. The PCR primers were about 50 bases before the start of the gene of interest with the 21 bases before the start of the FRT-KanR-FRT cassette, and about 50 bases after the stop of the gene of interest with 21 bases after the end of the FRT-KanR-FRT cassette. The gel purified PCR product was electroporated into the bacterial strain previously transformed with plasmid pKD46. L-arabinose was added to express λ Red recombinase from the pKD46. Kanamycin resistant colonies were screened for Ampicillin sensitivity, and used as a template in confirmation PCR with primers located in the chromosomal regions both upstream and downstream of the gene of interest. The PCR product was sequenced to confirm the chromosomal deletion. In the case of double deletions, the kan cassette of the first deletion was first removed (as per Datsenko and Wanner (Datsenko & Wanner, 2000)), followed by insertion of the second deletion.
EAW9 is a recG– strain. To construct EAW9, a plasmid containing the recG gene was digested with PmeI which cuts at base 1618 of recG. A Kanamycin gene flanked by HincII sites was excised from plasmid pKan and ligated into the PmeI site of the recG gene. The recG gene interrupted by the Kanamycin gene was excised from the plasmid by restriction digestion, and electroporated into a strain (MG1655 ΔrecA) containing the plasmid pKD46 (Datsenko & Wanner, 2000). A Kanamycin resistant colony was chosen. To confirm the recG chromosomal disruption, a PCR product was generated with primers for the chromosomal region upstream and downstream of the recG gene and sequenced.
EAW368 is founderΔe14ΔradD recG–. It was made by transduction of EAW232 to recG– with P1 grown on EAW9.
Plasmid construction
pEAW724 is the wt radD gene in the overproduction vector pET21a(Novagen). E. coli MG1655 genomic DNA was used as a template in a PCR with a primer consisting of a NdeI site followed by the first 27 bases of the radD gene. The ATG in the NdeI site is also the start codon for radD. The other primer consisted of a BamHI site followed by the last 25 bases of the radD gene. Changes were made for better codon usage in codons 4, 5, and 584. The PCR product was digested with NdeI and BamHI and inserted into pET21a digested with the same enzymes. The resulting plasmid, designated pEAW724, was directly sequenced to confirm the presence of the wt radD gene.
pEAW752 is the first 355 amino acids of radD in the overproduction vector pET21a(Novagen). pEAW724 was used as a template in a PCR with a primer consisting of a NdeI site followed by the first 27 bases of the radD gene. The ATG in the NdeI site is also the start codon for radD. The other primer consisted of a BamHI site followed by a stop codon, and bases 1065-1048 of the radD gene. Changes were made for better codon usage in codons 4, and 5. The PCR product was digested with NdeI and BamHI and inserted into pET21a digested with the same enzymes. The resulting plasmid, designated pEAW752, was directly sequenced to confirm the presence of radD aa 1-355.
pEAW755 is radD K37R mutant in the overproduction vector pET21a(Novagen). It was made by QuikChange site-directed mutagenesis (Agilent Technologies) of pEAW724 template using primers consisting of the radD gene bases 98-125, and their complement. The AAA bases coding for the Lys at aa 37 were changed to CGT to code for an Arg. The resulting plasmid, designated pEAW755, was directly sequenced to confirm the presence of the radD K37R mutant. pEAW977 is radD C437A mutant in the overproduction vector pET21a(Novagen). It was constructed in a fashion similar to pEAW755 except the QuikChange mutagenesis primers had the TGT bases coding for Cys at aa 437 changed to GCA to code for Ala.
pEAW915 is SuperGlo GFP(Qbiogene) under the control of the E. coli recN promoter, in the plasmid pACYC184. To clone the recN promoter, E. coli MG1655 genomic DNA was used as a template in a PCR with a primer consisting of a BglII site followed by bases 200-180 upstream of the start of the recN gene. The other primer consisted of a NheI site followed by a NdeI site and bases 1-21 upstream of the recN gene. The PCR product was digested with NheI and BglII, and ligated to Qbiogene's pQBI63 plasmid cut with the same enzymes. The recN promoter and SuperGlo GFP were excised from the resulting plasmid with BglII and HindIII, which cuts downstream of the end of SuperGlo GFP, and ligated to pACYC184 cut with BamHI and HindIII. The resulting plasmid was designated pEAW915.
Ionizing radiation resistance assays
Using an overnight culture, strains were inoculated into LB with appropriate antibiotic to an OD600 of approximately 0.02 and grown at 37°C to an OD600 of approximately 0.4. Cultures were then incubated on ice 10 min; 15 mL of culture was spun down in a tabletop centrifuge at 4°C, and cells were resuspended in 800 μl LB.
For the 0 Gy timepoint, 100 μl was removed prior to irradiation. Cells were irradiated in a Shepherd Mark I Model 30 irradiator with a Cesium 137 source at a rate of 662.37 rad/min to 1000 Gy and 2000 Gy, with 100 μl of sample removed at each point.
Irradiated and non-irradiated cells were serially diluted (100 μl into 900 μl) into M9 media, and 100 μl of appropriate dilutions were spread onto LB plates. Plates were incubated at 37°C overnight, and colonies counted the following morning.
Plating for UV, ciprofloxacin, induced double-strand break
For UV irradiation, cells were grown and serially diluted as above, and 100 μl of appropriate dilutions were spread onto LB plates. For the complementation experiments, 10 μl of each dilution (10−2 through 10−6) was spotted onto an LB plate. The plates were then exposed to UV in a Spectrolinker XL-1000 UV crosslinker (Spectronics Corp). Pictures or colony counts were taken after incubating at 37°C overnight.
For ciprofloxacin experiments, plates were poured with LB agar containing the ciprofloxacin (0.005 or 0.01 μg/mL). Cells were grown and serially diluted as above, and spot plated (10 μl, 10−2 through 10−6) on the ciprofloxacin-containing plates. Pictures were taken after growing overnight at 37°C.
For the induced double-strand break assay, strains with or without the palindrome sequence (Table 1) were grown as above, serially diluted, and spot plated (10 μl, 10−2 through 10−6) on LB plates containing either 0.5% glucose or 0.2% arabinose. Pictures were taken after growing overnight at 37°C.
Complementation assays
To test for complementation of the radD gene, cells lacking the radD gene (EAW 232 & EAW 370, Table 1) were made chemically competent. Plasmids containing the radD gene (wildtype or mutant; pEAW 724, 752, 755, 977) were individually transformed into the strains, and selected for on ampicillin plates. Strains were grown and plated as above, with the addition of ampicillin in the growth media.
SOS response assay
Overnight cultures were diluted 1:100 in fresh LB, and 200 μl was added to the wells of a black-walled, clear-bottom 96 well plate (Corning). For each sample, three overnights were grown from separate colonies, and each overnight filled three wells in the plate (three biological and three technical replicates, for 9 total wells per sample). The plate was inserted into a Tecan infinite M1000 Pro plate reader. A program was used to incubate the plate at 37°C with orbital shaking. Every 10 min, the plate was briefly shaken linearly, and the OD600 and 509 nm emission (with 474 nm excitation) were read.
Conjugational recombination assays
Donor EAW175 and recipient EAW174 strains were constructed by P1 transductions from several strains which were kindly provided from Steve Sandler. EAW175 was made by a consecutive P1 transduction of (1) the Δ(metA)::kan allele from SS6311 into CAG5052 (KL227 btuB3191::Tn10 metB1 relA1 89′→6′) to obtain an intermediate strain EAW173, checked by Tetr and Kanr phenotypes, then followed by flipping out the kan cassette; and (2) the ilvO::kan allele from SS4761 into the intermediate EAW173 strain, checking for both Tetr and Kanr phenotypes.
To make the recipient strain, the kan cassette was flipped out first from the SS338 (Δ(attB)::psulA-gfp δ(metE)100::kan) strain and strain EAW174 was made by P1 transduction of the Δ(aroB)::kan allele from SS2495 to SS3388. The ΔrecA::kan allele from EAW20 was then transferred to EAW174 by P1 transduction to make recipient EAW188. Additional recipient strains were constructed by using P1 transduction to delete the radD, radA, recG, and ruvB genes individually, as well as the combination of radD radA, from the EAW 174 strain.
Conjugation was carried out essentially as described previously (Miller, 1972, Experiments in molecular genetics, CSHL) with the following exceptions. Donor strain was grown at 37°C in Luria-Bertani (LB) broth with Tetracycline until an optical density (OD600) of 0.7 was reached. The recipient strains were grown with Chloramphenicol, Kanamycin and Streptomycin until an optical density (OD600) of 0.5 was reached. All strains were spun down and gently resuspended in the initial volume of fresh LB broth twice to remove antibiotics. Mating was carried out by mixing 200 μl of donor cells with 1800 μl of recipient cells and incubating 100 min at 37°C. For ease of colony counting, the mating mixture was diluted 1:100 in LB broth, and 100 μl (500 μl for the ruvB strain) was mixed with 3 mL of pre-warmed 0.7% Bacto agar solution to prevent additional mating and immediately poured onto a minimal media plate. The plate was rested for a few minutes at room temperature to allow the agar to set before being turned upside down and incubated for 40 hours at 37°C.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM32335 (to MMC). We thank Dr. David Leach for the kind gift of the double strand break-inducing strains, Dr. Steve Sandler for the original HFR strains, Dr. Bénédicte Michel for helpful discussion of results and for helpful comments on the manuscript, and Dr. Susan Lovett for sharing results prior to publication and commenting on early drafts of this paper.
References
- Adams DE, Tsaneva IR, West SC. Dissociation of RecA filaments from duplex DNA by the RuvA and RuvB DNA repair proteins. Proc Natl Acad Sci USA. 1994;91:9901–9905. doi: 10.1073/pnas.91.21.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Deib A, Mahdi AA, Lloyd RG. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Arrage AA, Phelps TJ, Benoit RE, Palumbo AV, White DC. Bacterial sensitivity to UV-light as a model for ionizing-radiation resistance. J Microbiol Meth. 1993;18:127–136. [Google Scholar]
- Asai T, Kogoma T. Roles of ruvA, ruvC and recG gene functions in normal and DNA damage-inducible replication of the Escherichia coli chromosome. Genetics. 1994;137:895–902. doi: 10.1093/genetics/137.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z, Babosan A, Mazel D. Identification of genes involved in low aminoglycoside-induced SOS response in Vibrio cholerae: a role for transcription stalling and Mfd helicase. Nuc Acids Res. 2014;42:2366–2379. doi: 10.1093/nar/gkt1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z, Petranovic M, Flores MJ, Michel B. RuvAB is essential for replication forks reversal in certain replication mutants. EMBO Journal. 2006;25:596–604. doi: 10.1038/sj.emboj.7600941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam CE, Saveson CJ, Lovett ST. Role for radA/sms in recombination intermediate processing in Escherichia coli. J Bacteriol. 2002;184:6836–6844. doi: 10.1128/JB.184.24.6836-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, West SC. RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc Natl Acad Sci USA. 1995;92:5635–5639. doi: 10.1073/pnas.92.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler SE, Noskin LA, Kuzovleva NA, Noskina IG. Nature of the damage to Escherichia coli DNA induced by gamma irradiation. Internat J Radiat Biol. 1979;36:289–300. doi: 10.1080/09553007914551061. [DOI] [PubMed] [Google Scholar]
- Byrne RT, Chen SH, Wood EA, Cabot EL, Cox MM. Surviving extreme exposure to ionizing radiation: Escherichia coli genes and pathways. J Bacteriol. 2014a;196:3534–3545. doi: 10.1128/JB.01589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RT, Klingele AJ, Cabot EL, Schackwitz WS, Martin JA, Martin J, Wang Z, Wood EA, Pennacchio C, Pennacchio LA, Perna NT, Battista JR, Cox MM. Evolution of extreme resistance to ionizing radiation via genetic adaptation of DNA Repair eLife. 2014b;3:e01322. doi: 10.7554/eLife.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ. SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nuc Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Leeson MC, Sandler SJ. UvrD303, a hyperhelicase mutant that antagonizes RecA-dependent SOS expression by a mechanism that depends on its C terminus. J Bacteriol. 2009;191:1429–1438. doi: 10.1128/JB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Sandler SJ. UvrD limits the number and intensities of RecA-Green fluorescent protein structures in Escherichia coli K-12. J Bacteriol. 2007;189:2915–2920. doi: 10.1128/JB.01777-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Margulies AD. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Zhou XF, Wang R, Barth MC, Jiang T, Coderre JA, Dedon PC. Differential oxidation of deoxyribose in DNA by gamma and alpha-particle radiation. Radiat Res. 2005;163:654–662. doi: 10.1667/rr3344. [DOI] [PubMed] [Google Scholar]
- Cox MM. Recombinational DNA repair in bacteria and the RecA protein. Prog Nuc Acids Res Mol Biol. 2000;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- Cox MM. The bacterial RecA protein: structure, function, and regulation. In: Rothstein R, Aguilera A, editors. Topics in Current Genetics: Molecular Genetics of Recombination. Springer-Verlag; Heidelberg: 2007. pp. 53–94. [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC. RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diver WP, Sargentini NJ, Smith KC. A mutation (radA100) in Escherichia coli that selectively sensitizes cells grown in rich medium to x-or u.v.-radiation, or methyl methanesulphonate. Internat J Radiat Biol Rel Stud Phys, Chem Med. 1982;42:339–346. doi: 10.1080/09553008214551251. [DOI] [PubMed] [Google Scholar]
- Drlica K, Malik M, Kerns RJ, Zhaol XL. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eykelenboom JK, Blackwood JK, Okely E, Leach DRF. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Finch PW, Chambers P, Emmerson PT. Identification of the Escherichia coli recN gene product as a major SOS protein. J Bacteriol. 1985;164:653–658. doi: 10.1128/jb.164.2.653-658.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville NC, Blankschien MD, Magner DB, Rosenberg SM. RecQ-dependent death-by-recombination in cells lacking RecG and UvrD. DNA Repair (Amst) 2010;9:403–413. doi: 10.1016/j.dnarep.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss JO, Tainer JA. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair. 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines NM, Kim YIT, Smith AJ, Savery NJ. Stalled transcription complexes promote DNA repair at a distance. Proc Natl Acad Sci USA. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Develop. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DR, Pollock SV, Wood EA, Goiffon RJ, Klingele AJ, Cabot EL, Schackwitz W, Martin J, Eggington J, Durfee TJ, Middle CM, Norton JE, Popelars M, Li H, Klugman SA, Hamilton LL, Bane LB, Pennacchio L, Albert TJ, Perna NT, Cox MM, Battista JR. Directed evolution of radiation resistance in Escherichia coli. J Bacteriol. 2009;191:5240–5252. doi: 10.1128/JB.00502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Flanders P. DNA repair. Annual Review of Biochemistry. 1968;37:175–200. doi: 10.1146/annurev.bi.37.070168.001135. [DOI] [PubMed] [Google Scholar]
- Khan SR, Kuzminov A. Replication Forks Stalled at Ultraviolet Lesions Are Rescued via RecA and RuvABC Protein-catalyzed Disintegration in Escherichia coli. J Biol Chem. 2012;287:6250–6265. doi: 10.1074/jbc.M111.322990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Cadwell GW, Barnard KG, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R, Fishel RA, Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad EB. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977;130:167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Eggleston AK. Homologous Pairing and DNA Strand-Exchange Proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leirmo S, Harrison C, Cayley DS, Burgess RR, Record MT., Jr. Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Porton MC, Buckman C. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol Gen Genet. 1988;212:317–324. doi: 10.1007/BF00334702. [DOI] [PubMed] [Google Scholar]
- Lombardo MJ, Rosenberg SM. radC102 of Escherichia coli is an allele of recG. J Bacteriol. 2000;182:6287–6291. doi: 10.1128/jb.182.22.6287-6291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi AA, Lloyd RG. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- Michel B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 2005;3:1174–1176. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22:5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: Protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- Ohta T, Sutton MD, Guzzo A, Cole S, Ferentz AE, Walker GC. Mutations affecting the ability of the Escherichia coli UmuD ' protein to participate in SOS mutagenesis. J Bacteriol. 1999;181:177–185. doi: 10.1128/jb.181.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Marr MT, Roberts JW. E-coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- Record MT, Jr., Anderson CF, Mills P, Mossing M, Roe JH. Ions as regulators of protein-nucleic acid interactions in vitro and in vivo. Adv Biophys. 1985;20:109–135. doi: 10.1016/0065-227x(85)90033-4. [DOI] [PubMed] [Google Scholar]
- Sancar A, Reardon JT. Nucleotide excision repair in E-coli and man. DNA Repair Replicat. 2004;69:43–71. doi: 10.1016/S0065-3233(04)69002-4. [DOI] [PubMed] [Google Scholar]
- Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Sargentini NJ, Smith KC. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat Res. 1986;107:58–72. [PubMed] [Google Scholar]
- Savery NJ. The molecular mechanism of transcription-coupled DNA repair. Trends in Microbiology. 2007;15:326–333. doi: 10.1016/j.tim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JHJ, Chambon P, Egly JM. DNA-Repair Helicase -A component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Senecoff JF, Rossmeissl PJ, Cox MM. DNA recognition by the FLP recombinasse of the yeast 2-mu plasmid -a mutational analysis of the FLP binding-site. J Mol Biol. 1988;201:405–421. doi: 10.1016/0022-2836(88)90147-7. [DOI] [PubMed] [Google Scholar]
- Shaner SL, Melancon P, Lee KS, Burgess RR, Record MT., Jr. Ion effects on the aggregation and DNA-binding reactions of Escherichia coli RNA polymerase. Cold Spring Harbor Symp Quant Biol. 1983;1:463–472. doi: 10.1101/sqb.1983.047.01.055. [DOI] [PubMed] [Google Scholar]
- Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. A collection of strains containing genetically linked alternating antibiotic-resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha RP, Hader DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J Biol Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- Smith AJ, Pernstich C, Savery NJ. Multipartite control of the DNA translocase, Mfd. Nuc Acids Res. 2012;40:10408–10416. doi: 10.1093/nar/gks775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Swarts SG, Gilbert DC, Sharma KK, Razskazovskiy Y, Purkayastha S, Naumenko KA, Bernhard WA. Mechanisms of direct radiation damage in DNA, based on a study of the yields of base damage, deoxyribose damage, and trapped radicals in d(GCACGCGTGC)(2). Radiat Res. 2007;168:367–381. doi: 10.1667/RR1058.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Methods. 2009;6:767–U721. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Delmas P, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert MR, Nguyen DC. Induction of specific Escherichia coli genes by sublethal treatments with alkylating agents. Proc Natl Acad Sci USA. 1984;81:4110–4114. doi: 10.1073/pnas.81.13.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GC, Smith BT, Sutton MD. The SOS response to DNA damage. In: Storz G, HenggeAronis R, editors. Bacterial Stress Responses. American Society of Microbiology; Washington, D.C.: 2000. pp. 131–144. [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells -identities, mechanisms of formation, and reparability. Prog Nuc Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.